Summary of recent advances
Recently developed techniques allow genomic DNA sequencing from single microbial cells [1]. Here, we focus on research strategies for putting these methods into practice in the laboratory setting. An immediate consequence of single cell sequencing is that it provides an alternative to culturing organisms as a prerequisite for genomic sequencing. The microgram amounts of DNA required as template are amplified from a single bacterium by a method called multiple displacement amplification (MDA) avoiding the need to grow cells. The ability to sequence DNA from a cell is likely to have an immense impact on microbiology considering the vast numbers of novel organisms which have been inaccessible unless culture-independent methods could be used. However, special approaches have been necessary to work with amplified DNA. MDA may not recover the entire genome from the single copy present in most bacteria. Also, some sequence rearrangements can occur during the DNA amplification reaction. Over the past 2 years many research groups have begun to use MDA, and some practical approaches to single cell sequencing have been developed. We review the consensus that is emerging on optimum methods, reliability of amplified template, and the proper interpretation of “composite” genomes which result from the necessity of combining data from several single cell MDA reactions in order to complete the assembly. Preferred laboratory methods are considered based on experience at several large sequencing centers where >70% of genomes are now often recovered from single cells. Methods are reviewed for preparation of bacterial fractions from environmental samples, single cell isolation, DNA amplification by MDA, and DNA sequencing.
Introduction
Single microbial cell isolation and propagation in axenic cultures has been the primary means of obtaining sufficient DNA for genomic sequencing. However, the vast majority of bacterial and archaeal taxa remain uncultivated [2,3]. The inaccessibility of genomic DNA from these organisms led to the development of cultivation-independent methods, based on the extraction and analysis of DNA from entire environmental microbial communities. The PCR amplification and sequencing of ribosomal RNA genes is widely used for taxonomic characterization of uncultured microbial assemblages [4]. Direct cloning and sequencing of environmental DNA, or metagenomics, has provided an enormous increase in our understanding of the genes, and their encoded proteins, present in the environment [5–7]. The metagenomic approach was recently used to nearly double the number of identified proteins [8] and to unveil community-wide patterns in gene and taxa distributions among various habitats [6,9]. Metagenomic sequencing of extremely simple microbial communities enabled genome reconstruction of their predominant members [10–12]. However, even very large sequencing efforts proved unsuitable for genome assemblies and metabolic reconstruction of the members of complex communities. The assembly and analysis of discrete microbial genomes from the environment thus remains a difficult yet fundamental requirement of biological research.
A new strategy has been developed that addresses these limitations by enabling study of single cells without culturing [1]. DNA from individual cells can be amplified in sufficient quantities for use as templates in genomic sequencing. The multiple displacement amplification (MDA) method [13–15] generates micrograms of DNA from the several femtograms present in a typical bacterium. MDA is based on isothermal (30°C) strand displacement synthesis in which the highly processive phi29 DNA polymerase repeatedly extends random primers on the template as it concurrently displaces previously synthesized copies [1]. The ability to sequence from single cells using the amplified DNA was demonstrated by Raghunathan et al. with flow sorted E. coli, Myxococcus xanthus, and B. subtilis [16].
Some sequence information may be lost during the process of cell lysis and single genome amplification. Extensive portions of the genome, however, have been obtained from previously inaccessible species including novel soil bacteria [17], a species of Crenarchaeota [18], the marine organism Prochlorococcus [19], TM7 from soil and human oral cavity, a candidate phylum for which no sequenced members had existed [20,21], and from uncultured marine Flavobacteria containing proteorhodopsins (TW, RS unpublished). An estimated 70% of the genome was recovered from a single filament (containing a few hundred cells) of an uncultivated species of Beggiatoa [22]. Even this partial draft has dramatically advanced the biological research by revealing the presence of enzymes for sulfur-oxidation, nitrate- and oxygen-respiration and CO2-fixation supporting a putative lithoautotrophic metabolism proposed for Beggiatoa in 1888. High throughput sorting and amplification of large arrays of novel environmental single cells enables large-scale screening of these single amplified genomes (SAGs) by PCR for multiple phylogenetic and metabolic marker genes [23]. Initial PCR screening allows sequencing efforts to be focused on taxa of interest and with some prequalification that the MDA reactions chosen for sequencing contain substantial coverage of the genome [1].
