Abstract
Background
Nemo-like kinase (NLK) is an evolutionarily conserved protein kinase involved in Wnt/beta-catenin signaling, which has been reported to be associated with gliomagenesis. In the present study, we aimed to identify a concrete mechanism of Wnt/beta-catenin pathway regulation by microRNAs (miRNAs) in glioma.
Methods
Quantitative reverse-transcription polymerase chain reaction and in situ hybridization were conducted to detect the expression of miR-92b. The cell proliferation rate and cell cycle kinetics were detected using 3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-di-phenytetrazoliumromide (MTT) assay and flow cytometry, cell invasion and migration were evaluated using Transwell assay and wound healing assay, and cell apoptosis was detected using annexin V staining. Furthermore, the relevant molecules regulating proliferation and invasion were examined using Western blot analysis, immunohistochemistry, and immunofluorescence staining. Luciferase reporter assay was used to identify the direct regulation of NLK by miR-92b and beta-catenin/TCF4 activity.
Results
We first showed that the expression of miR-92b was elevated in both glioma samples and glioma cells. Furthermore, down-regulation of miR-92b triggered growth inhibition, induced apoptosis, and suppressed invasion of glioma in vitro and in vivo. Luciferase assay and Western blot analysis revealed that NLK is a direct target of miR-92b. Restoring expression of NLK inhibited glioma proliferation and invasion. Mechanistic investigation revealed that miR-92b deletion suppressed beta-catenin/TCF-4 transcription activity by targeting NLK. Moreover, expression of NLK was inversely correlated with miR-92b in glioma samples and was predictive of patient survival in a retrospective analysis.
Conclusions
Our findings identify a role for miR-92b in glioma proliferation and invasion after activation of Wnt/beta-catenin signaling via NLK.
Keywords: beta-catenin, glioma, MiR-92b, NLK, Wnt
Glioblastoma is one of the most common forms of neural malignancy, with a median survival of 9–12 months. Despite new biological insights and advances in therapy, the prognosis of patients with gliomas has remained poor during the past 4 decades.1,2 Therefore, it is essential to investigate the mechanism involved in the development and progression of glioma.
Wnt signaling has proven to be associated with various disease pathologies, including an important role in gliomagenesis. The Wnt signaling pathway regulates many biological processes through a complex of beta-catenin and the T cell factor/lymphoid-enhancer factor 1 (TCF/LEF-1) family of high-mobility group transcription factors.3,4 Wnt stabilizes cytosolic beta-catenin, which then binds to TCF/LEF-1 and recruits transcription factors Brg1 and CREB-binding protein to initiate Wnt-targeted gene expression.5 NLK was originally identified by screening cDNAs coding for protein kinases6 and was recently characterized as a suppressor of the Wnt signaling pathway in mammalian cells.7–12 Another recent study showed that Wnt5a-Ca2t signaling, a known negative regulator of Wnt,13 can be an upstream activator of TAK1-NLK for suppression of canonical Wnt/beta-catenin signaling in the Xenopus system.14 Wnt/beta-catenin signaling is thought to play a critical role in human carcinogenesis;15 thus, it is possible that NLK acts as a tumor suppressor by regulating the Wnt/beta-catenin pathway. NLK reportedly induces apoptosis in DLD-1 human colon cancer cells and prostate cancer cells.16,17 Although NLK has recently been reported in glioma cells to induce apoptosis,18 the precise mechanisms underlying the progression of gliomagenesis are not well characterized.
MiRNAs regulate gene expression after recognition of sequence-specific binding sites, typically in the 3′-untranslated region of target mRNAs, causing mRNA deadenylation and degradation or translational repression.19,20 Alterations in the pattern of gene expression resulting from aberrant expression of miRNAs are associated with numerous diseases, including cancer.21,22 Recent studies have shown that the expression of many miRNAs is altered in gliomas and some miRNAs may function as oncogenes or tumor suppressor genes, including miR-21, miR-221/222, miR-124, and miR-27b.23–26
In our study, we showed increased expression of miR-92b in high-grade glioma, compared with low-grade glioma and normal brain tissue. We proved that NLK is a direct target of miR-92b. Transient transfection of miR-92b inhibitor into glioma cell lines inhibited cell proliferation and cell invasion. Furthermore, miR-92b inhibitor strongly inhibited in vivo glioma xenograft growth. Forced NLK expression partially rescued the effects of miR-92b on the glioma growth. We also identified that miR-92b functions, at least in part, by regulating beta-catenin/TCF-4 pathway. Moreover, miR-92b levels in human gliomas inversely correlated with NLK levels in the same tumors. These results identify a critical role for miR-92b in regulation of proliferation and apoptosis in glioma, suggesting that miR-92b could be critical therapeutic target for glioma intervention.
Materials and Methods
Cell Culture and Culture Conditions
The human U251, U87, LN229, SNB19, and A172 glioblastoma cell lines were purchased from the Institute of Biochemistry and Cell Biology, Chinese Academy of Science. All cells were maintained in a 37°C, 5% carbon dioxide incubator in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% fetal-bovine serum (Invitrogen).
Human Glioma Samples
Human glioma samples were obtained from the Department of Neurosurgery, Sir Run Run Shaw Hospital, Medical College, Zhejiang University, and Department of Neurosurgery, Tianjin Medical University General Hospital, after informed consent was obtainted from adult patients who had received a diagnosis of glioma. Samples were freshly resected during surgery and immediately frozen in liquid nitrogen for subsequent total RNA extraction. The tissue samples included 48 grade I-II gliomas (low grade), 45 grade III-IV gliomas (high grade), and 9 normal brain tissue samples.
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction
Total RNA was isolated from cultured cells, human glioma specimens, or normal brain tissue specimens with use of TRIzol reagent (Invitrogen), according to the manufacturer's instructions. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed in triplicate in ABI 7500HT fast real-time PCR System (Applied Biosystems) and normalized with U6 and glyceraldehyde 3-phosphate dehydrogenase endogenous control. Total RNA from normal brain tissue samples was used as a control. miR-92b levels were measured using the TaqMan microRNA assay kit, and endogenous mRNA levels of NLK were detected using SYBR Green PCR Master Mix kit in accordance with the manufacturer's instructions (Applied Biosystems).
