Abstract
The Tat (twin-arginine translocation) system is a protein targeting pathway utilized by prokaryotes and chloroplasts. Tat substrates are produced with distinctive N-terminal signal peptides and are translocated as fully folded proteins. In Escherichia coli, Tat-dependent proteins often contain redox cofactors that must be loaded before translocation. Trimethylamine N-oxide reductase (TorA) is a model bacterial Tat substrate and is a molybdenum cofactor-dependent enzyme. Co-ordination of cofactor loading and translocation of TorA is directed by the TorD protein, which is a cytoplasmic chaperone known to interact physically with the TorA signal peptide. In the present study, a pre-export TorAD complex has been characterized using biochemical and biophysical techniques, including SAXS (small-angle X-ray scattering). A stable, cofactor-free TorAD complex was isolated, which revealed a 1:1 binding stoichiometry. Surprisingly, a TorAD complex with similar architecture can be isolated in the complete absence of the 39-residue TorA signal peptide. The present study demonstrates that two high-affinity binding sites for TorD are present on TorA, and that a single TorD protein binds both of those simultaneously. Further characterization suggested that the C-terminal ‘Domain IV’ of TorA remained solvent-exposed in the cofactor-free pre-export TorAD complex. It is possible that correct folding of Domain IV upon cofactor loading is the trigger for TorD release and subsequent export of TorA.
Keywords: bacterial protein transport, Escherichia coli, molybdoenzyme, twin-arginine translocation protein transport pathway (Tat protein transport pathway), twin-arginine translocation proofreading chaperone (Tat proofreading chaperone)
Abbreviations: DTT, dithiothreitol; ICP-MS, inductively coupled plasma–MS; IMAC, immobilized metal-ion-affinity chromatography; LB, Luria–Bertani; MALLS, multi-angle laser light scattering; MGD, molybdopterin guanine dinucleotide; SAXS, small-angle X-ray scattering; SEC, size-exclusion chromatography; SPA, sequential peptide affinity; Tat, twin-arginine translocation; TMAO, trimethylamine N-oxide
INTRODUCTION
The Tat (twin-arginine translocation) protein transport pathway exports proteins across the cytoplasmic membranes of bacteria and archaea, and the thylakoid membranes of plant chloroplasts [1,2]. Protein substrates of the Tat pathway are folded before translocation and are targeted to the Tat machinery by cleavable N-terminal signal peptides containing an almost invariant pair of arginine residues [3]. Transport by the Tat pathway is energized solely by the protonmotive force [1,2].
The model bacterium Escherichia coli K-12 produces 28 proteins bearing twin-arginine signal peptides. Approximately two-thirds of these proteins are known or predicted to contain redox cofactors and some of them also form complexes with partner subunits that lack a signal peptide before export [4]. The complexity of these Tat substrates necessitates a pre-export ‘proofreading’ process to ensure that only folded and assembled proteins are presented for translocation. One hypothesis involves dedicated chaperone proteins that interact directly with twin-arginine signal peptides to prevent premature targeting of the substrate before cofactor insertion and binding of any protein partners [5].
The E. coli TMAO (trimethylamine N-oxide) reductase TorA is a soluble periplasmic enzyme containing the MGD (molybdopterin guanine dinucleotide) cofactor at its active site [6]. TorA is a well-characterized substrate of the Tat pathway; indeed, transport of TorA to the periplasm was found to be blocked in a mutant strain that was unable to synthesize MGD, which led to the original hypothesis that a transport pathway for folded proteins may exist in bacteria [7]. The TorA protein is encoded by the torCAD operon (Figure 1A) where TorC is a haem-containing quinol oxidase and TorD is a cytoplasmic protein [8]. Cellular levels of enzymatically active TorA are significantly reduced in a strain lacking the torD gene [9], and the TorD protein was found subsequently to interact with an unfolded (heat-denatured) form of the mature TorA enzyme [9]. This initial work therefore established that there was a clear binding site for TorD within the mature region of the protein and suggested a role for TorD in loading the MGD cofactor into the TorA apoenzyme [9,10]. In addition to this, a subsequent genetic screen identified TorD as a binding partner for the TorA twin-arginine signal peptide [11]. This was confirmed in vitro when recombinant TorD was shown to bind directly to the isolated TorA signal peptide with a 1:1 stoichiometry and an apparent Kd of ~59 nM [12,13]. The binding epitope for TorD has been mapped to the C-terminal end of the TorA signal peptide, close to its junction with the mature portion of the enzyme [12,14]. Interestingly, binding of the TorA signal peptide by TorD seems to be an activity not strictly connected with cofactor loading, since co-expression of torD can enhance the export of synthetic signal peptide fusion proteins on the Tat pathway [11,15]. The TorD protein itself is 22.5 kDa and is known to exist in equilibrium between monomeric and dimeric forms [16–18], have a low level of GTPase activity associated with the dimeric form [18], and have the ability to bind the MGD cofactor in vitro [19]. However, although it is clear that the monomeric form of TorD can bind the isolated TorA signal peptide [12,13], the physiological role and significance of TorD dimerization is not clear.
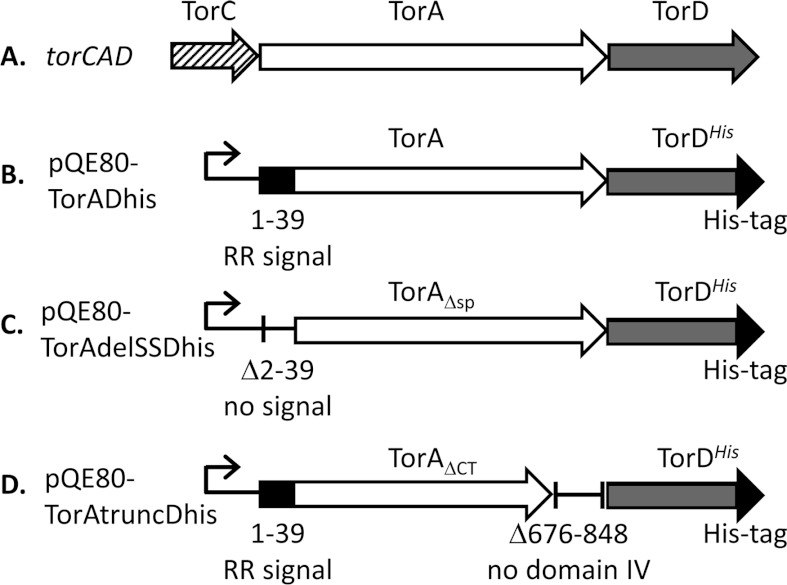
Figure 1. Tools for the isolation of TorAD complexes.
(A) A cartoon representing the structure of the torCAD operon located at 22.8 min on the E. coli chromosome. The names of the protein products of the genes are given above the arrows. (B) An overexpression vector based on pQE80 (Qiagen) encoding full-length TorA and TorDHis. The natural transcriptional and translational coupling between torA and torD is maintained. (C) An expression vector encoding TorA lacking its entire Tat signal peptide comprising Asn2–Ala39 (TorAΔsp, TorA Δ2–39) and TorDHis. (D) An expression vector encoding TorA lacking its entire C-terminal Domain IV comprising Ile676–Ser848 (TorAΔCT, TorA Δ676–848) and TorDHis.
Current models for TorA biosynthesis assume that TorD interacts with the TorA precursor by binding at two distinct sites: one being within the twin-arginine signal peptide with the other, uncharacterized, site lying elsewhere in the mature portion of the protein [5,20]. These two binding events are thought to not only delay the Tat transport event, but also actively facilitate efficient insertion of the MGD cofactor into the apoenzyme. This pre-export ‘Tat proofreading’ process would be completed by the controlled release of the bound chaperones and subsequent translocation of the folded active enzyme to the periplasm. However, it is not known whether each of the TorD-binding sites on the TorA apoenzyme are bound by separate TorD monomers or a single TorD dimer, or whether the TorD monomer itself contains two distinct TorA-binding sites. Moreover, the degree of folding of the TorA precursor when bound by TorD has not been explored, and it remains a mystery how release of the chaperones is ultimately triggered.
In the present study, a stable and cofactor-free complex of TorD with the TorA apoenzyme has been isolated, both with and without the twin-arginine signal peptide. Using a combination of biophysical and biochemical approaches, we have demonstrated that a single TorD monomer binds directly to a single TorA apoprotein regardless of the presence of the signal peptide. This suggests that a single TorD monomer recognizes two binding sites on the TorA polypeptide and that these epitopes are nearby or overlapping. Finally, we have demonstrated that binding of TorA by TorD changes the conformation of apo-TorA in relation to the mature active enzyme by inducing exposure of the C-terminal domain of the TorA protein.