Preparation of microbial cell fractions from the environment
Enrichment of a microbial fraction from environmental samples facilitates isolation of single cells. Soil was pretreated by density gradient centrifugation prior to single cell isolation [18,21]. For aquatic samples, tangential filtration should be considered if concentration of the biomass is necessary [24]. Collection of air samples for genomic analysis is a relatively new area. Sampling of microbes for culturing has been extensively researched [25–27]. Methods include impaction, liquid impingement, filtration, and electrostatic precipitation [28]. Each of these allows documentation of concentrations and compositions in air samples. Impaction, filtration, or electrostatic precipitation primarily collect samples on a semisolid or solid surface such as a culture plate or porous membrane filter and, therefore, are less suitable for isolation of viable single cells. Liquid impingement-type air collectors, which suspend cells in a solution that can be used to isolate single cells, allow for high efficiency particulate capture in ranges of 1–10 µm in a liquid medium [29,30]. In addition, bacteria associated with airborne particles in the requisite size range may also be captured.
Isolation of single cells
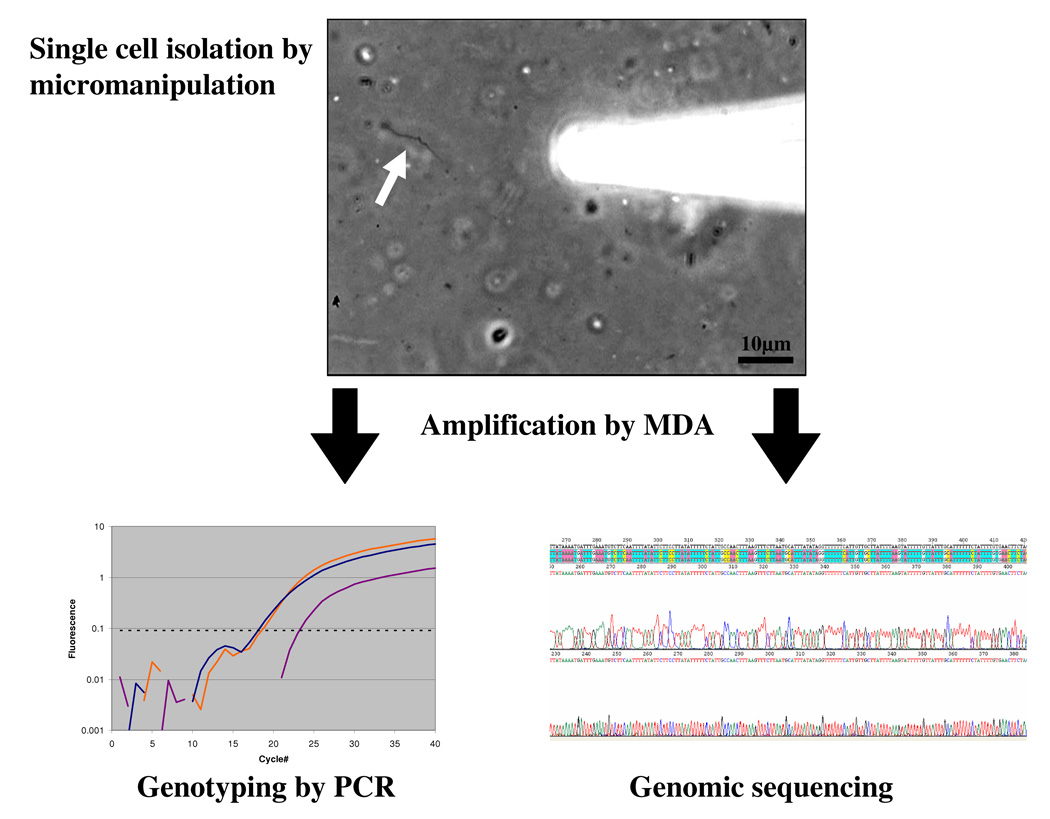
Depending on desired throughput and the environment and organisms targeted, single cells have been isolated for use in MDA reactions by dilution [17,19], fluorescence activated cell sorting (FACS) [16,23], micromanipulation [31] and microfluidics [20,32]. Cell sorting by FACS can isolate thousands of cells in minutes [16,21,23]. Potentially, single cell sorting can be combined with fluorescent in situ hybridization (FISH) to enrich for specific taxa [21,33,34]. Cells can be sorted into microplates (96 and 384 well format) facilitating automation of DNA amplification by MDA and downstream analyses. Micromanipulation methods can be divided in two categories. Optical tweezers have been described for single cell isolation and cultivation [35,36], but not in the context of whole genome amplification and sequencing. Mechanical micromanipulation (based on equipment used for in-vitro fertilization) in combination with modern research microscopy enables selection of single cells with high confidence [31]. It has been used in combination with FISH to select specific populations from environmental samples for single cell DNA amplification by MDA [17,18]. While micromanipulation is a relatively low throughput method compared to flow sorting, it is a powerful research tool that allows observation of cell morphology, documentation by imaging, and a high degree of certainty that a single cell was captured and delivered to the reaction vessel for amplification. Rinsing single cells in buffer allows the removal of free DNA or other contaminants. Cell densities of 50–1000 cells/µl are easily sufficient for micromanipulation enabling single cell studies of virtually any environmental sample. Single spirochetes of Borrelia burgdorferi (the causative agent of Lyme disease), for example, were identified by phase contrast microscopy and isolated by micromanipulation from infected tick midgut (TI, RL unpublished) in collaboration with the laboratory of Benjamin Luft (Stony Brook University, NY). Single cell MDA enabled phylogenetic analysis, genomic sequencing and genotyping by multi locus sequence typing (MLST) (Figure 1). By sequencing from single cells it is also possible to avoid the selectivity of culturing that can result in genotypic alterations.