Oligonucleotide, NLK Expression Plasmid Synthesis, and Transfection
miR-92b (5′-UAUUGCACUCGUCCCGGCCUCC-3′), miR-92b inhibitor (5′-GGAGGCCGGGACGAGUGCAAUA-3′), and negative control oligonucleotides were purchased from GenePharma (Shanghai, China). NLK expression plasmid was preserved in our laboratory. miRNA sequences/NLK expression plasmid were transfected into cultured cells with use of Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions.
Luciferase Reporter Assay
The human NLK 3′UTR was amplified and cloned into the XbaI site of the pGL3-control vector (Promega), downstream of the luciferase gene, to generate the plasmid pGL3-WT-NLK-3′UTR (NLK wild). pGL3-MUT-NLK-3′UTR (NLK mut) was generated from NLK wild by replacing the binding site of miR-92b with restriction enzyme cutting site GCUACGCAUAA. For the luciferase reporter assay, cells were cotransfected with luciferase reporter vectors and miR-92b I with use of Lipofectamine 2000.
To evaluate the beta-catenin/TCF-4 transcriptional activity, we used a pair of luciferase reporter constructs, TOP-FLASH and FOP-FLASH (Upstate). TOP-FLASH (with 3 repeats of the Tcf-binding site) or FOP-FLASH (with 3 repeats of a mutated Tcf-binding site) plasmids were transfected into cells treated with miR-92b I, as instructed by the suppliers.
After 48 h, luciferase activity was analyzed using the Dual-Luciferase Reporter Assay System according to the manufacturer's protocols (Promega, Madison).
Cell Proliferation Assay
Glioblastoma cells were seeded into 96-well plates at 2000 cells/well. After transfection as described previously, 20 μL of 3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-di-phenytetrazoliumromide (MTT) (0.5 mg/mL) was added into each well, each day for 7 consecutive days after treatment and incubated for 4 h, and the supernatant was then discarded. Finally, 200 μL of DMSO was added to each well to dissolve the precipitate. Optical density was measured at the wavelength of 570 nm. The data are presented as the mean ± standard error of the mean, derived from triplicate samples of at least 3 independent experiments.
Cell Cycle Assay
For cell cycle analysis by flow cytometry, transfected and NC cells in the log phase of growth were harvested, washed with phosphate-buffered saline, fixed with 90% ethanol overnight at 4°C, and then incubated with RNase at 37°C for 30 min. Nuclei of cells were stained with propidium iodide for an additional 30 min. A total of 10 000 nuclei were examined in a FACS Calibur flow cytometer (Becton-Dickinson), and DNA histograms were analyzed using Modifit software.
Apoptosis Assay
Forty-eight hours after transfection, apoptosis in cultured cells was evaluated using annexin V labeling. For the annexin V assay, an annexin V–FITC labeled Apoptosis Detection Kit (Abcam) was used according to the manufacturer's protocol. TUNEL assay was used to detect the apoptosis in tumor specimens and was performed as previously described.27
Transwell Assay
The transwell filters (Costar) were coated with matrigel (3.9 μg/μL, 60–80 μL) on the upper surface of polycarbonate membrane (diameter 6.5 mm, pore size 8 μm). After incubating at 37°C for 30 min, matrigel solidified and served as the extracellular matrix for tumor cell invasion analysis. A total of 600 μL of conditioned medium derived from tumor cells was used as a source of chemoattractant and was placed in the bottom compartment of the chamber. Harvested cells (1 × 105) in 100 μL of serum-free DMEM were added into the upper compartment of the chamber. After 24 h of incubation at 37°C with 5% carbon dioxide, the medium was removed from the upper chamber. The noninvaded cells on the upper side of the chamber were scraped off with a cotton swab. The cells that had migrated from matrigel into the pores of the inserted filter were fixed with 100% methanol, stained with hematoxylin, and mounted and dried at 80°C for 30 min. The number of cells invading through the matrigel was counted using 3 randomly selected visual fields from the central and peripheral portions of the filter by an inverted microscope at 200 × magnification. Each assay was repeated 3 times.
Wound Healing Assay
Cell culture and transfection conditions were optimized to ensure a homogeneous and viable cell monolayer before wounding. One day before transfection, equal numbers of glioma cells (2 × 105) were seeded in 6-well plates. When cell confluence reached about 90%, ∼24 h after transfection, an artifcial homogeneous wound was made on the monolayer with use of a sterile plastic 200 µL micropipette tip. After wounding, debris was removed by washing cells with phosphate-buffered saline. At different time points, cells that migrated into the wounded area or cells with extended protrusions from the wound border were photographed at 200× magnification under a light microscope.
Western Blot, miRNA Locked Nucleic Acid (LNA) In Situ Hybridization, Immunohistochemistry, and Immunofluorescence Staining
Western blot, miRNA-LNA in situ hybridization, and immunohistochemistry were performed as previously described.27 For immunofluorescence staining, control and transfected cells were seeded on coverslips and fixed with 4% paraformaldehyde (PFA, Sigma), treated with 3% H2O2 for 10 min and incubated with the antibodies described above overnight at 4°C. FITC- or TRITC-labeled secondary antibody (1:200 dilutions) was added for 2 h at 37°C. DAPI reagent was used to stain the glioma cell nuclei, and the cells were visualized using FV-1000 laser-scanning confocal microscopes and analyzed using IPP5.1 (Olympus).
Nude Mouse Tumor Xenograft Model and MiR-92b Inhibitor Treatment
SNB19 or LN229 glioma cells were subcutaneously injected to 5-week-old female nude mice (Cancer Institute of The Chinese Academy of Medical Science). When the tumor volume reached 100 mm3, the mice were randomly divided into 2 groups (10 mice per group), which were treated with 200 pmol scramble oligo or miR-92b inhibitor in 10 μL Lipofectamine 2000 through local injection of xenograft tumor in multiple sites. The treatment was performed once every 2 days for 14 days. The tumor volume was measured with a caliper every 2 days, using the formula: volume = length × width2/2.
