Abstract
Imaging traits are thought to have more direct links to genetic variation than diagnostic measures based on cognitive or clinical assessments and provide a powerful substrate to examine the influence of genetics on human brains. Although imaging genetics has attracted growing attention and interest, most brain-wide genome-wide association studies focus on voxel-wise single-locus approaches, without taking advantage of the spatial information in images or combining the effect of multiple genetic variants. In this paper we present a fast implementation of voxel- and cluster-wise inferences based on the random field theory to fully use the spatial information in images. The approach is combined with a multi-locus model based on least square kernel machines to associate the joint effect of several single nucleotide polymorphisms (SNP) with imaging traits. A fast permutation procedure is also proposed which significantly reduces the number of permutations needed relative to the standard empirical method and provides accurate small p-value estimates based on parametric tail approximation. We explored the relation between 448,294 single nucleotide polymorphisms and 18,043 genes in 31,662 voxels of the entire brain across 740 elderly subjects from the Alzheimer's Disease Neuroimaging Initiative (ADNI). Structural MRI scans were analyzed using tensor-based morphometry (TBM) to compute 3D maps of regional brain volume differences compared to an average template image based on healthy elderly subjects. We find method to be more sensitive compared with voxel-wise single-locus approaches. A number of genes were identified as having significant associations with volumetric changes. The most associated gene was GRIN2B, which encodes the N-methyl-d-aspartate (NMDA) glutamate receptor NR2B subunit and affects both the parietal and temporal lobes in human brains. Its role in Alzheimer's disease has been widely acknowledged and studied, suggesting the validity of the approach. The various advantages over existing approaches indicate a great potential offered by this novel framework to detect genetic influences on human brains.
Keywords: Imaging genetics, Association study, Random field theory, Voxel-wise inference, Cluster size inference, Nonstationarity, Least square kernel machines, Permutation, Pareto distribution, Parametric tail approximation, ADNI, MRI, Tensor-based morphometry, Alzheimer's disease, GRIN2B
Introduction
The past decade witnessed tremendous growth in both neuroimaging and genomics. Imaging genetics, as an interdisciplinary field, uses anatomical or functional imaging technologies as phenotypic assays to evaluate genetic variation. Quantitative imaging-derived traits are thought to have more direct links to genetic variation than diagnostic measures based on cognitive or clinical assessments and thus might offer more power to detect the association between specific genes, single nucleotide polymorphisms (SNPs) or copy number variations (CNVs) and various brain diseases (Gottesman and Gould, 2003). Therefore, a large amount of imaging and genetic data have been collected, such as the Alzheimer's Disease Neuroimaging Initiative (ADNI), with the hope of speeding up the studies in this area, and ultimately to improve human health care in the future (Akil et al., 2010).
In spite of the great investment and efforts poured into this area, imaging genetics is still a very young field. Statistical approaches are rudimentary compared to imaging-only or genetic-only methods. This is partly due to both the ultra-high dimension and complex noise structure of imaging and genetic data. Currently, most of the population-based association studies with neuroimaging phenotypes in the literature can be broadly categorized into candidate-phenotype candidate-SNP/gene association (e.g., Joyner et al., 2009), candidate-phenotype genome-wide association (e.g., Frayling et al., 2007; Potkin et al., 2009), or brain-wide candidate-SNP/gene association (e.g., Braskie et al., 2011; Filippini et al., 2009). All these studies reduce the dimensionality of the data using a priori knowledge, by either selecting specific, well-studied genetic variants that affect brain structure and function, or focusing on a specific brain area or a neuroimaging measure/contrast of interest.
It can be expected that the trend in imaging genetics is to embrace the brain-wide, genome-wide association paradigm, where both the entire genome and entire brain are searched for nonrandom associations (Hibar et al., 2011a). Several recent studies have been carried out in this category, e.g., a voxel-wise genome-wide association study (vGWAS) (Stein et al., 2010a) and a voxel-wise gene-wide association study (vGeneWAS) (Hibar et al., 2011b). Although these works are pioneering and important, showing that a completely unbiased search of the genome is feasible with imaging phenotypes, there are several limitations. Firstly, these mass-univariate analyses are based on statistics at each single voxel, focused only on the highest association, and therefore do not take full advantage of the spatial information in 3D imaging data. Such tests might miss spatially extended signals that do not “win” any voxel. Secondly, variants in the same haplotype block on the genome can be highly correlated due to linkage disequilibrium (LD) (Frazer et al., 2007). Although multivariate approaches based on principal components analysis (PCA) have been used to account for this colinearity (Hibar et al., 2011b), this linear approach may not detect interactions between SNPs and their nonlinear effect on the traits. Biologically-informed approaches that have the potential to capture epistatic effects are therefore needed (Schaid, 2010a, 2010b) to better model the interactions between genetic variants. Finally, image-wide genome-wide search is extremely computationally expensive, and, at the same time, lacks power. With millions or even billions of statistical tests to be performed, the colossal multiple testing problem can render no significant associations (Hibar et al., 2011b; Stein et al., 2010a). Other potential brain-wide, genome-wide association studies are based on penalized and sparse regression techniques, including ℓ2 regularization in ridge regression (Hoerl, 1985) and ℓ1 regularization in the least absolute shrinkage and selection operator (LASSO) (Tibshirani, 1996). These methods are able to localize both brain regions and genomic regions from high dimensional data, and have been reported to show boosted statistical power. Related studies are (Kohannim et al., 2011) using ridge regression, Kohannim et al. (in press) using ℓ1 + ℓ2 regularization, and a sparse reduced-rank regression (sRRR) method (Vounou et al., 2010). Sure independence screening (ISIS) (Fan and LV, 2008) is also a possible idea but needs to be tested with application to real imaging genetics datasets.