Figure 1.
Single cell MDA provides sufficient DNA to carry out PCR analysis of many loci. A single Borrelia cell (arrow) was isolated directly from tick midgut tissue by micromanipulation. The individual cells were amplified in 5μl MDA reactions yielding approximately 4.5μg DNA product which was used as template in PCR reactions with primers for loci previously employed for MLST analysis of cultured Borrelia isolates [49]. The PCR products were sequenced and confirmed the published genotypes of outer surface proteins (ospA, ospB, osp C). PCR using total DNA extracted from the tissue, an alternative approach, makes it impossible to determine if genotypes of different loci were genetically linked within individual cells, as the data obtained would be from all of the bacterial cells present. While culturing isolates is an alternative method that does provide linkage information, the organisms for which adequate culture conditions are available are very limited and there is often strong selection for fast growing strains biasing the results. Moreover, virulence factors associated with plasmids can be rapidly lost in culture [50].
Finally, microfluidic systems for combined cell sorting and DNA amplification are a promising new technology exhibiting decreased MDA bias compared to standard reactions [32]. E. coli cells isolated by an on-chip integrated procedure and subjected to a 60nl MDA reaction enabled highly accurate pyrosequencing (www.roche.com). Over 99% of contigs correctly mapped to the known E. coli genome and only 0.36% of sequence reads failed to map to E. coli boding well for the more challenging task of sequencing uncultured species.
Amplification of DNA by MDA
Amplification of a single copy microbial genome is a process highly susceptible to contamination. Purity of the MDA reagents is critical1. The level of care required is similar to that needed for PCR reactions from low template amounts (PCR Application Manual; www.roche-applied-science.com) and includes use of dedicated pipetters, standard methods to create work areas and instruments free of DNA contamination (e.g. use of bleach and UV light), and care in work flow design. Appropriate blank controls should be included for each experiment. While DNA artifacts [15], such as primer dimers and higher molecular weight DNAs derived from them, are synthesized in blank reactions (as measured by PicoGreen assay), sequence specific assays such as PCR should not generate amplicons. Fortunately, in the presence of even a single genome copy as template, specific amplification dominates and artifact synthesis should be low [16,32].
The quality of the amplified DNA should be evaluated prior to full-scale genomic sequencing with respect to DNA contamination and MDA bias. The first step in evaluating the purity of MDA amplicons can be fingerprinting (e.g. terminal restriction fragment length polymorphism, T-RFLP) [23] and sequencing of SSU rRNA PCR products [16,17]. Multiple broad-range primer sets, including those designed for bacteria, archaea, and eukaryotes, should be used. More challenging is the evaluation of MDA bias. If a reference genome is available, qPCR of multiple genomic loci can be performed to quantify loci representation [15,16,32]. However, this is more difficult for novel phylotypes, due to the lack of well-tested universal primers for genes other than the SSU rRNA. Nevertheless, the SSU rRNA-based phylogeny of a novel genome may help in designing narrow-range primers and probes for conserved single copy genes such as recA. Lastly, quality of the amplified DNA may be evaluated by low-level shotgun sequencing. The reads of such pre-sequencing could be analyzed for GC content distribution or other intrinsic DNA signatures [37], and BLAST [38] against GenBank, as a genome-wide DNA purity evaluation, while a read redundancy check enables MDA bias assessment.
