Abstract
Stem-cell functions require activation of stem-cell-intrinsic transcriptional programs and extracellular interaction with a niche microenvironment. How the transcriptional machinery controls residency of stem cells in the niche is unknown. Here we show that Id proteins coordinate stem-cell activities with anchorage of neural stem cells (NSCs) to the niche. Conditional inactivation of three Id genes in NSCs triggered detachment of embryonic and postnatal NSCs from the ventricular and vascular niche, respectively. The interrogation of the gene modules directly targeted by Id deletion in NSCs revealed that Id proteins repress bHLH-mediated activation of Rap1GAP, thus serving to maintain the GTPase activity of RAP1, a key mediator of cell adhesion. Preventing the elevation of the Rap1GAP level countered the consequences of Id loss on NSC–niche interaction and stem-cell identity. Thus, by preserving anchorage of NSCs to the extracellular environment, Id activity synchronizes NSC functions to residency in the specialized niche.
During mammalian development, cell-intrinsic transcriptional programs promote stem-cell self-renewal and the undifferentiated state1,2. As development proceeds, timely activation of lineage-specific transcription factors directs stem cells towards cell-type-specific gene expression patterns. Stem cells reside in a specialized microenvironment (niche) in which they establish intimate contacts with diverse cell types and the extracellular matrix3–5 (ECM). Depriving stem cells of anchorage to the niche precipitates loss of stem-cell identity and progression along specific differentiation programs6–8. Thus, cell-intrinsic transcriptional programs and the interaction with a specialized niche are essential to maintain stem-cell properties, but whether and how they are linked has remained unknown. The importance of the physical interactions between NSCs and the niche microenvironment has also been recognized by the discovery that a vascular niche supports NSCs in the subventricular zone (SVZ) of the postnatal brain9–12. After birth, GFAP-positive NSCs establish integrindependent contacts with the endothelial cells that provide the physical framework of a highly organized vascular network11. The close interactions between NSCs and blood vessels in the SVZ are essential to maintain stem-cell characters and prevent neurogenesis. However, the molecular mechanisms that control the interactions between NSCs and blood vessels in the SVZ remain largely unknown.
Id proteins are inhibitors of the bHLH class of transcription factors, a family of proteins that function as primary inducers of cell fate determination and differentiation in mammals13. Consistent with a redundant function of Id genes in the developing mouse brain, constitutive deletion of one Id family member does not significantly impact brain development, whereas constitutive Id1–Id3-knockout mice exhibit premature differentiation and major defects in the vasculature of the brain that lead to embryonic lethality between E10.5 and E13.5 (ref. 14). To elucidate the cell-intrinsic role of Id genes in NSCs and unravel the direct molecular targets engaged by Id proteins to preserve the NSC state, genetic models that target multiple Id genes selectively in the NSC compartment and eliminate the confounding effects resulting from Id deletion in other cell types are needed. Through the generation and analysis of mice carrying targeted deletions of three Id genes, we set out to determine the biological processes controlled by Id proteins to preserve the NSC state and identify the molecular network controlled by Id proteins in NSCs. An intriguing function of Id genes emerged from these studies whereby Id proteins operate in NSCs as transcriptional repressors of Rap1GAP, the inhibitor of RAP1–GTPase. The Id–Rap1GAP–RAP1 axis synchronizes stemness and anchorage of NSCs to the embryonic and postnatal niche.
RESULTS
Id proteins are multipotency factors for NSCs
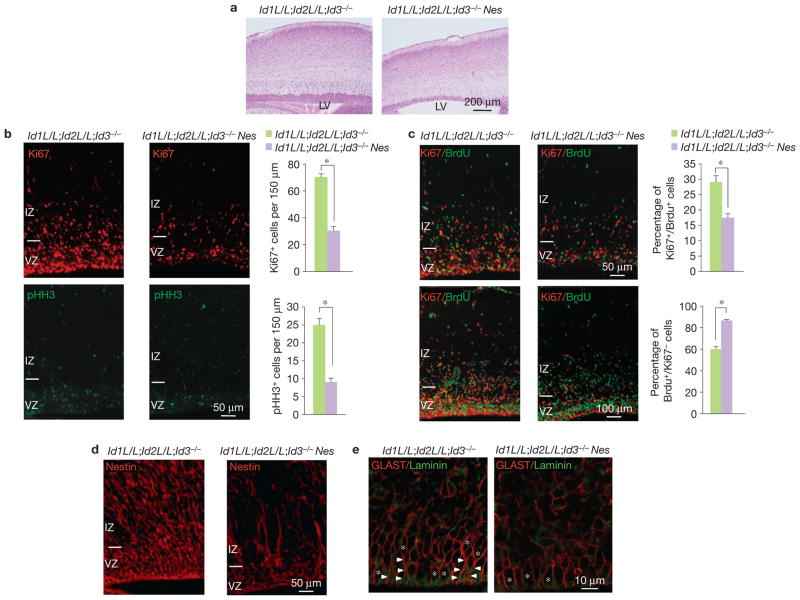
During development of the mouse brain, Id proteins are abundantly expressed with a broad and partially overlapping pattern particularly within the NSC-rich ventricular zone (VZ; Supplementary Fig. S1a,b). To ascertain the importance of Id proteins for NSC functions and identify key biological processes controlled by Id proteins in NSCs, we established a genetic model for Id ablation by generating compound mice carrying floxed Id1 and Id2 alleles and a constitutive Id3-knockout allele (Id1L/L;Id2L/L;Id3−/−, Id-conditional Triple knockout, Id-cTKO). NSC-specific Id-knockout mice were obtained by crossing Id-cTKO mice with a Nestin (Nes)–Cre transgenic line that has been widely used to delete genes in NSCs (Id-cTKO–Nes; ref. 15). Efficient deletion of Id1 and Id2 was obtained in vivo in the germinal layers and in vitro in NSCs from Id-cTKO mice following infection with a Cre-expressing lentivirus (Supplementary Figs S1 and S2a–d). Id-cTKO–Nes mice recovered at different embryonic stages and at birth were vital. However, >95% died within 24 h of postnatal life. Histological inspection of the mutant brains at embryonic day (E) 18.5 and postnatal day (P) 0 revealed enlarged ventricles, markedly reduced thickness of the germinal layers and an overall decreased brain cellularity (Fig. 1a). At E18.5, staining for the proliferation marker Ki67 and the mitotic marker phosphohistone H3 indicated that cells in the germinal areas of Id-cTKO–Nes embryonic brain had markedly reduced proliferative capacity (Fig. 1b). Next, we evaluated the impact of Id loss on cell-cycle length and rate of cell-cycle withdrawal of NSCs. Quantification of BrdU+ cells after a short pulse of BrdU (1 h) in the Ki67+ fraction of NSCs (‘cell-cycle timing’) and BrdU+ NSCs that had exited cell cycle 24 h after BrdU labelling (BrdU+/Ki67−) revealed that Id-cTKO–Nes NSCs had a significantly prolonged cell-cycle timing and increased probability of exiting from active cell cycle relative to controls (Fig. 1c). The perturbation of the cell cycle was associated with marked downregulation of cyclin D1 and increased expression of p27Kip1 in the VZ of Id-cTKO–Nes brains (Supplementary Fig. S3a,b). Immunostaining for the NSC marker Nestin depicted the striking reduction of the stem-cell compartment in Id-cTKO–Nes brains (Fig. 1d). Furthermore, high-magnification analysis of immunostaining using antibodies against laminin to label the VZ ECM and GLAST, another marker of NSCs, showed that loss of Id proteins disrupted the pseudo-stratified architecture of the VZ and altered the cell shape of the NSCs at the apical border (Fig. 1e). These effects occurred in the absence of noticeable changes of cell survival, as shown by immunostaining for the cleaved caspase-3 marker of apoptosis (Supplementary Fig. S3c).
Figure 1.
Deletion of Id genes in the developing neocortex decreases the size of the progenitor pool and alters the cell cycle. (a) Haematoxylin and eosin staining of the cerebral cortex of embryos at E18.5. (b) Sagittal sections from E18.5 embryonic brain were stained for Ki67 (upper panels) or pHH3 (lower panels). Quantification of Ki67+ or pHH3+ cells in the VZ–SVZ (150 μm) is shown in the histograms. Data are means ± s.d.; n = 3 both for Id-cTKO and Id-cTKO–Nes; P = 6.483 × 10−5. Asterisks indicate statistical significance. (c) Immunofluorescence micrographs for Ki67 (red) and BrdU (green) after a 1 h pulse of BrdU in E18.5 embryos (upper panels). The fraction of cycling progenitor cells (Ki67+) labelled with BrdU was scored. Data in the histogram are means ± s.d.; n = 3 for both Id-cTKO and Id-cTKO–Nes; P = 0.0013. The asterisk indicates statistical significance. Immunofluorescence micrographs for Ki67 (red) and BrdU (green) 24 h after a single pulse of BrdU in E18.5 embryos (lower panels). The fraction of cells labelled only with BrdU (BrdU+/Ki67−, no longer dividing) was scored. Data in the histogram are means ± s.d.; n = 3 for both Id-cTKO and Id-cTKO–Nes; P = 5.981× 10−5. The asterisk indicates statistical significance. (d) Immunofluorescence micrographs for the NSC marker Nestin show a significant depletion of NSC bodies and radial fibres in the Id-cTKO–Nes. (e) High-magnification immunostaining of the VZ/SVZ for laminin (green) and GLAST (red) in E18.5 brains. Wild-type progenitors in the VZ are embedded in a laminin-rich environment (green) that anchors cell bodies (asterisks) and end-feet, highlighted by GLAST immunostaining at the ventricular border. Arrowheads indicate apical processes. Id-cTKO–Nes progenitors at the ventricular surface present with relaxed cell bodies. Id-cTKO–Nes cells in the more basal areas of the VZ lack apical processes and anchored end-feet. LV, lateral ventricle; VZ, ventricular zone; IZ, intermediate zone.