Random field theory is a core inferential tool in neuroimaging, especially in the analysis of fMRI and PET data (Friston et al., 2007), in order to detect the effect or activation at an unknown spatial location. In particular, it assesses whether the test statistic value or the spatial extent of a set of contiguous voxels exceeding some predefined threshold is unusually large by chance alone, known as voxel-wise inferences and cluster size inferences respectively. Based on early work on Gaussian random field theory (Adler, 1981; Aldous, 1989), the theoretical basis for voxel-wise inferences (Worsley et al., 1996), stationary cluster size inferences (Cao and Worsley, 2001; Friston et al., 1994; Poline and Mazoyer, 1993; Roland et al., 1993), and nonstationary cluster size inferences (Worsley, 2002; Worsley et al., 1999) have been well established and extended to various types of random fields (Cao, 1999). Cluster-wise inference is known to have increased sensitivity compared to tests based on voxel intensity when the signal is spatially distributed (Friston et al., 1996; Poline et al., 1997), though they have never been used to imaging genetics, perhaps due to the computational demands or the number of assumptions that are required to make parametric approximation valid, as shown in many nonparametric simulations and tests (Hayasaka and Nichols, 2003; Hayasaka et al., 2004; Holmes, 1994).
To model the interaction of genes or SNPs in the same genetic path-way or at nearby loci, a semiparametric regression model has been proposed where covariate effects are modeled parametrically and the pathway effect/interaction of multiple gene expressions/SNPs is modeled parametrically or nonparametrically using least-squares kernel machines (LSKM)1 (Kwee et al., 2008; Liu et al., 2007). This approach has been shown to be flexible by modeling high-dimensional interactions while allowing for covariates, and to have superior performance for association mapping of quantitative traits.
Nonparametric permutation methods have been introduced to neuroimaging and found successful applications (Holmes et al., 1996; Nichols and Holmes, 2002). They are particularly useful when parametric distributional assumptions cannot be met. However, when faced with the ultra-high dimensional data comprising of thousands of millions of SNPs or genes, standard permutation procedures are normally too computationally expensive, which impedes their wide implementation in imaging genetics. Furthermore, the resolution of obtainable p-values as well as the smallest p-value are bounded by inverse of the number of permutations. Since large multiple testing corrections rely on precise approximation of small p-values, standard permutation procedures would seem to be impractical. Pooling and parametric fitting of permutation distributions offer the potential to make permutation practical in this setting.
The contributions of this paper are three-fold. Firstly, to our knowledge, this is the first paper to apply random field theory to imaging genetics on a whole genome basis. Our fast implementation of random field theory not only takes advantage of the spatial information in images, which increases sensitivity, but also significantly reduces the computational burden. Secondly, we introduce to neuroimaging a multi-locus method based on least square kernel machines to model interactions among SNPs in the same gene so that a gene-based association test can be performed. Thirdly, we provide a tail approximation procedure which makes permutation-based inference feasible for brain-wide genome-wide data. All these methods were intensively tested on both simulated Gaussian images and real tensor based morphometry (TBM) data as a part of the Alzheimer's disease Neuroimaging Initiative (ADNI). Several genes were detected as significantly associated with the volumetric changes in human brains of elderly subjects. The most associated gene was GRIN2B, which encodes the N-methyl-d-aspartate (NMDA) glutamate receptor NR2B subunit and affects both the parietal and temporal lobes in human brains. Its role in Alzheimer's disease has been widely acknowledged and studied (see Discussion section), suggesting the validity of the approach. Our proposed methods show various advantages over existing approaches and indicate a great potential to detect genetic influences on the brain.
The rest of the paper is organized as follows. In the Materials and methods section, the fast implementation of random field theory, the LSKM approach and the pooling and parametric fitting of permutation distributions are introduced. In the Results section, null simulation results on both simulated Gaussian images and real TBM data are shown, in order to validate the proposed approaches. The methods are then evaluated by a detailed comparison with the existing brain-wide genome-wide studies using the same dataset. The pros and cons of the approaches as well as the significance of the biological findings are summarized in the Discussion section. The details of the LSKM approach and voxel- and cluster-wise inferences using random field theory are provided in the Appendix.