Single cell genomic sequencing
For many laboratories, sequencing the complete genome of a single cell is a major objective. Some practical experience is now emerging from major sequencing centers on best procedures and performance expectations. The amplified DNA is suitable for Sanger sequencing, pyrosequencing (www.roche.com) [39] or Illumina sequencing (www.illumina.com) [40]. The dramatic increase in pyrosequence read length and the availability of paired-end libraries for these newer and significantly less expensive high throughput sequencing platforms, makes them increasingly popular. The formation of about one chimeric DNA rearrangement (largely inversion-deletion events) per 20 Kb by the MDA reaction [41] poses problems for large insert libraries like BACs since most paired ends would be compromised by the inversions. Occasional sequence rearrangements in small to mid-size insert clone libraries are not expected to impact the sequence assembly at sufficient sequencing coverage.
Pyrosequencing of single uncultured TM7 cells led to a fairly fragmented assembly of 1,825 scaffolds totaling 2.86 Mb [20]. At the DOE Joint Genome Institute, a single amplified genome of a marine Flavobacterium [23] was used to generate 18 Mb of Sanger sequence and 95 Mb of pyrosequence (TW, RS, unpublished). Pyrosequencing provided a less labor intensive, less expensive, high throughput sequence output free of cloning bias. The addition of Sanger sequences provided high-quality sequence coverage of homopolymeric regions and improved the assembly by bridging regions through paired end information of larger insert clones. The sequence was assembled resulting in a more fragmented genome than typically expected (183 contigs, 1.85 Mb), presumably due to the amplification bias generated by MDA. Primer walking on medium and small insert size clones and PCR/adapter PCR on the diluted MDA products eliminated some of the gaps, indicative of underrepresentation but not lack of these sequences in the MDA reaction [19]. This generated a consensus sequence of 1.9 Mb in 17 contigs, with the largest contig being approximately 700 Kb, indicating the great potential of these methods to achieve a single cell genome reconstruction.
