Abstract
Background
The objective of this study was to examine the predictive value of parameters of 3D 1H magnetic resonance spectroscopic imaging (MRSI) prior to treatment with radiation/chemotherapy (baseline) and at a postradiation 2-month follow-up (F2mo) in relationship to 6-month progression-free survival (PFS6) and overall survival (OS).
Methods
Sixty-four patients with newly diagnosed glioblastoma multiforme (GBM) being treated with radiation and concurrent chemotherapy were involved in this study. Evaluated were metabolite indices and metabolite ratios. Logistic linear regression and Cox proportional hazards models were utilized to evaluate PFS6 and OS, respectively. These analyses were adjusted by age and MR scanner field strength (1.5 T or 3 T). Stepwise regression was performed to determine a subset of the most relevant variables.
Results
Associated with shorter PFS6 were a decrease in the ratio of N-acetyl aspartate to choline-containing compounds (NAA/Cho) in the region with a Cho-to-NAA index (CNI) >3 at baseline and an increase of the CNI within elevated CNI regions (>2) at F2mo. Patients with higher normalized lipid and lactate at either time point had significantly worse OS. Patients who had larger volumes with abnormal CNI at F2mo had worse PFS6 and OS.
Conclusions
Our study found more 3D MRSI parameters that predicted PFS6 and OS for patients with GBM than did anatomic, diffusion, or perfusion imaging, which were previously evaluated in the same population of patients.
Keywords: glioblastoma, magnetic resonance spectroscopic imaging, survival
Glioblastoma multiforme (GBM) is the most common and most malignant type of glioma. Despite advances in multimodal treatments that combine surgery, radiation therapy (RT), and chemotherapy, patients with GBM have a limited prognosis, with a median survival of 15 months.1 Conventional MRI can delineate structural abnormalities such as regions where the blood–brain barrier has been compromised, but it fails to distinguish among edema, gliosis, inflammation, and active tumor. The Macdonald criteria that have been used for many years to assess tumor progression and response to therapy are based upon changes in cross-sectional diameters of the contrast-enhancing lesion in combination with clinical worsening.2 Recent studies have shown that some patients who are treated with standard therapies can suffer from a temporary increase in the contrast-enhancing lesion that later subsides without further treatment.3,4 This phenomenon, which is termed pseudoprogression, complicates clinical decision making and leads to ambiguities in evaluation of the effectiveness of new therapies. While the newer Response Assessment for Neuro-Oncology criteria5 also take into account changes in the size of regions with abnormal signal on T2-weighted images, they are unable to differentiate tumor from nonspecific treatment effects.
Proton magnetic resonance spectroscopic imaging (1H-MRSI) is a powerful noninvasive tool for characterizing the spatial extent and metabolic properties of lesions for patients with brain tumors.6 Markers that can be detected include choline-containing compounds (Cho), N-acetyl aspartate (NAA), creatine (Cr), lactate (Lac), and lipid (Lip). Elevations in the level of Cho, due to increased membrane synthesis or cell proliferation, and reductions in levels of the neuronal marker NAA have been used as biomarkers for distinguishing regions of tumor from normal brain tissue. Cr and phosphocreatine, which both resonate as singlets at 3.0 ppm, are involved in ATP metabolism. The level of total Cr is a marker of energy transfer, and storage and has been reported to increase7,8 or decrease9 in gliomas. Lac is a marker of anaerobic metabolism, and its CH3 component (1.3 ppm) often overlaps with CH2 groups in long alkyl chains of Lip that arise from necrosis or subcutaneous Lip. Spectral editing using J-difference methods have been applied to separate Lac from Lip10 for assessing the malignancy of tumors.11 Previous work has shown that the parameters obtained from 3D MRSI data may provide useful information for diagnosis,12,13 directing image-guided surgery and14 radiation planning,15–17 and predicting survival18–22 in patients with glioma.
In an earlier study, we showed that integrated anatomic and diffusion- and perfusion-weighted imaging examinations obtained postsurgery but pretreatment and at posttreatment follow-ups could provide parameters that predicted progression-free survival (PFS) and overall survival (OS) in patients with GBM.23 We found that larger volumes of the region with T2 hyperintensity at baseline (pre-RT) and at post-RT were associated with worse OS, while higher blood volumes, peak height, and recirculation factors at pre-RT and larger blood volumes at post-RT, in the T2 hyperintensity lesion, were associated with shorter PFS.23 In the current study, we evaluated data from baseline and post-RT scans for the predictive values of MRSI parameters in relation to 6-month PFS (PFS6) and OS in the same population. Parameters evaluated were the metabolite indices24 and ratios between spectroscopic voxels containing anatomic and/or metabolic lesions and those within normal appearing white matter (NAWM). We hypothesized that metabolic imaging parameters from the pre-/post-RT examinations were more likely to identify the true spatial extent and malignancy of tumors than would conventional MRI. The results of this analysis were also compared with the findings from anatomic, diffusion, and perfusion imaging in order to evaluate the importance of these modalities in understanding the clinical course of the disease.
Materials and Methods
Study Population
The study population comprised 66 patients who were newly diagnosed with GBM, were recruited for treatment at the University of California, San Francisco, and gave informed consent to participate in serial MR scans. Two patients were subsequently excluded from analysis: 1 died of hepatitis B virus infection before progression, and 1 had surgical complications. The remaining 64 patients (26 females, 38 males; median age = 53 y; KPS ≥ 60) were treated with radiation and concurrent temozolomide alone (n = 23 patients), temozolomide with erlotinib (n = 29), poly-ICLC (polyinosinic-polycytidylic acid stabilized with polylysine and carboxymethylcellulose) (n = 10), or R115777 (n = 2). This population had been studied previously in terms of the patterns of changes in diffusion and perfusion imaging parameters.23 In the current study, we examined the predictive values of MRSI parameters at baseline (after surgical resection but prior to RT and chemotherapy) and at post-RT in relation to PFS6 and OS.
Half of the patients received gross total resection, and the others had either a subtotal resection or a biopsy alone due to the location of the lesion. RT was administered with a total dose of 60 Gy in 2-Gy fractions over a 6-week period. Of the 57 patients for whom the exact start date of RT was recorded, baseline examinations were acquired at a median of 5 days before RT (ranging from 19 days pre- to 8 days post-RT). Post-RT MRSI data were acquired at times varying 2–4 months after baseline, with a total of 53 MRSI datasets available at the 2-month follow-up scan (F2mo). When tumor progression was suspected, patients received an additional scan at a short time interval (∼1 mo) to help distinguish true from pseudoprogression. Criteria for defining true progression were according to Macdonald et al2 and were in many cases confirmed by a second surgery. Survival information was collected up to 6.8 years after the first scan.
Forty-seven of the patients progressed during the study period, with 44 recurring locally/regionally (within 10 mm from the margin of the resection cavity, residual contrast enhancement, and/or T2 hyperintensity lesions at baseline) and 3 showing distant recurrence. The recurrence pattern was not a significant factor in OS. Of these patients, 20 switched to a new type of chemotherapy, 24 had new therapy in combination with a second surgical resection, and 3 had no treatment information available. Patients who received a combination of reoperation and new chemotherapy had significantly longer survival than those who received new chemotherapy alone (median survival, 558 days vs 342 days; P < .0001, log-rank test).23 Only 2 of the patients who progressed received anti-angiogenic therapy at the time of recurrence. Age, sex, KPS, steroids, extent of resection, and type of chemotherapy were not significantly correlated with OS or PFS6 in this population.23
MR Examination
MR data were acquired using a standard transmit/receive quadrature head coil or an 8-channel receive-only phased-array coil from either 1.5-T or 3-T scanners (GE Healthcare Technologies). The examinations included the acquisition of anatomic, perfusion, diffusion, and spectroscopic imaging data. As the perfusion and diffusion imagings have been described previously,23 we provide details of only the standard MRI and MRSI data acquisition parameters.