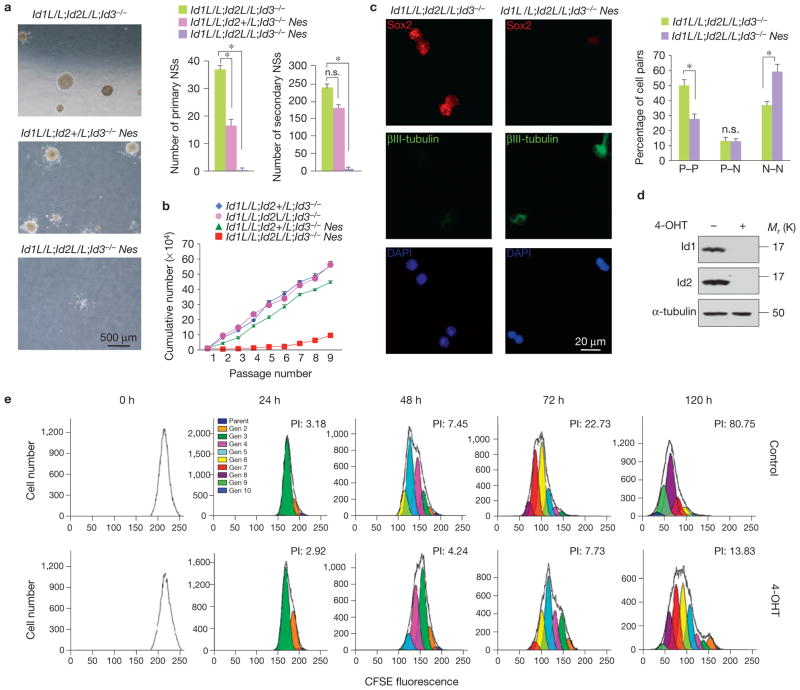
To investigate the NSC-intrinsic role of Id proteins, we isolated NSCs from the telencephalon of E13.5 Id-cTKO–Nes embryos and littermate controls and cultured them at clonal densities (<1 cell per microlitre) to assess neurosphere formation. Both primary and secondary neurosphere formation was lost in cultures derived from Id-cTKO–Nes brains (Fig. 2a). The self-renewal profile tracked with severely impaired Id-cTKO–Nes NSC proliferation measured in consecutive passages (Fig. 2b). Notably, preservation of one wild-type Id2 allele was sufficient to maintain self-renewal and proliferation at levels similar to those of wild-type cells (Fig. 2a,b). To determine whether Id proteins direct the mode of division (symmetric versus asymmetric) and/or daughter cell fate specification (progenitor versus neuron), we performed the clonal pair-cell assay16–18. In this assay, NSCs are identified by the expression of the transcription factor Sox2, and newly born neurons by the neuronal marker βIII-tubulin. Whereas the fraction of asymmetric divisions (progenitor–neuron, P–N) was similar in Id-cTKO–Nes and control cells, the generation of Sox2-positive, paired progenitors (P–P) was significantly reduced (approximately twofold) and correspondingly the production of βIII-tubulin-positive, paired neurons (N–N) was enhanced in cells carrying Id recombined alleles (Fig. 2c). Thus, in the absence of Id, shifting cell division from a proliferative to a differentiation mode compromises NSC cycling and self-renewal capacity.
Figure 2.
Id proteins regulate self-renewal and proliferation of NSCs. (a) The micrographs show the neurosphere formation potential of NSCs isolated from the brains of the indicated genotypes. The total number of neurospheres generated in medium containing EGF and FGF2. Data in the histograms represent the means ± s.d. of the number of primary and secondary neurospheres (NSs) obtained from E13.5 telencephalon of the indicated genotypes; n = 3 for each genotype; P = 6.525×10−6 for Id1L/L;Id2L/L;Id3−/− Nes and Id1L/L;Id2L/L;Id3−/−; P = 3.412×10−5 for Id1L/L;Id2 +/L;Id3−/−Nes and Id1L/L;Id2L/L;Id3−/−; P = 1.624×10−5 for secondary neurospheres. Asterisks indicate statistical significance; n.s., not significant. (b) Proliferation kinetics of NSCs by passage and Id status. Data are the means ± s.d.; n = 3 for each genotype. (c) Id regulates cell fate of NSCs in clonal culture. Left, representative images of prevalent types of daughter-cell pairs originating from individual Id-cTKO or Id-cTKO–Nes NSCs: two progenitor cells (P–P) and two post-mitotic neurons (N–N). Sibling cell pairs derived from individual cells were immunostained with antibodies against Sox2 (red), a neural progenitor restricted transcription factor and βIII-tubulin (green), a neuronal marker, and counterstained with DAPI (blue). Right, quantification of the percentage of P–P, P–N and N–N daughter cell pairs for each genotype. Data are the means ± s.d.; n = 3; (Id-cTKO, 750 cells; Id-cTKO–Nes, 635 cells). P = 5.249 × 10−5 for P–P; P = 2.687 × 10−5 for N–N. Asterisks indicate statistical significance; n.s., not significant. (d) Western blot analysis of Id1 and Id2 in Id-cTKO–Rosa-Cre-ER NSCs demonstrates efficient ablation of Id genes following treatment with 4-OHT for 72 h. (e) Proliferation kinetics of vehicle- (upper panels) and 4-OHT treated (lower panels) Id-cTKO–Rosa-Cre-ER NSCs by CFSE dilution. The fluorescence histogram plots at increasing time intervals show the ModFit-generated CFSE profile. Each Gaussian peak represents one generation. The plots at 0 h demonstrate equal loading of CFSE in both 4-OHT- and vehicle-treated cells. Id-deleted NSCs exhibit decreased cell divisions relative to wild-type cells, as indicated by higher CFSE fluorescence signal at each time interval. Proliferation index (PI) measures the proliferative activity of NSCs and indicates the total expansion of the original cell population. Uncropped images of blots are shown in Supplementary Fig. S6.
Rap1GAP is a direct bHLH–Id target gene in NSCs
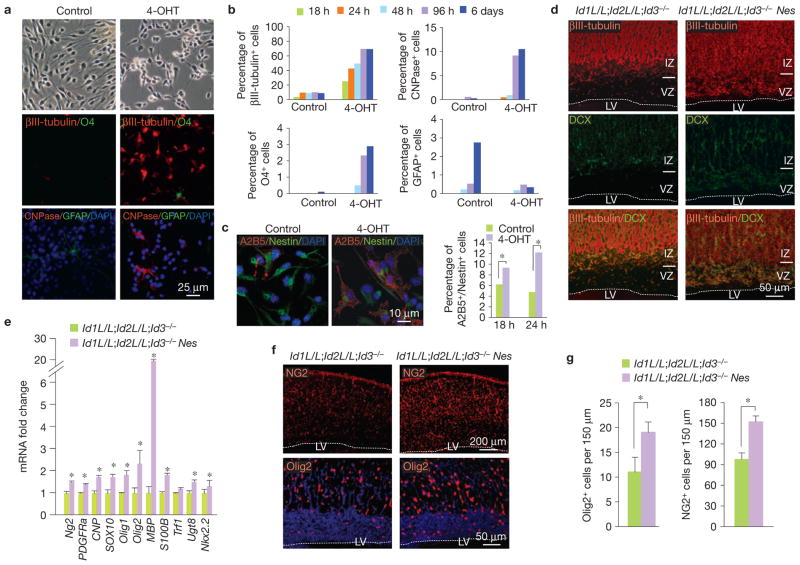
To uncover the early phenotypic modifications and the molecular consequences instigated by loss of Id activity, we generated Id-cTKO mice carrying a Cre-recombinase-oestrogen-receptor-T2 (Cre-ER) allele targeted to the ROSA26 locus (Id-cTKO–Rosa-Cre-ER). Efficient deletion of Id1 and Id2 with complete loss of Id protein expression was detectable after treatment of NSCs with 4-OHT for 72 h (Fig. 2d). We used this system to determine the kinetics of cell division of Id-deleted NSCs in proliferative conditions in the presence of EGF and FGF2, which preserve self-renewal and prevent differentiation using carboxyfluorescein diacetate succinimidyl ester (CFSE). When compared with vehicle-treated cells, 4-OHT-treated Id-cTKO–Rosa-Cre-ER NSCs showed higher intracellular levels of CFSE fluorescence intensity measured by flow cytometry at each sequential time analysed after the initial loading. This finding is indicative of a progressive decline in the rate of cell division of Id-deleted cells resulting in a sixfold reduction in the proliferation index at 120 h (13.83 in 4-OHT-treated versus 80.75 in vehicle-treated cells, Fig. 2e). 4-OHT-mediated deletion of Id genes initiated a swift morphological and molecular differentiation along the neuronal and oligodendrocyte lineages (Fig. 3a,b). In fact, an early event precipitated by Id loss was the commitment of NSCs towards a Nestin–A2B5 double-positive oligodendrocyte-restricted progenitor in the absence of apoptotic changes (Fig. 3c and Supplementary Fig. S3d). When recombination of Id genes was induced in concert with removal of the growth factors EGF and FGF2 from the culture media, NSCs differentiated prematurely along the neuronal and oligodendrocyte lineages at the expense of the astrocytic fate (Supplementary Fig. S4a,b). Accordingly, sphere differentiation assay of NSCs showed that 100% of the clones derived from wild-type NSCs contained astrocytes. Conversely, 67% of the clones generated by Id-knockout NSCs lacked astrocytes but contained neurons and oligodendrocytes in an advanced state of maturation (Supplementary Fig. S4c,d). Immunohistochemical analysis of E18.5 Id-cTKO–Nes embryonic brains demonstrated expansion of the βIII-tubulin/doublecortin (DCX)-positive compartment into the VZ/SVZ (Fig. 3d). Both messenger RNA and protein levels for transcription regulators and markers of the oligodendrocyte lineage were increased in Id-cTKO–Nes brains (Fig. 3e–g).