The uneven representation of the genomic information (Figure 2a) due to amplification bias adds cost and labor to the process of obtaining high-quality assembly and finishing. The bias is a random process [16] and thus sequences underrepresented in MDA products from one cell may be obtained by MDA from another cell of the same taxon. Using single cell MDAs has the advantage that these can be pre-screened by PCR for any known or suspected sequences to confirm their identity. Alternatively, multiple identical cells may be pooled prior to the MDA to reduce bias [16]. Moreover, several strategies have been proposed to increase uniform amplification, including the decrease in MDA reaction volumes [32] and supplementing amplification reactions with polyethylene glycol or single-strand binding proteins and spermidine [42,43].
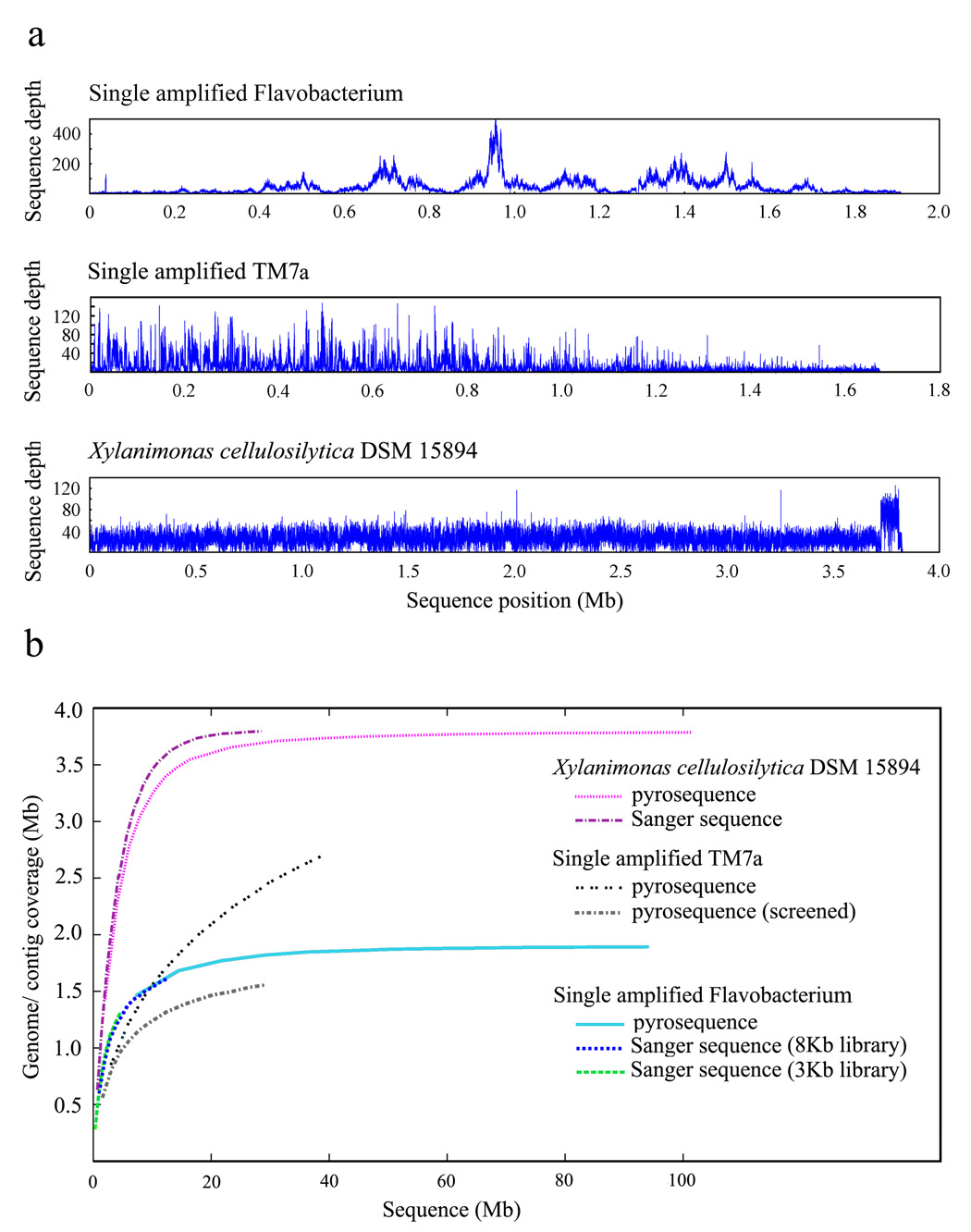
Figure 2.
Characterization of single cell MDA DNA sequence. (a) Pyrosequence depth distribution for a single amplified Flavobacterium (top panel) (TW, RS unpublished) and the single amplified TM7a (middle panel) [20] demonstrate MDA-originated high regional fluctuations in sequence coverage, while the sequence depth for the unamplified sample Xylanimonas cellulosilytica DSM 15894 exhibits a low degree of variation (bottom panel). Sequence positions were determined via arranging contigs by length. TM7a pyrosequence was screened to exclude reads contributing to contigs <300 bp and human reads. The mean sequence depth is 85.7 (± 73.7) for the flavobacterial SAG, 14.4 (± 16.2) for the TM7a SAG, and 31.9 (± 13.9) for X. cellulosilytica. A certain degree of sequence depth fluctuation may be attributed to repeats and mis-assemblies. (b) Genome coverage as function of the genome sequencing effort. Additional shotgun sequencing would not be effective in recovering the under-amplified genomic regions in the single amplified Flavobacterium (estimated genome size of ~2.3Mb) or TM7a (no genome estimate available). The X. cellulosilytica genome is estimated to be ~3.8Mb.
Recently, the complete genome was reported for an uncultivable bacteria derived from protists inhabiting the termite gut [44]. The genome was reconstructed by carrying out MDA on multiple bacterial cells derived from a single protist host cell so that individual cells were expected to have only slight genome variations (genomovars). While exciting progress on the biology and biochemistry of these bacteria resulted from this work, it will be important to remember that the current genome is a composite from multiple individuals. Similarly, the first reported human genome was a composite of individuals [45,46] and even the recently completed genome of a single individual [47] was derived from many cells differing by somatic mutations.