Statistical Analysis
Statistics were determined using the Student’s t-test, χ2-test and analysis of variance. Overall survival curves were plotted according to the Kaplan-Meier method, with the log-rank test applied for comparison. All differences were considered to be statistically signifcant at the level of P < .05. Statistics were performed using the SPSS Graduate Pack, version 11.0, statistical software (SPSS).
Results
miR-92b Is Overexpressed in Human Glioma
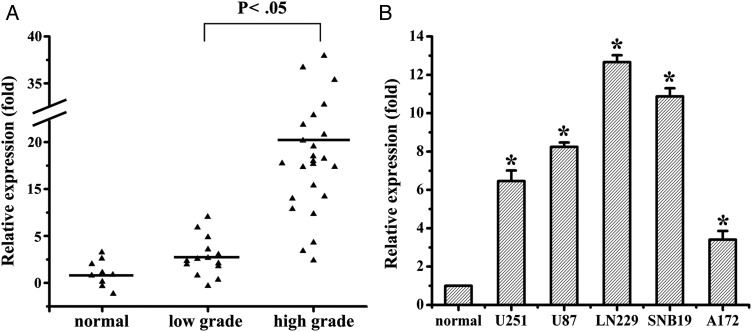
To determine the levels of miR-92b in glioma samples and cell lines, total RNAs were extracted from glioma tissues at grades I, II, III, and IV and glioma cell lines, and the expression levels of miR-92b were analyzed using qRT-PCR. As shown in Fig. 1A, the levels of miR-92b expression in glioma tissues were significantly increased, compared with normal brain tissues. There was no significant difference of miR-92b expression levels in glioma tissues between 14 low-grade gliomas (grade I and II) and 9 normal brain tissues, which was significantly lower than that in 24 high-grade gliomas (grade III and IV). It was also shown that miR-92b was up-regulated in 5 glioma cell lines, compared with 5 normal brain tissues (Fig. 1B). Taken together, our results revealed that miR-92b was abnormally overexpressed both in human glioma samples and in cell lines.
Fig. 1.
miR-92b is upregulated in glioma samples and glioma cells. (A) miR-92b expression in different grade glioma and normal brain tissues by qRT-PCR. (B) qRT-PCR analysis showed that U251, U87, LN229, SNB19, and A172 glioma cells express high levels of miR-92b, compared with normal brain tissues. *P < .05.
miR-92b Inhibitor Suppresses Glioma Malignancy
To examine biological significance of miR-92b in glioma, MiR-92b I or NC was transiently transfected into the glioma cells. Cell proliferation, cell cycle, cell death, and cell invasion were analyzed using MTT, propidium iodide flow cytometry, annexin V assay, and transwell invasion assays, respectively.
qRT-PCR results determined that the relative expression level of miR-92b in miR-92b I–treated SNB19 cells was 21.23% ± 0.98% (P < .05) and 16.76% ± 0.78% for LN229 cells (P < .05), compared with NC group, respectively (Fig. 2A). After miR-92b was inhibited, the expression of proliferation- and invasion-related proteins, including PCNA, Cyclin D1, Bcl2, and MMP9 was downregulated (Fig. 2B).
Fig. 2.
miR-92b I suppresses glioma cell proliferation and invasion. (A) miR-92b expression was quantified by qRT-PCR analysis. miR-92b inhibitor significantly suppressed miR-92b expression in both SNB19 and LN229 cells (P < .05), relative to the NC. (B) Expression of cell proliferation and invasion-related molecules was analyzed by Western blot 48 h after transfection with miR-92b inhibitor and NC. (C and D) Representative cartogram showing the proliferation, cell cycle, apoptosis, and invasion regulated by miR-92b inhibitor or NC in glioma cells. *P < .05.
The MTT results (Fig. 2C) revealed that miR-92b I–treated cells showed a significant decrease in viability, than NC group cells (P < .05, from day 2 to 5). Suppression of miR-92b also induced cell cycle arrest and increased the G0/G1 fraction from 50.1% ± 2.4% to 64.5% ± 2.1% in miR-92b I–treated SNB19 cells and 48.7% ± 2.0% to 63.8% ± 2.8% in miR-92b I–treated LN229 cells (P < .05) (Fig. 2D). Treatment with miR-92b I resulted in a decrease in the population of cells that were in S phase. It was shown in a representative experiment in which 27.3% ± 2.2% of NC SNB19 cells were in S phase, whereas miR-92b I–treated cultures had 18.2% ± 3.2% S-phase cells. Similarly, in LN229 cells, miR-92b I caused a decrease in S phase from 31.3% ± 3.1% of NC to 21.8% ± 3.5% of miR-92b I group.
miR-92b I also inhibited glioma cell survival. As shown in Fig. 2E, compared with NC group (2.67% ± 0.32% and 3.04% ± 0.62%) in SNB19 and LN229 cells, the downregulation of miR-92b caused a significant (P < .05) increase in (16.89% ± 1.63% and 20.18% ± 2.36%) apoptotic death, showing an induction of apoptosis in the cells transfected with the miR-92b I.
To test whether miR-92b regulates tumor cell migration, wound scraping assay was performed. Approximately 12 h after wound scraping of a confluent monolayer, nearly full wound closure was observed in the NC cells. In contrast, wounds scraped in confluent cultures of miR-92b I–transfected cells closed only partially (Fig. 2F). Similarly, transwell assays showed that miR-92b I markedly inhibited transwell invasion of glioma cells (P < .05) (Fig. 2G). The number of cells invading through the matrigel in the miR-92b I group was (21.2 ± 2.1) significantly decreased from those of the NC group (110.4 ± 10.5) in SNB19 cells. In LN229 cells, invasive activity was also inhibited in the miR-92b I group (16.1 ± 3.3), compared with that of the NC group (92.7 ± 8.6). The above data show that miR-92b inhibitor suppresses several malignancy parameters in human glioma cells.