Anatomic MRI included a T1-weighted sagittal scout (repetition time [TR]/echo time [TE] = 54/2 ms), axial T2-weighted fluid attenuated inversion recovery (FLAIR; TR/TE/inversion time [TI] = 10 002/127–157/2200 ms, 48 slices, slice thickness = 3 mm, in-plane resolution = 0.9–1.0 mm), and pre- and postcontrast T1-weighted spoiled gradient echo (SPGR) (TR/TE = 26/2–8 ms, 180 × 240mm2 field of view, 64 slices, slice thickness = 1.5–3 mm, in-plane resolution = 0.9–1.0 mm). For data obtained with the 8-channel coil, proton density-weighted gradient echo images were acquired using the manufacturer-provided parallel imaging calibration sequence in order to obtain estimates of coil sensitivities (TR/TE = 150/2 ms) for the combination of the spectral data from each channel.
The 3D 1H-MRSI data were obtained using chemical shift selective (CHESS) water suppression, very selective suppression (VSS), outer volume suppression,25 and point resolved spectral selection (PRESS), volume selection with TE/TR of 144/1000–1100 ms. Of these 117 examinations, 47 MRSI datasets were acquired using a Lac-edited sequence at 1.5 T10 (29 at baseline; 18 at F2mo), which offered peaks of Lac and Lip separately. The remaining exams were obtained with a non-Lac-edited sequence at 3 T. An overpress factor of 1.2 was utilized to reduce chemical shift artifact at 3 T.26 The spectral data were acquired with 512 dwell points and 1000 Hz sweepwidth at 1.5 T and with 1024 dwell points and 2000 Hz sweepwidth at 3 T. Spectral array sizes were 12 × 12 × 8 or 16 × 16 × 8, acquired with sampling restricted to an elliptical region of k-space and with fields of view corresponding to 120 × 120 × 80 mm3 or 160 × 160 × 80 mm3.27
Postprocessing and Analysis
Postprocessing was performed using a previously published methodology.28 Spectral arrays from the 8-channel coil were processed individually, and the signals were combined using software developed in-house that weights the data by their coil sensitivities. The spectral data were apodized with a 2-Hz and a 4-Hz Lorentzian filter at 1.5 T and 3 T, respectively, Fourier transformed, and processed with phase and frequency corrections. The peak parameters, including individual peak heights (Cho, Cr, NAA, and Lac/Lip) and the Cho-to-NAA index (CNI), were computed automatically for each voxel within the excited region using in-house software.24 Since not all MRSI was acquired using a Lac-edited sequence, an index of combined Lac and Lip (LL) was defined as the absolute peak intensity at 1.3 ppm for the non-Lac-edited case, and by combining Lac and Lip peak heights for Lac-edited data.21
The 3D 1H-MRSI data were referenced to the postcontrast 3D SPGR image under the assumption that there was no movement between the acquisition of imaging and spectral data. The FLAIR image was aligned to the corresponding postcontrast 3D SPGR images.29 Segmentation of the brain was performed automatically on the precontrast 3D SPGR images using a program based on a Markov random field model.30 The segmented white-matter mask was then used to identify voxels that had at least 75% NAMW. The T2ALL region of interest (ROI), the T2 hyperintensity defined from the FLAIR images, was downsampled to the resolution of MRSI, and only voxels containing >50% of the T2ALL ROI were defined as T2L. The other ROIs included regions within the PRESS volume that had a CNI value >2 (CNI2) or >3 (CNI3) and in the intersection of the T2L and CNI2 (CNI2T; see Fig. 1). All ROIs excluded the resection cavity.
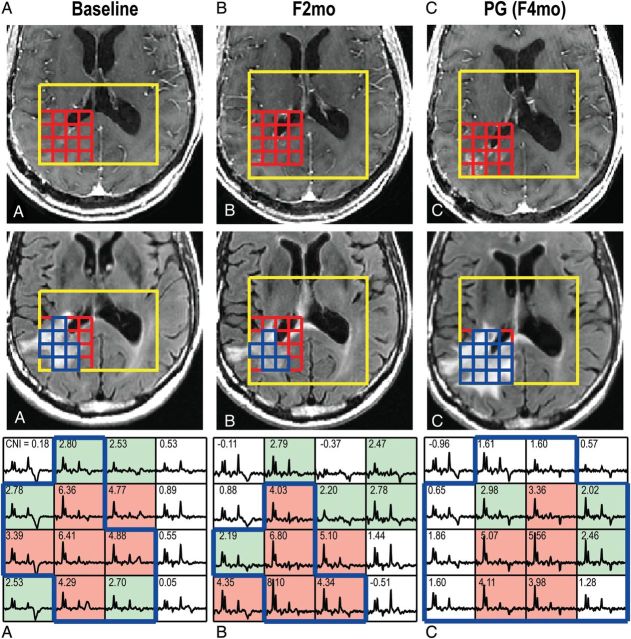
Fig. 1.
(A) Images and spectra at baseline, (B) 2-month follow-up after baseline (F2mo), and (C) progression (PG) from a patient with newly diagnosed GBM. The postcontrast and FLAIR images display the PRESS selected volume and the corresponding arrays of spectra. The corresponding Cho-to-NAA index (CNI) values are shown in the spectral array. Voxels with CNI > 3.0 are highlighted in light red, voxels in light green had CNI between 2 and 3, and voxels in the T2 lesion within the PRESS volume (T2L) are outlined in blue. The patient had PFS of 4 mo and OS of 8 mo. CNI values varied in the T2L at PG (C).
Metabolite peak heights were normalized by median peak heights in NAWM for Cho and Cr (nCho and nCr), and normalized Lac, Lip, and LL (nLac, nLip, and nLL) were estimated by metabolite peak heights divided by median peak NAA intensity in NAWM. Excess Cho (exCho; Fig. 2) or excess Cr (exCr) was calculated using the formula (Cho-[Cho/NAA]NAWM *NAA)/ChoNAMW or (Cr-[Cr/NAA]NAWM *NAA)/CrNAMW,31 respectively. The medians and maximums of the metabolite ratios (normalized metabolite values, exCho, exCr, and CNI) were analyzed within these abnormalities, but maximum Cho/NAA and NAA/Cho were not evaluated, to avoid these voxels being dominated by voxels with extremely low NAA or low Cho. The CNI is a z-score that measures the deviation of Cho and NAA in the spectrum relative to levels in spectra from normal-appearing brain. Also considered in the analysis were the sum of nLac, nLip, and nLL, the volumes of the T2ALL, and the number of voxels within the lesions, including T2L, CNI2, CNI3, and CNI2T.