The implications of using reconstructed genomes for bacteria will be affected by the genetic diversity of the species which has proven to be exceedingly variable in many cases [8]. However, single cell genomics may offer some advantages over other methods. Metagenomic shotgun sequencing provides gene frequencies at the community level but rarely reveals the genetic linkage of sequences within individuals. Metagenomics can fail to distinguish where highly variable sequences are present in one species or highly homologous sequences are derived from divergent groups. In contrast, single cell sequencing resolves evolutionary distance between two cells, although the genome from each may be incomplete. Of critical importance, single cell sequencing also provides the biological context of observing what is really possible in individuals where genetic linkage has been driven by natural selection. Finally, single cell sequencing may provide a new means for addressing the highly intractable problem of genetic diversity even where the traditional terminology of “species” is not applicable. In these cases of extreme variation the concept of a “pan-genome” (the range of sequences present) and “core-genome” (the defining sequences essential to the group) have been used instead of relying on the more limiting terminology of species [8,48]. Single cell sequencing can contribute a measurement of the diversity between multiple cells and the sequence context and genetic linkage needed to define boundaries of the pan-genome.
What can one expect from sequencing data of a novel single amplified genome? Current MDA protocols enable the reconstruction of major portions of the genome from one cell of an uncultured microorganism obtained from a complex community, a goal that is currently not attainable with other methods. Single cell MDA products from Prochlorococcus [19,21], E. coli [32] and TM7 [20] recovered up to 70% of their genomes. For the marine Flavobacterium we recovered approximately 80% of the genome, with genome size estimate based on the presence of single copy genes. Evaluation of the genome recovery as a function of our sequencing effort indicates near-saturation, i.e. additional shotgun sequencing would mostly result in repeated sampling of the over-amplified genomic regions (Figure 2b). Recovery of the remaining 20% of the genome and genome closure may require a combination of approaches including use of multiple cells, integration of single cell and metagenomic data [2] or, potentially, use of information in the sequence drafts to guide successful development of culture methods, still a paramount goal of the biological research. At present, partial genomes and completed composite genomes are sufficient for many exciting applications.
Conclusions
Methods for the analysis of single cells from environmental samples have matured over the past several years and are now ready to be employed for discoveries in basic research and biotechnology. Efforts continue to improve the MDA reaction enzymology to reduce bias and chimeric rearrangements. However, even with current limitations, single cell sequencing should enable rapid progress identifying metabolic properties and ecological adaptations in the great numbers of uncultivated microorganisms. In addition, it provides a new method to examine patterns in inter- and intra-species genetic variation in evolutionary, phylogenetic, and epidemiological studies. Single cell sequencing, combined with metagenomics, will be a powerful tool for addressing the complexity when species encompass a broad range of sequences and distinct boundaries between species are unclear. Finally, analysis of the microbial cell provides an indispensable view of the living organism. As in the case of the human genome, there is much to learn at the level of the individual.
Acknowledgements
We would like to thank Alex Copeland for helpful discussions and generation of Figure 2. RS was supported by NSF grants MCB 0738232, MCB-0741329, and OCE-0623288, and DOE grant DOEM-78201.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
MDA reagent-derived exogenous DNA contamination has been suggested by various laboratories and thus stringent quality control of each reagent lot is crucial.
References