Figure 3.
Enforced differentiation of NSCs following acute loss of Id genes. (a) Upper panels, bright-field images of Id-cTKO–Rosa-Cre-ER NSCs cultured for 18 h in the absence or presence of 4-OHT in EGF- and FGF2-containing medium. Middle panels, immunofluorescence staining for the neuronal marker βIII-tubulin (red) and the oligodendrocyte marker O4 (green). Lower panels, immunofluorescence staining for the astrocytic marker GFAP (green) and the oligodendrocyte marker CNPase (red). Nuclei were counterstained with DAPI. (b) Quantification of the percentage of βIII-tubulin+, O4+, CNPase+ and GFAP+ cells at the indicated times of 4-OHT treatment. Data represent the means; n = 6 from two independent experiments, each carried out in triplicate. (c) Left, double immunofluorescence staining for the progenitor markers Nestin (green) and A2B5 (red). Nuclei were counterstained with DAPI. Right, quantification of the percentage of A2B5+/Nestin+ cells. Data represent the means; n = 6 from two independent experiments, each carried out in triplicate. P = 0.0011 for 18 h; P = 1.275 × 10−5 for 24 h. Asterisks indicate statistical significance. (d) Immunofluorescence staining for βIII-tubulin (red) and DCX (green) in E18.5 embryo brains. VZ, ventricular zone; IZ, intermediate zone; LV, lateral ventricle. (e) Gene expression analysis by qRT-PCR for oligodendrocyte-specific genes in E18.5 embryo cortex. Gene expression is normalized to the expression of 18S ribosomal RNA. Data are the means ± s.d.; n = 3 for both Id-cTKO and Id-cTKO–Nes. Asterisks indicate statistical significance. (f) Immunofluorescence staining for NG2 (upper panels) and Olig2 (lower panels) in E18.5 Id-cTKO and Id-cTKO–Nes embryos. Nuclei were counterstained with DAPI. LV, lateral ventricle. (g) Left, the fraction of cells labelled with Olig2 in VZ/IZ was scored. Bars indicate means ± s.d.; n = 3 for both Id-cTKO and Id-cTKO–Nes; P = 0.0007. The asterisk indicates statistical significance. Right, the fraction of cells labelled with NG2 was scored. Bars indicate the means ± s.d. n = 3 for both Id-cTKO and Id-cTKO–Nes; P = 0.0016. The asterisk indicates statistical significance.
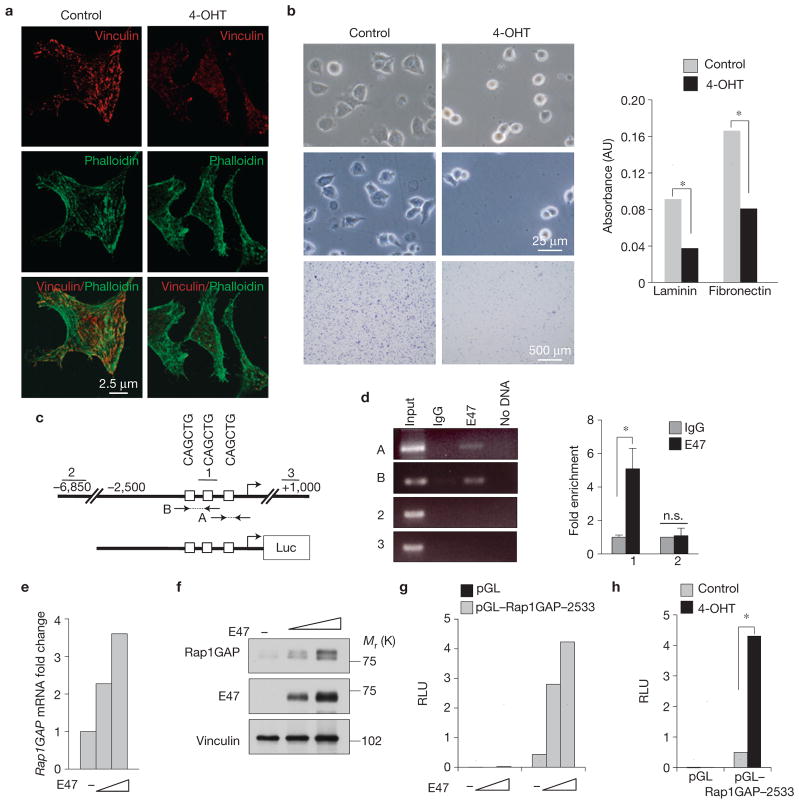
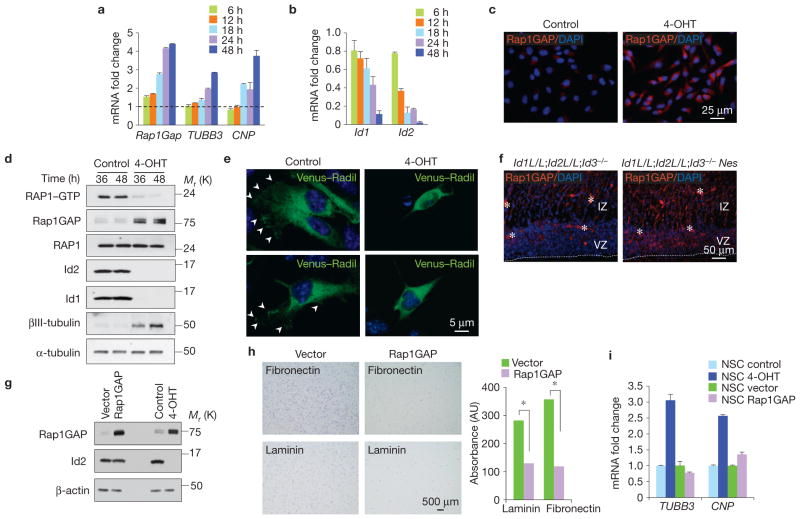
Having established that Id-cTKO–Rosa-Cre-ER NSCs represent a suitable system to interrogate the impact of acute loss of Id activity in NSCs, we obtained transcriptomic profiles of Id-cTKO–Rosa-Cre-ER NSCs treated for short times with 4-OHT or vehicle. Microarray experiments were analysed using Bayesian analysis of time series (BATS), an algorithm that identifies and ranks differentially expressed genes from time-course microarray experiments19 (Supplementary Table S1). Surprisingly, knowledge-based pathway analysis applied to the 300 most significant BATS-inferred variant genes revealed convergent regulation by Id genes of functional gene categories linked to cell membrane, cell adhesion and ECM (ranked first, second and third, respectively, Supplementary Tables S2 and S3). Gene categories associated with the Nes–Cre-driven phenotype of Id deletion (oligodendrocyte differentiation, neuronal differentiation and cell division) were also modified but with notably lower rankings. Thus, we explored whether acute loss of Id genes perturbs cell adhesion. Id recombination triggered profound changes in cell shape, as shown by severe loss of focal adhesion contacts and actin stress fibres as early as 12–18 h of 4-OHT treatment of NSCs cultured on laminin (Fig. 4a). Furthermore, acute loss of Id genes disrupted the ability of NSCs to adhere to laminin and fibronectin (Fig. 4b). Next, we sought to identify direct Id targets that might operate as master regulators of cell adhesion in NSCs. To do this, we processed the BATS-derived global output of 3,176 differentially expressed genes in Id-cTKO–Rosa-Cre-ER NSCs using a reverse engineering algorithm designed for the identification of direct targets of transcription factors (TSNI, Time-Series Network Identification)20. Remarkably, two known direct targets of Id proteins—transcriptionally activated by bHLH transcription factors21,22—ranked sixth (Cdkn1c) and ninth (Id4) in the TSNI-predicted direct Id target list. The gene encoding Rap1GAP, the GTPase activating protein for RAP1, ranked twelfth (Supplementary Tables S4 and S5; ref. 23). By acting as an essential intracellular mediator of integrin signalling, RAP1 is a key regulator of cellular adhesion in multiple biological settings23–25. Chromatin immunoprecipitation (ChIP) assays revealed that the primary Id-targeted bHLH transcription factor, the ubiquitously expressed protein E47, binds to three high-affinity E-boxes in the Rap1GAP promoter (Fig. 4c,d). Expression of E47 in NSCs elevated Rap1GAP mRNA and protein levels, and enhanced the activity of a Rap1GAP promoter/luciferase reporter containing the E47-bound E-boxes (Fig. 4e–g). The Rap1GAP promoter was also activated by deletion of Id genes triggered by 4-OHT in Id-cTKO–Rosa-Cre-ER NSCs (Fig. 4h).
Figure 4.