Materials and methods
The tensor based morphometry data and the SNP data are the same as those used in the previous studies (Shen et al., 2010). The mapping from SNPs to genes is the same as used in Hibar et al. (2011b). However, we describe all imaging and genetic data preprocessing for completeness.
Study design and subjects
Data used in this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.ucla.edu). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public–private partnership. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer's disease (AD). Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials. The Principal Investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California — San Francisco. ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 adults, ages 55 to 90, to participate in the research, approximately 200 cognitively normal older individuals to be followed for 3 years, 400 people with MCI to be followed for 3 years and 200 people with early AD to be followed for 2 years. For up-to-date information, see www.adni-info.org.
818 subjects were genotyped as part of the ADNI study. However, only 740 unrelated Caucasian subjects identified by self-report and confirmed by MDS analysis (Stein et al., 2010a) were included to reduce population stratification effects. Volumetric brain differences were assessed in 173 AD patients (78 female/95 male; 75.54 ± 7.66 years old), 361 MCI subjects (130 female/231 male; 75.16 ± 7.29 years old), and 206 healthy elderly subjects (94 female/112 male; 76.13 ± 4.94 years old).
Data preprocessing
MRI images
High-resolution structural brain MRI scans were acquired at 58 ADNI sites with 1.5 T MRI scanners using a sagittal 3D MP-RAGE sequence developed for consistency across sites (Jack et al., 2008) (TR = 2400 ms, TE = 1000 ms, flip angle = 8°, field of view = 24 cm, final reconstructed voxel resolution = 0.9375 × 0.9375 × 1.2 mm3). Images were calibrated with phantom-based geometric corrections to ensure consistency across scanners. Additional image corrections included (Jack et al., 2008): (1) correction of geometric distortions due to gradient non-linearity, (2) adjustment for image intensity inhomogeneity due to B1 field non-uniformity using calibration scans, (3) reducing residual intensity inhomogeneity, and (4) geometric scaling according to a phantom scan acquired for each subject to adjust for scanner- and session-specific calibration errors. Images were linearly registered with 9 parameters to the International Consortium for Brain Imaging template (ICBM-53) (Mazziotta et al., 2001) to adjust for differences in brain position and scaling.
For TBM analysis, a minimal deformation template (MDT) was first created for the healthy elderly group to serve as an unbiased average template image to which all other images were warped using a non-linear inverse-consistent elastic intensity-based registration algorithm (Leow et al., 2005). Volumetric tissue differences were assessed at each voxel in all individuals by calculating the determinant of the Jacobian matrix of the deformation, which encodes local volume excess or deficit relative to the mean template image. The maps of volumetric tissue differences were then down-sampled using trilinear interpolation to 4 × 4 × 4 mm3 isotropic voxel resolution for computational efficiency. The percentage volumetric difference relative to a population-based brain template at each voxel served as a quantitative measure of brain tissue volume difference for genome-wide association.
Genetic analysis
Genome-wide genotype information was collected at 620,901 markers. Only Single Nucleotide Polymorphisms (SNPs) were included in this analysis. Alleles on the forward strand are reported. Individual markers were excluded from the analysis that did not satisfy the following quality criteria based on previous genome-wide association studies (Cardon et al., 2007): genotype call rate <95% (42,670 SNPs removed), significant deviation from Hardy–Weinberg equilibrium p<5.7 × 10−7 (871 markers removed), minor allele frequency (MAF) <0.10 (161,354 SNPs removed), and a platform-specific recommended quality control score of <0.15 (variable number of SNPs removed across subjects). The minor allele frequency cut-off of 0.10 (10%) was used to ensure that sufficient numbers of subjects would be found in our sample in each genotypic group (homozygous major allele, heterozygous, homozygous minor allele) using an additive genetic model. If alleles are in Hardy−Weinberg equilibrium, a minor allele frequency cut off of 0.10 ensures that at least 7 subjects are in the smallest genotypic category on average. NSNP = 448,294 SNPs remained for analysis after quality control. See Table 1 for the final number of SNPs on each chromosome considered. Missing data still occurs over these remaining SNPs, but we require that at least 95% of the subjects have a successfully genotyped SNP for that SNP to be included. (Our multi-locus method allows for missing SNPs, so no further subjects or SNPs are dropped when considering a set of SNPs).
Table 1.
Final number of SNPs on each chromosome analyzed.
| Chr | # of SNPs | Chr | # of SNPs | Chr | # of SNPs | Chr | # of SNPs |
|---|---|---|---|---|---|---|---|
| 1 | 33,850 | 8 | 24,768 | 15 | 13,355 | 22 | 6757 |
| 2 | 36,384 | 9 | 21,283 | 16 | 13,460 | X | 10,637 |
| 3 | 30,765 | 10 | 23,089 | 17 | 11,721 | Y | 12 |
| 4 | 27,073 | 11 | 21,796 | 18 | 13,198 | XY | 25 |
| 5 | 27,396 | 12 | 21,461 | 19 | 7895 | Mitochondrial | 12 |
| 6 | 30,054 | 13 | 16,605 | 20 | 11,169 | – | – |
| 7 | 24,446 | 14 | 14,501 | 21 | 6582 | Total | 448,294 |
Gene grouping
SNPs were grouped by gene, where “gene” is defined by the gene transcript region including both introns and exons. SNPs upstream/downstream from the gene region were not included. SNPs that were not located in a gene were excluded. All splice variants were considered as belonging to the same gene. After gene grouping, Ngene = 18,043 genes were left for analysis out of the estimated 20,000–25,000 protein coding genes in the human genome (International Human Genome Sequencing Consortium, 2004).