- 1.Lasken RS. Single-cell genomic sequencing using Multiple Displacement Amplification. Curr Opin Microbiol. 2007;10:510–516. doi: 10.1016/j.mib.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Rappe MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 3.Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olsen GJ, Lane DJ, Giovannoni SJ, Pace NR, Stahl DA. Microbial Ecology and Evolution: A Ribosomal RNA Approach. Annual Review of Microbiology. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- 5.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 6.Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 7.Beja O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, Jovanovich SB, Gates CM, Feldman RA, Spudich JL, et al. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science. 2000;289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- 8.Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU, Martinez A, Sullivan MB, Edwards R, Brito BR, et al. Community genomics among stratified microbial assemblages in the ocean's interior. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 10.Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y-i, Sugahara J, Preston C, de la Torre J, Richardson PM, DeLong EF. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. PNAS. 2006;103:18296–18301. doi: 10.1073/pnas.0608549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 12.Woyke T, Teeling H, Ivanova NN, Hunteman M, Richter M, Gloeckner FO, Boffelli D, Anderson IJ, Barry KW, Shapiro HJ, et al. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature. 2006 doi: 10.1038/nature05192. [DOI] [PubMed] [Google Scholar]
- 13.Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, Sun Z, Zong Q, Du Y, Du J, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc.Natl.Acad.Sci.U.S.A. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosono S, Faruqi AF, Dean FB, Du Y, Sun Z, Wu X, Du J, Kingsmore SF, Egholm M, Lasken RS. Unbiased whole-genome amplification directly from clinical samples. Genome Res. 2003;13:954–964. doi: 10.1101/gr.816903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raghunathan A, Ferguson HR, Jr, Bornarth CJ, Song W, Driscoll M, Lasken RS. Genomic DNA amplification from a single bacterium. Appl.Environ.Microbiol. 2005;71:3342–3347. doi: 10.1128/AEM.71.6.3342-3347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lasken RS, Raghunathan A, Kvist T, Ishoey T, Westermann P, Ahring BK, Boissy R. Multiple displacement amplification from single bacterial cells. In: Hughes S, Lasken RS, editors. Whole Genome Amplification. Scion Publishing Ltd; 2005. pp. 119–147. Methods Express. [Google Scholar]
- 18.Kvist T, Ahring BK, Lasken RS, Westermann P. Specific single-cell isolation and genomic amplification of uncultured microorganisms. Appl Microbiol Biotechnol. 2006 doi: 10.1007/s00253-006-0725-7. [DOI] [PubMed] [Google Scholar]
- 19.Zhang K, Martiny AC, Reppas NB, Barry KW, Malek J, Chisholm SW, Church GM. Sequencing genomes from single cells by polymerase cloning. Nat Biotechnol. 2006;24:680–686. doi: 10.1038/nbt1214. [DOI] [PubMed] [Google Scholar]
- 20.Marcy Y, Ouverney C, Bik EM, Losekann T, Ivanova N, Martin HG, Szeto E, Platt D, Hugenholtz P, Relman DA, et al. Dissecting biological "dark matter" with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. PNAS. 2007 doi: 10.1073/pnas.0704662104. 0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Podar M, Abulencia CB, Walcher M, Hutchison D, Zengler K, Garcia JA, Holland T, Cotton D, Hauser L, Keller M. Targeted access to the genomes of low-abundance organisms in complex microbial communities. Appl Environ Microbiol. 2007;73:3205–3214. doi: 10.1128/AEM.02985-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mussmann M, Hu FZ, Richter M, de Beer D, Preisler A, Jorgensen BB, Huntemann M, Glockner FO, Amann R, Koopman WJ, et al. Insights into the Genome of Large Sulfur Bacteria Revealed by Analysis of Single Filaments. PLoS Biol. 2007;5:e230. doi: 10.1371/journal.pbio.0050230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stepanauskas R, Sieracki ME. Matching phylogeny and metabolism in the uncultured marine bacteria, one cell at a time. PNAS. 2007;104:9052–9057. doi: 10.1073/pnas.0700496104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giovannoni SJ, DeLong EF, Schmidt TM, Pace NR. Tangential flow filtration and preliminary phylogenetic analysis of marine picoplankton. Appl Environ Microbiol. 1990;56:2572–2575. doi: 10.1128/aem.56.8.2572-2575.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiomori T, Miyamoto H, Makishima K, Yoshida M, Fujiyoshi T, Udaka T, Inaba T, Hiraki N. Evaluation of bedmaking-related airborne and surface methicillin-resistant Staphylococcus aureus contamination. J Hosp Infect. 2002;50:30–35. doi: 10.1053/jhin.2001.1136. [DOI] [PubMed] [Google Scholar]
- 26.Groschel DHM. Air sampling in hospitals. Annals of the New York Academy of Sciences. 1980;353:230–240. doi: 10.1111/j.1749-6632.1980.tb18926.x. [DOI] [PubMed] [Google Scholar]
- 27.Andersen AA. New sampler for the collection, sizing, and enumeration of viable airborne particles. J Bacteriol. 1958;76:471–484. doi: 10.1128/jb.76.5.471-484.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Omberg K, Stentzenbach L. Indoor and Outdoor Air Sampling. In: Emanuel P, Roos JW, Niyogi K, editors. Sampling for Biological Agents in the Environment. ASM Press; 2008. pp. 133–164. [Google Scholar]