EXPERIMENTAL
Plasmid and strain construction
The torAD genes form a transcription unit on the E. coli chromosome with overlapping stop and start codons. To co-overproduce TorA and TorDHis, this natural translational coupling was retained. DNA encoding SPA (sequential peptide affinity)-tagged TorD along with TorA was amplified from the chromosome of E. coli strain DY330 torD-SPA [21,22] using oligonucleotides TorAMfeIup (5′-GCGCCAATTGCGATAAGAAGGAAGAAAAATAATG-3′) and SPASphIdown (5′-GCGCGCATGCCTACTTGTCATCGTCATCC-3′). The resulting PCR product was digested with MfeI and SphI and cloned into pQE80 digested previously with EcoRI and SphI. To replace DNA encoding the SPA-tagged variant of TorD with a His-tagged variant, the pQE80torADSPA plasmid was digested with NheI (which removes DNA covering the 3′-end of torD, the SPA-tag-coding sequence and a small region of the vector). Plasmid pQE60torD [13] which codes for a C-terminally His-tagged TorD was also digested with NheI and the fragment covering the 3′-end of torD along with the His-tag-coding sequence was ligated into the digested pQE80torADSPA vector. Resultant clones were analysed by sequencing to ensure that the NheI fragment had been cloned into the correct orientation, and the construct was designated pQE80TorADhis (Figure 1B).
To overproduce TorDHis along with TorA lacking its N-terminal signal peptide, the encoding DNA was amplified using oligonucleotides TorAMfeIdelSS (5′-GCGCCAATTGCGATAAGAAGGAAGAAAAATAATGGCGCAAGCGGCGACTGACGCTGTCATCTCG-3′) and QEreverse (5′-GTTCTGAGGTCATTACTGGAT-3′), with pQE80 TorADhis as a template. The resultant PCR product was digested with MfeI and HindIII and cloned into pQE80 that had been pre-digested with EcoRI and HindIII, to give construct pQE80TorAdelSSDhis (Figure 1C).
To produce a C-terminally truncated TorA along with TorDHis, a fragment of torA terminating at codon 675 and followed by a stop codon was amplified with oligonucleotides TorAfor (5′-GACCTCAACCTGCCGCTTTGC-3′) and TorA675STOPHindIII (5′-GCGCAAGCTTATTTCTCAAACCACATCGGATGCCC-3′) and digested with NruI (the recognition site for which is present in the torA gene) and HindIII. A second PCR product covering the torD ribosome-binding site and a fragment of the torD gene was amplified with oligonucleotides TorDHindRBS (5′-GCGCAAGCTTGCAGGTGAAATCATGACCACG-3′) and TorDrev (5′-GGTCATCAGAAACAGGCCGC-3′) and digested with HindIII and SacII (the recognition site for SacII occurs naturally in torD). A three-way ligation was carried out with these two fragments and pQE80TorADhis that had been digested previously with NruI and SacII. The resultant clone was designated pQE80TorAtruncDhis (Figure 1D).
Strain FTH007 encodes a C-terminally hexahistidine-tagged TorA produced from the native chromosomal location and was constructed as follows. A 582 bp fragment of torA DNA finishing immediately before the torA stop codon was amplified with oligonucleotides TorAHis1 (5′-GCGCCCATGGCTCGAAAACTATCGCCGATATGAAC-3′) and TorAHis2 (5′-GCGCAGATCTTGATTTCACCTGCGACGCGGGAAC-3′), digested with NcoI and BglII and cloned into similarly digested pFAT210 [23]. DNA covering the torD ribosome-binding site and approximately 600 nucleotides of downstream DNA was amplified with oligonucleotides TorAHis3 (5′-GCGCAAGCTTTGTTCCCGCGTCGCAGGTGAAATC-3′) and TorAHis4 (5′-GCGCTCTAGAGACGTTATCTGTTTTGGTGGTCGC-3′), digested with XbaI and HindIII and cloned into the above construct. DNA covering the torAhis allele was subsequently excised by digestion with XbaI and KpnI and cloned into similarly digested pMAK705 [24]. The torAhis allele was then moved on to the chromosome of strain DSS401 (as MC4100, ΔdmsABC::KanR) [25] to give strain FTH007.
All constructs were fully sequenced to ensure that no mistakes had been introduced inadvertently during the cloning procedure.
TMAO reductase assays
TMAO reductase assays were performed using strain GB426 (as MC4100, ΔtorCAD::AprR, ΔdmsABC::KanR [22]), which contained either pQE80, pQE80TorADhis, pQE80TorAdelSSDhis or pQE80TorAtruncDhis. Cells were cultured anaerobically overnight in LB (Luria–Bertani) medium supplemented with 0.5% glycerol and 0.4% TMAO, along with the antibiotics apramycin (50 μg/ml), kanamycin (25 μg/ml) and ampicillin (100 μg/ml). Cells were harvested, washed and lysed by passage through a French press pressure cell as described previously [26]. After removal of unbroken cells and debris by a short centrifugation step, the resultant crude cell extract was assayed for TMAO reductase activity essentially as described previously [6,27]. Protein concentration was determined using the Lowry assay [28].
Preparation of recombinant proteins
All plasmid-encoded TorA/TorDHis proteins were overproduced and purified in an identical manner. A single colony of freshly transformed E. coli BL21(DE3) [F− ompT hsdS(rB− mB−) gal dcm λ(DE3)] was cultured overnight in 5 ml of LB medium supplemented with 100 μg/ml ampicillin. This provided the inoculum for a 500 ml culture, which was grown aerobically at 37°C and at 140 rev./min in a baffled 2 litre flask to a D600 of 0.6 before induction of recombinant protein production by addition of 2 mM IPTG (isopropyl β-d-thiogalactopyranoside), followed by a temperature shift to 18°C. After 16 h, cells were harvested and resuspended (10 ml per 1 g of cells) in buffer A [50 mM Tris/HCl (pH 7.5), 200 mM KCl, 1 mM DTT (dithiothreitol) and 25 mM imidazole]. Protease inhibitor [EDTA-free Complete™ protease inhibitor cocktail (Roche)], lysozyme and DNase were added to the resuspension and cells were lysed using an Emulsiflex C3 high-pressure homogenizer. Cell debris was removed by centrifugation at 18850 g for 15 min, followed by removal of membranes by ultracentifugation at 45000 rev./min for 1 h using a Beckman Ti70 rotor. All centrifugation steps were conducted at 4°C. The supernatant obtained following ultracentrifugation was filtered through a 0.22-μm-pore-size membrane filter (Millipore) before being subjected to IMAC (immobilized metal-ion-affinity chromatography) as follows. The extract was loaded on to a 5 ml HisTrap column (GE Healthcare) equilibrated with buffer A. After extensive washing with buffer A, hexahistidine-tagged proteins were subsequently eluted using a gradient of 25–500 mM imidazole in buffer A. TorA/TorDHis-containing fractions were identified by SDS/PAGE (12% gels), pooled and concentrated to 2 ml using a Vivaspin 20 spin concentrator (10 kDa cut-off, Sartorius). The concentrated sample was then loaded on to a calibrated HiLoad 16/60 Superdex 200 Prep Grade (GE Healthcare) size-exclusion column equilibrated with buffer A (50 mM Tris/HCl, pH 7.5, 200 mM KCl and 1 mM DTT), with TorA/TorDHis-containing fractions again identified by SDS/PAGE (12% gels).
Chromosomally encoded TorAHis was purified from cells that had been cultured anaerobically in 5 litre capped bottles that had been filled with LB medium supplemented with 0.4% TMAO, 0.5% glycerol and 50 μg/ml kanamycin. After inoculation with a 5 ml pre-culture, bottles were incubated for 16 h without shaking at 37°C. Cells were harvested, washed and resuspended to a final concentration of 10 ml/g of cells in 50 mM Tris/HCl (pH 7.5) and 40% (w/v) sucrose. The suspension was then supplemented with EDTA (5 mM final concentration) and 0.6 mg/ml lysozyme and incubated for 30 min at 37°C to release the periplasmic fraction. Sphaeroplasts were removed by centrifugation at 12500 rev./min for 20 min using a Beckman JA-25.50 rotor at 4°C and the supernatant was retained. The EDTA was removed by dialysis in buffer A before loading the supernatant on to a 5 ml HisTrap column and purifying as above.
SAXS (small-angle X-ray scattering)
SAXS data using purified TorDHis, TorA/TorDHis and TorAΔSP/TorDHis samples were recorded on the ID14-3 BioSAXS beamline at the European Synchrotron Radiation Facility (ESRF Grenoble, France). Sample-detector distance was 2.43 m for an X-ray wavelength of 0.931 Å (1 Å=0.1 nm). Then, 50 μl sample and buffer volumes were loaded in a flowthrough quartz capillary cell at 25°C. The sample volume exposed to the X-ray beam was approximately 10 μl. Samples were checked for radiation damage by using ten successive exposure times of 10 s each. Final exposure time was 100 s for all samples/buffers. The two-dimensional diffraction patterns were normalized to an absolute scale and azimuthally averaged to obtain the intensity profiles I(Q), within BSxCuBE (ESRF beamline data collection software). Solvent contributions (buffer backgrounds collected before and after every protein sample) were averaged and subtracted from the associated protein sample using the program PRIMUS [29].