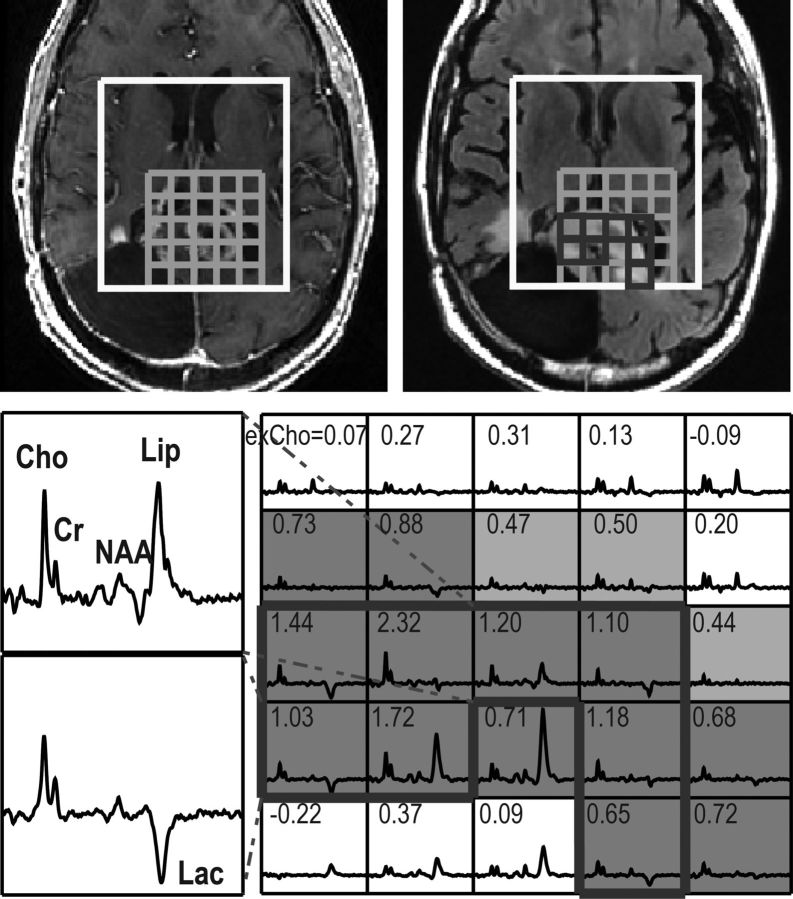
Fig. 2.
3D (non-Lac-edited) MRSI acquired from a patient with PFS of 2 mo and OS of 7 mo at F2mo. The individual spectra show peaks corresponding to Cho, Cr, NAA, Lac, and Lip in the lesions. Voxels with a Cho-to-NAA index (CNI) > 3.0 are highlighted in dark grey, voxels in light grey had CNI between 2 and 3, and voxels in the T2 lesion within the PRESS volume (T2L) were outlined in black thick line. Numbers in the spectral array represent values of excess Cho (exCho), the differences in Cho compared with those in NAWM adjusted by the changes in NAA. Spectrum from the top left corner with a relatively high NAA peak had exCho of 0.07, whereas that in the opposite corner with a low NAA was 0.72. Elevated exCho values are seen in the regions of the T2L with increased CNI.
Statistical analysis was performed using R version 2.9.2 (www.r-project.org). A nonparametric Wilcoxon signed-rank test was used to access the difference in volumes at baseline, F2mo, and progression. Variables influencing early progression (Y = 1 if PFS was ≤6 mo and Y = 0 if PFS was >6 mo) and OS were identified by applying multivariable logistic regression and Cox proportional hazards models, respectively. The same model development procedure was applied for assessing both outcomes at the time of baseline and F2mo. Each variable was assessed in a multivariate model with field strength and age. At each time point, all estimates significant at P ≤ .05 within an ROI were included as potential variables in a forward stepwise procedure, where age and field strength were forced into the model. Selected variables within each ROI were then evaluated together in this same stepwise procedure to obtain a single overall model. The difference between the 2 time points (F2mo and baseline) was included in the model development at F2mo if both scans were performed on the same field strength. Patients who progressed at F2mo were excluded from the F2mo PFS6 analysis. Due to the exploratory nature of this study, adjustment for multiple comparisons was not made.
Results
Volumes of ROIs
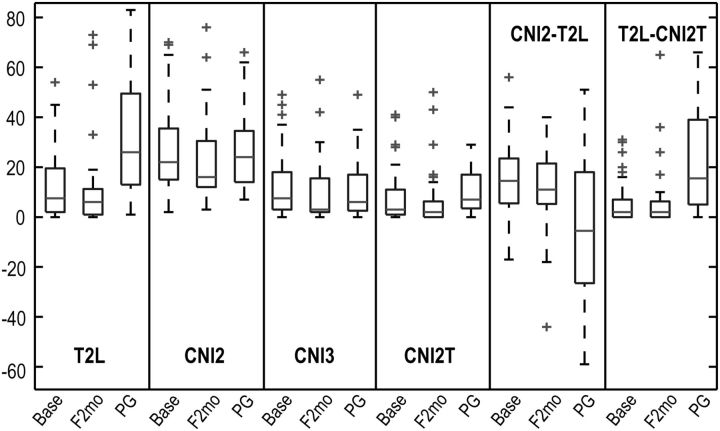
The mean ± SD of the T2 lesion coverage by MRSI PRESS selected volume was 0.73±0.17. The numbers of voxels in the various abnormalities are plotted in Fig. 3. The median differences in the numbers of voxels between CNI2 and T2L (CNI2-T2L) and between T2L and CNI2T (T2L-CNI2T) were 15 and 1 at baseline (P < .0001, P < .0001), while they were –1 and 13.5 at progression (PG in Fig. 3) (P = .8699, P < .0001). There was a trend for larger metabolic lesions (CNI2) than anatomic lesions (T2L) at baseline, while the CNI values were more variable in T2L at progression. Serial 3D MRSI data in Fig. 1 show that the patient (PFS of only 4 mo) had new areas of contrast enhancement that originated from the metabolic lesion at baseline and F2mo. Note that voxels in the T2L at progression had elevated CNI but with more heterogeneity than at baseline and F2mo.
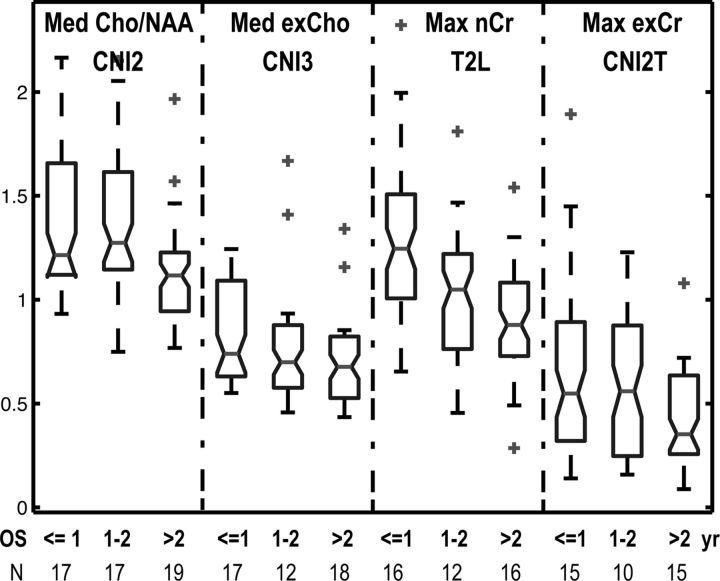
Fig. 3.