Acute loss of Id genes in NSCs impairs cell adhesion and induces bHLH-mediated transcription of Rap1GAP. (a) Immunofluorescence staining for vinculin (red) and F-actin (phalloidin, green) in NSCs treated with 4-OHT or vehicle for 18 h. (b) Cell adhesion on ECM components of NSCs treated with 4-OHT for 48 h. Left, upper panels, bright-field images of NSCs treated with vehicle or 4-OHT 1 h after plating on fibronectin-coated plates. Left, middle panels, bright-field images of NSCs remaining adherent after washes. Left, lower panels, crystal violet staining of NSCs remaining adherent after washes. Right, colorimetric quantification of NSCs adherent to laminin or fibronectin. Data represent the means; n = 8 from two independent experiments, each performed in quadruplicate. P = 5.401 × 10−8 (adhesion to laminin); P = 2.383 × 10−7 (adhesion to fibronectin). Asterisks indicate statistical significance. (c) Rap1GAP promoter (upper scheme) and Rap1GAP promoter/luciferase construct (lower scheme). Squares indicate E-boxes. Arrows indicate primers used to amplify the E-box-containing regions in ChIP assay. 2 and 3 are regions in the Rap1GAP gene that were not bound by E47. (d) Crosslinked chromatin prepared from NSCs transfected with E47 was incubated with IgG or anti-E47 antibody. Immunoprecipitates were amplified by PCR and analysed by agarose gel electrophoresis (left panel) or qPCR (right panel). Fold enrichment is relative to background chromatin pulled down from IgG immunoprecipitation. Data represent the means ± s.e.m.; n = 3 independent experiments, each carried out in triplicate; P = 1.400× 10−5. The asterisk indicates statistical significance; n.s., not significant. (e) Rap1GAP mRNA induction as a function of E47 in NSCs. Data represent the means; n = 6 from two independent experiments, each performed in triplicate. (f) Western blot using antibodies against Rap1GAP, E47 and vinculin in NSCs transfected with pcDNA–E47 or the empty vector. (g) NSCs were transfected with pGL vector or pGL–Rap1GAP–2533 in the absence or in the presence of increasing amounts of pcDNA3.1–E47. Data represent the means; n = 6 from two independent experiments, each performed in triplicate. (h) NSCs were transfected with pGL vector or pGL–Rap1GAP–2533 and treated with 4-OHT for 72 h. Data represent the means; n = 6 from two independent experiments, each performed in triplicate. P = 0.0029. RLU is relative Firefly luminescence units normalized to Renilla luminescence. The asterisk indicates statistical significance. Uncropped images of blots are shown in Supplementary Fig. S6.
Id proteins repress Rap1GAP and maintain active RAP1 in NSCs
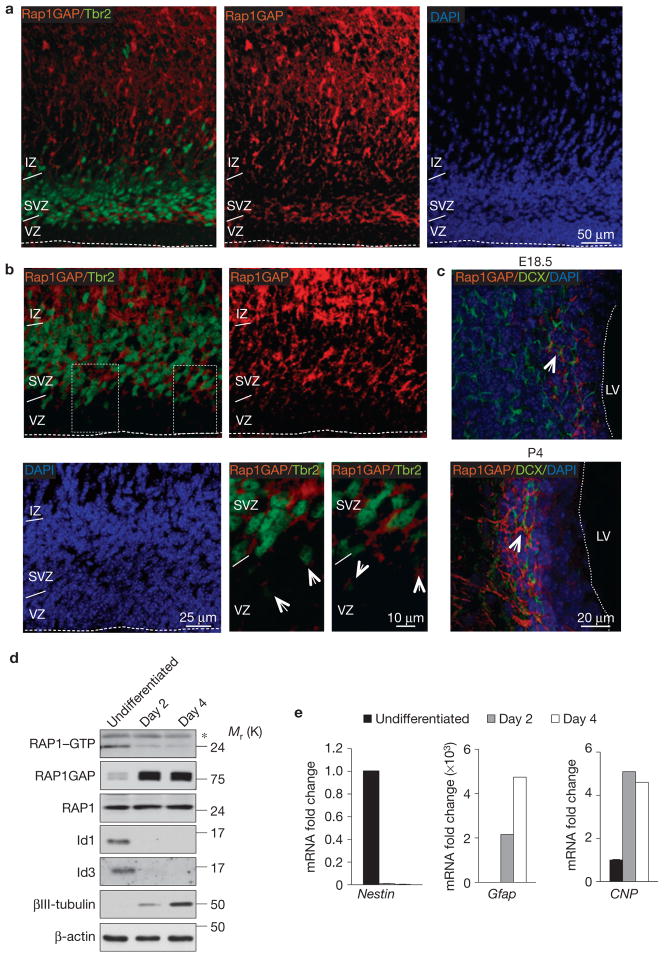
Multipotent NSCs in the VZ retain an apical membrane that is anchored to the ventricular surface26. Conversely, Tbr2-positive progenitors lack apical contact with the ventricular surface27. Double immunostaining experiments for Tbr2 and Rap1GAP in the developing brain revealed that the two proteins are largely absent in the VZ but are co-induced as apical progenitors give rise to basal progenitors in the SVZ (Fig. 5a,b). We identified scattered cells in the outer VZ that co-express low levels of Tbr2 and Rap1GAP, indicating that progenitors acquire Rap1GAP as they delaminate from the ventricular surface (Fig. 5b, arrows). Co-expression analysis of Rap1GAP with DCX shows that Rap1GAP is induced in concert with DCX, a protein marking the commitment towards a migrating neuroblastic fate (Fig. 5c). Next, we investigated whether expression of Rap1GAP and the RAP1 GTPase are regulated when Id proteins are downregulated in an in vitro model of NSC differentiation. When growth factors are removed from the culture, NSCs downregulate Id proteins, lose neural stem cell markers (for example, Nestin) and induce lineage-specific differentiation markers. Rap1GAP was strongly upregulated and RAP1 activity was inhibited (Fig. 5d,e). Mimicking the loss of self-renewal of NSCs initiated by removal of growth factors, the acute deletion of Id genes in Id-cTKO–Rosa-Cre-ER NSCs in the presence of growth factors resulted in a rapid, sustained and strong induction of Rap1GAP mRNA and protein with consequent extinction of RAP1–GTP (Fig. 6a–d). As a direct consequence of RAP1 inactivation, we observed the loss of localization on the plasma membrane of Radil, a RAP1 effector whose membrane localization is dependent on the intracellular activity of RAP1 and is essential for the biological effects of RAP1 on cell adhesion28,29 (Fig. 6e). Deletion of Id genes in the Id-cTKO–Nes brain triggered ectopic expression of Rap1GAP in the VZ, indicating that Id proteins repress Rap1GAP in vivo (Fig. 6f). To dissect the significance of Rap1GAP in NSCs, we expressed Rap1GAP at similar levels to Id-cTKO–Rosa-Cre-ER NSCs after Id deletion and analysed NSCs at the earliest time of Rap1GAP accumulation after lentiviral transduction (72 h, Fig. 6g). The results in Fig. 6g–i show that acute expression of Rap1GAP impaired adhesion of NSCs to laminin and fibronectin to the same extent manifested by NSCs depleted of Id genes (compare Fig. 6h with Fig. 4b). However, under these experimental conditions, Rap1GAP did not induce either neuronal or oligodendroglial differentiation (Fig. 6i). Thus, the primary consequence of Rap1GAP expression in NSCs is to disrupt cell adhesion but this effect is invariably associated with initiation of differentiation in the context of Id deletion.
Figure 5.
Expression of Rap1GAP in the normal cerebral cortex and NSC differentiation. (a) Immunofluorescence staining for Rap1GAP (red) and Tbr2 (green) in the telencephalon of E18.5 mouse brain. (b) Higher-magnification image of the VZ/SVZ showing that Rap1GAP (red) and Tbr2 (green) are co-expressed by progenitors in the SVZ in the telencephalon. Arrows in the lower right panels (higher-magnification images of the outlined areas in upper left panel) point to the cells in the outer VZ that exhibit weak co-expression of Tbr2 and Rap1GAP. (c) Immunofluorescence staining for Rap1GAP and DCX in the telencephalon of E18.5 and P4 mouse brain. Arrows indicate clusters of Rap1GAP–DCX double-positive cells. The dashed line marks the ventricular surface. VZ, ventricular zone; SVZ, subventricular zone; IZ, intermediate zone; LV, left ventricle. (d) Analysis of RAP1 activity in undifferentiated NSCs or NSCs cultured in differentiation medium for the indicated times using the RAP1 pulldown assay. Protein samples were analysed by SDS–PAGE and western blotting using RAP1 antibody (RAP1–GTP). Total cell lysates were analysed by SDS–PAGE and western blotting using the indicated antibodies. β-actin is shown as a loading control. The asterisk denotes a nonspecific band. (e) Gene expression analysis by qRT-PCR for Nestin, Gfap and CNP in NSCs treated as in d. Data are the means; n = 6 from two independent experiments, each performed in triplicate. Gene expression is normalized to the expression of 18S ribosomal RNA. Uncropped images of blots are shown in Supplementary Fig. S6.
Figure 6.