The single locus model and statistical parametric mapping
A general linear model was used which can be written as
| (1) |
where v is an index for voxels, Y(v) = [Y1(v),⋯,YN(v)]T is a vector of volumetric tissue differences at voxel v across N subjects, and X is a known N × q design matrix. In this study, age, sex and an intercept are included as covariates, which comprise the first three columns of X. The fourth column of X is the number of minor allele for a particular SNP ℓ, which is the effect of interest. β(v) is a q-dimensional vector of unknown parameters, and ε(v) = [ε1(v),⋯,εN(v)]T is a vector of random errors with standard deviation σ(v).
Let β̂(v) denote an unbiased estimate of β(v), then the residuals are
| (2) |
and an estimate of the residual variance is
| (3) |
where v = N − q is the error degrees of freedom. Assuming εi(v), i = 1,⋯,N are independent and identically normally distributed, the t-statistic at voxel v is defined as
| (4) |
where c is a contrast vector of interest, which can be designed as [0 0 0 1]T in the present study to test the association between a particular SNP and the volumetric difference at a particular voxel. T(v) follows a t-distribution with v degrees of freedom, and T(v) at all voxels together form a Statistical Parametric Map.
The uncorrected p-value for observed statistic t(v) is , where is the null hypothesis for SNP ℓ, and we use a two-sided alternative since the direction of a SNP's effect cannot be anticipated in general. The Family-wise error (FWE) corrected p-value for a voxel-wise statistic t(v) searching over the brain is ; for a cluster of size s, , where max S is over all clusters in the brain. We compute these FWE-corrected p-values with either the random field theory or permutation, obtaining two-sided p-values as twice the usual one-sided p-values.2 The FWE-corrected p-value for our single-locus models searching over both the brain and all SNPs is found with Bonferroni, . The multilocus model introduced below produces an approximate χ2 test statistic that can similarly be converted into a FWE-corrected p-value. The FWE-corrected p-value for searching over brain and genes is . All methods considered below are fitted for each voxel, so going forward we suppress the voxel index v unless needed for clarity.
For cluster size inference, each cluster is formed as a set of contiguous voxels with test statistic values exceeding a pre-defined cluster forming threshold, where contiguity is defined by an order-18 neighborhood (voxels need at least a common edge to be connected). The cluster-forming threshold is always set corresponding to an uncorrected p-value threshold of 0.001. We use stat_threshold from the fmristat toolbox3 for FWE-corrected voxel-wise inference, and stat_thresh from the NS toolbox4 for cluster size p-values, as both of them implement χ2 results (unlike SPM).
A multi-locus approach — least square kernel machines
To model the complex interaction of SNPs, we consider a semi-parametric regression model (Liu et al., 2007) that relates the imaging traits to a number of SNPs, where the covariate effects, e.g., gender, age, etc., are modeled parametrically (i.e. linearly) and the interaction of SNPs is modeled nonparametrically using a least-squares kernel machines (LSKM) approach. This unified framework allows a flexible function of the joint effect of multiple SNPs on the imaging traits by specifying a kernel function. It also allows for the possibility that nearby SNPs are likely to interact with each other and the overall effect might be nonlinear.
Assume that there are N unrelated subjects. For each subject i, i = 1,⋯,N, let Yi denote the quantitative imaging trait at a particular voxel. x0,i is a q × 1 vector of the (non-SNP) covariates and Gi,s is the genotype for the SNP s of subject i, which is coded to be the number of copies of the minor allele that subject i possesses for SNP s, and takes the values of 0, 1, or 2. Let Gi = (Gi,1, Gi,2,⋯,Gi,S)T be a S × 1 vector of all the genotypes for subject i. The semi-parametric model for a given voxel is:
| (5) |
where βK is a q × 1 vector of the regression coefficients, h(Gi) denotes a nonparametric function of the SNPs in some function space ℋK, and the errors εK,i are assumed to be normally distributed with mean 0 and standard deviation σK. Eq. (5) models covariate effects parametrically and the joint effect of SNPs nonparametrically. The function space ℋK is determined by a N × N kernel matrix K which is a function of the genotype data and must be positive definite. In particular, the element (j,k) of K is a scalar kernel function k(Gj, Gk) of the SNP genotypes of subjects j and k. There are a variety of choices for the kernel function. In this study, the kernel function proposed by Kwee et al. (2008) is used.
| (6) |
where IBS(Gj,s, Gk,s) denotes the number of alleles shared identical by decent by subjects j and k at the SNP s, and takes values 0, 1, or 2. In this study, we assume IBS = 0 if one of the subjects has missing genotype. We have also tested IBS = 1 and IBS = 2 for missing genotypes and found that this has little effect on the results.