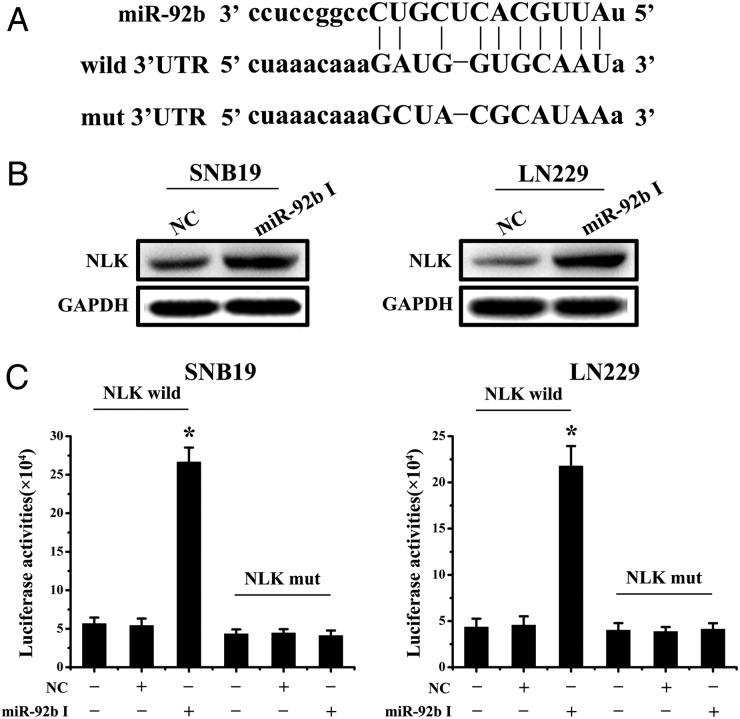
miR-92b Binds to 3′UTR of NLK and Inhibits Wnt/beta-catenin Signaling
On the basis of the Sanger microRNA database, miR-92b has the predicted seed matches in the 3′UTR of NLK (Fig. 3A). Moreover, NLK has been reported to act as a negative regulator of Wnt signaling by interacting with and phosphorylating TCF/LEF-1 family proteins on 2 serine/threonine residues located in the central region recently.17 This phosphorylation by NLK inhibits DNA binding by the beta-catenin-TCF complex. Therefore, we considered that miR-92b may play an important role in regulating the activity of the Wnt/beta-catenin pathway via NLK.
Fig. 3.
miR-92b directly targets NLK. (A) Bioinformatic analysis identifed NLK as a potential target for miR-92b. NLK wild and NLK mut luciferase report plasmids were constructed. (B) Western blotting for NLK at 48 h after transfection of miR-92b inhibitor in SNB19 and LN229 cells. (C) Luciferase reporter assays confirmed that NLK was the direct target of miR-92b in SNB19 and LN229 cells. *P < .05.
To determine whether NLK is regulated by miR-92b, we knocked down miR-92b in SNB19 and LN229 cells and evaluated NLK protein levels at 48 h after transfection. Western blotting analysis showed that miR-92b I obviously increased NLK protein levels in SNB19 and LN229 glioma cells (Fig. 3B). To assess whether NLK is a direct target of miR-92b, we created NLK wild and NLK mut (Fig. 3A), which were cotransfected with miR-92b I or NC, respectively, into glioma cells for 48 h, followed by measurement of luciferase activity in transfected cells. Our results showed that the reporter plasmid with wild-type 3′-UTR of NLK caused a significant increase in luciferase activity in cells transfected with miR-92b I, whereas mut-type 3′-UTR of NLK produced no change in luciferase activity (Fig. 3C). Taken together, these data indicate that miR-92b directly modulates NLK expression by binding to 3′UTR of NLK.
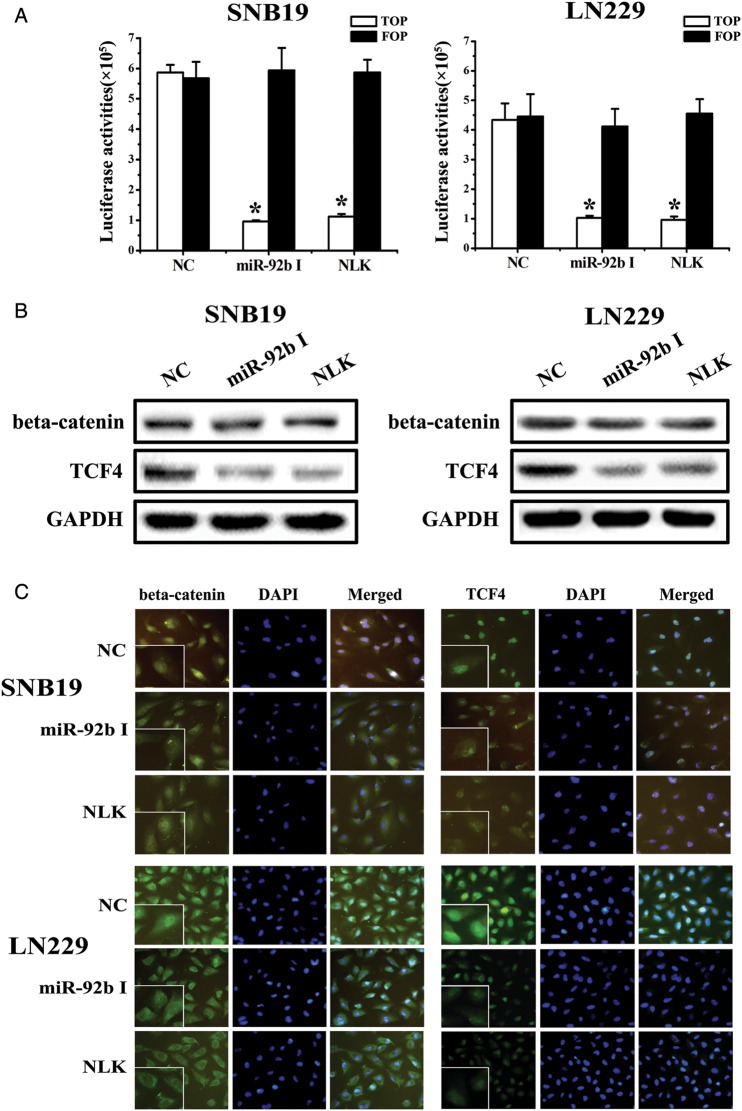
To determine the relationship between the expression of miR-92b/NLK and the activity of the beta-catenin/Wnt pathway, we first used TOPflash and FOPflash reporters, which are widely used to evaluate beta-catenin–dependent signaling activity to evaluate the effects of miR-92b and NLK on Wnt/beta-catenin signaling in SNB19 and LN229 cells. The luciferase activity of the cells changed, as we hypothesized (Fig. 4A). Wnt/beta-catenin signaling was inhibited when the level of miR-92b was down-regulated or NLK was upregulated. We then used Western blot assays to investigate the expression levels of beta-catenin and TCF-4 proteins (Fig. 4B). Fluorescence microscopy of beta-catenin and TCF-4 (Fig. 4C) showed that the location of beta-catenin in cells shifts from nucleus to cytoplasm when the expression of miR-92b decreased or NLK was overexpressed. At the same time, TCF-4 levels decreased in the nucleus. Together, these data show that miR-92b binds to NLK 3′UTR and miR-92b or NLK expression are correlated with Wnt/beta-catenin/TCF4 signaling activity.