Expression of Rap1GAP inhibits RAP1 activity and impairs adhesion of NSCs. (a) Gene expression analysis by qRT-PCR for Rap1GAP, TUBB3 and CNP in NSCs at the indicated times of 4-OHT treatment. Data represent the means ± s.d.; n = 3 independent experiments, each carried out in triplicate. Gene expression is normalized to the expression of 18S ribosomal RNA. The dashed line indicates the normalized value of the untreated control for each time point. (b) Gene expression analysis by qRT-PCR for Id1 and Id2 in NSCs at the indicated times of 4-OHT treatment. Gene expression is normalized to the expression of 18S ribosomal RNA. Data represent the means ± s.d.; n = 3 independent experiments, each performed in triplicate. (c) Immunofluorescence staining for Rap1GAP in NSCs treated for 24 h with vehicle or 4-OHT. (d) Analysis of RAP1 activity in NSCs treated with vehicle or 4-OHT using the RAP1 pulldown assay. Proteins were analysed by western blotting using RAP1 antibody (RAP1–GTP). Total cell lysates were analysed by western blotting using the indicated antibodies. α-tubulin is shown as a loading control. (e) Fluorescence microscopy of Id-cTKO–Rosa-Cre-ER NSCs infected with pLOC–Venus–Radil in the absence or presence of 4-OHT. Arrows indicate accumulation of Venus–Radil at the plasma membrane in control cells. 4-OHT-treated NSCs show diffuse cytoplasmic fluorescence signal. (f) Immunofluorescence staining for Rap1GAP in the brain of Id-cTKO and Id-cTKO–Nes E18.5 embryos shows de-repression of Rap1GAP in the mutant VZ/SVZ. VZ, ventricular zone; IZ, intermediate zone. (g) Analysis of Rap1GAP expression in NSCs following lentiviral transduction (pLOC–GFP–vector, pLOC–GFP–Rap1GAP) or treatment with vehicle or 4-OHT for 72 h. Total cell lysates were analysed by western blotting using the indicated antibodies. β-actin is shown as a loading control. (h) Cell adhesion on ECM components of NSCs transduced with vector or Rap1GAP lentivirus as in g. Left, bright-field images of adherent NSCs 30 min and 90 min after plating on fibronectin or laminin, respectively. Right, colorimetric quantification. Data represent the means; n = 8 from two independent experiments each performed in quadruplicate; P = 0.0011 (adhesion to fibronectin); P = 0.0018 (adhesion to laminin). Asterisks indicate statistical significance. (i) Gene expression analysis by qRT-PCR for TUBB3 and CNP in NSCs treated as in g. Data represent the means; n = 3 independent experiments, each performed in triplicate. Gene expression is normalized to the expression of 18S ribosomal RNA. Uncropped images of blots are shown in Supplementary Fig. S6.
The Id–RAP1 pathway directs adhesion and residency of NSCs in the neural niche
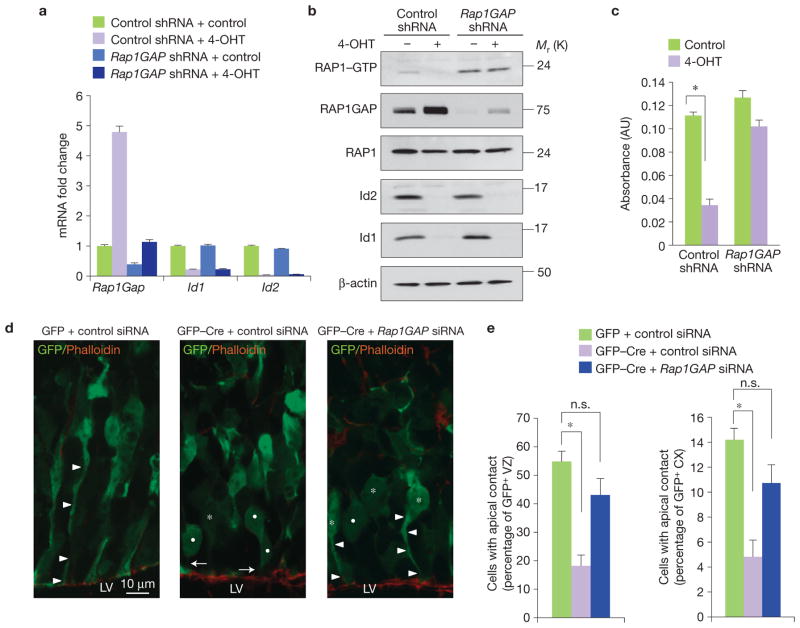
The premature increase of Rap1GAP in Id-deficient NSCs and brains, combined with concurrent inactivation of RAP1, prompted us to perform complementary studies in purified NSCs and the whole brain to functionally interrogate the significance of Rap1GAP accumulation precipitated by Id deletion for: RAP1 activity; adhesion to the ECM; and anchorage of NSCs to the niche. To determine whether accumulation of Rap1GAP in NSCs that are being depleted of Id proteins is directly responsible for the observed inactivation of RAP1 (loss of RAP1–GTP), we countered elevation of the Rap1GAP level in NSCs undergoing acute deletion of Id genes by short hairpin RNA (shRNA)-mediated silencing of the Rap1GAP gene (Fig. 7a). Preventing Rap1GAP accumulation reversed the inhibition of RAP1–GTP induced by Id loss (Fig. 7b). Concurrently, NSCs adhesiveness to the ECM was restored (Fig. 7c).
Figure 7.
Silencing of Rap1GAP restores adhesion of NSCs carrying deletion of Id genes in vitro and in vivo. (a) Gene expression analysis by qRT-PCR for Rap1GAP, Id1 and Id2 in control-shRNA- or Rap1GAP-shRNA-expressing NSCs treated with vehicle or 4-OHT for 48 h. Data indicate the means ± s.d.; n = 3 independent experiments, each performed in triplicate. (b) Analysis of RAP1 activity and protein expression in replicate cultures treated as in a. (c) Analysis of cell adhesion to fibronectin-coated plates in replicate cultures treated as in a. Data represent the means ± s.e.m.; n = 3 independent experiments, each performed in quadruplicate. P = 5.401 × 10−8. The asterisk indicates statistical significance. (d) Ex vivo electroporation of MSCV–GFP or MSCV–GFP–Cre plasmid together with control or Rap1GAP siRNA into E14.5 mouse cortices followed by organotypic slice culture for 36 h. Cortical sections were stained for GFP to identify transfected cells and F-actin/phalloidin (red) to define the ventricular surface. Arrowheads and asterisks indicate GFP+ apical processes and cell bodies, respectively, which make contact with F-actin; arrows and dots indicate GFP+ processes and cell bodies, respectively, which are disconnected from F-actin. LV, lateral ventricle. (e) Quantification of GFP+ cells with apical contacts. Results represent the means ± s.d.; n = 6 from three electroporated embryos per group from two independent experiments (P = 0.008 for the left panel and P = 0.0096 for the right panel). Left, the y axis reports the percentage of the total number of GFP+ cells in the VZ whose apical process is in contact with the ventricular surface (determined by co-localization with phalloidin staining). Right, the y axis reports the percentage of the total number of GFP+ cells in the cerebral cortex (CX) whose apical process is in contact with the ventricular surface (determined by co-localization with phalloidin staining). Asterisks indicate statistical significance; n.s., not significant. Uncropped images of blots are shown in Supplementary Fig. S6.
In the developing cortex, ECM components and integrin receptors represent the molecular glue that anchors NSCs to the ventricular surface and provides crucial signals to maintain the NSC identity30. RAP1 is a key intracellular mediator of integrin-based cell–ECM adhesion and has been implicated in retention of Drosophila germ stem cells inside the niche8,25. To explore in vivo the functional significance of ectopic elevation of Rap1GAP triggered by Id deletion for the ability of NSCs to retain anchorage to the ventricular surface, we analysed the consequences of acute loss of Id function in the NSC compartment of the embryonic brain. Deletion of floxed genes in the cortical germinal layers of the developing mouse brain can be achieved by Cre recombinase31,32. Electroporation of Id-cTKO cortices at E14.5 with a Cre–GFP-expressing plasmid efficiently recombined Id genes in the electroporated, GFP-positive cells in the VZ (data not shown). We determined the number of GFP-positive cells that retained apical contact with the ventricular surface, which was marked by phalloidin staining. When compared with GFP-electroporated cells, expression of Cre–GFP triggered detachment of the apical processes of VZ cells from the ventricular surface. However, the expression of Cre–GFP in the presence of siRNA oligonucleotides targeting Rap1GAP, but not control siRNAs, restored anchorage of the apical processes of NSCs to the ventricular surface (Fig. 7d,e). Thus, the disruption of apical contacts precipitated by Id loss in the developing brain requires accumulation of Rap1GAP. Preventing Rap1GAP upregulation by siRNA-mediated silencing also rescued cell-cycle withdrawal and premature neuronal differentiation induced by Id ablation in the embryonic VZ (Supplementary Fig. S5a,b).
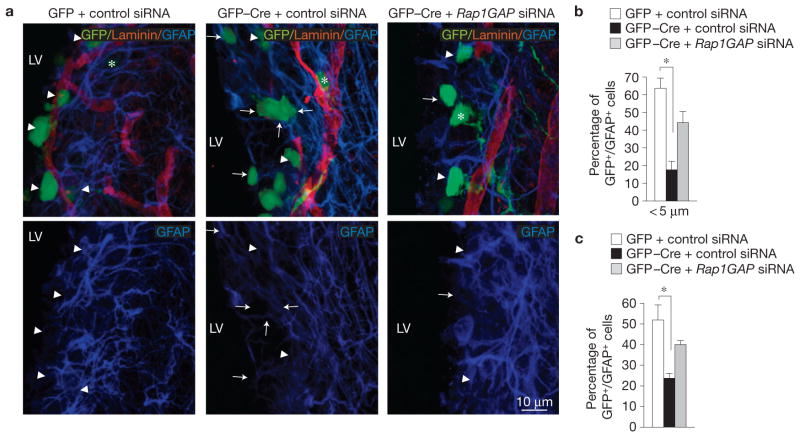
After birth, NSCs are retained in a vascular niche in the SVZ (ref. 9). Recent analysis of the postnatal NSC niche revealed a prominent network of blood vessels and showed that NSCs, which express GFAP, lie in close contact with the vascular surface10–12. We investigated whether Id deletion elicits a Rap1GAP-dependent detachment of NSCs from the specialized niche even in this temporal context. Towards this goal, the brain of Id-cTKO mice at P5 was electroporated with: GFP control plasmid plus siRNA control oligonucleotides; GFP–Cre plasmid plus siRNA control oligonucleotides; or GFP–Cre plasmid plus siRNA oligonucleotides targeting Rap1GAP. After 5 days, the mice were killed and the integrity of the vascular niche–NSC interaction was examined by scoring the fraction of GFP/GFAP-positive NSCs that lie within 5 μm of the laminin-positive endothelium. In Id-deleted NSCs, the proportion of GFP/GFAP-positive cells less than 5 μm away from the endothelium was reduced by more than threefold when compared with the control and silencing Rap1GAP prevented this effect (Fig. 8a,b). Moreover, we found that the proportion of GFAP-positive NSCs was halved by Cre-mediated deletion of Id genes and this effect was significantly rescued by Rap1GAP silencing (Fig. 8c).