Surprisingly, to make inference on the effect of h(Gi), the nonparametric function h(·) need not be estimated. By using a connection to Linear Mixed Models, a score statistic based on the null (no-SNP) model can be used to test the effect of multiple SNPs on the traits (Liu et al., 2007):
| (7) |
where Y = (Y1,⋯,YN)T, X0 = (x0,1,⋯,x0,N)T, β̂0 and σ̂0 are the maximum-likelihood estimates from the linear regression model Y = X0β0 + ε0. The test statistic Qτ is a quadratic function of Y and follows a mixture of chi-squares under the null hypothesis. See Liu et al. (2007) and Appendix A for more details on the development of the score statistic and the Satterthwaite approximation of Qτ by matching its first two moments with a scaled chi-squared distribution .
For the purpose of inference with random field theory, we assume that the Qτ statistic image behaves as a random field after scaling, where ν̃ = max{1,[ν]}, and [ν] denotes the round function. We estimate the χ2 random field smoothness to be (Worsley, 1994), where FWHM is the error smoothness estimated in the usual way from the regression model Y = X0β0 + ε0 (see Appendix C for the definition of FWHM and details on smoothness estimation). In brief, Resels Per Voxel (RPV) image was estimated using SPM's spm_est_smoothness function in SPM 8, setting defaults.stats.maxres Inf instead of 64 to use all images.
Fast implementation of random field theory — the “small effect” assumption
Random field theory (RFT) finds FWE-corrected p-values for voxel and cluster statistics by accounting for the volume and smoothness of the statistic image (see Appendix C for a review of voxel- and cluster-wise inference). The smoothness is estimated from the standardized residuals and this is a time consuming process, often taking more time than model fitting itself. For single-locus models, the smoothness estimation would have to be repeated NSNP times, once for each SNP. However, we expect the overall fit of the model – and hence the residuals – to be quite similar between different single-locus models. Thus we propose smoothness estimation under a “small effect” assumption. Under this assumption a SNP has such a small impact on the residuals that the single-locus model's smoothness estimate will be well-approximated by the smoothness estimate from the no-SNP model. We evaluate this assumption in the Results section.
Fast permutation and parametric tail approximation
FWE-corrected voxel-wise and cluster-wise inferences are generated for each SNP or gene's statistic image using random field theory. This parametric method is based on a series of assumptions and approximations, and in our previous work we have found that the random field theory can be inaccurate even when the sample size is large (Silver et al., 2011). In these situations, nonparametric methods such as permutation might be a good substitution.
There are different ways to conduct permutation in the presence of nuisance variables (Anderson and Robinson, 2001). For a single-locus model, we randomly permutated the column of X containing the SNP covariate (Kennedy and Cade, 1996). For our multi-locus model, we permute all SNPs together (effectively permuting the rows and columns of K), to preserve the correlation between SNPs while breaking the relationship between SNPs and subjects. To compute FWE-corrected p-values the maximal statistic (voxel or cluster) over the brain is saved for each permutation, Mb = maxv | Tb (v)|, b = 1,…,Nperm (Holmes et al., 1996; Nichols and Hayasaka, 2003); for an observed statistic t then
| (8) |
However, standard permutation procedures are normally too computationally expensive. Specifically, we cannot afford the recommended Nperm = 10,000, nor even 100 permutations on each single SNP. Hence, in the present study, instead of constructing a null maximum statistic distribution (voxel or cluster) for each single SNP, we pooled maximum distributions over SNPs from the whole genome and construct only one null distribution. This is based on the assumption that the null distribution for a single SNP or single gene is similar to the pooled distribution. Again, we evaluate this assumption in the Results section.
As detailed above, image-wise FWE-corrected p-values require a further Bonferroni correction over SNPs or genes, performed by multiplying the p-value by the number of SNPs or genes. Such a severe multiple testing correction relies heavily on precise approximation of small p-values, yet permutation p-values can only be multiples of the inverse of the number of permutations, requiring a large number of permutation samples. For example, for 500,000 SNPs, a of 10−7 is just significant (0.05/500,000 = 10−7); yet pooling 10 permutations per SNP (5 million total!) cannot produce p-values smaller than 10−6.7 (1/5,000,000 = 2 × 10−7). Hence, we introduce a parametric tail approximation method proposed by Knijnenburg et al. (2009) which provides accurate small p-value approximation based on a moderate number of permutation samples.
