Abstract
Introduction
Recent discoveries of inherited glioma risk loci and acquired IDH mutations are providing new insights into glioma etiology. IDH mutations are common in lower grade gliomas and secondary glioblastomas and uncommon in primary glioblastomas. Because the inherited variant in 11q23 has been associated with risk of lower grade glioma and not with glioblastomas, we hypothesized that this variant increases susceptibility to IDH-mutated gliomas, but not to IDH-wild-type gliomas.
Methods
We tested this hypothesis in patients with glioma and controls from the San Francisco Adult Glioma Study, the Mayo Clinic, and Illumina controls (1102 total patients, 5299 total controls). Case-control additive associations of 11q23 risk alleles (rs498872, T allele) were calculated using logistic regression, stratified by tumor IDH status (mutated or wild-type) and by histology and grade. We also adjusted for the recently discovered 8q24 glioma risk locus rs55705857 G allele.
Results
The 11q23 glioma risk locus was associated with increased risk of IDH-mutated gliomas of all histologies and grades (odds ratio [OR] = 1.50; 95% confidence interval [CI] = 1.29–1.74; P = 1.3X10−7) but not with IDH-wild-type gliomas of any histology or grade (OR = 0.91; 95% CI = 0.81–1.03; P = 0.14). The associations were independent of the rs55705857 G allele.
Conclusion
A variant at the 11q23 locus increases risk for IDH-mutated but not IDH-wild-type gliomas, regardless of grade or histology.
Keywords: adult glioma, IDH1 and IDH2 mutation, rs498872, rs55705857, single-nucleotide polymorphism
We and others first reported several inherited variants that increase glioma risk in 2009.1,2 Additional confirmation and new risk loci have been identified since then.3–8 We9 and Simon et al.10 showed that the glioma risk loci in 8q24 and 11q23 first identified by Shete et al.1 were associated with risk of lower grade infiltrating gliomas but not with glioblastomas.
It is now well established that mutation in the isocitrate dehydrogenase (IDH) 1 and 2 genes leads to genome-wide histone and DNA methylation changes that result in abnormal gene expression and gliomagenesis, defining a distinct subclass of gliomas.11,12IDH mutations occur in ∼50%–80% of grade II-III gliomas and secondary glioblastomas but in fewer than 10% of primary glioblastomas.13–17 Tumor IDH mutations are associated with younger age of onset and better overall survival among patients with gliomas of all grades and histologies18,19 and are also associated with other somatic, genetic, and epigenetic alterations.17,20
Given these observations, we hypothesized that the 8q24 and 11q23 glioma risk loci might be specific to IDH-mutated, but not to IDH-wild-type gliomas. In a separate recent publication,21 we showed that the 8q24 locus is specifically associated with risk for oligodendroglial tumors and IDH-mutated astrocytomas of all grades. Here, we report that the 11q23 glioma risk variant rs498872 T allele is associated with risk of IDH-mutated gliomas but not with risk of IDH-wild-type gliomas, regardless of grade or histology.
Materials and Methods
Subjects
The Institutional Review Boards at University of California San Francisco (UCSF) and Mayo Clinic approved the methods for this study, and informed consent was obtained from each study subject. For this study, only patients with no previous diagnosis of glioma were included.
Subjects from UCSF included case and control participants in the San Francisco Bay Area Adult Glioma Study (AGS) and additional controls obtained from Illumina, as previously described.2 In brief, patients aged ≥20 years with newly diagnosed and histologically confirmed incident glioma (International Classification of Diseases for Oncology, morphology codes 9380–9481) were recruited from the local population-based registry, the Northern California Rapid Case Ascertainment program, and the UCSF Neuro-Oncology Clinic. All patients were diagnosed from 1991 through 2010. UCSF AGS controls were ascertained through random-digit dialing, had no history of brain tumor at time of recruitment, and were frequency matched to population-based patients on age, sex, and ethnicity; 74% of patients and 83% of controls who were contacted consented to participate. We obtained tumor tissue samples from 72% of participants who received a diagnosis from 1991 through 2005, and acquisition is ongoing for more recent series.