Figure 8.
Id proteins are required for adhesion of NSCs to the vasculature in the postnatal SVZ. (a) In vivo electroporation of MSCV–GFP or MSCV–GFP–Cre plasmid together with control or Rap1GAP siRNA into P5 mouse cortices, followed by immunostaining five days later. Cortical sections were stained for GFP to identify successfully transduced cells, GFAP (blue) to label NSCs and laminin (red) to define blood vessel surface. Arrowheads and arrows indicate GFP–GFAP-positive cells that lie <5 μm or >5 μm from laminin-positive structures, respectively. Asterisks indicate GFP-positive cells that are negative for GFAP. LV, lateral ventricle. (b), Quantification of GFP–GFAP-positive cells <5 μm from laminin+ structures. The asterisk indicates statistical significance. (c) Quantification of GFP–GFAP-positive cells. Results represent the means ± s.d.; n = 6 from 3 electroporated brains per group from two independent experiments. The asterisk indicates statistical significance. P = 0.0024 for b; P = 0.009 for c.
Together, these findings support the conclusion that Rap1GAP is a physiologically relevant direct target of the bHLH–Id transcriptional pathway, whose de-repression in Id-deleted NSCs is essential to release them from the embryonic ventricular niche and the postnatal vascular niche.
DISCUSSION
Id proteins are expressed by embryonic and somatic stem cells, in which they act redundantly to drive stemness by enhancing proliferation and inhibiting differentiation33–36. Here we provide evidence that besides maintaining the multipotent and self-renewing state35, Id proteins impart to NSCs the competence of adhering to the extracellular niche microenvironment of the embryonic and postnatal brain. In the absence of Id genes, NSCs cannot sustain stemness and undergo premature differentiation along the neuronal and oligodendroglial lineages at the expense of the astrocytic lineage. Using unbiased computational methods, we uncovered an Id-based network that protects the RAP1–GTPase from the inhibitory activity of Rap1GAP, the latter emerging as the product of a bHLH-target gene. RAP1 is a key mediator of cell adhesion that functions by positively regulating integrin signalling and its inactivation impairs cell–ECM and cell–cell adhesion in different cell types23–25,37. From multiple experimental systems in vitro and in vivo, our work established that, by preserving high RAP1 activity in NSCs, Id proteins operate as essential master regulators of the anchorage of embryonic and postnatal NSCs to the extracellular niche microenvironment, a feature linked to stem-cell identity.
The finding that maintenance of active RAP1 through Id-mediated repression of Rap1GAP is critical to hold NSCs in the ECM niche of the embryonic and postnatal brain is consistent with the notion that α6-integrin was identified as the sole common element of different types of embryonic and adult stem cells38. Resembling the Rap1GAP-mediated inactivation of RAP1 after Id ablation in NSCs, blockage of α6β1-integrin with neutralizing antibodies caused NSCs to move away from the ventricular and the vascular surfaces of the embryonic and postnatal neural niche, respectively11,30. The fact that markers of apoptosis are not induced after Id deletion in NSCs in vitro and in vivo is consistent with the notion that the association of NSCs with the niche is through adhesion mechanisms, rather than survival selection11. Our results are also coherent with work in the Drosophila testis where RAP1 activity controls stem-cell anchoring to the stromal niche8, thereby indicating that the master regulatory function of RAP1 for adhesion to the stem-cell niche may be an evolutionarily conserved mechanism.
Interactions between stem cells, blood vessels and the ECM are found in most stem-cell niches. These interactions are frequently mediated by integrins and integrin receptors. Therefore, by Id-mediated negative control of Rap1GAP, a NSCs-intrinsic transcriptional program synchronizes self-renewal, inhibition of differentiation and anchorage to the embryonic and postnatal NSC niche. The broad activity of Id proteins as self-renewing factors and inhibitors of differentiation in embryonic and somatic stem cells, together with the general maintenance of stem cells in a niche microenvironment, indicate that the Id–RAP1 network may be a common mechanism used by stem cells to synchronize self-renewal with adhesion to the niche.
METHODS
Generation of conditionally mutant Id2 mice
The Id2 conditional allele (Id2Flox) was created by inserting an Frt-flanked pGKNeoR cassette into intron 2 and loxP sites 5′ of exon 1 and intron 2. As the coding region of Id2 is in the first and second exon, the entire coding region of Id2 is floxed, and thus a target for deletion by the Cre recombinase. NeoR-targeted embryonic stem cell lines were produced, and appropriate integration was verified by genomic PCR (the position of primers is indicated in Supplementary Fig. S2). Breeding male chimaeras with C57Bl/6 female mice generated F1 animals in a 129/Svj;C57Bl/6 hybrid background. Mice were screened for germline transmission of the floxed Id2 allele (Id2+/L) by genomic PCR and crossed with mice carrying an flp transgene under the control of the β-actin promoter to delete the PGK–neo cassette in vivo. Id1LoxP/LoxP (Id1L/L), Id3LoxP/LoxP (Id3L/L) and Id3−/− mice have been described previously35,39,40. Id1+/L, Id2+/L and Id3−/− mice were crossed to obtain Id1L/L;Id2L/L;Id3−/− mice. To generate Id1L/L;Id2L/L;Id3−/−;Nestin–Cre animals, homozygous females were bred to Nestin–Cre males. For embryonic analysis, Id1L/L;Id2L/L;Id3−/− females were crossed with Id1L/L; Id2+/L;Id3−/−; Nestin–Cre males, which were viable. All animal experiments were approved by and performed in accordance with the guidelines of the International Agency for Research on Cancer’s Animal Care and Use Committee.
Histology and immunostaining
Tissue preparation and immunohistochemistry on mouse embryo brain and immunofluorescence staining of NSCs were performed as previously described31,32,41. Antibodies used in immunostaining are listed in Supplementary Table S6. In the histograms, values represent the mean values; error bars are s.d. or s.e.m. as indicated in the figure legends. Statistical significance was determined by t-test (two-tailed).
Ex vivo and in vivo electroporation and immunostaining
Ex vivo electroporation was performed as described previously31,32. Briefly, Id-cTKO embryos were recovered at E14.5. Endotoxin-free plasmids (1 mg ml−1, MSCV–GFP or MSCV–Cre–GFP) mixed with siRNA were injected into lateral ventricles using a Femtojet microinjector (Eppendorf). Electroporated cerebral hemispheres were sectioned coronally into 300-μm-thick slices using a vibratome (VT1000S, Leica). siRNAs were used at 10 μM and were Rap1GAP Smart Pool (M-050831-01, Dharmacon, sequence: 5′-CAGGAUGGAUGAACAGCGA-3′, 5′-GUUGAUCAAUGCCGAGUAU-3′, 5′-CAAUGUGGAUCGAUUCUAC-3′, 5′-GCACUAAACCUGUUACUGG-3′) or siGENOME Non-Targeting siRNA Pool targeting firefly luciferase (U47296, D-001206-13, Dharmacon). The slices were cultured for 36 h in Neurobasal medium containing 1% N2 and B27 supplements (Invitrogen), 2 mM l-glutamine, penicillin–streptomycin and amphotericin B. Slices were labelled with 10 μM BrdU for 2 h followed by fixation in 4% paraformaldehyde (PFA) at 4 °C for 14 h. Slices were incubated for 24 h in PBS containing 15% sucrose and 30% sucrose sequentially and embedded in OCT. Confocal images were acquired with a Zeiss Axioscop2 FS MOT microscope and used to score double-positive cells. Each experiment was performed with samples (mid-rostral sections) from at least three animals from two independent experiments. Quantification was performed using six to eight sections from each embryo. In the histograms, values represent the mean values; error bars are s.d. Different subregions of the cerebral cortex were identified on the basis of cell density and visualized with DAPI nuclear staining and neuronal marker (βIII-tubulin) expression as described previously42,43. Briefly, the VZ/SVZ is identified by high cell density and absent/faint βIII-tubulin staining, IZ with low cell density and strong βIII-tubulin staining, and cortical plate with high cell density and strong βIII-tubulin staining.
Electroporation of postnatal mouse brain was performed as described previously44. Briefly, following anaesthesia by hypothermia, P5 pups were fixed on a custom-made support plate placed in a stereotaxic rig. Injections were performed at the midpoint of a virtual line connecting the eye with the cranial landmark Lambda as visualized by a light source. Plasmid solution (1.2 μl) was injected at a depth of 3 mm from the skull surface using a Hamilton syringe equipped with a 34G needle.