CD analysis
CD analysis of protein samples was provided as a service by the Scottish Circular Dichroism Facility, University of Glasgow. Spectra of protein solutions were obtained using a JASCO J-810 spectropolarimeter. Far-UV CD measurements (185–260 nm) were collected in quartz cells of 0.02 cm pathlength at 25°C with a scan speed of 10 nm/min, bandwidth of 1 nm, response of 2 s and data pitch of 0.2 nm. Before collection of CD data, proteins were buffer-exchanged into CD-appropriate buffer (50 mM Tris/H2SO4, pH 7.5, 200 mM K2SO4 and 1 mM DTT). Protein concentrations were estimated using a NanoDrop spectrophotometer before the experiment. Near-UV CD measurements (250–320 nm) were obtained in a 0.2-cm-pathlength quartz cuvette using the following parameters: temperature, 25°C; scan speed, 10 nm/min; bandwidth, 1 nm; response, 2.0 s; and data pitch, 0.2 nm. Protein was analysed in appropriate buffer (50 mM Tris/HCl, pH 7.5, 200 mM KCl and 1 mM DTT). The spectra were corrected for protein concentration and cell pathlength before being analysed by DichroWeb (http://dichroweb.cryst.bbk.ac.uk), an online server which hosts the various algorithms used to estimate protein secondary structures.
Metal analysis
Metal content was analysed by ICP-AES (inductively coupled plasma–atomic emission spectrometry)/ICP-MS (inductively coupled plasma–MS) and was provided as a service by the School of Chemistry at the University of Edinburgh.
Limited trypsinolysis of TorA–TorDHis and TorAΔSP–TorDHis complexes
Protein samples (1 mg/ml in 50 mM Tris/HCl, pH 7.5, 200 mM KCl and 1 mM DTT; 100 μl final volume) were incubated with 1% (w/w) trypsin (porcine pancreas, proteomics grade, Sigma–Aldrich) at 37°C. Aliquots of 5 μl were withdrawn at various intervals and the reaction stopped by addition of 20 μl of Laemmli buffer (Bio-Rad Laboratories) followed by immediate boiling for 10 min. Once the time course was completed, samples were analysed by SDS/PAGE (12% gels).
Proteins were identified by tryptic peptide mass fingerprinting of gel slices performed by the FingerPrints Proteomic Service (College of Life Sciences, University of Dundee).
Analysis of TorA–TorDHis complexes by SEC (size-exclusion chromatography)–MALLS (multi-angle laser light scattering)
Size estimates of TorA–TorDHis complexes were obtained by SEC–MALLS, which was provided as a service by the University of York Bioscience Technology Facility. Measurements were conducted on a system comprising a Wyatt HELEOS-II multi-angle light-scattering detector and a Wyatt rEX refractive index detector linked to a Shimadzu HPLC system (SPD-20A UV detector, LC20-AD isocratic pump system, DGU-20A3 degasser and SIL-20A autosampler). Work was conducted at room temperature (20±2°C). Solvent was filtered through a 0.2-μm-pore-size filter before use and a further 0.1-μm-pore-size filter was present in the flow path. Filtered (0.2 μm pore size) TorA–TorDHis samples (100 μl, corresponding to 100 μg of protein) were applied to a Superdex 200 10/300 GL column (GE Healthcare) equilibrated with 50 mM Tris/HCl (pH 7.5), 200 mM KCl and 1 mM DTT at 0.5 ml/min; Shimadzu LC Solutions software was used to control the HPLC, and Astra V software was used for the HELEOS-II and rEX detectors. BSA was used for molecular mass calibration. The Astra data collection was 1 min shorter than the LC solutions run to maintain synchronization. Blank buffer injections were used as appropriate to check for carry-over between sample runs. Data were analysed using the Astra V software. MWs were estimated using the Zimm fit method [30] with degree 1. A value of 0.19 was used for protein refractive index increment (dn/dc).
RESULTS
Isolation of a TorA–TorDHis complex
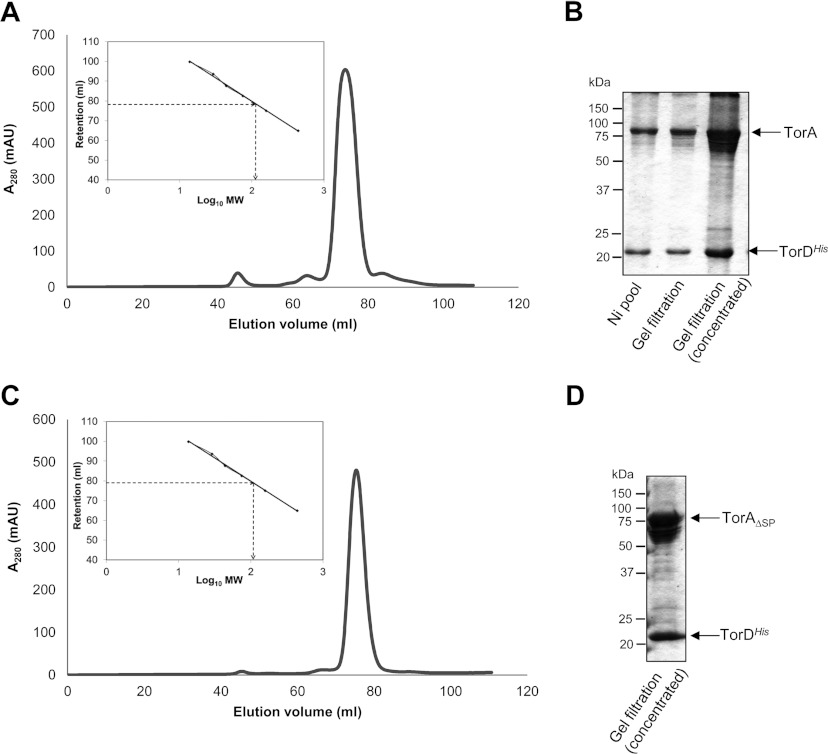
Isolation of a complex between untagged TorA and affinity-tagged TorD was originally demonstrated on a small scale using a C-terminally SPA-tagged form of TorD [22]. In order to scale up the co-expression system to allow detailed analysis of the complex, the original torADSPA allele was cloned into expression vector pQE80 and the DNA coding for the SPA tag at the C-terminus of TorD replaced with a hexahistidine tag (Figure 1B). The soluble cell extract containing overproduced TorA and TorDHis was subjected to IMAC and fractions containing proteins of the expected size of TorA and TorDHis were identified in a single peak at 110 mM imidazole. The IMAC samples were pooled, concentrated and subjected to SEC using a HiLoad 16/60 Superdex 200 Prep Grade column. As shown in Figure 2(A), protein was eluted at 78.5 ml, corresponding to an estimated molecular mass of 114 kDa. Analysis of the peak fraction by SDS/PAGE revealed the presence of two major proteins (Figure 2B), which were confirmed by tryptic peptide mass fingerprinting to be TorA and TorDHis. The overall protein yield from this process was approximately 1.5 mg/g of bacterial cells.
Figure 2. Isolation of a TorA–TorDHis and TorAΔSP–TorDHis complex.
(A and C) TorDHis-containing fractions after metal chelate chromatography of cell extracts overproducing (A) TorA and TorDHis or (C) TorAΔSP and TorDHis were pooled, concentrated and applied to a HiLoad 16/60 Superdex 200 Prep Grade size-exclusion column. Eluted protein was monitored by measuring absorbance at 280 nm. The column was calibrated with the standard proteins ribonuclease (14 kDa), carbonic anhydrase (29 kDa), ovalbumin (44 kDa), conalbumin (75 kDa), aldolase (158 kDa) and ferritin (440 kDa), and the linear regression analysis is shown as inset boxes. R2=0.9976, y=−23.5x+126.83. MW, molecular mass. (B and D) SDS/PAGE analysis (12% gels) of (B) the concentrated fractions after metal chelate chromatography (Ni pool), and the non-concentrated and concentrated peak fractions from SEC of the TorA–TorDHis complex and (D) the concentrated peak fraction from SEC of the TorAΔSP–TorDHis complex. Molecular masses are indicated in kDa.
It was notable during the purification of TorADHis, particularly following concentration of the samples, that more than one electrophoretic form of TorA was evident by SDS/PAGE. A low-percentage acrylamide gel was therefore used to separate these forms, which were excised and analysed by tryptic peptide mass fingerprinting. The analysis showed that, whereas the majority of the TorA was present as full-length intact polypeptide, there was a proportion of TorA that had undergone limited proteolysis and was lacking up to 150 amino acids from its C-terminal end (Supplementary Figure S1 at http://www.biochemj.org/bj/452/bj4520057add.htm). In addition, in some instances, up to 12 amino acid residues could be lost from the N-terminus of TorA. It is concluded that, whereas a complex of TorADHis is stable to purification, the extremities of TorA can be subjected to variable levels of proteolysis during isolation.
Removal of the entire Tat signal peptide from the TorA protein (TorA Δ2–39) still allows isolation of a stable complex with TorDHis
As well as signal peptide binding, previous biochemical and genetic experiments pointed to a binding site for TorD within the mature region of the TorA enzyme (e.g. [9,10]). To investigate this further, a vector was designed that would co-overproduce a version of TorA lacking its entire 39-residue Tat signal peptide (TorAΔSP) with TorDHis (Figure 1C). When TorDHis was isolated from the soluble cell fraction by IMAC, a protein of the expected size of TorAΔSP was also co-purified. The TorDHis-containing fractions were pooled, concentrated and applied to a Superdex 200 size-exclusion column. The TorDHis eluted at 78.8 ml, corresponding to an estimated molecular mass of 110 kDa (Figure 2C) and analysis of the concentrated peak fraction by SDS/PAGE revealed the presence of two major proteins (Figure 2D), which were confirmed by tryptic peptide mass fingerprinting to be TorAΔSP and TorDHis. Thus it is clear that a stable complex of TorDHis and TorA can be isolated in the absence of the twin-arginine signal peptide.