The Mayo Clinic cases consisted of patients ≥18 years of age who had surgical resection or biopsy of a glioma from 2001 through 2009. Patients were identified at diagnosis for those initially cared for at the Mayo Clinic and at the time of pathologic confirmation for those who initially received a diagnosis elsewhere and were subsequently cared for at Mayo. Pathologic diagnosis was confirmed by review of the primary surgical material for all cases by 2 Mayo Clinic neuropathologists. The control group consisted of consenting individuals who underwent a general medical examination at the Mayo Clinic and have been previously described.9 Individuals <18 years of age and those with a history of a brain tumor were not eligible to be controls. The participation rates were ∼70% for patients and 50% for controls, and tumor tissue samples were available from ∼67% of patients.
Analyses for this study were restricted to white participants, and histological glioma definitions were based on World Health Organization (WHO) criteria.22 Histological categories included glioblastomas, grades 2 and 3 astrocytomas, oligodendrogliomas, and oligoastrocytomas.
Assays for IDH Mutation
UCSF AGS tumor specimens were sequenced to identify IDH1 and IDH2 mutations with use of previously described methods.13 In brief, the region spanning the R132 codon of IDH1 and the region spanning the R172 codon of IDH2 were amplified by polymerase chain reaction with M13 tagged primers to facilitate amplification and sequencing. Products were run on a 1.5% agarose gel and subsequently sequenced in both directions at the UCSF Genomics Core Facility according to the manufacturer's protocol. Sequences were analyzed with Applied Biosystems Sequence Scanner Software, version 1.0. At the Mayo Clinic, IDH1 mutation detection was performed using pyrosequencing. IDH2 mutation detection was performed using both pyrosequencing and Sanger sequencing as previously described.23 Primer sequences are available upon request.
Genotyping Germline Single-Nucleotide Polymorphisms (SNPs)
Genotype data for SNP rs498872 in the 11q23 region from UCSF AGS, Mayo Clinic, and Illumina controls came from Illumina genome-wide and custom panels with use of previously described genotyping methods and quality-control measures.2,9,21 Genotype data for SNP rs55705857 in the 8q24 region subjects came from custom panels of previously described UCSF AGS and Mayo Clinic patients and controls.22
Statistical Analysis
Initial analyses included additive logistic regression models for 0, 1, or 2 copies of the T risk allele of rs498872 to obtain unadjusted single point associations. Models were run separately for (1) patients with tumor IDH mutation versus controls and (2) patients without tumor IDH mutation versus controls. Analyses were conducted for 7 nondiscrete grade and histology groupings: (1) all gliomas, (2) glioblastomas, (2) grade 2 gliomas, (4) grade 3 gliomas, (5) grade 2/3 oligodendrogliomas, (6) grade 2/3 oligoastrocytomas, and (7) grade 2/3 astrocytomas. Analyses were first performed separately for UCSF AGS and Mayo Clinic subjects and then pooled and analyzed using logistic regression models adjusted for study site. We also examined genetic heterogeneity between sites by including a site-by-SNP interaction term. To determine whether the association of glioma with rs498872 is independent of the association of glioma with rs55705857 G allele, we also conducted analyses in which both rs498872 and rs55705857 were included in the logistic model along with study site.