Box plots of the numbers of voxels in the T2 lesion within the PRESS volume (T2L), regions that had CNI value >2 (CNI2), regions that had CNI value >3 (CNI3), intersection of the T2L and CNI2 (CNI2T), CNI2-T2L and T2L-CNI2T at baseline, F2mo, and progression (PG). Number of patients are 64, 41, and 36, respectively, at these 3 time points. CNI2-T2L and T2L-CNI2T represent the difference in number of voxels between CNI2 and T2L and between T2L and CNI2T, respectively. Metabolic lesions were larger than the T2L, and the percent of the metabolic lesions within the T2L varied at PG.
MRSI and 6-month Progression-Free Survival
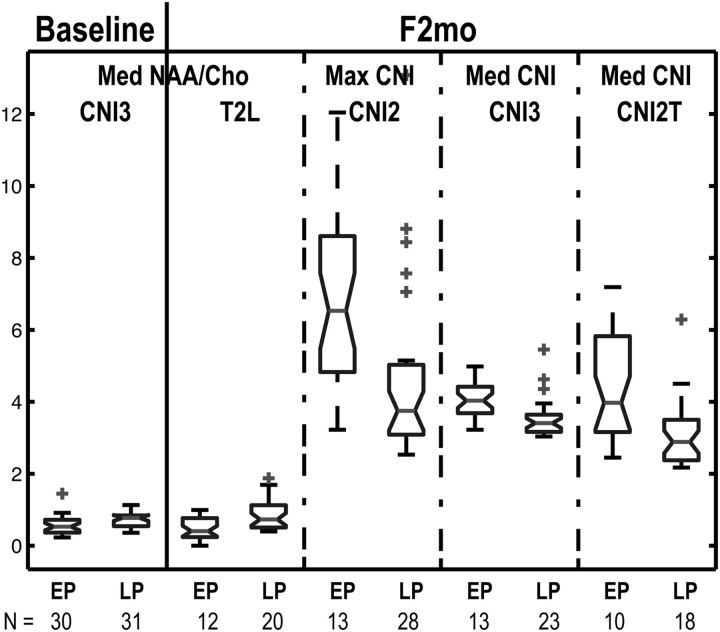
Median PFS was 6 months (95% confidence interval [CI], 5–7 mo), with 17 patients being censored after 1-year follow-up. Of the 64 patients, 32 had progressed at 6 months and were designated as early progressors. The metabolite parameters that were significantly associated with PFS6 are given in Table 1. The only baseline parameter that was related to PFS6 was median NAA/Cho in CNI3 (odds ratio [OR] of early progression = 0.0678, P = .0301). At F2mo, not only NAA/Cho but also CNI and exCho were significant predictors of PFS. Patients with lower median NAA/Cho in T2L, higher maximum CNI in CNI2, higher median CNI in CNI3, and CNI2T at F2mo had worse PFS6 (OR = 0.0233, 1.4175, 5.6299, and 2.1531, respectively; Ps = .0229, .0193, .0196, and .0271). Of the volumetric parameters, the number of voxels in CNI2 (OR = 1.0458, P = .0431) at F2mo was a significant predictor of progression at 6 months. These predictors are illustrated in Fig. 4 for early and late progressors. Of the patients who had T2L, CNI2, CNI2T, and CNI3 lesions, those with higher NAA/Cho in T2L at F2mo were associated with late progression (n = 26; OR = 0.0123, P = .0312). However, changes in metabolites between these 2 time points were not related to PFS6.
Table 1.
Odds ratios and P-values for metabolite parameters that were significant predictors of PFS6 (P ≤ .05) after controlling for age and field strength
| Time | ROI | n | Logistic Linear Regression |
Stepwise Output | ||
|---|---|---|---|---|---|---|
| Parameter | P | OR | ||||
| Baseline | CNI3 | 61 | Median NAA/Cho | .0301 | 0.0678 | Median NAA/Cho |
| F2mo | T2L | 32 | Median NAA/Cho | .0229 | 0.0233 | Median NAA/Cho |
| CNI2 | 41 | Max CNI | .0193 | 1.4175 | Max CNI | |
| Median CNI | .0291 | 6.3427 | ||||
| Max exCho | .0335 | 3.3471 | ||||
| CNI2T | 28 | Median CNI | .0271 | 2.1531 | Median CNI | |
| Max CNI | .0441 | 1.3459 | ||||
| CNI3 | 35 | Median CNI | .0196 | 5.6299 | Median CNI | |
| Median NAA/Cho | .0450 | 0.0159 | ||||
| Volume | 41 | CNI2 | .0431 | 1.0458 | CNI2 vol | |
Metabolite parameters in the final column represent the selected parameter from the stepwise analysis within each ROI. Parameters in bold were selected over the variables from different ROIs that were significant at that time point.
Fig. 4.
Box plots of median NAA/Cho in the regions that had CNI value >3 (CNI3) at baseline, median NAA/Cho in the T2 lesion within the PRESS volume (T2L), maximum Cho-to-NAA index (CNI) in the regions that had CNI value >2 (CNI2), median CNI in the CNI3 and intersection of the T2L and CNI2 (CNI2T) at F2mo in the groups of early progressor (EP) and late progressor (LP). N represents the number of patients. Smaller CNI values and higher NAA/Cho are seen in patients who progressed later.
MRSI and Overall Survival
Median OS was 20 months (95% CI, 16–24 mo), with 11 patients being censored. Treatment failure or progression by 6 months was associated with significant survival disadvantage (P = .0010). Table 2 summarizes the significant factors predicting OS. Overall, the normalized Lac and Lip values were the best OS predictors at all time points. Fig. 2 (non-Lac-edited MRSI) and Fig. 5 (Lac-edited MRSI) demonstrate that patients with elevated median nLL in T2L at F2mo had poor OS (7 and 11 mo, respectively). Maximum exCr, maximum nCr, median Cho/NAA, maximum CNI, and maximum exCho at F2mo were also found to be significantly associated with OS (see Fig. 6), but only maximum nCr in T2L was selected with median nLL in the stepwise regression analysis (hazard ratios [HRs] = 2.2613, 1.9338+ [increased HR per 0.1 unit]; Ps = .0956, .0040). Figure 7 shows that a patient with higher maximum nCr and elevated nLL in T2L had relatively shorter OS (11 vs 61 mo). Patients with small volumes of T2ALL at baseline and CNI2T at F2mo or a decreased number of voxels in CNI2T from pre-RT to post-RT (F2mo-baseline, delta) had a significant benefit in OS (HRs = 1.0163, 1.0442, 1.0755; Ps = .0201, .0036, .0034). The stepwise regression models based on these metabolite and volumetric parameters for the patients who had T2L, CNI2, CNI3, and CNI2T lesions indicated significant association with OS of higher sum nLL in T2L, CNI2, and CNI2T at baseline (n = 46; HRs = 1.2894, 1.1025, 0.7394, respectively; Ps = .0020, .0757, .0119); larger maximum nCr in T2L, maximum nLL in CNI2T, and number of voxels in CNI2T at F2mo (n = 37; HRs = 5.9858, 8.8522, 0.9524, respectively; Ps = .0120, .0346, .1707); and elevated maximum nLL in T2L and number of voxels in CNI2T post-RT (n = 42; HRs = 2.6992, 1.0531, respectively; Ps = .0575, .0702).