Fig. 4.
Effect of miR-92b on the activity the Wnt/beta-catenin signaling pathway. (A) The luciferase reporter assay using TopFlash and FopFlash vectors was used to study beta-catenin TCF/LEF promoter activity. The TopFlash reporter vector has 3-TCF binding sites, and FopFlash has mutated TCF binding sites. SNB19 and LN229 cells were cotransfected with different expression vectors as indicated. MiR-92b inhibitor and NLK treatment decreased beta-catenin TCF/LEF promoter activity. (B) Down-regulation of miR-92b or upregulation of NLK did not affect beta-catenin expression, but decreased TCF-4 expression. (C) Immunofluorescence assay for beta-catenin indicated that the location of beta-catenin in cells shifted from nucleus to cytoplasm when the expression of miR-92b was decreased or NLK was overexpressed and that TCF-4 expression decreased in the nucleus, compared with NC. *P < .05.
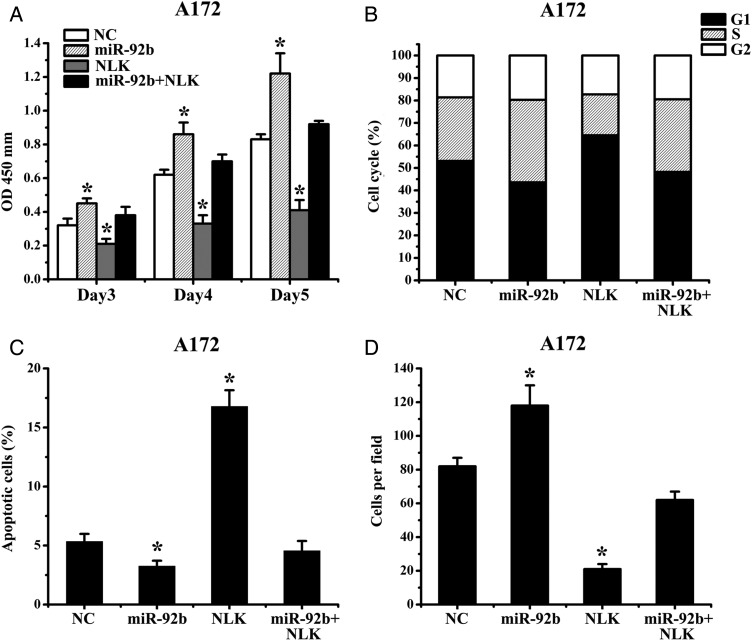
Expression of NLK Overrides miR-92b Survival Function
We next assessed the importance of NLK in miR-92b–mediated cell survival. Because A172 cells had relatively low expression of miR-92b, we transfected miR-92b or NC oligonucleotide 24 h after transfection with NLK-expression plasmid lacking 3′-UTR. The results show that ectopic expression of NLK abrogated miR-92b effects on cell proliferation and invasion (Fig. 5A–D). Together, these data suggest that the effects of miR-92b on the glioma growth are partially mediated by NLK.
Fig. 5.
Expression of NLK abrogates miR-92b survival function. (A) Proliferation of A172 cells after Bim transfection with or without miR-92b was significantly reduced, compared with NC. (B) A172 cells were transfected with NLK (not including the 3′UTR) and miR-92b, and cell proliferation and invasion were measured by MTT, flow cytometry, annexin V, and Transwell assay. *P < .05.
miR-92b Inhibitor Suppresses In Vivo Glioblastoma Xenograft Growth
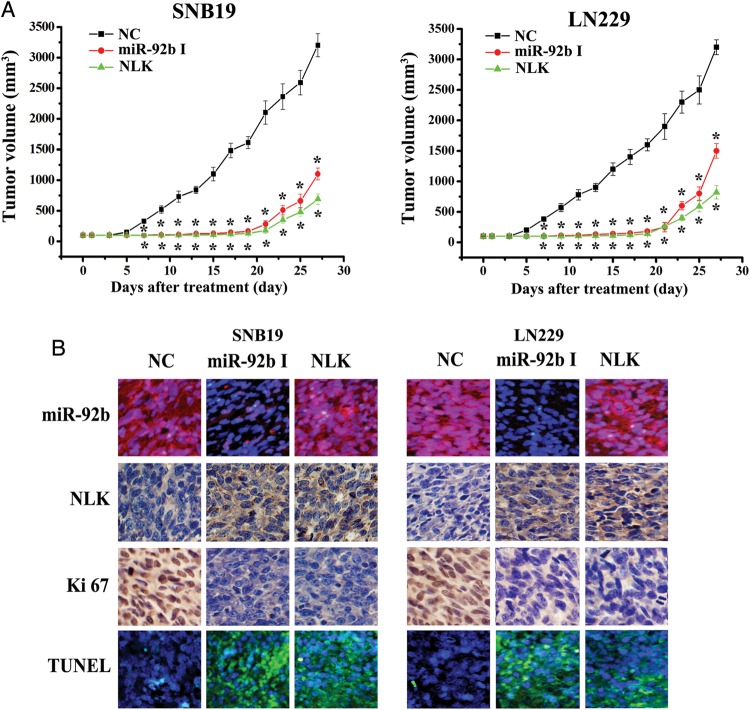
We assessed the effects of miR-92b expression on in vivo glioblastoma xenograft growth. MiR-92b I or NC was transfected into SNB19 or LN229 cells. The cells were subsequently subcutaneously injected to 5-week-old female nude mice, and tumor sizes were measured after 4 weeks (Fig. 6A). As shown in Fig. 6A, miR-92b I or NLK significantly reduced tumor growth, compared with NC group (P < .05).