- 29.Lin X, Reponen T, Willeke K, Grinshpun SA, Foarde KK, Ensor DS. Long-term sampling of airborne bacteria and fungi into a non-evaporating liquid. Atmos. Environ. 1999;33:4291–4298. [Google Scholar]
- 30.Lin X, Reponen T, Willeke K, Wang Z, Grinshpun SA, Trunov M. Survival of airborne microorganisms during swirling aerosol collection. Aerosol Sci. Technol. 2000;32:184–196. [Google Scholar]
- 31.Ishoy T, Kvist T, Westermann P, Ahring BK. An improved method for single cell isolation of prokaryotes from meso-, thermo- and hyperthermophilic environments using micromanipulation. Appl Microbiol Biotechnol. 2006;69:510–514. doi: 10.1007/s00253-005-0014-x. [DOI] [PubMed] [Google Scholar]
- 32.Marcy Y, Ishoey T, Lasken RS, Stockwell TB, Walenz BP, Halpern AL, Beeson KY, Goldberg SMD, Quake SR. Nanoliter Reactors Improve Multiple Displacement Amplification of Genomes from Single Cells. PLoS Genetics. 2007;3:e155. doi: 10.1371/journal.pgen.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sekar R, Fuchs BM, Amann R, Pernthaler J. Flow sorting of marine bacterioplankton after fluorescence in situ hybridization. Appl Environ Microbiol. 2004;70:6210–6219. doi: 10.1128/AEM.70.10.6210-6219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wallner G, Fuchs B, Spring S, Beisker W, Amann R. Flow sorting of microorganisms for molecular analysis. Appl Environ Microbiol. 1997;63:4223–4231. doi: 10.1128/aem.63.11.4223-4231.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huber R, Huber H, Stetter KO. Towards the ecology of hyperthermophiles: biotopes, new isolation strategies and novel metabolic properties. FEMS Microbiol Rev. 2000;24:615–623. doi: 10.1111/j.1574-6976.2000.tb00562.x. [DOI] [PubMed] [Google Scholar]
- 36.Jahn U, Gallenberger M, Paper W, Junglas B, Eisenreich W, Stetter KO, Rachel R, Huber H. Nanoarchaeum equitans and Ignicoccus hospitalis: new insights into a unique, intimate association of two archaea. J Bacteriol. 2008;190:1743–1750. doi: 10.1128/JB.01731-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karlin S, Campbell AM, Mrazek J. Comparative DNA analysis across diverse genomes. Annu Rev Genet. 1998;32:185–225. doi: 10.1146/annurev.genet.32.1.185. [DOI] [PubMed] [Google Scholar]
- 38.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 39.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5:433–438. doi: 10.1517/14622416.5.4.433. [DOI] [PubMed] [Google Scholar]
- 41.Lasken RS, Stockwell TB. Mechanism of chimera formation during the Multiple Displacement Amplification reaction. BMC Biotechnol. 2007:7–19. doi: 10.1186/1472-6750-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Inoue J, Shigemori Y, Mikawa T. Improvements of rolling circle amplification (RCA) efficiency and accuracy using Thermus thermophilus SSB mutant protein. Nucleic Acids Res. 2006;34:e69. doi: 10.1093/nar/gkl350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu L, Liu X, Schadt CW, Zhou J. Microarray-Based Analysis of Subnanogram Quantities of Microbial Community DNAs by Using Whole-Community Genome Amplification. Appl. Environ. Microbiol. 2006;72:4931–4941. doi: 10.1128/AEM.02738-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hongoh Y, Sharma VK, Prakash T, Noda S, Taylor TD, Kudo T, Sakaki Y, Toyoda A, Hattori M, Ohkuma M. Complete genome of the uncultured Termite Group 1 bacteria in a single host protist cell. Proceedings of the National Academy of Sciences. 2008;105:5555–5560. doi: 10.1073/pnas.0801389105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 46.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 47.Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, et al. The Diploid Genome Sequence of an Individual Human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coleman ML, Sullivan MB, Martiny AC, Steglich C, Barry K, Delong EF, Chisholm SW. Genomic islands and the ecology and evolution of Prochlorococcus. Science. 2006;311:1768–1770. doi: 10.1126/science.1122050. [DOI] [PubMed] [Google Scholar]
- 49.Attie O, Bruno JF, Xu Y, Qiu D, Luft BJ, Qiu WG. Co-evolution of the outer surface protein C gene (ospC) and intraspecific lineages of Borrelia burgdorferi sensu stricto in the northeastern United States. Infect Genet Evol. 2007;7:1–12. doi: 10.1016/j.meegid.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 50.Glockner G, Schulte-Spechtel U, Schilhabel M, Felder M, Suhnel J, Wilske B, Platzer M. Comparative genome analysis: selection pressure on the Borrelia vls cassettes is essential for infectivity. BMC Genomics. 2006;7:211. doi: 10.1186/1471-2164-7-211. [DOI] [PMC free article] [PubMed] [Google Scholar]