Injected mice were subjected to 5 electrical pulses (100 V, 50 ms, separated by 950 ms intervals) using the ECM 830 BTX electroporator (Harvard Apparatus) and 10 mm plate electrodes coated with conductive gel (Signa gel, Parker Laboratories). Five days later, animals were deeply anaesthetized and killed by transcardial perfusion of saline followed by 4% PFA. After 24 h, 50-μm vibratome sections were immunostained with the antibodies listed in Supplementary Table S6. Eight z-stacks (1 μm) were captured using a Zeiss LSM510Meta-NLO confocal microscope. z-stacks were taken along the medial and dorsal side of the SVZ using a ×40 objective. GFP–GFAP double-positive NSCs in the postnatal SVZ (B1 cells) were identified as reported in recent studies10–12 and according to the following criteria: close contact to the ventricular surface; distinctive morphology (radial glia-like morphology versus the multipolar, branched and small soma morphology of parenchymal astrocytes); and close association (<5 μm) to laminin-positive endothelium. The distance of GFP–GFAP double-positive DAPI-stained cell nuclei from laminin-positive blood vessels was quantified using ImageJ64 software. At least 200 cells per section and four to six sections from each embryo were included in the analysis. Statistical significance was determined by t-test (with Welch’s correction).
Cell culture conditions and cell-culture-based assays
Human embryonic kidney 293T and bEnd3 cells (ATCC) were grown in DMEM containing 10% fetal bovine serum (Invitrogen). Primary murine NSCs isolated from E13.5 Id1L/L;Id2L/L;Id3−/−; Nes–Cre or Id1L/L;Id2L/L;Id3−/−; Rosa-Cre-ER mouse telencephalon were cultured in the presence of FGF2 and EGF (20 ng ml−1 each). For Id gene deletion, cells cultured on laminin-coated dishes in the presence of FGF2 and EGF were treated with 250 nM 4-OHT dissolved in 100% ethanol or vehicle. Differentiation of NSCs was induced by culturing cells on laminin-coated dishes in NSC medium lacking growth factors. Primary and secondary mouse neurosphere assay was performed by plating 5,000 and 2,000 cells, respectively, in 35 mm dishes in collagen containing NSC medium in triplicates. The number of colonies was scored 14 days later.
For the sphere differentiation assay, passage-7 single neurospheres (of ~150 μm diameter) were transferred to laminin-coated coverslips in proliferation medium. After 24 h, the medium was replaced with growth-factor-deprived medium. Coverslips were processed five days later for triple-labelled immunofluorescence staining to detect neurons, oligodendrocytes and astrocytes, using anti-βIII-tubulin, anti-O4 and anti-GFAP antibodies, respectively. The phenotype of the progenitor was determined on the basis of the cell types (neuron, astrocyte and/or oligodendrocyte) that were present in the clone, regardless of the cell number. Eighteen spheres per condition were examined in two independent experiments.
For cell adhesion assay, Id1L/L;Id2L/L;Id3−/−;Rosa-Cre-ER+/− NSCs were cultured as neurospheres in the presence of 4-OHT or vehicle for 48 h. Dissociated neurospheres were plated on laminin- or fibronectin-coated plates in quadruplicates. At the times indicated in the figure legends, plates were vigorously washed with PBS/0.1% BSA twice to remove non-adherent cells. Cells were fixed with 4% PFA for 10 min and stained with 5% crystal violet. Crystal violet was solubilized in 2% SDS and absorbance was measured at 550 nm. For cell adhesion assay following Rap1GAP overexpression, NSCs were infected with pLOC–GFP or pLOC–GFP–Rap1GAP lentiviral particles and assayed as described above after 72 h. RNA and proteins from parallel cultures were analysed by real-time quantitative PCR (qRT-PCR) and western blot, respectively.
Lentiviral production
pLKO.1 lentiviral expression vectors carrying shRNAs were purchased from Sigma. The hairpin sequence targeting the Rap1GAP gene is 5′-CCTGGTATTCTCGCTCAAGTA-3′. pLOC–GFP–Rap1GAP lentivirus was obtained from Open Biosystems. Lentivirus preparation and infections were performed as described previously41.
CFSE proliferation assay
Cell proliferation analysis of NSCs was done using CFSE (ref. 45,46; Invitrogen). Cells were incubated with 20 μM CFSE in PBS/0.1% BSA at 37 °C for 15 min in the dark, washed with NSC medium, counted and plated in the presence of 4-OHT or vehicle. At 24, 48, 72 and 120 h after CFSE labelling, single cells were fixed with 1% PFA and analysed on a FACSCalibur flow cytometer (BD Biosciences). One aliquot of cells was analysed immediately after staining, as a reference for CFSE loading. Unlabelled cells were used as a control for background autofluorescence signal. Fluorescence-activated cell sorting (FACS) data were analysed using the Proliferation Wizard algorithm of the ModFit LT 3.0 software (Verity Software House). Cells were gated on the basis of forward and side scatter profiles. ModFit deconvoluted the raw histograms of fluorescence intensity into Gaussian peaks, where each peak represented a generation, to determine the proportion of cells at each generation. The proliferation index is the ratio of the total number of cells in all generations including the parental to the computed number of original parent cells present at the start of the experiment. Goodness of fit of a model was assessed by the reduced χ2 value.
Microarrays and computational analyses
Total RNA was extracted from triplicate samples of Id-cTKO–Rosa-Cre-ER NSCs treated for different times (6 h, 12 h, 18 h, 24 h, 48 h, 96 h or 144 h) with 4-OHT or vehicle and used for analysis on the Illumina MouseWG-6 expression BeadChip array. The raw array data were normalized using the Bioconductor package Lumi using quantile normalization. To identify and rank the differentially expressed genes between NSCs treated with 4-OHT or vehicle control from the time-course experiments, we used the BATS algorithm as described previously19. This method ranks the differentially expressed genes between the two time series on the basis of a Bayes factor (Supplementary Table S1). Significant functional annotation clusters enriched in the 300 most significant BATS-inferred variant genes were predicted using DAVID (Database for Annotation, Visualization and Integrated Discovery, Bioinformatics Resources at the National Institute of Allergy and Infectious diseases, NIH). The expression data for 3,176 significant genes were processed using the TSNI algorithm20. TSNI identifies the targets of experimentally perturbed genes, given a time series obtained after the perturbation has been applied. In our experiments the perturbation was the suppression of Id gene expression. The output of TSNI is a vector (‘B vector’), where the k th value is proportional to the estimated effect of the perturbation on gene k. Therefore, genes associated with a larger (absolute) value represent candidate targets of the perturbation; these values are ranked and reported in Supplementary Tables S4 and S5. The sign of the B element is an indication of whether the corresponding gene has experienced a concordant or discordant effect on its expression when compared with the sign of the perturbation. Given that the sign of the perturbation in this experiment is negative (repression of Id gene expression), a large negative value represents a transcriptional enhancing action, and a positive value represents a repressive action on the specific gene by Id proteins. The microarray expression data have been deposited in the GEO database (accession number: GSE36488).
Biochemical methods
The levels of active GTP-bound RAP1 were determined using the Active RAP1 Pull-Down and Detection Kit (Pierce) according to the manufacturer’s instruction using Id1L/L;Id2L/L;Id3−/−;Rosa-Cre-ER NSCs cultured in medium containing vehicle or 4-OHT for the indicated times. Proteins were analysed by immunoblotting using anti-RAP1 antibody. For Rap1GAP-shRNA-mediated silencing experiments, Id1L/L;Id2L/L;Id3−/−;Rosa-Cre-ER NSCs were infected with a lentivirus expressing shRNA control or shRNA targeting Rap1GAP and processed as described above.
RNA preparation and qRT-PCR
RNA preparation and qRT-PCR were performed as described previously32,41. Primers used in qRT-PCR are listed in Supplementary Table S7a.
ChIP and luciferase reporter studies
ChIP was carried out as described previously41 using anti-E47 antibody (sc-649, Santa Cruz) or rabbit immunoglobulins. DNA was eluted in 100 μl of water and 1 μl was analysed by PCR or SYBR-green-based qRT-PCR using the DNA pool from E47 and IgG ChIP and input control. Melting curve analysis was performed for each reaction to ensure a single peak. Results are presented as the mean ± s.d. Experiments were performed in triplicate and were repeated three times. Primers used in the ChIP assay are listed in Supplementary Table S7b. For the Rap1GAP reporter assay, a fragment of ~2550 bp corresponding to the mouse Rap1gap promoter was amplified by PCR using the following primers (forward: 5′-TGCACCTTGGTACCCATGGAAACCAGAGGAGGTCTTT-3′; reverse: 5′-GGGGACGCACCTACCCTCGAGAGGCACCGAAGGTGAAG-3′) and cloned into the KpnI–XhoI sites of the pGL3 vector (Promega) to generate pGL3–2533. Transfection of mouse NSCs was performed using the Amaxa Mouse Neural Stem Cell Nucleofector Kit (Lonza) according to the manufacturer’s instructions. Firefly and Renilla luciferase activities were assayed using the Dual-Luciferase Reporter Assay System (Promega). Results are mean values of Firefly normalized to Renilla.
Supplementary Material
Acknowledgments
This work was supported by National Cancer Institute grants R01CA101644 and R01CA131126 (A.L.), R01CA085628 and R01CA127643 (A.I.) and National Institute of Neurological Disorders and Stroke R01NS061776 (A.I.). F.N. is supported by a fellowship from the Italian Ministry of Welfare/Provincia di Benevento. We thank S. Angers for the Venus–Radil expression plasmid.