Fig. 6.
Knock-down of miR-92b inhibits glioma growth in vivo. (A) When subcutaneous tumors were established, miR-92b inhibitor was injected in a multisite injection manner every 2 days for 14 days. Tumor volumes were measured every 2 days during treatment. (B) Fluorescence in situ hybridization showed that miR-92b inhibitor effectively inhibited the expression of miR-92b; NLK and Ki67 expressions were detected by immunohistochemistry assay in xenograft tumor sections; TUNEL assay in xenograft tumor sections revealed that miR-92b inhibitor induced cell apoptosis.
LNA-ISH analysis confirmed that miR-92b levels were considerably reduced in the miR-92b I group (Fig. 6B). Immunohistochemical staining analysis revealed that NLK levels were up-regulated in miR-92b I group (Fig. 6B), confirming the data in vitro that NLK is a direct target of miR-92b. In addition, Ki-67 staining showed that miR-92b I–treated tumors had a lower proliferation index, compared with the NC group (Fig. 6B). TUNEL assay analysis of xenograft tumor taken at 28 days after treatment revealed more apoptosis in the miR-92b I group, when compared with tumors from NC group (Fig. 5B). Therefore, miR-92b inhibitor suppresses the growth of human glioma experimental tumors in vivo.
Inverse Correlation of Expression of miR-92b and NLK in Glioma Samples
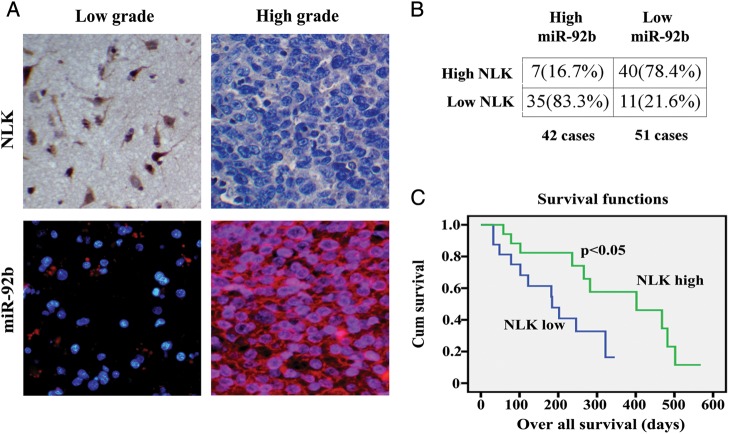
Having shown NLK as a major target of miR-92b, we further investigated the correlation between miR-92b and NLK expression in glioma samples. We examined 93 human glioma specimens with LNA-ISH and immunohistochemical staining. Representative images of miR-92b and NLK are shown in Fig. 7A. Upregulation of miR-92b was detected in 42 gliomas (Fig. 7B). Of the 42 tumors with elevated miR-92b, 35 (83.3%) had low levels of NLK (P < .05). Forty (78.4%) of 51 specimens with downregulated miR-92b presented high levels of NLK. In addition, we found that miR-92b expression increased significantly in high-grade gliomas, compared with low-grade gliomas.
Fig. 7.
NLK expression inversely correlated with miR-92b and low NLK expression correlated with poor survival in GBM. (A) Expression of NLK in resected glioma specimen was assessed by IHC assay. ISH assay showed the expression of miR-92b in the homologous specimens. (B) χ2-test analysis of miR-92b and NLK expressions. The inverse correlation is significant. (C) Kaplan-Meier survival curves indicating cumulative survival as a function of time for those patients with low expression of NLK versus high expression of NLK. The patients with low NLK expression experienced significantly worse outcome.
Furthermore, retrospective analysis of the clinical outcome of our patients revealed that decreased immune detection of NLK correlated with poor survival (Fig. 7C). In sum, these data indicate that miR-92b regulation of NLK expression has significant clinical impact in glioma.
Discussion
Extensive studies have shown that aberrant Wnt/beta-catenin signaling has a key role in the development of glioblastoma,28 including cell proliferation,29 cell apoptosis,30 and cell invasion.31 NLK is an evolutionarily conserved MAP kinase–like kinase that regulates diverse signaling processes via phosphorylation of several transcription factors.15,32–34 The negative regulation of TCF/LEF by NLK has been observed in human embryonic kidney 293 (HEK293) cells and the cervical epithelioid carcinoma cell line HeLa.9,32 In these cell lines, overexpression of NLK inhibits beta-catenin-TCF/LEF complex-mediated transcription via phosphorylation of TCF/LEF.
miR-92 has been involved in the progression of tumorigenesis in several types of tumors. H Al-Nakhle et al35 reported that levels of ERβ1 expression are negatively regulated by the microRNA miR-92 in a cohort of primary breast tumors. Haug36 et al showed that MYCN regulates miR-92 expression, and knockdown of MYCN in MYCN-amplified (MNA) neuroblastoma cell lines increases secretion of endogenous DKK3 to the culture media. Antagomir and miRNA-mimic transfections in neuroblastoma cell lines confirmed that DKK3 secretion to the culture media is regulated by miR-92. Therefore, MYCN-regulated miR-92 inhibits secretion of the tumor suppressor DICKKOPF-3 (DKK3) in neuroblastoma. Patients with multiple myeloma with miR-17, miR-20, and miR-92 high-expression had shorter progression-free survival, compared to those with miR-17, miR-20, and miR-92 low expression.37 Of note, miR-92b was reported previously to be over-expressed in neuronal-specific stem cells and to exhibit dynamic expression patterns in the developing brain.38–41 Thus, it appears that elevated expression of this microRNA is a feature common to brain stem cells and brain tumor cells. Thus far, only one study has investigated the role of miR-92b in glioma.42 Sun et al42 specifically examined a Notch signaling pathway subnetwork. They first constructed a comprehensive glioblastoma-specific miRNA-TF–mediated regulatory network. Then, from the network, they extracted a composite glioblastoma-specific regulatory network. Their topological and functional analyses of the subnetwork revealed that 6 miRNAs (miR-124, miR-137, miR-219–5p, miR-34a, miR-9, and miR-92b) might play important roles in glioblastoma. Therefore, miR-92b has been rarely reported to be involved in the progression of gliomagenesis.