Results and Discussion
Gliomas are a heterogeneous class of tumors. In each subgroup defined by grade and histology, there are distinct clinical and molecular profiles.24 The discovery of IDH mutations has altered our understanding of gliomagenesis and may lead to the eventual inclusion of IDH status in the WHO glioma classification scheme.18,25,26 Numerous reports have demonstrated that IDH mutation segregates gliomas into clinically relevant subgroups; patients with IDH-mutated tumors tend to have much better prognoses.14,16,27
We identified tumor IDH mutations in 379 (34%) of 1102 patients with glioma (874 from UCSF and 228 from the Mayo Clinic) for whom both IDH mutation and rs498872 genotype data were available. The IDH mutation rate was 8% in glioblastomas, 84% in grade 2 gliomas, 63% in grade 3 gliomas, 83% in grade 2/3 oligodendrogliomas, 90% in grade 2/3 oligoastrocytomas, and 61% in grade 2/3 astrocytomas (Table 1). Patient characteristics, including age, sex, and median survival, are also shown in Table 1. The prevalence of IDH mutations in our study patients by histology and grade was comparable to that found in other studies16,17,28 and in the remaining UCSF and Mayo samples without constitutional SNP genotyping. The control group included 5299 controls (1116 from UCSF, 3389 Illumina controls, and 794 from the Mayo Clinic) (Table 2).
Table 1.
Demographic and survival characteristics of UCSF Adult Glioma Study and Mayo Clinic glioma patients stratified by IDH1/2 mutation status
| Histology | Total |
IDH1/2-wild-type |
IDH1/2-mutated |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number | % IDH mutated | Number | % men | Median age at diagnosis | Number deceased | Median survival years | Number | % men | Median age at diagnosis | Number deceased | Median survival years | |
| All gliomas | 1102 | 34 | 723 | 63 | 58 | 631 | 1.3 | 379 | 62 | 40 | 133 | 11.5 |
| Glioblastomas | 663 | 8 | 607 | 64 | 58 | 551 | 1.2 | 56 | 66 | 44 | 43 | 2.1 |
| Gr 2 Gliomas | 224 | 84 | 36 | 56 | 51 | 21 | 6.4 | 188 | 63 | 39 | 53 | 13.6 |
| Gr 3 Gliomas | 215 | 63 | 80 | 53 | 54 | 59 | 1.7 | 135 | 59 | 41 | 37 | 14.5 |
| Oliodendrogliomas Gr 2/3 | 134 | 83 | 23 | 39 | 51 | 12 | 12.6 | 111 | 57 | 43 | 21 | NA |
| Oliogoastrocytomas Gr 2/3 | 87 | 90 | 9 | 33 | 33 | 7 | 2.3 | 78 | 63 | 39 | 22 | 13.1 |
| Astrocytomas Gr 2/3 | 218 | 61 | 84 | 60 | 55 | 61 | 1.7 | 134 | 64 | 37 | 47 | 10.3 |
NA - The median survival has not yet been reached.
Table 2.
Associations of 11q23 rs498872T allele with glioma risk stratified by tumor IDH1/2 mutation status in UCSF Adult Glioma and Mayo Clinic Studies
| Histology |
IDH1/2-wild-type gliomas |
Test for heterogeneity by study site |
IDH1/2-mutated gliomas |
Test for heterogeneity by study site | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| case-control association |
case-control association |
|||||||||
| Number | RAF | OR (95% CI) | P value | P value | Number | RAF | OR (95% CI) | P value | P value | |
| Controls | 5299 | 0.31 | 5299 | 0.31 | ||||||
| All gliomas | 723 | 0.29 | 0.91 (0.81–1.03) | 0.14 | 0.32 | 379 | 0.41 | 1.50 (1.29–1.74) | 1.3E-07 | 0.11 |
| Glioblastomas | 607 | 0.29 | 0.91 (0.80–1.04) | 0.17 | 0.24 | 56 | 0.43 | 1.65 (1.14–2.40) | 0.008 | 0.28 |
| Gr 2 Gliomas | 36 | 0.29 | 0.93 (0.56–1.53) | 0.77 | 0.69 | 188 | 0.39 | 1.38 (1.12–1.71) | 0.002 | 0.35 |
| Gr 3 Gliomas | 80 | 0.29 | 0.90 (0.64–1.27) | 0.54 | 0.74 | 135 | 0.43 | 1.59 (1.25–2.03) | 1.8E-04 | 0.27 |
| Oliodendrogliomas Gr 2/3 | 23 | 0.24 | 0.72 (0.37–1.40) | 0.33 | 0.95 | 111 | 0.41 | 1.52 (1.16–1.99) | 0.002 | 0.31 |
| Oliogoastrocytomas Gr 2/3 | 9 | 0.28 | 0.86 (0.31–2.39) | 0.78 | NA* | 78 | 0.38 | 1.29 (0.93–1.79) | 0.134 | 0.17 |
| Astrocytomas Gr 2/3 | 84 | 0.30 | 0.97 (0.70–1.34) | 0.84 | 0.93 | 134 | 0.41 | 1.53 (1.20–1.96) | 5.5E-04 | 0.80 |
*All IDH-wild-type oligoastrocytomas are from UCSF.