Specifically, the tail of the permutation distribution is modeled as a generalized Pareto distribution (GPD). It has been demonstrated that the GPD approximates the distribution of the extreme values of a set of independent and identically distributed random variables, i.e. those values that exceed a particular high threshold, using extreme value theory (Gumbel, 2004; Pickands, 1975). The cumulative distribution function (CDF) of GPD takes the form:
| (9) |
where ξ is the shape parameter, τ is the scale parameter and θ is the threshold — only values greater than θ are modeled. The range of t is θ<t<∞ when ξ ≥ 0 and θ<t< − τ/ξ when ξ < 0. If ξ = 0 or ξ = 1, the GPD becomes the exponential and uniform distribution respectively. When ξ > 0, the GPD is equivalent to Pareto distribution which has a long tail. The threshold parameter is set according to the observed tail of the permutation distribution. Knijnenburg et al. (2009) proposed to select θ such that the 250 most extreme samples are included, as this balances the bias and variance of the distribution fit. The other two parameters are fitted using Maximum likelihood (ML) estimation, denoted ξ̂ and τ̂. It has been shown that when ξ > −1/2 the ML estimates exist and are asymptotically efficient (Smith, 1984). In this study, ξ always fell in this range.
For an observed voxel-wise or cluster size statistic t we compute as follows. Create a pooled permutation distribution of the maximal statistics {Mb}, consisting of NPerm elements. If #{Mb ≥ t} ≥ 10 compute the p-value in the conventional way, i.e.,
| (10) |
Otherwise, use a GPD tail approximation
| (11) |
.
Hence, the p-value for an observed statistic that exceeds the largest value in {Mb} can be estimated by extrapolating the fitted GPD.
Simulation-based evaluations of p-values
Monte Carlo simulations based on smooth Gaussian images were carried out for both voxel- and cluster-wise inferences (brain-wise FWE-corrected), based on both T maps generated from the single-locus model and the χ2 maps generated from the multi-locus LSKM approach. Each Gaussian image was generated from a 104 × 104 × 104 white noise image convolved with a Gaussian smoothing kernel. The FWHM was 4.5 voxels to match the smoothness of the real TBM data analyzed in this study. The outer 36 voxels in every direction were truncated after smoothing to avoid nonstationarity at the edge. 10,000 realizations were created, where each realization consisted of 740 scans to match our actual sample size.
For null simulations of the single-locus method, a SNP was randomly selected from real data with the subject ID shuffled. For the multi-locus approach, a set of contiguous SNPs was randomly selected from the real data with the number of SNPs to combine given by a random integer between 1 and 20, reflecting the varying number of SNPs per gene or kb region in the real data analysis; the subject ID was shuffled once for the set of SNPs to reflect the null hypothesis of no association and to retain any LD structure. The cluster forming threshold was set at 0.001 for all cluster size inferences.
Previous work has characterized RFT results for linear regression with structural data (Silver et al., 2011), hence we focused our real data null simulations on LSKM which has not had extensive validation work. To assess the accuracy of LSKM's scaled χ2 approximation on real data, univariate null simulations were conducted. For each of 10,000 realizations, data was obtained from a randomly-selected in-mask voxel, and the multi-locus test run based on, again, between 1 and 20 SNPs with subject ID shuffled (synchronized permutation over SNPs); the puc based on the χ2 approximation was recorded for each realization.
The same approach used for null simulations on Gaussian images was also taken to assess brain-wise FWE-corrected voxel and cluster inferences on real TBM data; again 10,000 realizations were computed, and minimum based on random field theory was recorded for voxel and cluster inferences on each image.
Accurate p-values are uniformly distributed under the null hypothesis, and hence we plot observed ordered p-values versus expected ordered uniform variates to judge their validity. We also plot the 95% confidence interval for each ordered p-value, using the result that the k-th ordered uniform variate follows a Beta distribution ℬ(k, Nsim − k + 1), where Nsim is the number of independent simulations (David and Nagaraja, 1970).
Simulation-based evaluations of the “small effect” assumption
All the RFT-based depend on the “small effect” assumption, i.e., that noise smoothness is accurately estimated by residuals of the no-SNP, demographic-only regression model. Voxel-wise RFT-corrected p-values only depend on the overall FWHM (and the search volume), and thus the accuracy of the voxel-wise RFT inferences depend on this assumption. Thus we compare the FWHM estimated from 1000 randomly selected single-locus models to the FWHM of the no-SNP model.
Cluster size RFT p-values accounting for nonstationarity depend on accurate smoothness estimates at each and every voxel. Verifying the accuracy of the “small effect” assumption for cluster inference is involved, because these local estimates are so noisy. We have relegated the details of the cluster “small effect” validations to Appendix B, but summarized the findings in the Results section below.
Simulation-based evaluations of parametric tail approximation
The fast permutation procedure is based on two assumptions that we need to verify. The first one is that after parametric fitting the tail of a moderate number of permutation samples, small p-values can be well estimated. The second key assumption is that the distribution of the maximum peak or cluster size under null hypothesis pooled from all SNPs is very similar to the null distribution for each single SNP. These two assumptions ensure that we only need to construct one null distribution by pooling SNPs from the whole genome with a moderate number of permutation samples.