TorDHis binds the full-length TorA precursor, and the TorAΔsp species lacking its entire signal peptide (Δ2–39), at a 1:1 stoichiometry
SEC indicates that the complex of TorDHis with TorA elutes at a similar molecular mass regardless of whether the signal peptide was present on TorA (Figure 2). A more accurate technique is SEC–MALLS, which uses HPLC-linked SEC coupled with static laser light scattering to provide a direct measure of molecular mass. Using this technique, the TorA–TorDHis complex was observed to exhibit a mass range of 100–120 kDa and a peak mass of 114.7 kDa (Table 1), which is very close to the calculated mass (117.9 kDa) for a 1:1 complex of the two proteins (Table 1). The TorAΔSP–TorDHis complex exhibited a mass range of 95–115 kDa and a peak mass of 108.2 kDa (Table 1), which is close to the calculated theoretical mass (113.8 kDa) for a 1:1 complex of the two proteins (Table 1). The relatively wide mass range seen for both samples reflects the observed proteolytic events leading to heterogeneity of the TorA sample observed by SDS/PAGE (Figure 2). Overall, the SEC–MALLS analyses point to complexes containing one TorA protein and one TorD protein, and suggest strongly that TorD can remain stably bound to TorA in the absence of the twin-arginine signal peptide.
Table 1. The apparent molecular masses of TorAD complexes.
Parameters obtained from SEC–MALLS analysis of each of the indicated complexes.
| Sample | Major peak elution range (min) | Molecular mass at peak (kDa) | Range across peak (kDa) | Predicted molecular mass of a 1:1 complex (kDa) |
|---|---|---|---|---|
| TorA–TorDHis | 26.4–30.4 | 114.7 | 100–120 | 117.9 |
| TorAΔSP–TorDHis | 26.3–30.0 | 108.2 | 95–115 | 113.8 |
| TorAΔCT–TorDHis | 27.6–30.2 | 99.7 | 97–107 | 99.2 |
The TorA C-terminal domain is protease-accessible in TorAD complexes
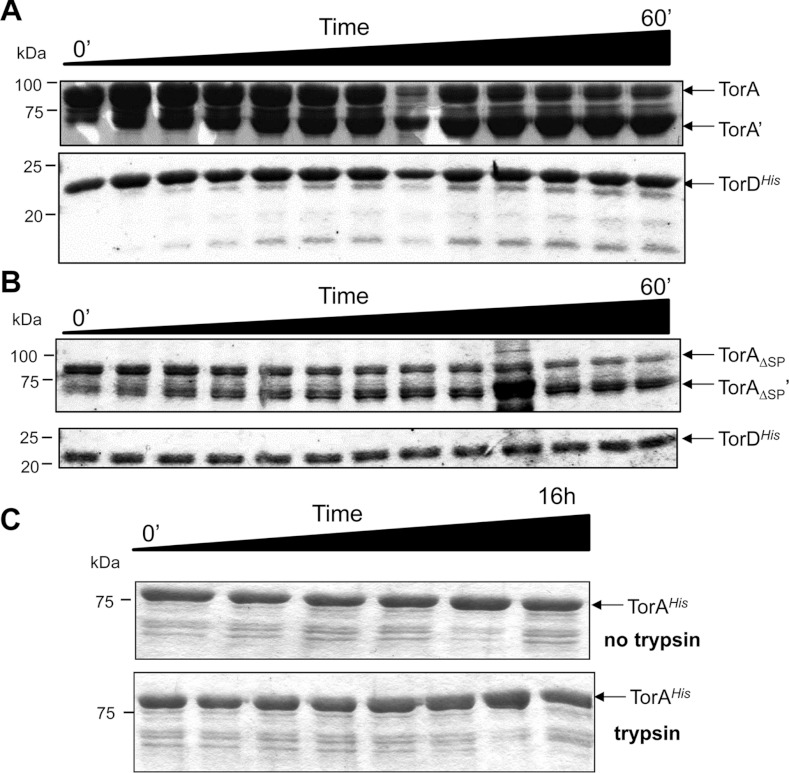
To gain more insight into the architecture of the TorAD complexes under investigation in the present study, limited proteolysis experiments were undertaken. First, the purified TorA–TorDHis complex was incubated with a low concentration of trypsin over a time course of 60 min. As shown in Figure 3(A), TorDHis was almost completely resistant to trypsinolysis under these conditions. In contrast, TorA was cleaved to yield a stable proteolytic fragment of approximately 70 kDa that was apparently resistant to further digestion by trypsin. Peptide mass fingerprinting of the stable 70 kDa TorA proteolysis product revealed that the initial 675 amino acid residues from the N-terminus of the protein were intact and that the trypsin treatment had therefore removed a short section of polypeptide from the C-terminal end of TorA (Supplementary Figure S1).
Figure 3. Limited trypsinolysis of the TorA–TorDHis and TorAΔSP–TorDHis complexes.
The TorA–TorDHis (A) and TorAΔSP–TorDHis complexes (B) were isolated. Each purified complex was incubated with trypsin and 5 μl aliquots removed at 5 min intervals between 0 and 60 min, mixed with Laemmli buffer and immediately boiled to prevent any further digestion. (C) As a control, periplasmic mature TorAHis was purified and incubated with trypsin before 5 μl aliquots were removed at the following eight time points (0 min, 10 min, 20 min, 30 min, 1 h, 2 h, 4 h and 16 h). Concomitantly, mature TorAHis incubated without trypsin before aliquots were removed at the following six time points (0 min, 10 min, 30 min, 1 h, 4 h and 16 h). Samples were mixed with Laemmli buffer and immediately boiled. At the end of the time courses, the samples were analysed by SDS/PAGE (12% gels). Molecular masses are indicated in kDa.
Next, the TorAΔSP–TorDHis complex, where TorA had already had its 39-residue signal peptide deliberately removed, was incubated with trypsin in a similar manner. As shown in Figure 3(B), TorDHis was almost completely resistant to protease digestion, whereas the TorAΔSP was degraded to a smaller stable product. Analysis of this protease-resistant fragment by tryptic peptide mass fingerprinting showed that it was cleaved at Lys675, which was the identical position where the full-length TorA precursor was cleaved. As a control, a C-terminally His-tagged, but otherwise native, form of TorA was purified from the periplasm of E. coli strain FTH007. The fully folded mature form of TorA was completely resistant to digestion with trypsin, even after a 16-h incubation period (Figure 3C). These results are consistent with the suggestion that the TorA–TorDHis and TorAΔSP–TorDHis complexes have a similar overall architecture, both being susceptible to limited trypsinolysis in an identical manner. In both cases, a C-terminal stretch of immature TorA is obviously exposed to the aqueous phase in a conformation that is clearly not adopted by the mature fully assembled enzyme.
The C-terminal domain of TorA is not involved in stable TorAD complex formation
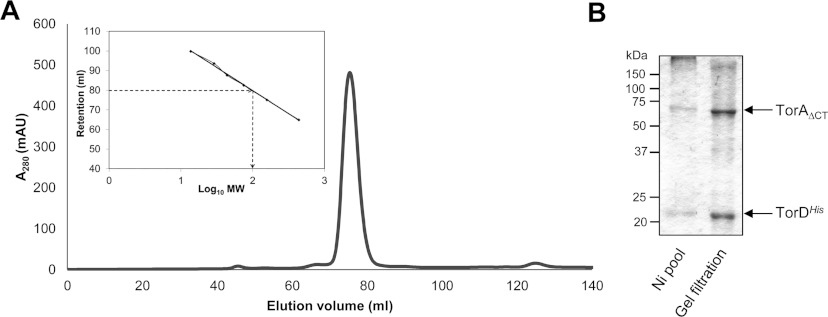
TorA is a member DMSO reductase family of molybdoenzymes. Proteins in this family adopt a fold that can be divided into four structurally distinct domains [31], and, of these, Domain IV is formed by the extreme C-terminal part of the protein and is the only domain in this fold that comprises a clear contiguous stretch of polypeptide chain (Supplementary Figure S2 at http://www.biochemj.org/bj/452/bj4520057add.htm). Although the crystal structure of the E. coli TorA protein has not been solved, structure predictions based on the Shewanella massilia TorA crystal structure [32] suggest the position of the trypsin cleavage site in the E. coli TorA–TorDHis complexes lies within an unstructured loop region that connects Domain III to Domain IV (Supplementary Figure S2). To investigate this further, a construct was designed that would co-overproduce TorDHis together with the first 675 amino acids of TorA, including the signal peptide (designated TorAΔCT). First, the construct was expressed in a strain lacking endogenous TMAO reductase activity under strict anaerobic conditions, which would induce expression of the MGD biosynthetic machinery. Then a crude cell extract was prepared and the TMAO reductase activity assayed. The resultant data showed that the C-terminally truncated form of TorA was completely devoid of enzymatic activity (Supplementary Table S1 at http://www.biochemj.org/bj/452/bj4520057add.htm), establishing that this domain of the protein is absolutely critical for function. Next, TorDHis was isolated from the soluble cell fraction by IMAC, which resulted in a protein of the expected size of TorAΔCT co-purifying (Figure 4). The two proteins remained fully associated during SEC through a Superdex 200 column (Figure 4A), with the complex eluting with an estimated molecular mass of 100 kDa (Figure 4A). Analysis of the peak fractions by SDS/PAGE confirmed that both proteins were present in the sample, but this time very little degradation of the deliberately truncated TorA was apparent (Figure 4B). SEC–MALLS analysis of the purified complex (Table 1) confirmed that the TorAΔCT–TorDHis complex had a narrow mass range of 97–107 kDa, reflecting the relative homogeneity of the sample, and a peak mass of 99.7 kDa, which is close to the calculated mass (99.2 kDa) for a 1:1 complex of the two proteins (Table 1). It can be concluded that, although the C-terminal 173 amino acids are essential for the ultimate enzymatic activity of TorA, they do not influence the formation or stability of the pre-export complex between TorA and TorD.