ORs from additive model for 0, 1, or 2T alleles in rs498872 are adjusted for study site. P values ≤.05 are in bold. RAF = risk allele frequency.
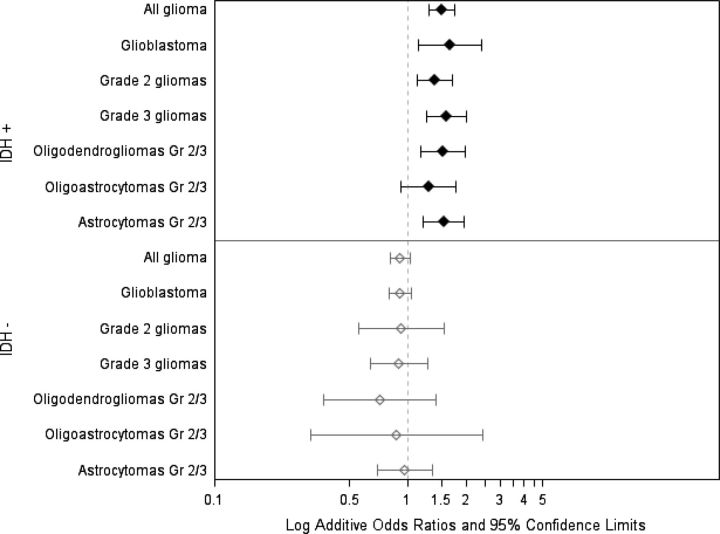
In a recent report,21 we showed that a novel variant on 8q24 is strongly associated with risk of all oligodendroglial gliomas and with IDH-mutated astrocytic gliomas. In this report, we show that the T allele of rs498872 on 11q23 confers increased risk for IDH-mutated gliomas of all grades and histological groups but not for IDH-wild-type gliomas of any grade or histology (Table 2 and Fig. 1). These associations were statistically significant for all histological groups of IDH-mutated gliomas, with the exception of grade 2/3 oligoastrocytomas. Associations did not display significant heterogeneity across study site (Table 2). Sanson et al.5 reported a similar result in their analysis of all glioma grades and histologies. By stratifying analyses by histology and grade, we revealed that the association between rs498872 and glioma risk in IDH-mutated gliomas is independent of the known associations between low-grade glioma and both IDH mutation and the 11q23 variant.9,27,28
Fig. 1.
Association of 11q23 variant with glioma risk, stratified by histology, grade, and IDH mutation status. Case-control ORs and 95% CIs for each category are from additive logistic models of 0, 1, or 2 rs498872 T alleles and adjusted for study site.
We further showed that the specific and significant association of the inherited T allele in rs498872 is independent of the association of the 8q24 variant, rs55705857 (Table 3), which we had recently found to be associated with IDH-mutated astrocytic gliomas and with both IDH-mutated and IDH-wild-type oligodendroglial tumors.21 Thus, the 11q23 variant appears to be even more specifically associated with IDH mutation status than the 8q24 locus. We have now identified 2 independent risk loci that confer inherited susceptibility to IDH-mutated gliomas, which comprise a distinct glioma subclass.11,12
Table 3.