In the Sanger microRNA database, miR-92b is shown to have multiple predicted targets among which are many tumor suppressors. We chose to focus on NLK and Wnt/beta-catenin pathways because they are dysregulated in glioma and because it is the subject of intensive research in our laboratory. NLK is frequently downregulated in gliomas, but the cause of its downregulation is not well understood.26 Our study provides several new findings on this microRNA. In our study, we further confirmed overexpression of miR-92b both in glioma tissues and cell lines. We found, for the first time to our knowledge, that miR-92b inhibits NLK expression in glioma cells via binding to the 3′UTRs. We showed, for the first time to our knowledge, that downregulation of miR-92b expression in glioma cells strongly inhibits cell proliferation, cell cycle progression, cell survival, cell invasion, and in vivo glioma xenograft growth by targeting NLK and further decreases the beta-catenin/TCF4 complex activities. Moreover, we found a significant inverse correlation between miR-92b expression and NLK expression in glioma tissues. Furthermore, we found that decreased immune detection of NLK correlated with poor survival. We therefore present the new mechanism involving miR-92b in human gliomas.
Together, this study provides new insights into the role of miR-92b in human gliomas. It shows that miR-92b is upregulated in gliomas and that miR-92b inhibitor potently inhibits glioma growth by targeting a tumor suppressor-NLK. The study also suggests that miR-92b inhibitor might serve as a glioma therapeutic agent.
Funding
This work was supported by Natural Science Foundation of Zhejiang Province of China (Y2080024).
Acknowledgments
We thank Prof. Pu for her critical review of the manuscript.
Conflict of interest statement. None declared.
References
- 1.Hadjipanayis CG, Van Meir EG. Tumor initiating cells in malignant gliomas: biology and implications for therapy. J Mol Med. 2009;87:363–374. doi: 10.1007/s00109-009-0440-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Purow B, Schiff D. Advances in the genetics of glioblastoma: are we reaching critical mass? Nat Rev Neurol. 2009;5:419–426. doi: 10.1038/nrneurol.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 4.Giles RH, van Es JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta. 2003;1653:1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- 5.Willert K, Jones KA. Wnt signaling: is the party in the nucleus? Genes Dev. 2006;20:1394–1404. doi: 10.1101/gad.1424006. [DOI] [PubMed] [Google Scholar]
- 6.Brott BK, Pinsky BA, Erikson RL. Nlk is a murine protein kinase related to Erk/MAP kinases and localized in the nucleus. Proc Natl Acad Sci USA. 1998;9:963–968. doi: 10.1073/pnas.95.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishitani T, Ninomiya-Tsuji J, Nagai S, et al. The TAK1-NLK-MAPK-related pathway antagonizes signalling between beta-catenin and transcription factor TCF. Nature. 1999;399:798–802. doi: 10.1038/21674. [DOI] [PubMed] [Google Scholar]
- 8.Ishitani T, Kishida S, Hyodo-Miura J, et al. The TAK1-NLK mitogen-activated protein kinase cascade functions in the Wnt-5a/Ca(2+) pathway to antagonize Wnt/beta-catenin signaling. Mol Cell Biol. 2003;23:131–139. doi: 10.1128/MCB.23.1.131-139.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishitani T, Ninomiya-Tsuji J, Matsumoto K. Regulation of lymphoid enhancer factor 1/T-cell factor by mitogen-activated protein kinase-related Nemo-like kinase-dependent phosphorylation in Wnt/beta-catenin signaling. Mol Cell Biol. 2003;23:1379–1389. doi: 10.1128/MCB.23.4.1379-1389.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goold RG, Owen R, Gordon-Weeks PR. Glycogen synthase kinase 3b phosphorylation of microtubule-associated protein 1B regulates the stability of microtubules in growth cones. J Cell Sci. 1999;112:3373–3384. doi: 10.1242/jcs.112.19.3373. [DOI] [PubMed] [Google Scholar]
- 11.Gould TD, Zarate CA, Manji HK. Glycogen synthase kinase-3: a target for novel bipolar disorder treatments. J Clin Psychiatry. 2004;65:10–21. [PubMed] [Google Scholar]
- 12.Terry S, Yang X, Chen MW, et al. Multifaceted interaction between the androgen and Wnt signaling pathways and the implication for prostate cancer. J Cell Biochem. 2006;99(2):402–410. doi: 10.1002/jcb.20983. [DOI] [PubMed] [Google Scholar]
- 13.Kuhl M, Sheldahl LC, Park M, et al. The Wnt/Ca2+ pathway: a new vertebrate Wnt signaling pathway takes shape. Trends Genet. 2000;16:279–283. doi: 10.1016/s0168-9525(00)02028-x. [DOI] [PubMed] [Google Scholar]
- 14.Meneghini MD, Ishitani T, Carter JC, et al. MAP kinase and Wnt pathways converge to downregulate an HMG-domain repressor in Caenorhabditis elegans. Nature. 1999;399:793–797. doi: 10.1038/21666. [DOI] [PubMed] [Google Scholar]
- 15.Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 16.Yasuda J, Tsuchiya A, Yamada T, et al. Nemo-like kinase induces apoptosis in DLD-1 human colon cancer cells. Biochem Biophys Res Commun. 2003;308:227–233. doi: 10.1016/s0006-291x(03)01343-3. [DOI] [PubMed] [Google Scholar]