Table 2.
Significant metabolic predictors for OS (*P < .01; **P < .001) after controlling for age and field strength for all data (upper values) and for Lac-edited data alone (below)
| Time | ROI | n | Cox Regression Parameter | Stepwise Output |
||
|---|---|---|---|---|---|---|
| Parameter | P | HR | ||||
| Baseline | T2L | 52 | Sum nLL* | Sum nLL | .0025 | 1.1017 |
| CNI2 | 64 | Sum nLL | Sum nLL | .0137 | 1.0561 | |
| CNI2T | 49 | Sum nLL | Sum nLL | .0344 | 1.0799 | |
| CNI3 | 61 | Sum nLL | Sum nLL | .0159 | 1.0673 | |
| Volume | 64 | T2ALL | T2ALL vol | .0201 | 1.0163 | |
| F2mo | T2L | 44 | Max exCr,* max nCr,* max/median*/sum nLL | Median nLL | .0398 | 1.9338+ |
| Max nCr | .0956 | 2.2613 | ||||
| CNI2 | 53 | Median Cho/NAA,* max*/median/sum** nLL | Max nLL | .0028 | 4.5151 | |
| CNI2T | 40 | Max CNI, max exCho, max exCr, max nCr, max**/median*/sum nLL | Max nLL | .0004 | 6.8927 | |
| CNI3 | 47 | Median Cho/NAA, max exCho, max*/median/sum** nLL | Max nLL | .0038 | 4.6228 | |
| Vol | 53 | T2ALL,* T2L,* CNI2, CNI2T,* CNI3 | CNI2T vol | .0036 | 1.0442 | |
| Delta | CNI3 | 42 | Median CNI, max*/sum nLL | Max nLL | .0084 | 3.8105 |
| Volume | 48 | CNI2T* | CNI2T vol | .0034 | 1.0755 | |
| Baseline | T2L | 24 | Sum nLac | Sum nLac | .0294 | 1.1634 |
| CNI2 | 28 | Max/sum nLac | Max nLac | .0225 | 1.6600+ | |
| CNI3 | 26 | Max/median/sum nLac, sum nLip | Sum nLac | .0112 | 1.1473 | |
| F2mo | T2L | 15 | Max/sum nLac | Sum nLac | .0467 | 1.1137 |
| CNI2 | 17 | Max/sum* nLac, sum nLip | Sum nLac | .0098 | 1.1708 | |
| CNI2T | 15 | Max/sum nLac | Max nLac | .0228 | 1.8158+ | |
| CNI3 | 15 | Max/sum nLac | Max nLac | .0166 | 1.7394+ | |
| Delta | CNI2 | 17 | Max/sum nLac | Sum nLac | .0195 | 1.6136 |
| CNI2T | 13 | Max nLac | Max nLac | .03091 | 2.2599+ | |
| CNI3 | 15 | Max/sum nLac, sum nLip | Max nLac | .0888 | 2.0651+ | |
| Sum nLac | .0958 | 1.6146 | ||||
| Sum nLip | .0967 | 1.6361 | ||||
Parameters in column 5 (“Parameter” under “Stepwise Output”) were selected from the stepwise procedure within each ROI. Metabolite parameters in bold are the outcomes of the stepwise analyses from the significant variables across all ROIs at each time point. HR values with + stand for the amount of increased hazard per 0.1 unit.
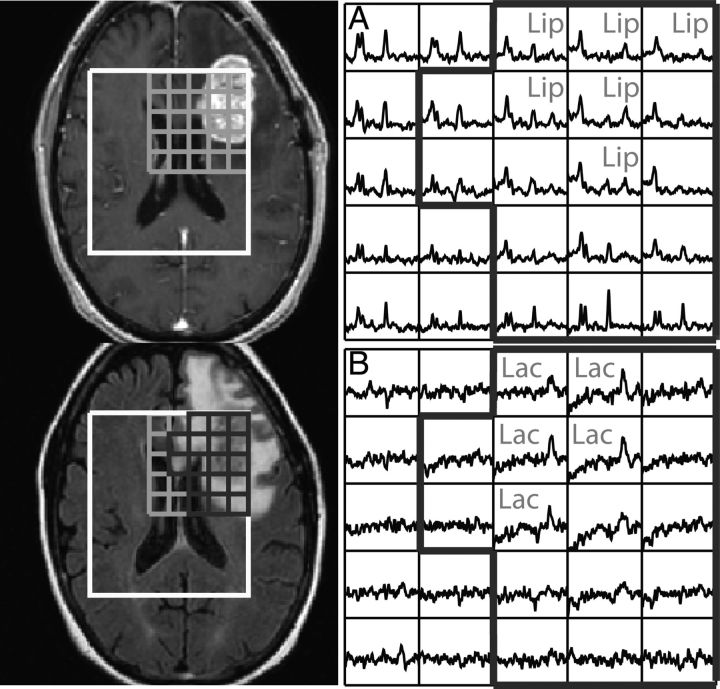
Fig. 5.
3D (Lac-edited) MRSI acquired from a patient with PFS of 2 mo and OS of 11 mo at F2mo. Spectral array corresponded to (A) summed and (B) difference of the Lac-edited spectra. Voxels having Lac or Lip with signal-to-noise ratio ≥ 5.0 are labeled, and voxels within the T2 lesion within the PRESS volume (T2L) are outlined in black thick line.
Fig. 6.
Box plots of median Cho/NAA in the regions that had CNI value >2 (CNI2), median exCho in the regions that had CNI value >3 (CNI3), maximum nCr in the T2 lesion within the PRESS volume (T2L) or maximum excess creatine (exCr) in the intersection of T2L and CNI2 (CNI2T) at F2mo in 3 groups of patients, who died within a year, between 1 and 2 years, and lived longer than 2 years. N represents the number of patients. Patients who had elevated metabolic values had relatively shorter OS.
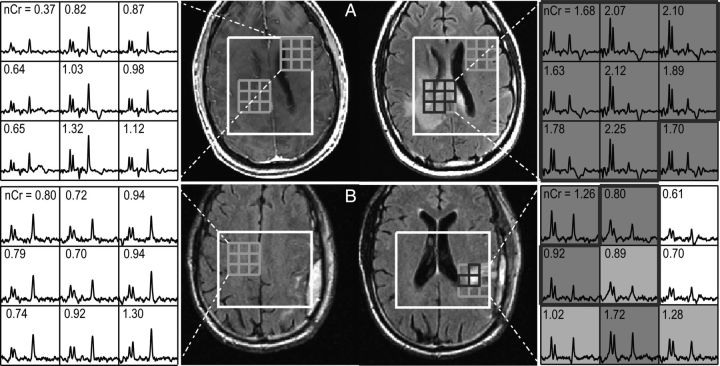
Fig. 7.
Postcontrast T1-weighted FLAIR images and 3D (non-Lac-edited) MRSI from 2 patients (A, B) with GBM at F2mo. Voxels with Cho-to-NAA index (CNI) >3.0 are highlighted in dark grey, voxels in light grey had CNI between 2 and 3, and voxels in the T2 lesion within the PRESS volume (T2L) were outlined in black thick line. Numbers in the spectral array represent values of normalized creatine (nCr). Patient A had a short OS of 11 mo and patient B died at 61 mo.