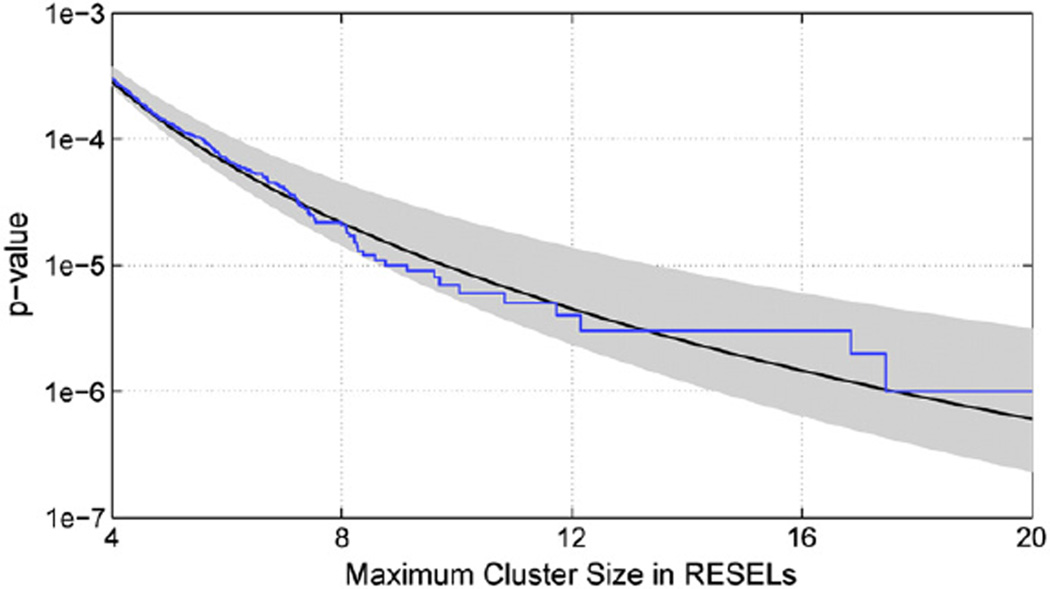
We validated both assumptions with the nonstationarity cluster test. For the first assumption, the distribution of maximum cluster size in RESELs was collected to compute . As a gold standard, we generated a 1,000,000-element permutation distribution of the maximum cluster size, randomly sampling over SNPs (and of course shuffling subject ID). We considered approximating this gold standard with GPD fits to permutation distributions of size 100,000, either (1) drawn randomly from the gold standard distribution, or (2) as an independent sample of permutations. For case (1), we repeated the sampling 1000 times, creating 1000 GPD fits, allowing us to characterize the sampling variability of the GPD fits with confidence intervals.
To validate the second assumption, 5 SNPs with MAF 0.1, 0.2,⋯,0.5 were randomly selected and the null maximum cluster size distribution for each SNP was constructed based on 100,000 permutation samples each. Each of these 5 distributions was compared to the 1,000,000-element gold standard pooled from all SNPs. Considering only the tails (including at least the 250 most extreme permutation samples), a Kolmogorov-Smirnov test was used to detect any differences between the distributions. The tails of the 5 distributions were also assessed by the Anderson-Darling 5-sample procedure to test the hypothesis that the distributions from which the samples were drawn are identical.
Real data application
The TBM ADNI data was analyzed with both single- and multi-locus models, with voxel- and cluster-based inferences. As our null simulations of RFT cluster size inferences showed conservativeness on Gaussian images and dramatic anti-conservativeness on real TBM data for either T or χ2 statistics (see below, Fig. 1), we used our permutation approach for cluster size inferences. Specifically, for the single-locus model, 100,000 permutations were computed, randomly sampling SNPs and permuting subject ID, to create a SNP-pooled permutation distribution, and the GPD tail fit used to find for extreme cluster sizes. Multi-locus models were defined based on SNP groupings described above in the Gene grouping section. A cluster-forming threshold corresponding to univariate puc = 0.001 was used.
Fig. 1.