- 17.Emami KH, Brown LG, Pitts TEM, et al. Nemo-like kinase induces apoptosis and inhibits androgen receptor signaling in prostate cancer cells. Prostate. 2009;69:1481–1492. doi: 10.1002/pros.20998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui G, Li Z, Shao B, et al. Clinical and biological significance of nemo-like kinase expression in glioma. J Clin Neurosci. 2011;18:271–275. doi: 10.1016/j.jocn.2010.05.037. [DOI] [PubMed] [Google Scholar]
- 19.Standart N, Jackson RJ. MicroRNAs repress translation of m7Gppp-capped target mRNAs in vitro by inhibiting initiation and promoting deadenylation. Genes Dev. 2007;21:1975–1982. doi: 10.1101/gad.1591507. [DOI] [PubMed] [Google Scholar]
- 20.de Moor CH, Meijer H, Lissenden S. Mechanisms of translational control by the 3′UTR in development and differentiation. Semin Cell Dev Biol. 2005;16:49–58. doi: 10.1016/j.semcdb.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 21.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 22.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 23.Gwak HS, Kim TH, Jo GH, et al. Silencing of MicroRNA-21 Confers Radio-Sensitivity through Inhibition of the PI3K/AKT Pathway and Enhancing Autophagy in Malignant Glioma Cell Lines. PLoS One. 2012;7:e47449. doi: 10.1371/journal.pone.0047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quintavalle C, Garofalo M, Zanca C, et al. miR-221/222 overexpession in human glioblastoma increases invasiveness by targeting the protein phosphate PTPμ. Oncogene. 2012;31:858–868. doi: 10.1038/onc.2011.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie YK, Huo SF, Zhang G, et al. CDA-2 induces cell differentiation through suppressing Twist/SLUG signaling via miR-124 in glioma. J Neurooncol. 2012;110:179–186. doi: 10.1007/s11060-012-0961-x. [DOI] [PubMed] [Google Scholar]
- 26.Chen L, Li H, Han L, et al. Expression and function of miR-27b in human glioma. Oncol Rep. 2011;26:1617–1621. doi: 10.3892/or.2011.1458. [DOI] [PubMed] [Google Scholar]
- 27.Zhang CZ, Zhang JX, Zhang AL, et al. MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Mol Cancer. 2010;9:229. doi: 10.1186/1476-4598-9-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palos TP, Zheng S, Howard BD. Wnt signaling induces GLT-1 expression in rat C6 glioma cells. J Neurochem. 1999;73:1012–1023. doi: 10.1046/j.1471-4159.1999.0731012.x. [DOI] [PubMed] [Google Scholar]
- 29.Pulvirenti T, Van Der Heijden M, Droms LA, et al. Dishevelled 2 signaling promotes self-renewal and tumorigenicity in human gliomas. Cancer Res. 2011;71:7280–7290. doi: 10.1158/0008-5472.CAN-11-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Satoh J, Kuroda Y. Beta-catenin expression in human neural cell lines following exposure to cytokines and growth factors. Neuropathology. 2000;20:113–123. doi: 10.1046/j.1440-1789.2000.00293.x. [DOI] [PubMed] [Google Scholar]
- 31.Roth W, Wild-Bode C, Platten M, et al. Secreted Frizzled-related proteins inhibit motility and promote growth of human malignant glioma cells. Oncogene. 2000;19:4210–4220. doi: 10.1038/sj.onc.1203783. [DOI] [PubMed] [Google Scholar]
- 32.Ishitani T, Hirao T, Suzuki M, et al. Nemo-like kinase suppresses Notch signalling by interfering with formation of the Notch active transcriptional complex. Nat Cell Biol. 2010;12:278–285. doi: 10.1038/ncb2028. [DOI] [PubMed] [Google Scholar]
- 33.Kanei-Ishii C, Ninomiya-Tsuji J, Tanikawa J, et al. Wnt-1 signal induces phosphorylation and degradation of c-Myb protein via TAK1, HIPK2, and NLK. Genes Dev. 2004;18:816–829. doi: 10.1101/gad.1170604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohkawara B, Shirakabe K, Hyodo-Miura J, et al. Role of the TAK1-NLK-STAT3 pathway in TGF-b-mediated mesoderm induction. Genes Dev. 2004;18:381–386. doi: 10.1101/gad.1166904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Al-Nakhle H, Burns PA, Cummings M, et al. ERβ1 expression is regulated by miR-92 in breast cancer. Cancer Res. 2010;70:4778–4784. doi: 10.1158/0008-5472.CAN-09-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haug BH, Henriksen JR, Buechner J, et al. MYCN-regulated miRNA-92 inhibits secretion of the tumor suppressor DICKKOPF-3 (DKK3) in neuroblastoma. Carcinogenesis. 2011;32:1005–1012. doi: 10.1093/carcin/bgr073. [DOI] [PubMed] [Google Scholar]
- 37.Chen L, Li C, Zhang R, et al. miR-17–92 cluster microRNAs confers tumorigenicity in multiple myeloma. Cancer Lett. 2011;309:62–70. doi: 10.1016/j.canlet.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 38.Kapsimali M, Kloosterman WP, de Bruijn E, et al. MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system. Genome Biol. 2007;8:R173. doi: 10.1186/gb-2007-8-8-r173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krichevsky AM, King KS, Donahue CP, et al. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe T, Takeda A, Mise K, et al. Stage-specific expression of microRNAs during Xenopus development. FEBS Lett. 2005;579:318–324. doi: 10.1016/j.febslet.2004.11.067. [DOI] [PubMed] [Google Scholar]
- 41.Zhao JJ, Hua YJ, Sun DG, et al. Genome-wide microRNA profiling in human fetal nervous tissues by oligonucleotide microarray. Childs Nerv Syst. 2006;22:1419–1425. doi: 10.1007/s00381-006-0173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun J, Gong X, Purow B, et al. Uncovering MicroRNA and Transcription Factor Mediated Regulatory Networks in Glioblastoma. PLoS Comput Biol. 2012;8:e1002488. doi: 10.1371/journal.pcbi.1002488. [DOI] [PMC free article] [PubMed] [Google Scholar]