Combined Imaging and Spectroscopic Predictors
Significant anatomic, diffusion, and perfusion imaging23 as well as spectroscopic predictors of OS and PFS or PFS6 are summarized in Table 3. Only those MRSI parameters calculated from stepwise regression models are included in the table. Note that none of the diffusion parameters is associated with OS or PFS and that the perfusion values are associated with only PFS. The T2L and non-enhancing lesion volumes are associated with OS at both baseline and F2mo, but the volume of the contrast-enhancing lesion is associated with OS at only F2mo. By contrast, there are MRSI parameters at both baseline and F2mo, which relate to PFS6 and OS.
Table 3.
Combined anatomic, diffusion, and perfusion imaging21 and spectroscopic imaging predictors
| OS | PFS/PFS6 | ||
|---|---|---|---|
| Baseline | MRI | T2ALL vol, NEL vol | T2ALL median nPH*/nCBV/nRF |
| NEL median nPH/nCBV/nRF | |||
| MRSI | T2L*/CNI2/CNI2T/CNI3 sum nLL | CNI3 median NAA/Cho | |
| F2mo | MRI | T2ALL vol,* NEL vol,* CEL vol* | T2ALL/NEL median nCBV |
| MRSI | T2L median nLL (+ max nCr), CNI2*/CNI2T**/CNI3* max nLL, CNI2T vol* | CNI2 vol | |
| CNI2 max CNI | |||
| CNI2T/CNI3 median CNI | |||
| T2L median NAA/Cho | |||
| Delta | MRI | None | T2ALL/NEL 90th nRF |
| MRSI | CNI3 max nLL,* CNI2T vol* | None |
Abbreviations: NEL, non-enhancing lesion; nPH, normalized peak height; nCBV, normalized cerebral blood volumes; nRF, normalized recirculation factor. *P < .01; **P < .001. All the tests were adjusted by age and field strength. Italics represent the parameters that had HR or OR < 1.
Discussion
While the prognosis for patients with GBM is relatively poor, variability of survival among patients who are given the same aggressive treatment suggests that there are additional underlying factors that influence how the tumor responds to treatment. Although conventional MRI is a powerful tool to visualize changes in morphological abnormalities, it is not a direct reflection of biochemical changes in the tumor and surrounding tissue. In this study, we used 3D MRSI to assess changes in metabolism between NAWM, anatomic, and metabolic lesions during RT and demonstrated that parameters, which described these changes, could be used to predict PFS and OS.
Radiation is one of the standard treatments for patients with GBM. Normal tissue that surrounds the tumor lesion has a limited tolerance to radiation but is unavoidably subjected to this treatment. Reduced NAA/Cr and Cho/Cr have been reported in normal brain that has been irradiated.32,33 In this study, we took this into account by using the median peak height in the NAWM to calculate the normalized peak height in the lesions. However, the local field inhomogeneities in the lesion caused by necrosis and/or hemorrhage may broaden the linewidths of metabolite peaks differently from patient to patient. Treatment-induced necrosis could also be present in the tumor lesion. The presence of Lip within the lesion immediately after the completion of radiation may reflect treatment-induced necrosis. The fact that the portion of the lesion being considered also had elevated Cho relative to NAA indicates that it comprised a combination of tumor and necrosis. Another factor that influences radiation sensitivity is the hypoxic environment that ensues due to tumor cells outgrowing their blood supply and potentially becoming more resistant than normal cells. MRSI is able to assess the levels of Lac in the tumor that are associated with reduced oxygenation.
The assessment of treatment response is further complicated by pseudoprogression, which can result from an RT-induced increase in vascular permeability.3 Of the 32 patients in this study who progressed within 6 months after completion of concomitant RT and chemotherapy, 50% were confirmed as having true progression by reoperation. For the patients who did not get a resection, the assessment of progression was based upon longer-term clinical and imaging follow-up, which minimized any confounding effects associated with pseudoprogression. Although the patients have been followed by serial MRI/MRSI for maximally 1 year, the OS analysis was carried out for more than 5 years after the first enrollment, leaving relatively few censored observations.
Multivoxel 3D MRSI provides an assessment of the spatial distribution of metabolic lesions in patients with glioma. The data provide parameters reflecting cellular metabolism, and CNI has been shown to be more sensitive in differentiating tumor from nontumor than have levels of individual metabolites.24,34 ROIs in this study included the morphological abnormality that was covered by the PRESS box (T2L), the metabolic abnormality (CNI2 and CNI3), and regions containing both anatomic and metabolic abnormalities (CNI2T). Due to the limited spatial resolution of 3D MRSI, regions within the smaller contrast-enhancing lesions were not evaluated separately in this study. Even though data acquisition did not always cover the entire spatial extent of the lesion, it has been shown here and in previous studies that the number of voxels within the metabolic abnormality could be used to predict OS.15,17,18
A previously published analysis of anatomic imaging in the same population reported that larger volumes of the T2 lesion and non-enhancing lesion at pre-RT and increased anatomic volumes at post-RT were significantly correlated with worse OS but not worse PFS.23 In this study, we were interested in assessing the metabolic changes that may assist in predicting outcome in a prospective group of patients. A large number of 3D MRSI exams were acquired from patients with GBM, with an average coverage of the T2 lesion by 3D MRSI of 73%. Metabolic heterogeneity within the T2 lesion and metabolic abnormalities outside anatomic lesions were observed. The number of voxels within the T2 and metabolic lesions were significantly correlated with both PFS6 and OS.18,20,21 At the F2mo scan, the number of voxels with elevated CNI within the T2 hyperintensity played the most important role in predicting OS compared with the other volumetric parameters based on the stepwise regression model. This implies that the CNI lesion may be a more reliable measure of tumor burden.
Disease progression in this study was determined from the size of the contrast-enhancing lesion in conjunction with neurologic function and corticosteroid use.2 Patients who progressed by 6 months had a significantly shorter median survival time than those who did not, P = .001. As previously reported, PFS6 has been proposed as an alternative endpoint for evaluating patients with GBM receiving temozolomide.35 The consistent metabolite observations for predicting PFS6 in our study were changes in the relative levels of NAA and Cho in the metabolic lesions. Correlated with early PFS6 were decreased median NAA/Cho in CNI3 at baseline, lower median NAA/Cho in T2L, and higher median/maximum CNI in CNI2/CNI2T/CNI3 at F2mo. This suggests that increased tumor cellularity and decreased neuronal function promote tumor progression. Incorporating what we reported previously (see Table 3), the presence of regions with elevated blood volumes within the T2 lesion at the pre-RT and post-RT time points were also associated with shorter PFS.23 These results suggest that high tumor cellularity causes regional hypoxia and promotes the formation of new blood vessels.36,37 As a result, patients with a large volume of residual tumor cells and increased blood volume were more likely to resist RT and progress at an early time point.