Figure 4. Domain IV of TorA is not required for complex formation with TorD.
(A) TorDHis-containing fractions after metal chelate chromatography of cell extracts overproducing TorAΔCT and TorDHis were pooled, concentrated and applied to a HiLoad 16/60 Superdex 200 Prep Grade size-exclusion column. Eluted protein was monitored by measuring absorbance at 280 nm. MW, molecular mass. (B) SDS/PAGE analysis (12% gels) of the pooled fractions after metal chelate chromatography (Ni pool), and the non-concentrated peak fraction following gel-filtration chromatography. Molecular masses are indicated in kDa.
The TorAD complexes are devoid of MGD, but retain most of their secondary structure
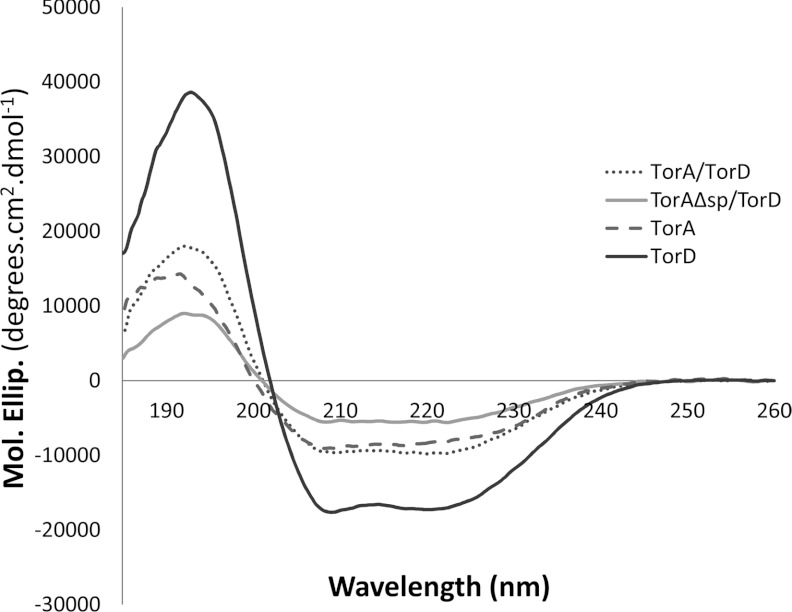
Whereas it has generally been assumed that a Tat substrate protein is maintained in an ‘unfolded conformation’ when in complex with its pre-export chaperone, the degree of folding of such Tat precursors has not been examined in detail. In the present study, recombinant TorDHis, active TorAHis, and the TorA–TorDHis and TorAΔSP–TorDHis complexes were analysed by CD spectroscopy. The CD spectra recorded for each sample are shown in Figure 5 and the calculated percentages of helix, strand, turn and disorder are given in Supplementary Table S2 (at http://www.biochemj.org/bj/452/bj4520057add.htm). Consistent with the highly α-helical structure of TorD [17], the CD spectrum of purified TorDHis shows a high proportion of helix (Figure 5 and Supplementary Table S2). Enzymes of the DMSO reductase family are mixed α/β proteins [31–34] and this is reflected in the CD spectrum of purified TorAHis (Figure 5 and Supplementary Table S2). Interestingly, CD analysis of the TorA–TorDHis and TorAΔSP–TorDHis complexes showed that both had a significant degree of secondary structure and that the observed percentage of disorder was no higher than that of isolated fully assembled TorAHis. Using the parameters determined for TorAHis and TorDHis (Supplementary Table S2), the predicted percentages of helix, strand, turn and disorder that would be expected for a simple 1:1 mixture of these two proteins can be calculated (Supplementary Table S2). It is notable that these predicted values are a close match to those experimentally determined for the purified TorA–TorDHis and TorAΔSP–TorDHis complexes, with the values particularly close for the TorA–TorDHis complex (Supplementary Table S2). Taken together, these results indicate that the TorA component of the TorA–TorDHis complex has probably adopted its native secondary structure. Moreover, whereas the TorAHis protein was isolated from an anaerobic culture and exhibited a straw colour in solution, the TorA–TorDHis and TorAΔSP–TorDHis complexes examined were produced from cultures grown under aerobic conditions without added molybdate. As a result, they would be expected to be largely devoid of cofactor and indeed were colourless and devoid of enzymatic activity. Metal content in the complexes by ICP-MS showed that the molar ratio of molybdenum to TorA/D complex was less than 1:1000. Taken together with the trypsinolysis experiments (Figure 3), these data suggest that the cofactor-free TorD-bound species of the TorA apoenzyme adopts a fold very similar to that of its fully assembled counterpart, save for an exposed C-terminal Domain IV.
Figure 5. CD spectra of purified TorAHis, TorDHis, and the TorA–TorDHis and TorAΔSP–TorDHis complexes.
CD spectra (185–260 nm) were collected in quartz cells of 0.02 cm pathlength at 25°C with a scan speed of 10 nm/min, bandwidth of 1 nm, response of 2 s and data pitch of 0.2 nm. The buffer used was 50 mM Tris/H2SO4 (pH 7.5), 200 mM K2SO4 and 1 mM DTT, and protein concentrations were: TorA, 0.7 mg/ml; TorD, 1 mg/ml; TorA–TorDHis, 1 mg/ml; and TorAΔsp–TorDHis, 0.5 mg/ml.
SAXS analysis of full-length TorA–TorDHis and signal peptide-less TorAΔSP–TorDHis complexes reveals shared structural features
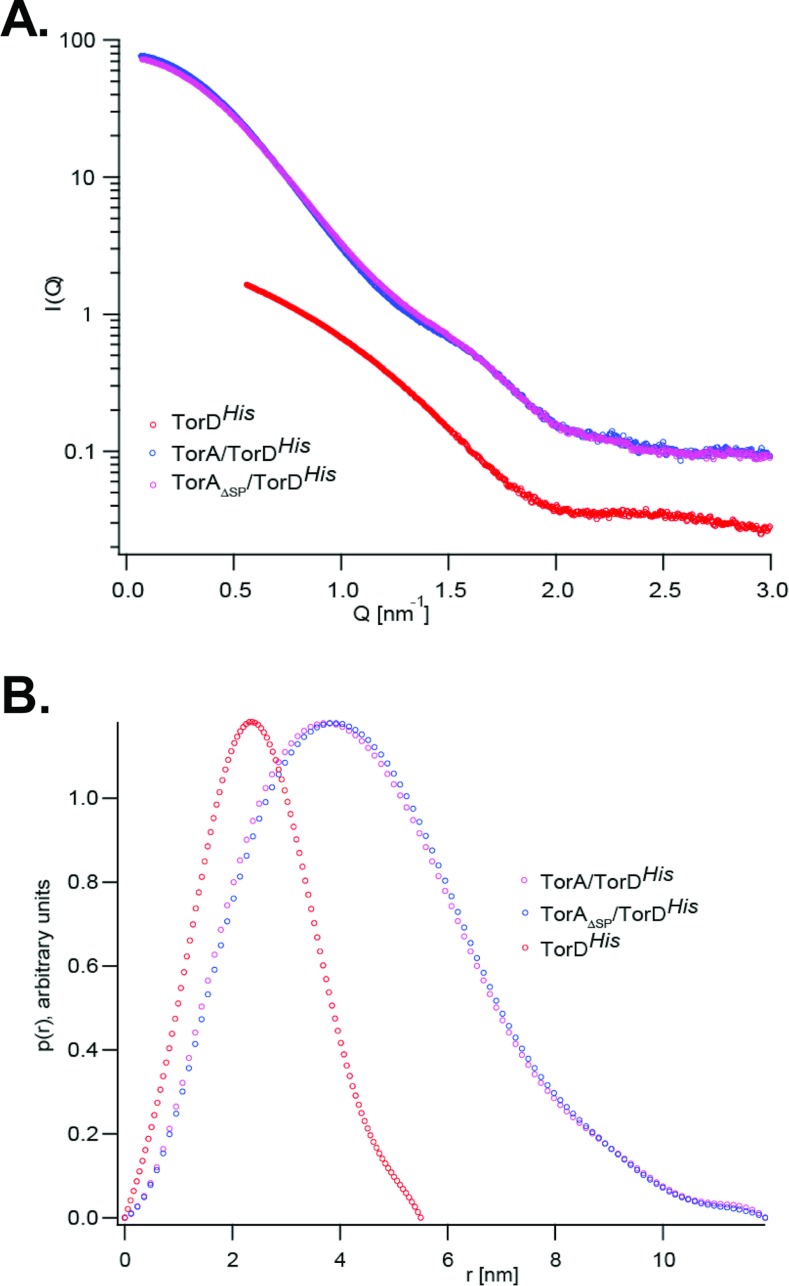
SAXS is a powerful technique that can be used to compare the overall shapes and sizes of proteins in solution. We analysed the TorA–TorDHis and TorAΔSP–TorDHis complexes, together with isolated TorDHis, since the high-resolution structure of TorD from S. massilia suggested that the protomer could adopt a highly extended conformation [17]. Representative SAXS scattering curves from each of the three samples are shown in Figure 6. It is clear that the scattering curves for the TorA–TorDHis and TorAΔSP–TorDHis complexes are almost indistinguishable, indicating that they have an identical overall shape. An overview of the general parameters extracted from the SAXS data including the radii of gyration (Rg; obtained from the Guinier approximation [35]), the distance distribution function, P(r) and the maximum particle size (Dmax; obtained from the GNOM [36] analysis) are given in Table 2.