Footnotes
AUTHOR CONTRIBUTIONS
A.L. and A.I. designed and supervised this study, analysed data and wrote the manuscript. X.Z. generated the conditional Id2-knockout mice. F.N., X.Z., D.S., A.C., R.S. and A.L. carried out experiments. H-s.N. and R.B. generated the conditional Id1-knockout mice. Y.Z. generated the constitutive and conditional Id3-knockout mice. M.L. and D.D.B. performed the computational analysis of the microarray data. All authors discussed the results and commented on the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at www.nature.com/reprints
Note: Supplementary Information is available on the Nature Cell Biology website
References
- 1.He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annu Rev Cell Dev Biol. 2009;25:377–406. doi: 10.1146/annurev.cellbio.042308.113248. [DOI] [PubMed] [Google Scholar]
- 2.Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuchs E, Tumbar T, Guasch G. Socializing with the neighbors: stem cells and their niche. Cell. 2004;116:769–778. doi: 10.1016/s0092-8674(04)00255-7. [DOI] [PubMed] [Google Scholar]
- 4.Marthiens V, Kazanis I, Moss L, Long K, Ffrench-Constant C. Adhesion molecules in the stem cell niche–more than just staying in shape? J Cell Sci. 2010;123:1613–1622. doi: 10.1242/jcs.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellis SJ, Tanentzapf G. Integrin-mediated adhesion and stem-cell-niche interactions. Cell Tissue Res. 2010;339:121–130. doi: 10.1007/s00441-009-0828-4. [DOI] [PubMed] [Google Scholar]
- 7.Raymond K, Deugnier MA, Faraldo MM, Glukhova MA. Adhesion within the stem cell niches. Curr Opin Cell Biol. 2009;21:623–629. doi: 10.1016/j.ceb.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Wang H, et al. Rap-gef signaling controls stem cell anchoring to their niche through regulating de-cadherin-mediated cell adhesion in the Drosophila testis. Dev Cell. 2006;10:117–126. doi: 10.1016/j.devcel.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Currle DS, Gilbertson RJ. The niche revealed. Cell Stem Cell. 2008;3:234–236. doi: 10.1016/j.stem.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 10.Mirzadeh Z, Merkle FT, Soriano-Navarro M, Garcia-Verdugo JM, Alvarez-Buylla A. Neural stem cells confer unique pinwheel architecture to the ventricular surface in neurogenic regions of the adult brain. Cell Stem Cell. 2008;3:265–278. doi: 10.1016/j.stem.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen Q, et al. Adult Svz stem cells lie in a vascular niche: a quantitative analysis of niche cell–cell interactions. Cell Stem Cell. 2008;3:289–300. doi: 10.1016/j.stem.2008.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tavazoie M, et al. A specialized vascular niche for adult neural stem cells. Cell Stem Cell. 2008;3:279–288. doi: 10.1016/j.stem.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perk J, Iavarone A, Benezra R. Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer. 2005;5:603–614. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- 14.Lyden D, et al. Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature. 1999;401:670–677. doi: 10.1038/44334. [DOI] [PubMed] [Google Scholar]
- 15.Tronche F, et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- 16.Bultje RS, et al. Mammalian Par3 regulates progenitor cell asymmetric division via notch signaling in the developing neocortex. Neuron. 2009;63:189–202. doi: 10.1016/j.neuron.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanada K, Tsai LH. G protein βγ subunits and AGS3 control spindle orientation and asymmetric cell fate of cerebral cortical progenitors. Cell. 2005;122:119–131. doi: 10.1016/j.cell.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Shen Q, Zhong W, Jan YN, Temple S. Asymmetric numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. Development. 2002;129:4843–4853. doi: 10.1242/dev.129.20.4843. [DOI] [PubMed] [Google Scholar]
- 19.Angelini C, Cutillo L, De Canditiis D, Mutarelli M, Pensky M. BATS: a Bayesian user-friendly software for analyzing time series microarray experiments. BMC Bioinform. 2008;9:415. doi: 10.1186/1471-2105-9-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Della Gatta G, et al. Direct targets of the Trp63 transcription factor revealed by a combination of gene expression profiling and reverse engineering. Genome Res. 2008;18:939–948. doi: 10.1101/gr.073601.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pagliuca A, Cannada-Bartoli P, Lania L. A role for Sp and helix-loop-helix transcription factors in the regulation of the human Id4 gene promoter activity. J Biol Chem. 1998;273:7668–7674. doi: 10.1074/jbc.273.13.7668. [DOI] [PubMed] [Google Scholar]
- 22.Rothschild G, Zhao X, Iavarone A, Lasorella A. E proteins and Id2 converge on P57kip2 to regulate cell cycle in neural cells. Mol Cell Biol. 2006;26:4351–4361. doi: 10.1128/MCB.01743-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caron E. Cellular functions of the Rap1 GTP-binding protein: a pattern emerges. J Cell Sci. 2003;116:435–440. doi: 10.1242/jcs.00238. [DOI] [PubMed] [Google Scholar]
- 24.Boettner B, Van Aelst L. Control of cell adhesion dynamics by Rap1 signaling. Curr Opin Cell Biol. 2009;21:684–693. doi: 10.1016/j.ceb.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bos JL, et al. The role of Rap1 in integrin-mediated cell adhesion. Biochem Soc Trans. 2003;31:83–86. doi: 10.1042/bst0310083. [DOI] [PubMed] [Google Scholar]
- 26.Fietz SA, Huttner WB. Cortical progenitor expansion, self-renewal and neurogenesis-a polarized perspective. Curr Opin Neurobiol. 2011;21:23–35. doi: 10.1016/j.conb.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Pontious A, Kowalczyk T, Englund C, Hevner RF. Role of intermediate progenitor cells in cerebral cortex development. Dev Neurosci. 2008;30:24–32. doi: 10.1159/000109848. [DOI] [PubMed] [Google Scholar]
- 28.Ahmed SM, Daulat AM, Meunier A, Angers S. G protein βγ subunits regulate cell adhesion through Rap1a and its effector radil. J Biol Chem. 2010;285:6538–6551. doi: 10.1074/jbc.M109.069948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smolen GA, et al. A rap GTPase interactor, RADIL, mediates migration of neural crest precursors. Genes Dev. 2007;21:2131–2136. doi: 10.1101/gad.1561507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loulier K, et al. β1 integrin maintains integrity of the embryonic neocortical stem cell niche. PLoS Biol. 2009;7:e1000176. doi: 10.1371/journal.pbio.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao X, et al. The N-Myc-Dll3 cascade is suppressed by the ubiquitin ligase Huwe1 to inhibit proliferation and promote neurogenesis in the developing brain. Dev Cell. 2009;17:210–221. doi: 10.1016/j.devcel.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao X, et al. The hect-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat Cell Biol. 2008;10:643–653. doi: 10.1038/ncb1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jankovic V, et al. Id1 restrains myeloid commitment, maintaining the self-renewal capacity of hematopoietic stem cells. Proc Natl Acad Sci USA. 2007;104:1260–1265. doi: 10.1073/pnas.0607894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakashima K, et al. Bmp2-mediated alteration in the developmental pathway of fetal mouse brain cells from neurogenesis to astrocytogenesis. Proc Natl Acad Sci USA. 2001;98:5868–5873. doi: 10.1073/pnas.101109698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nam HS, Benezra R. High levels of Id1 expression define B1 type adult neural stem cells. Cell Stem Cell. 2009;5:515–526. doi: 10.1016/j.stem.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ying QL, Nichols J, Chambers I, Smith A. Bmp induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with stat3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 37.Kooistra MR, Dube N, Bos JL. Rap1: a key regulator in cell–cell junction formation. J Cell Sci. 2007;120:17–22. doi: 10.1242/jcs.03306. [DOI] [PubMed] [Google Scholar]
- 38.Fortunel NO, et al. Comment on “‘Stemness’: transcriptional profiling of embryonic and adult stem cells” and “a stem cell molecular signature”. Science. 2003;302:393. doi: 10.1126/science.1086384. [DOI] [PubMed] [Google Scholar]
- 39.Guo Z, et al. Modeling Sjogren’s syndrome with Id3 conditional knockout mice. Immunol Lett. 2011;135:34–42. doi: 10.1016/j.imlet.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan L, Sato S, Frederick JP, Sun XH, Zhuang Y. Impaired immune responses and B-cell proliferation in mice lacking the Id3 gene. Mol Cell Biol. 1999;19:5969–5980. doi: 10.1128/mcb.19.9.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carro MS, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heng JI, et al. Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature. 2008;455:114–118. doi: 10.1038/nature07198. [DOI] [PubMed] [Google Scholar]
- 43.Nguyen L, et al. P27kip1 independently promotes neuronal differentiation and migration in the cerebral cortex. Genes Dev. 2006;20:1511–1524. doi: 10.1101/gad.377106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boutin C, Diestel S, Desoeuvre A, Tiveron MC, Cremer H. Efficient in vivo electroporation of the postnatal rodent forebrain. PLoS One. 2008;3:e1883. doi: 10.1371/journal.pone.0001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lyons AB, Parish CR. Determination of lymphocyte division by flow cytometry. J Immunol Methods. 1994;171:131–137. doi: 10.1016/0022-1759(94)90236-4. [DOI] [PubMed] [Google Scholar]
- 46.Quah BJ, Warren HS, Parish CR. Monitoring lymphocyte proliferation in vitro and in vivo with the intracellular fluorescent dye carboxyfluorescein diacetate succinimidyl ester. Nat Protoc. 2007;2:2049–2056. doi: 10.1038/nprot.2007.296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.