Case-control ORs of IDH1/2–mutated gliomas with rs498872 T allele and rs55705857 G allele in UCSF Adult Glioma and Mayo Clinic Studies
| Variable | Number | rs498872 (T allele) |
rs55705857 (G allele) |
||||
|---|---|---|---|---|---|---|---|
| RAF | OR (95% CI) | P value | RAF | OR (95% CI) | P value | ||
| Controls | 1300 | 0.32 | 0.05 | ||||
| IDH mutated tumors | |||||||
| All gliomas | 359 | 0.42 | 1.52 (1.27–1.83) | 5.1E-06 | 0.19 | 5.21 (3.90–6.96) | 6.8E-29 |
| Glioblastomas | 48 | 0.46 | 1.84 (1.19–2.85) | 6.2E-03 | 0.22 | 7.55 (4.12–13.9) | 6.5E-11 |
| Gr 2 Gliomas | 180 | 0.39 | 1.42 (1.12–1.80) | 4.3E-03 | 0.21 | 5.56 (3.93–7.88) | 4.5E-22 |
| Gr 3 Gliomas | 131 | 0.44 | 1.60 (1.23–2.07) | 4.4E-04 | 0.16 | 4.25 (2.83–6.38) | 2.8E-12 |
| Oliodendrogliomas Gr 2/3 | 107 | 0.42 | 1.60 (1.18–2.16) | 2.3E-03 | 0.20 | 5.89 (3.83–9.07) | 7.6E-16 |
| Oliogoastrocytomas Gr 2/3 | 74 | 0.39 | 1.33 (0.95–1.88) | 0.10 | 0.16 | 3.73 (2.30–6.04) | 9.0E-08 |
| Astrocytomas Gr 2/3 | 130 | 0.42 | 1.54 (1.18–2.02) | 1.7E-03 | 0.19 | 5.37 (3.59–8.04) | 3.4E-16 |
Note: Fewer subjects had data available for rs55705857 than for rs498872 because only a recently completed custom genotyping panel22 contained rs55705857.
ORs for each glioma subtype are from an additive model that includes both 0, 1, or 2 T alleles in rs498872 and 0, 1, or 2 G alleles in rs55705857 and is adjusted for study site. P values ≤ .05 are in bold.
RAF = risk allele frequency.
Because germline risk SNPs, by definition, precede tumor development, our results may help elucidate the mechanism behind the pathogenesis of these tumors after the causal variant in 11q23 is identified. At present, we can only speculate whether the 11q23 variant actively promotes development of IDH mutations, whether individuals without the risk alleles more readily suppress cells that develop these mutations, or whether some other mechanism explains the specific association of this SNP with risk of IDH-mutated gliomas. At this time, there are no known biologic functions associated with the 11q23 SNP to explain our observed associations, and further functional studies are needed.
Genome-wide association studies (GWAS) have greatly advanced our knowledge of the genetic mechanisms underlying carcinogenesis but typically have not focused on associations between inherited variants and acquired somatic mutations. As knowledge of cancer biology improves, the resolution at which tumors are meaningfully classified will continue to become finer. Although GWAS of cancer have long restricted cases to only those subjects whose tumor is of a particular histologic subtype, there is a growing trend to restrict cases to only those tumors that carry specific somatic mutations.29,30 Both the present finding and our recently identified novel variant on 8q24, rs55705857, that was strongly associated with risk of IDH-mutated astrocytic gliomas21 showed that IDH mutation stratifies gliomas into relevant etiologic subgroups. This demonstrates that acquired genetic alterations in the tumor can be used to restrict analyses to phenotypically homogeneous cases that are likely to share similar underlying genetic risk factors. The identification of 2 germline alterations that impact the development of IDH-mutated gliomas may bring us closer to understanding the causal mechanisms underlying the development of these tumors. In addition, our results may help to improve disease classification and prognostic accuracy in future studies.