Figure 6. SAXS characterization of TorDHis alone and TorDHis in complex with different forms of TorA.
(A) SAXS results for TorDHis, and the TorA–TorDHis and TorAΔSP–TorDHis complexes. Q is the scattering vector and I represents intensity. (B) Distance distributions P(r) for TorDHis, TorA–TorDHis and TorAΔSP–TorDHis. All curves were normalized.
Table 2. SAXS parameters for the TorA and TorD proteins.
This output was obtained from the scattering data and GNOM modelling for each of the indicated protein samples.
| Sample | Rg (Guinier) | Rg [P(r)] | Dmax [P(r)] |
|---|---|---|---|
| TorA–TorDHis | 35.0 Å | 35.2 Å | 119 Å |
| TorAΔSP–TorDHis | 35.6 Å | 35.1 Å | 119 Å |
| TorDHis | 20.0 Å | 19.2 Å | 55 Å |
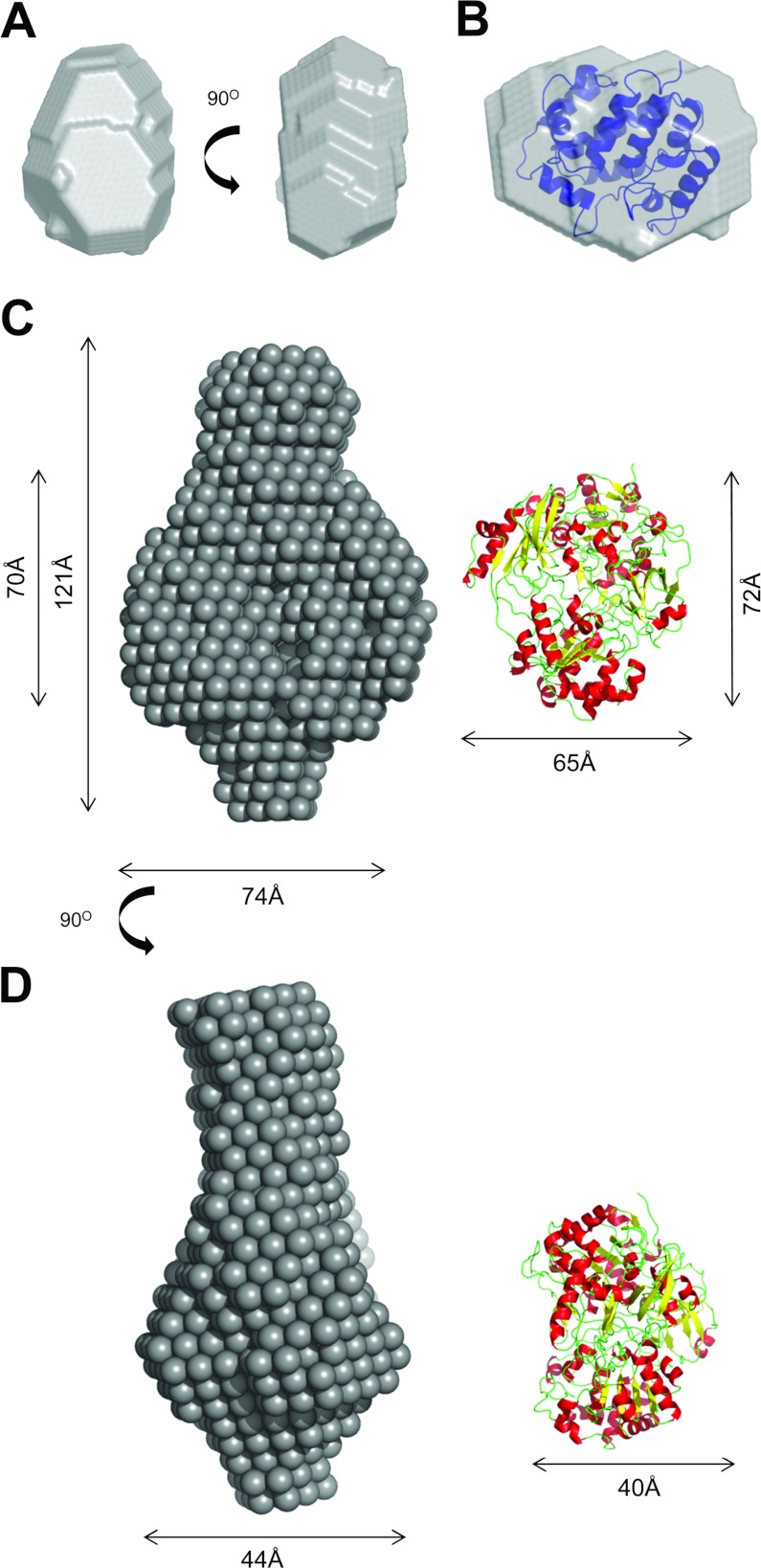
A low-resolution model of TorDHis, obtained using the program DAMMIN [37], is shown in Figure 7(A). The overall shape of TorDHis is compact and elliptical with dimensions of approximately 36 Å×50 Å×34 Å. From the crystal structures of S. massilia TorD [17] and the TorD family protein DmsD from E. coli [38–40], scattering envelopes can be calculated [41]. It is clear from Supplementary Figure S3 (http://www.biochemj.org/bj/452/bj4520057add.htm) that neither the S. massilia TorD domain-swapped dimer nor one of the protomers involved in forming that dimer would give scattering curves similar to that measured for TorDHis in solution. By contrast, the scattering curve predicted from the X-ray structure of DmsD is very similar in shape, and the electron density of monomeric DmsD, which can be readily docked into the TorDHis SAXS envelope (Figure 7B).
Figure 7. Ab initio modelling of TorDHis and the TorA–TorDHis complex.
(A) Two different views of the ab intio model of TorD, generated using DAMMIN [37]. (B) The electron density for E. coli DmsD (PDB code 3CW0 [39]) docked into the SAXS-derived shape of TorDHis. (C) and (D) represent two different views of the ab intio model of the TorA–TorDHis complex. The X-ray structure of S. massilia TorA (PDB code 1TMO [32]) is shown to the right.
Since the scattering curves indicated that the TorA–TorDHis and TorAΔSP–TorDHis complexes were indistinguishable, a single SAXS-derived shape corresponding to either the TorA–TorDHis or the TorAΔSP–TorDHis complex is shown in Figures 7(C) and 7(D). The complex is highly elongated, with an overall length of approximately 120 Å. There is a large central area of density with one relatively large lobe of density protruding from one side of the central region and a smaller lobe of density protruding from the opposite side. The large central area has approximate dimensions of 70 Å×74 Å×44 Å and is a reasonably close match to the dimensions of the crystal structure of S. massilia TorA (Figures 7C and 7D).
DISCUSSION
The process of co-ordinated assembly (‘proofreading’ or ‘quality control’) is an important consideration in the biosynthesis of all complex cofactor-containing Tat-dependent enzymes [5]. The simplest model system used to study this process is the E. coli TMAO reductase, which has a single MGD cofactor and a single biosynthetic chaperone, TorD. Whereas early research has seen analyses of the TorD–signal peptide interaction in isolation [12,42], the present study has focused on the characterization of the entire pre-export TorAD complex.
Design of co-overproduction vectors established that untagged TorA formed a tight and stable complex with hexahistidine-tagged TorD. Surprisingly, however, despite the strong evidence for two separate TorD-binding sites on TorA [9,11], we have clearly demonstrated using the accurate SEC–MALLS technique in conjunction with other experiments that TorD binds TorA with a 1:1 stoichiometry. Even more surprisingly, given the well-characterized interaction between TorD and the TorA signal peptide [11–15,42], a truncated TorA enzyme completely lacking its entire 39-residue signal peptide retained the ability to interact in a stable manner with TorD. Whereas the present study clearly establishes that there is a high-affinity TorD-binding site within the mature portion of TorA, it also raises questions about the role and importance of the signal peptide interaction.