The regions with elevated levels of Lac and Lip were the best predictors for poor OS in this study. Results from the Lac-edited data, which were consistent with the previous findings,20,21 indicate that this was mostly due to increased Lac. As the end product of anaerobic respiration, increased Lac may indicate tumor hypoxia or unregulated tumor growth. Higher Lip peak within the metabolic abnormalities, the region with elevated CNI (>2), could arise from tumor necrosis and/or treatment-induced necrosis (post-RT), along with increased tumor cells. Another interesting finding was that increased nCr in T2L was significantly correlated with worse OS by the end of radiation (F2mo). This resonance corresponds to a combination of Cr and phosphocreatine. The latter serves as a short-term energy reserve in the brain. Changes in total Cr are possibly associated with altered synthesis in Cr because astroglial cells of the normal brain are able to synthesize Cr.38 Increased nCr levels may reveal the infiltrative characteristics of the tumor cells and thus could explain the relationship to tumor progression. Increased Cr was also found in a grade II astrocytoma,39 and gliomatosis8 and was reported to have prognostic implication on malignant transformation in low-grade glioma.40 Another interpretation is that the elevation of nCr in T2L is associated with decreased Cr in NAWM due to the effect of RT.
Although a previous study has shown that nCho at the post-RT follow-up scan was correlated with PFS and OS in a group of 26 patients with high-grade glioma using single-slice multivoxel MRS with a nominal voxel size of 1.0 × 1.1 × 2.0 cm,3,22 it was not found to be a significant predictor in our study. The nominal spatial resolution in our study was 1 cm3, and we achieved more extensive coverage of the lesion for a larger patient population. Besides evaluating metabolite ratios and peak heights normalized relative to NAWM, we also evaluated 2 parameters that we have termed exCho and exCr.31 These parameters took into account the differences in Cho or Cr relative to their levels in NAWM. They were found to be significantly correlated with OS and were retained in the stepwise regression analysis.
Overall, our study examined the potential predictive value of the pre-RT and post-RT in vivo 3D MRSI for PFS6 and OS in patients with newly diagnosed primary GBM being treated with radiation and concurrent chemotherapy. It was observed that the metabolic abnormality frequently extends beyond that of the anatomic lesions. Relative to conventional and diffusion- and perfusion-weighted MRI, in which only the volumes of the anatomic lesions are predictors of OS,21,23,41 increased CNI in the metabolic lesions were observed in patients who progressed earlier, and elevated nLac and nLip levels were associated with worse OS. This suggests that 3D MRSI may be a more reliable measure for tumor burden in predicting OS. The majority (81%) of patients in this study received temozolomide and radiation, which is the current standard of care for patients with GBM. Future studies will use similar analytical methods for 3D MRSI to follow patients participating in clinical trials that include novel treatment strategies, such as inhibiting angiogenesis or blocking cell proliferation.41–43
Funding
This work was supported by UC Discovery grant no. ITL-BIO04-10148, NIH grant nos R01 CA127612, P01 CA11816, and NIH P50 CA97257, and the St Louis Fall Festival Committee American Brain Tumor Association Fellowship.
Acknowledgments
The authors thank Annette Molinaro, Wei Bian, and Mekhail Anwar for useful discussion and assistance.
References
- 1.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Macdonald DR, Cascino TL, Schold SC, Jr, Cairncross JG. Response criteria for phase II studies of supratentorial malignant glioma. J Clin Oncol. 1990;8:1277–1280. doi: 10.1200/JCO.1990.8.7.1277. [DOI] [PubMed] [Google Scholar]
- 3.Taal W, Brandsma D, de Bruin HG, et al. Incidence of early pseudo-progression in a cohort of malignant glioma patients treated with chemoirradiation with temozolomide. Cancer. 2008;113:405–410. doi: 10.1002/cncr.23562. [DOI] [PubMed] [Google Scholar]
- 4.Wasserfallen JB, Ostermann S, Leyvraz S, Stupp R. Cost of temozolomide therapy and global care for recurrent malignant gliomas followed until death. Neuro Oncol. 2005;7:189–195. doi: 10.1215/S1152851704000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wen PY, Macdonald DR, Reardon DA, et al. Updated response assessment criteria for high-grade gliomas: Response Assessment in Neuro-Oncology working group. J Clin Oncol. 2010;28:1963–1972. doi: 10.1200/JCO.2009.26.3541. [DOI] [PubMed] [Google Scholar]
- 6.Martinez-Bisbal MC, Celda B. Proton magnetic resonance spectroscopy imaging in the study of human brain cancer. Q J Nucl Med Mol Imaging. 2009;53:618–630. [PubMed] [Google Scholar]
- 7.Urenjak J, Williams SR, Gadian DG, Noble M. Proton nuclear magnetic resonance spectroscopy unambiguously identifies different neural cell types. J Neurosci. 1993;13:981–989. doi: 10.1523/JNEUROSCI.13-03-00981.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galanaud D, Chinot O, Nicoli F, et al. Use of proton magnetic resonance spectroscopy of the brain to differentiate gliomatosis cerebri from low-grade glioma. J Neurosurg. 2003;98:269–276. doi: 10.3171/jns.2003.98.2.0269. [DOI] [PubMed] [Google Scholar]
- 9.Li X, Lu Y, Pirzkall A, McKnight T, Nelson SJ. Analysis of the spatial characteristics of metabolic abnormalities in newly diagnosed glioma patients. J Magn Reson Imaging. 2002;16:229–237. doi: 10.1002/jmri.10147. [DOI] [PubMed] [Google Scholar]
- 10.Star-Lack J, Spielman D, Adalsteinsson E, et al. In vivo lactate editing with simultaneous detection of choline, creatine, NAA, and lipid singlets at 1.5T using PRESS excitation with applications to the study of brain and head and neck tumors. J Magn Reson. 1998;133:243–254. doi: 10.1006/jmre.1998.1458. [DOI] [PubMed] [Google Scholar]
- 11.Park I, Chen AP, Zierhut ML, et al. Implementation of 3T lactate-edited 3D 1H MR spectroscopic imaging with flyback echo-planar readout for glioma patients. Ann Biomed Eng. 2011;39:193–204. doi: 10.1007/s10439-010-0128-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Catalaa I, Henry R, Dillon WP, et al. Perfusion, diffusion and spectroscopy values in newly diagnosed cerebral gliomas. NMR Biomed. 2006;19:463–475. doi: 10.1002/nbm.1059. [DOI] [PubMed] [Google Scholar]
- 13.Law M, Yang S, Wang H, et al. Glioma grading: sensitivity, specificity, and predictive values of perfusion MR imaging and proton MR spectroscopic imaging compared with conventional MR imaging. AJNR Am J Neuroradiol. 2003;24:1989–1998. [PMC free article] [PubMed] [Google Scholar]
- 14.Chang SM, Nelson S, Vandenberg S, et al. Integration of preoperative anatomic and metabolic physiologic imaging of newly diagnosed glioma. J Neurooncol. 2009;92:401–415. doi: 10.1007/s11060-009-9845-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson SJ, Graves E, Pirzkall A, et al. In vivo molecular imaging for planning radiation therapy of gliomas: an application of 1H MRSI. J Magn Reson Imaging. 2002;16:464–476. doi: 10.1002/jmri.10183. [DOI] [PubMed] [Google Scholar]