Funding
Work at University of California, San Francisco was supported by the National Institutes of Health (grant numbers R01CA52689, P50CA097257, R01CA126831, and CA112355) and the National Brain Tumor Foundation, the UCSF Lewis Chair in Brain Tumor Research, and by donations from families and friends of John Berardi, Helen Glaser, Elvera Olsen, Raymond E. Cooper, and William Martinusen. Work at the Mayo Clinic was supported by the National Institutes of Health (grant numbers P50CA108961 and P30 CA15083), National Institute of Neurological Disorders and Stroke (grant number RC1NS068222Z), and the Ting Tsung and Wei Fong Chao Family Foundation and a gift from Bernie and Edith Waterman.
Acknowledgments
The collection of cancer incidence data used in this study was supported by the California Department of Public Health as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885; the National Cancer Institute's Surveillance, Epidemiology and End Results Program under contract HHSN261201000036C awarded to the Cancer Prevention Institute of California, contract HHSN261201000035C awarded to the University of Southern California, and contract HHSN261201000034C awarded to the Public Health Institute; and the Centers for Disease Control and Prevention's National Program of Cancer Registries, under agreement #1U58 DP000807-01 awarded to the Public Health Institute.
The ideas and opinions expressed herein are those of the author(s) and endorsement by the State of California Department of Public Health, the National Cancer Institute, and the Centers for Disease Control and Prevention or their Contractors and Subcontractors is not intended nor should be inferred.
We thank the study participants, the clinicians, and research staff at participating medical centers; Dr. Kenneth Aldape; deCODE genetics; Dr. Bernd Scheithauer; and the Mayo Clinic Comprehensive Cancer Center Biospecimens and Processing and Genotyping Shared Resources.
Conflict of interest statement. M. B. has consulted with Ivivi health sciences and Pharmacokinesis. All other authors: no conflicts.
References
- 1.Shete S, Hosking FJ, Robertson LB, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41(8):899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wrensch M, Jenkins RB, Chang JS, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41(8):905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson U, Schwartzbaum J, Wiklund F, et al. A comprehensive study of the association between the EGFR and ERBB2 genes and glioma risk. Acta Oncol. 2010;49(6):767–775. doi: 10.3109/0284186X.2010.480980. [DOI] [PubMed] [Google Scholar]
- 4.Egan KM, Thompson RC, Nabors LB, et al. Cancer susceptibility variants and the risk of adult glioma in a US case-control study. J Neurooncol. 2011;104(2):535–542. doi: 10.1007/s11060-010-0506-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanson M, Hosking FJ, Shete S, et al. Chromosome 7p11.2 (EGFR) variation influences glioma risk. Hum Mol Genet. 2011;20(14):2897–2904. doi: 10.1093/hmg/ddr192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartzbaum JA, Xiao Y, Liu Y, et al. Inherited variation in immune genes and pathways and glioblastoma risk. Carcinogenesis. 2010;31(10):1770–1777. doi: 10.1093/carcin/bgq152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang TH, Kon M, Hung JH, Delisi C. Combinations of newly confirmed Glioma-Associated loci link regions on chromosomes 1 and 9 to increased disease risk. BMC Med Genomics. 2011;4:63. doi: 10.1186/1755-8794-4-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stacey SN, Sulem P, Jonasdottir A, et al. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat Genet. 2011;43(11):1098–1103. doi: 10.1038/ng.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenkins RB, Wrensch MR, Johnson D, et al. Distinct germ line polymorphisms underlie glioma morphologic heterogeneity. Cancer Genet. 2011;204(1):13–18. doi: 10.1016/j.cancergencyto.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simon M, Hosking FJ, Marie Y, et al. Genetic risk profiles identify different molecular etiologies for glioma. Clin Cancer Res. 2010;16(21):5252–5259. doi: 10.1158/1078-0432.CCR-10-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prensner JR, Chinnaiyan AM. Metabolism unhinged: IDH mutations in cancer. Nat Med. 2011;17(3):291–293. doi: 10.1038/nm0311-291. [DOI] [PubMed] [Google Scholar]