These findings can be interpreted in two possible ways. First, that the signal peptide-binding activity of TorD is of a much lower affinity than that of the second binding site within the mature TorA sequence, so much so that there is a strong bias towards the higher-affinity species during the purification protocol. Although this scenario is not impossible, recent studies place the apparent Kd for TorD binding to the isolated TorA signal peptide at 59–330 nM [12,42], which is indicative of very tight binding such that it is likely that a detectable portion of TorA would not fail to have two TorD molecules bound. The second possibility is that the TorD-binding site within the mature portion of TorA is very close to the signal peptide-binding epitope such that a single TorD protein could interact with both simultaneously. This hypothesis would require there to be two different TorA-binding sites on a single TorD protein. Although the location of the TorA signal peptide-binding site on TorD has not yet been reported, there is a larger body of evidence available on the location of the signal peptide-binding site on another member of the TorD family, DmsD. The E. coli DmsD protein was the first twin-arginine signal peptide-binding chaperone to be described [43]. It is essential for the assembly of molybdenum-dependent DMSO reductase and selenate reductase enzymes in E. coli and binds to the signal peptides of those enzymes with apparent Kd values in the range 10–100 nM [43–45]. Genetic, biochemical and structural analyses predict the signal peptide-binding site to occupy only one face of DmsD (primarily involving residues of the N-terminal helices) and so suggests that a significant proportion the DmsD protein is not involved in signal peptide recognition [39,45,46]. This leaves the possibility, at least, that a second protein- or peptide-recognition site may be present on this type of chaperone. Indeed, variants of E. coli TorD have been described (C79R and L83P) that retain signal peptide-binding activity, but are impaired in their binding to mature TorA [10].
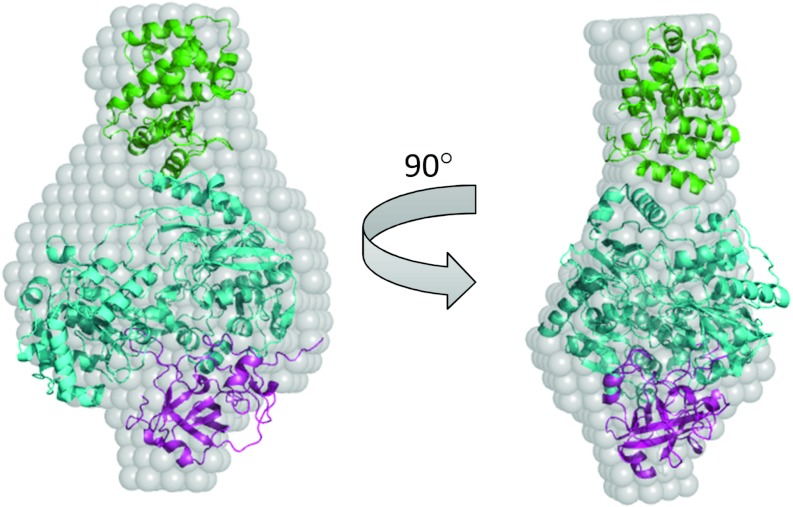
Further evidence for overlapping TorD-binding sites near the N-terminus of TorA come from analysis of the SAXS data obtained in the present study. To investigate the possible arrangement of TorA and TorD in the TorAD complex, rigid body modelling was undertaken using the SASREF program [47] and the structures of S. massilia TorA and E. coli DmsD as building blocks. To facilitate this analysis, TorA was split into two parts to give the proteolytically stable N-terminal part (Domains I–III) and the proteolytically sensitive Domain IV identified in the present study. A constraint was then applied that the last amino acid of N-terminal part should be within 10 Å of the first amino acid of Domain IV, in order to incorporate flexibility into the TorA component of the complex, again consistent with the protease accessibility results of the present study. The results from the rigid body modelling show a similar overall shape to that obtained during ab initio analysis (the fit of the model with the experimental data is shown in Supplementary Figure S4 at http://www.biochemj.org/bj/452/bj4520057add.htm). The rigid body model is shown docked into the ab initio SAXS-derived envelope of TorAD in Figure 8. When docked into the ab initio model, the density for TorD fits very well into the larger protruding lobe, whereas the remainder of the ab initio envelope is of an appropriate volume to accommodate TorA. The C-terminal Domain IV of TorA distal from TorD fits into the smaller protruding lobe, which is consistent with the biochemical data of the present study that Domain IV is not required for interaction of TorD with TorA. Bearing in mind that the SAXS scattering curves for both signal peptide-containing and signal peptide-free TorAD complexes are identical, this must be taken as good evidence that the binding epitope for TorD on the mature portion of TorA must lie near the N-terminus and thus also near the second epitope within the signal peptide.
Figure 8. Rigid body modelling of the TorA–TorDHis complex.
Rigid body modelling was conducted using the homologous component parts of the complex, i.e. TorD homologue DmsD (PDB code 3CW0, shown in green), and TorA (PDB code 1TMO) split into two parts: Domains I–III (i.e. amino acids 5–629; shown in cyan) and Domain IV (amino acids 631–798; shown in magenta). Modelling was conducted using SASREF [47]. The generated model is docked into the TorA–TorDHis ab intio model for comparison.
The CD and SAXS analyses suggest that there is a very high degree of folding by the TorA protein even in the absence of the MGD cofactor. However, it is clear that the TorA–TorDHis and TorAΔSP–TorDHis complexes are not in the native conformation and that the C-terminal Domain IV of the TorA protein is exposed. This could suggest that the locking of Domain IV into position represents the final act in cofactor insertion. One possible role of TorD could be to prevent premature closing of the Domain IV ‘flap’ before MGD has bound, or, alternatively, that closing of Domain IV after cofactor loading is the trigger that releases TorD from the now mature enzyme. The molecular mechanism of how TorD senses the events surrounding cofactor loading remains to be unearthed.
Online data
AUTHOR CONTRIBUTION
Jennifer Dow, Frank Gabel, Frank Sargent and Tracy Palmer performed the research and analysed data; Jennifer Dow, Frank Sargent and Tracy Palmer designed the research; Tracy Palmer supervised the research; and Jennifer Dow, Frank Gabel, Frank Sargent and Tracy Palmer wrote the paper.
ACKNOWLEDGEMENTS
We acknowledge Dr Bérengère Ize, Dr Grant Buchanan and Dr Dave Guymer for useful discussions and preliminary studies. We also thank Dr Cyril Dian for technical assistance at the ID14-3 BioSAXS beamline at the ESRF.
FUNDING
This work was funded via a Wellcome Trust Ph.D. Studentship Award to J.M.D. [grant number WT089692/Z09/z] and a Royal Society Wolfson Research Merit Award to T.P.
References
- 1.Celedon J. M., Cline K. Intra-plastid protein trafficking: how plant cells adapted prokaryotic mechanisms to the eukaryotic condition. Biochim. Biophys. Acta. 2013;1833:341–351. doi: 10.1016/j.bbamcr.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmer T., Berks B. C. The twin-arginine translocation (Tat) protein export pathway. Nat. Rev. Microbiol. 2012;10:483–496. doi: 10.1038/nrmicro2814. [DOI] [PubMed] [Google Scholar]
- 3.Berks B. C. A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 4.Palmer T., Sargent F., Berks B. C. The Tat protein export pathway. In: Böck A., Curtiss R., Kaper J. B., Karp P. D., Neidhardt F. C., Nyström T., Slauch J. M., Squires C. L., Ussery D., editors. EcoSal–Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: ASM Press; 2010. Chapter 4.3.2, http://www.ecosal.org. [Google Scholar]
- 5.Sargent F. Constructing the wonders of the bacterial world: biosynthesis of complex enzymes. Microbiology. 2007;153:633–651. doi: 10.1099/mic.0.2006/004762-0. [DOI] [PubMed] [Google Scholar]
- 6.Silvestro A., Pommier J., Giordano G. The inducible trimethylamine-N-oxide reductase of Escherichia coli K12: biochemical and immunological studies. Biochim. Biophys. Acta. 1988;954:1–13. doi: 10.1016/0167-4838(88)90049-0. [DOI] [PubMed] [Google Scholar]
- 7.Santini C. L., Ize B., Chanal A., Müller M., Giordano G., Wu L. F. A novel sec-independent periplasmic protein translocation pathway in Escherichia coli. EMBO J. 1998;17:101–112. doi: 10.1093/emboj/17.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mejean V., Iobbi-Nivol C., Lepelletier M., Giordano G., Chippaux M., Pascal M. C. TMAO anaerobic respiration in Escherichia coli: involvement of the tor operon. Mol. Microbiol. 1994;11:1169–1179. doi: 10.1111/j.1365-2958.1994.tb00393.x. [DOI] [PubMed] [Google Scholar]
- 9.Pommier J., Mejean V., Giordano G., Iobbi-Nivol C. TorD, a cytoplasmic chaperone that interacts with the unfolded trimethylamine N-oxide reductase enzyme (TorA) in Escherichia coli. J. Biol. Chem. 1998;273:16615–16620. doi: 10.1074/jbc.273.26.16615. [DOI] [PubMed] [Google Scholar]
- 10.Genest O., Neumann M., Seduk F., Stocklein W., Mejean V., Leimkuhler S., Iobbi-Nivol C. Dedicated metallochaperone connects apoenzyme and molybdenum cofactor biosynthesis components. J. Biol. Chem. 2008;283:21433–21440. doi: 10.1074/jbc.M802954200. [DOI] [PubMed] [Google Scholar]