- 16.Park I, Tamai G, Lee MC, et al. Patterns of recurrence analysis in newly diagnosed glioblastoma multiforme after three-dimensional conformal radiation therapy with respect to pre-radiation therapy magnetic resonance spectroscopic findings. Int J Radiat Oncol Biol Phys. 2007;69:381–389. doi: 10.1016/j.ijrobp.2007.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laprie A. [Proton magnetic resonance spectroscopic imaging and other types of metabolic imaging for radiotherapy planning in adult and pediatric high-grade gliomas] Cancer Radiother. 2009;13:556–561. doi: 10.1016/j.canrad.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 18.Kuznetsov YE, Caramanos Z, Antel SB, et al. Proton magnetic resonance spectroscopic imaging can predict length of survival in patients with supratentorial gliomas. Neurosurgery. 2003;53:565–574. doi: 10.1227/01.neu.0000079331.21178.8e. discussion 574–566. [DOI] [PubMed] [Google Scholar]
- 19.Oh J, Henry RG, Pirzkall A, et al. Survival analysis in patients with glioblastoma multiforme: predictive value of choline-to-N-acetylaspartate index, apparent diffusion coefficient, and relative cerebral blood volume. J Magn Reson Imaging. 2004;19:546–554. doi: 10.1002/jmri.20039. [DOI] [PubMed] [Google Scholar]
- 20.Crawford FW, Khayal IS, McGue C, et al. Relationship of pre-surgery metabolic and physiological MR imaging parameters to survival for patients with untreated GBM. J Neurooncol. 2009;91:337–351. doi: 10.1007/s11060-008-9719-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saraswathy S, Crawford FW, Lamborn KR, et al. Evaluation of MR markers that predict survival in patients with newly diagnosed GBM prior to adjuvant therapy. J Neurooncol. 2009;91:69–81. doi: 10.1007/s11060-008-9685-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quon H, Brunet B, Alexander A, et al. Changes in serial magnetic resonance spectroscopy predict outcome in high-grade glioma during and after postoperative radiotherapy. Anticancer Res. 2011;31:3559–3565. [PubMed] [Google Scholar]
- 23.Li Y, Lupo JM, Polley MY, et al. Serial analysis of imaging parameters in patients with newly diagnosed glioblastoma multiforme. Neuro Oncol. 2011;13:546–557. doi: 10.1093/neuonc/noq194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKnight TR, Noworolski SM, Vigneron DB, Nelson SJ. An automated technique for the quantitative assessment of 3D-MRSI data from patients with glioma. J Magn Reson Imaging. 2001;13:167–177. doi: 10.1002/1522-2586(200102)13:2<167::aid-jmri1026>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 25.Tran TK, Vigneron DB, Sailasuta N, et al. Very selective suppression pulses for clinical MRSI studies of brain and prostate cancer. Magn Reson Med. 2000;43:23–33. doi: 10.1002/(sici)1522-2594(200001)43:1<23::aid-mrm4>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Osorio JA, Ozturk-Isik E, et al. Considerations in applying 3D PRESS H-1 brain MRSI with an eight-channel phased-array coil at 3 T. Magn Reson Imaging. 2006;24:1295–1302. doi: 10.1016/j.mri.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Vigneron DB, Cha S, et al. Relationship of MR-derived lactate, mobile lipids, and relative blood volume for gliomas in vivo. AJNR Am J Neuroradiol. 2005;26:760–769. [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson SJ. Analysis of volume MRI and MR spectroscopic imaging data for the evaluation of patients with brain tumors. Magn Reson Med. 2001;46:228–239. doi: 10.1002/mrm.1183. [DOI] [PubMed] [Google Scholar]
- 29.Nelson SJ, Nalbandian AB, Proctor E, Vigneron DB. Registration of images from sequential MR studies of the brain. J Magn Reson Imaging. 1994;4:877–883. doi: 10.1002/jmri.1880040621. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 31.Park I, Elkhaled A, Kadambi A, et al. Finding of early prognostic marker from 3D 1H-MRSI and diffusion tensor imaging for newly-diagnosed GBM patients receiving radiation, temozolomide and PKC Inhibitor. May 2010 Paper presented at: International Society for Magnetic Resonance in Medicine annual meeting; Stockholm, Sweden. [Google Scholar]
- 32.Sundgren PC, Nagesh V, Elias A, et al. Metabolic alterations: a biomarker for radiation-induced normal brain injury—an MR spectroscopy study. J Magn Reson Imaging. 2009;29:291–297. doi: 10.1002/jmri.21657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esteve F, Rubin C, Grand S, Kolodie H, Le Bas JF. Transient metabolic changes observed with proton MR spectroscopy in normal human brain after radiation therapy. Int J Radiat Oncol Biol Phys. 1998;40:279–286. doi: 10.1016/s0360-3016(97)00714-1. [DOI] [PubMed] [Google Scholar]
- 34.McKnight TR, von dem Bussche MH, Vigneron DB, et al. Histopathological validation of a three-dimensional magnetic resonance spectroscopy index as a predictor of tumor presence. J Neurosurg. 2002;97:794–802. doi: 10.3171/jns.2002.97.4.0794. [DOI] [PubMed] [Google Scholar]
- 35.Polley MY, Lamborn KR, Chang SM, et al. Six-month progression-free survival as an alternative primary efficacy endpoint to overall survival in newly diagnosed glioblastoma patients receiving temozolomide. Neuro Oncol. 2010;12:274–282. doi: 10.1093/neuonc/nop034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wesseling P, Ruiter DJ, Burger PC. Angiogenesis in brain tumors; pathobiological and clinical aspects. J Neurooncol. 1997;32:253–265. doi: 10.1023/a:1005746320099. [DOI] [PubMed] [Google Scholar]
- 37.Damert A, Machein M, Breier G, et al. Up-regulation of vascular endothelial growth factor expression in a rat glioma is conferred by two distinct hypoxia-driven mechanisms. Cancer Res. 1997;57:3860–3864. [PubMed] [Google Scholar]
- 38.Dringen R, Verleysdonk S, Hamprecht B, et al. Metabolism of glycine in primary astroglial cells: synthesis of creatine, serine, and glutathione. J Neurochem. 1998;70:835–840. doi: 10.1046/j.1471-4159.1998.70020835.x. [DOI] [PubMed] [Google Scholar]
- 39.Londono A, Castillo M, Armao D, Kwock L, Suzuki K, Unusual MR. Spectroscopic imaging pattern of an astrocytoma: lack of elevated choline and high myo-inositol and glycine levels. AJNR Am J Neuroradiol. 2003;24:942–945. [PMC free article] [PubMed] [Google Scholar]
- 40.Hattingen E, Raab P, Franz K, et al. Prognostic value of choline and creatine in WHO grade II gliomas. Neuroradiology. 2008;50:759–767. doi: 10.1007/s00234-008-0409-3. [DOI] [PubMed] [Google Scholar]
- 41.Lund EL, Spang-Thomsen M, Skovgaard-Poulsen H, Kristjansen PE. Tumor angiogenesis—a new therapeutic target in gliomas. Acta Neurol Scand. 1998;97:52–62. doi: 10.1111/j.1600-0404.1998.tb00609.x. [DOI] [PubMed] [Google Scholar]
- 42.Franceschi E, Tosoni A, Bartolini S, et al. Treatment options for recurrent glioblastoma: pitfalls and future trends. Expert Rev Anticancer Ther. 2009;9:613–619. doi: 10.1586/era.09.23. [DOI] [PubMed] [Google Scholar]
- 43.Hsu JY, Wakelee HA. Monoclonal antibodies targeting vascular endothelial growth factor: current status and future challenges in cancer therapy. BioDrugs. 2009;23:289–304. doi: 10.2165/11317600-000000000-00000. [DOI] [PubMed] [Google Scholar]