- 13.Christensen BC, Smith AA, Zheng S, et al. DNA methylation, isocitrate dehydrogenase mutation, and survival in glioma. J Natl Cancer Inst. 2011;103(2):143–153. doi: 10.1093/jnci/djq497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houillier C, Wang X, Kaloshi G, et al. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology. 2010;75(17):1560–1566. doi: 10.1212/WNL.0b013e3181f96282. [DOI] [PubMed] [Google Scholar]
- 15.van den Bent MJ, Dubbink HJ, Marie Y, et al. IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tumors: a report of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Clin Cancer Res. 2010;16(5):1597–1604. doi: 10.1158/1078-0432.CCR-09-2902. [DOI] [PubMed] [Google Scholar]
- 16.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai A, Kharbanda S, Pope WB, et al. Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J Clin Oncol. 2011;29(34):4482–4490. doi: 10.1200/JCO.2010.33.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartmann C, Meyer J, Balss J, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118(4):469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 19.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Labussiere M, Idbaih A, Wang XW, et al. All the 1p19q codeleted gliomas are mutated on IDH1 or IDH2. Neurology. 2010;74(23):1886–1890. doi: 10.1212/WNL.0b013e3181e1cf3a. [DOI] [PubMed] [Google Scholar]
- 21.Jenkins RB, Xiao Y, Sicotte H, et al. A low-frequency variant at 8q24.21 is strongly associated with risk of oligodendroglial tumors and astrocytomas with IDH1 or IDH2 mutation. Nat Genet. 2012;44(10):1122–1125. doi: 10.1038/ng.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kipp BR, Voss JS, Kerr SE, et al. Isocitrate dehydrogenase 1 and 2 mutations in cholangiocarcinoma. Human Pathology. 2012;43(10):1552–1558. doi: 10.1016/j.humpath.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Huse JT, Phillips HS, Brennan CW. Molecular subclassification of diffuse gliomas: seeing order in the chaos. Glia. 2011;59(8):1190–1199. doi: 10.1002/glia.21165. [DOI] [PubMed] [Google Scholar]
- 25.Brennan C. Genomic profiles of glioma. Curr Neurol Neurosci Rep. 2011;11(3):291–297. doi: 10.1007/s11910-011-0198-7. [DOI] [PubMed] [Google Scholar]
- 26.Korshunov A, Meyer J, Capper D, et al. Combined molecular analysis of BRAF and IDH1 distinguishes pilocytic astrocytoma from diffuse astrocytoma. Acta Neuropathol. 2009;118(3):401–405. doi: 10.1007/s00401-009-0550-z. [DOI] [PubMed] [Google Scholar]
- 27.Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27(25):4150–4154. doi: 10.1200/JCO.2009.21.9832. [DOI] [PubMed] [Google Scholar]
- 28.Gupta R, Webb-Myers R, Flanagan S, Buckland ME. Isocitrate dehydrogenase mutations in diffuse gliomas: clinical and aetiological implications. J Clin Pathol. 2011;64(10):835–844. doi: 10.1136/jclinpath-2011-200227. [DOI] [PubMed] [Google Scholar]
- 29.Haiman CA, Chen GK, Vachon CM, et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor-negative breast cancer. Nat Genet. 2011;43(12):1210–1214. doi: 10.1038/ng.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kilpivaara O, Mukherjee S, Schram AM, et al. A germline JAK2 SNP is associated with predisposition to the development of JAK2(V617F)-positive myeloproliferative neoplasms. Nat Genet. 2009;41(4):455–459. doi: 10.1038/ng.342. [DOI] [PMC free article] [PubMed] [Google Scholar]