- 11.Jack R. L., Buchanan G., Dubini A., Hatzixanthis K., Palmer T., Sargent F. Coordinating assembly and export of complex bacterial proteins. EMBO J. 2004;23:3962–3972. doi: 10.1038/sj.emboj.7600409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buchanan G., Maillard J., Nabuurs S. B., Richardson D. J., Palmer T., Sargent F. Features of a twin-arginine signal peptide required for recognition by a Tat proofreading chaperone. FEBS Lett. 2008;582:3979–3984. doi: 10.1016/j.febslet.2008.10.049. [DOI] [PubMed] [Google Scholar]
- 13.Hatzixanthis K., Clarke T. A., Oubrie A., Richardson D. J., Turner R. J., Sargent F. Signal peptide–chaperone interactions on the twin-arginine protein transport pathway. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8460–8465. doi: 10.1073/pnas.0500737102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genest O., Seduk F., Ilbert M., Mejean V., Iobbi-Nivol C. Signal peptide protection by specific chaperone. Biochem. Biophys. Res. Commun. 2006;339:991–995. doi: 10.1016/j.bbrc.2005.11.107. [DOI] [PubMed] [Google Scholar]
- 15.Li S. Y., Chang B. Y., Lin S. C. Coexpression of TorD enhances the transport of GFP via the TAT pathway. J. Biotechnol. 2006;122:412–421. doi: 10.1016/j.jbiotec.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 16.Tranier S., Mortier-Barriere I., Ilbert M., Birck C., Iobbi-Nivol C., Mejean V., Samama J. P. Characterization and multiple molecular forms of TorD from Shewanella massilia, the putative chaperone of the molybdoenzyme TorA. Protein Sci. 2002;11:2148–2157. doi: 10.1110/ps.0202902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tranier S., Iobbi-Nivol C., Birck C., Ilbert M., Mortier-Barriere I., Mejean V., Samama J. P. A novel protein fold and extreme domain swapping in the dimeric TorD chaperone from Shewanella massilia. Structure. 2003;11:165–174. doi: 10.1016/s0969-2126(03)00008-x. [DOI] [PubMed] [Google Scholar]
- 18.Guymer D., Maillard J., Agacan M. F., Brearley C. A., Sargent F. Intrinsic GTPase activity of a bacterial twin-arginine translocation proofreading chaperone induced by domain swapping. FEBS J. 2010;277:511–525. doi: 10.1111/j.1742-4658.2009.07507.x. [DOI] [PubMed] [Google Scholar]
- 19.Genest O., Neumann M., Seduk F., Stocklein W., Mejean V., Leimkuhler S., Iobbi-Nivol C. Dedicated metallochaperone connects apoenzyme and molybdenum cofactor biosynthesis components. J. Biol. Chem. 2008;283:21433–21440. doi: 10.1074/jbc.M802954200. [DOI] [PubMed] [Google Scholar]
- 20.Genest O., Mejean V., Iobbi-Nivol C. Multiple roles of TorD-like chaperones in the biogenesis of molybdoenzymes. FEMS Microbiol. Lett. 2009;297:1–9. doi: 10.1111/j.1574-6968.2009.01660.x. [DOI] [PubMed] [Google Scholar]
- 21.Butland G., Peregrin-Alvarez J. M., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 22.Ize B., Coulthurst S. J., Hatzixanthis K., Caldelari I., Buchanan G., Barclay E. C., Richardson D. J., Palmer T., Sargent F. Remnant signal peptides on non-exported enzymes: implications for the evolution of prokaryotic respiratory chains. Microbiology. 2009;155:3992–4004. doi: 10.1099/mic.0.033647-0. [DOI] [PubMed] [Google Scholar]
- 23.de Leeuw E., Granjon T., Porcelli I., Alami M., Carr S. B., Müller M., Sargent F., Palmer T., Berks B. C. Oligomeric properties and signal peptide binding by Escherichia coli Tat protein transport complexes. J. Mol. Biol. 2002;322:1135–1146. doi: 10.1016/s0022-2836(02)00820-3. [DOI] [PubMed] [Google Scholar]
- 24.Hamilton C. M., Aldea M., Washburn B. K., Babitzke P., Kushner S. R. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambasivarao D., Weiner J. H. Dimethyl sulfoxide reductase of Escherichia coli: an investigation of function and assembly by use of in vivo complementation. J. Bacteriol. 1991;173:5935–5943. doi: 10.1128/jb.173.19.5935-5943.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sargent F., Bogsch E. G., Stanley N. R., Wexler M., Robinson C., Berks B. C., Palmer T. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 1998;17:3640–3650. doi: 10.1093/emboj/17.13.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmer T., Berks B. C., Sargent F. Analysis of Tat targeting function and twin-arginine signal peptide activity in Escherichia coli. Methods Mol. Biol. 2010;619:191–216. doi: 10.1007/978-1-60327-412-8_12. [DOI] [PubMed] [Google Scholar]
- 28.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 29.Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., Svergun D. I. PRIMUS: a Windows-PC based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 30.Zimm B. H. The scattering of light and the radial distribution function of high polymer solutions. J. Chem. Phys. 1948;16:1093–1099. [Google Scholar]
- 31.Schindelin H., Kisker C., Hilton J., Rajagopalan K. V., Rees D. C. Crystal structure of DMSO reductase: redox-linked changes in molybdopterin coordination. Science. 1996;272:1615–1621. doi: 10.1126/science.272.5268.1615. [DOI] [PubMed] [Google Scholar]
- 32.Czjzek M., Dos Santos J. P., Pommier J., Giordano G., Mejean V., Haser R. Crystal structure of oxidized trimethylamine N-oxide reductase from Shewanella massilia at 2.5 Å resolution. J. Mol. Biol. 1998;284:435–447. doi: 10.1006/jmbi.1998.2156. [DOI] [PubMed] [Google Scholar]
- 33.Schneider F., Lowe J., Huber R., Schindelin H., Kisker C., Knablein J. Crystal structure of dimethyl sulfoxide reductase from Rhodobacter capsulatus at 1.88 Å resolution. J. Mol. Biol. 1996;263:53–69. doi: 10.1006/jmbi.1996.0555. [DOI] [PubMed] [Google Scholar]
- 34.McAlpine A. S., McEwan A. G., Bailey S. The high resolution crystal structure of DMSO reductase in complex with DMSO. J. Mol. Biol. 1998;275:613–623. doi: 10.1006/jmbi.1997.1513. [DOI] [PubMed] [Google Scholar]
- 35.Guinier A. La diffraction des rayons X aux tres faibles angles: applications a l’etude des phenomenes ultra-microscopiques. Ann. Phys. 1939;12:161–236. [Google Scholar]
- 36.Svergun D. I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- 37.Svergun D. I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stevens C. M., Winstone T. M., Turner R. J., Paetzel M. Structural analysis of a monomeric form of the twin-arginine leader peptide binding chaperone Escherichia coli DmsD. J. Mol. Biol. 2009;389:124–133. doi: 10.1016/j.jmb.2009.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramasamy S. K., Clemons W. M., Jr Structure of the twin-arginine signal-binding protein DmsD from Escherichia coli. Acta Crystallogr., Sect. F: Struct. Biol. Crystal. Commun. 2009;65:746–750. doi: 10.1107/S1744309109023811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qiu Y., Zhang R., Binkowski T. A., Tereshko V., Joachimiak A., Kossiakoff A. The 1.38 Å crystal structure of DmsD protein from Salmonella typhimurium, a proofreading chaperone on the Tat pathway. Proteins. 2008;71:525–533. doi: 10.1002/prot.21828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Svergun D., Barberato C., Koch M. H. J. CRYSOL: a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- 42.Shanmugham A., Bakayan A., Voller P., Grosveld J., Lill H., Bollen Y. J. The hydrophobic core of twin-arginine signal sequences orchestrates specific binding to Tat-pathway related chaperones. PLoS ONE. 2012;7:e34159. doi: 10.1371/journal.pone.0034159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oresnik I. J., Ladner C. L., Turner R. J. Identification of a twin-arginine leader-binding protein. Mol. Microbiol. 2001;40:323–331. doi: 10.1046/j.1365-2958.2001.02391.x. [DOI] [PubMed] [Google Scholar]
- 44.Guymer D., Maillard J., Sargent F. A genetic analysis of in vivo selenate reduction by Salmonella enterica serovar Typhimurium LT2 and Escherichia coli K12. Arch. Microbiol. 2009;191:519–528. doi: 10.1007/s00203-009-0478-7. [DOI] [PubMed] [Google Scholar]
- 45.Chan C. S., Winstone T. M., Chang L., Stevens C. M., Workentine M. L., Li H., Wei Y., Ondrechen M. J., Paetzel M., Turner R. J. Identification of residues in DmsD for twin-arginine leader peptide binding, defined through random and bioinformatics-directed mutagenesis. Biochemistry. 2008;47:2749–2759. doi: 10.1021/bi702138a. [DOI] [PubMed] [Google Scholar]
- 46.Stevens C. M., Okon M., McIntosh L. P., Paetzel M. 1H, 13C and 15N resonance assignments and peptide binding site chemical shift perturbation mapping for the Escherichia coli redox enzyme chaperone DmsD. Biomol. NMR Assign. 2012 doi: 10.1007/s12104-012-9408-8. doi:10.1007/s12104-012-9408-8. [DOI] [PubMed] [Google Scholar]
- 47.Petoukhov M. V., Svergun D. I. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.