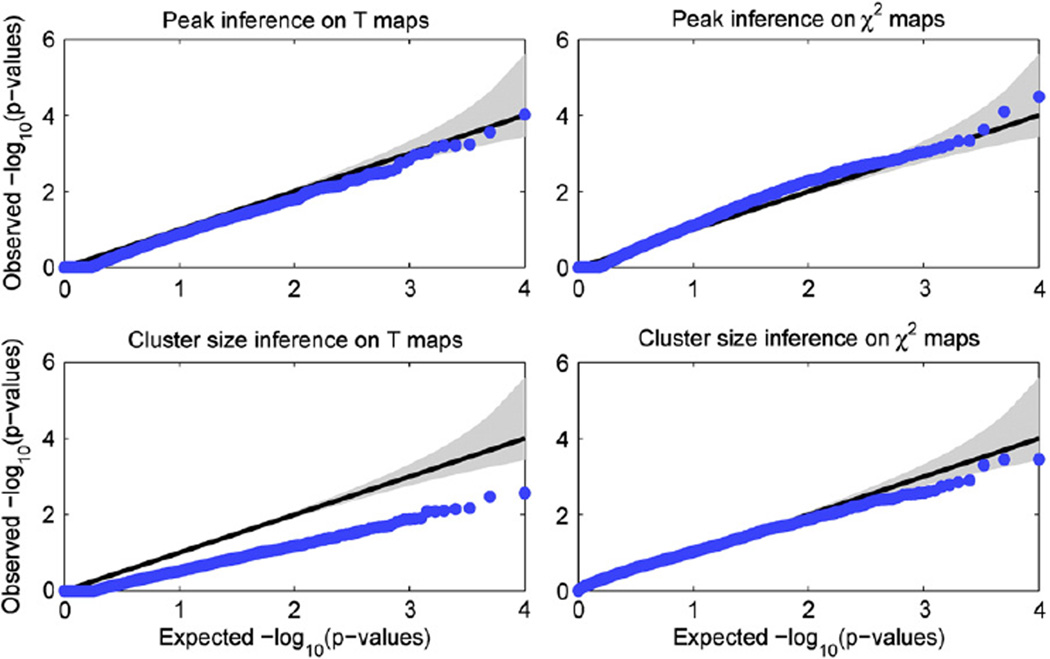
Gaussian null simulations to evaluate validity of brain-wise FWE-corrected p-values. Q–Q plots of for the four combinations of voxel vs. cluster and single-locus T vs. multi-locus χ2 tests, showing that voxel-wise inferences are valid, while cluster size inferences are also valid but somewhat conservative. For each case 10,000 simulations were carried out, with SNPs randomly selected and their subject IDs shuffled; cluster forming threshold was set at 0.001. Gray region shows 95% confidence bands (see text for details).
Results
Null simulations on generated Gaussian images
Fig. 1 shows the Q–Q plots for the four combinations of voxel vs. cluster and single-locus T vs. multi-locus χ2 tests, with the confidence intervals based on the Beta distribution shown by the gray region. It can be seen that for voxel-wise inferences, small p-values are well within the confidence intervals while for cluster size inferences the results are conservative but valid.
Null simulations on real TBM images
Voxel-wise least square kernel machines
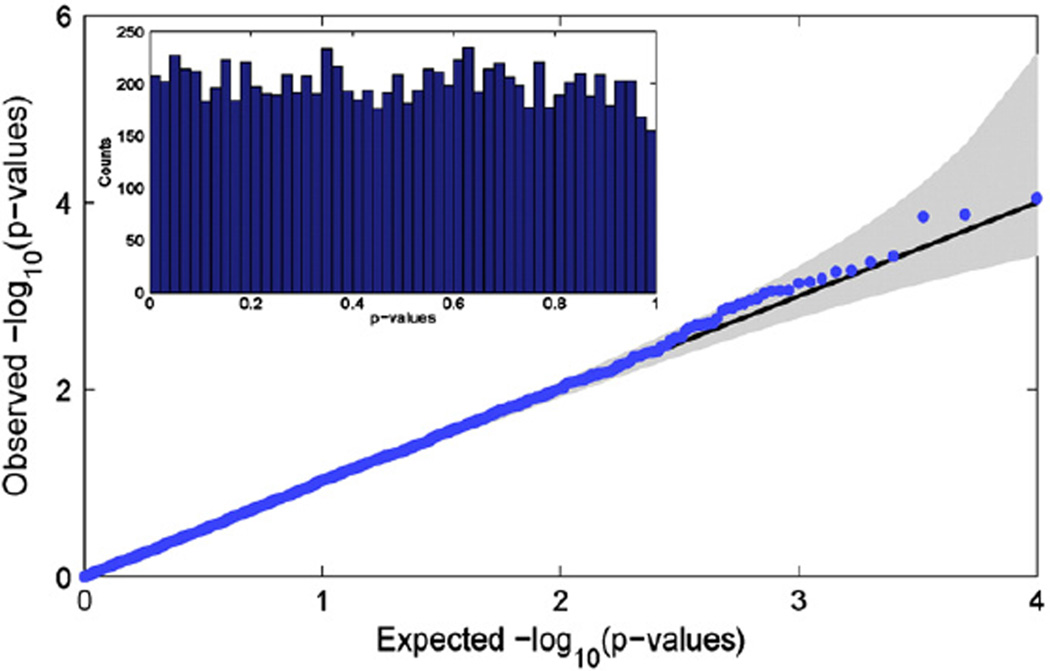
Fig. 2 shows a Q–Q plot for the observed distribution of null real data p-values against a uniform distribution. The embedded panel of Fig. 2 shows a histogram of the p-values. Both plots clearly show that the null p-values approximately follow a uniform distribution on the interval [0,1], providing evidence that the LSKM produces accurate uncorrected inferences based on the possibly non-Gaussian TBM data (Leow et al., 2007).
Fig. 2.
Real data null simulations to evaluate LSKM uncorrected p-values. Q–Q plot (main figure) and histogram (inset) of puc from the LSKM multi-locus model, showing that approximate χ2 distribution appears accurate. 10,000 realizations were created from randomly selected voxels and 1–20 randomly selected SNPs with their IDs shuffled.
Null simulations on real image data
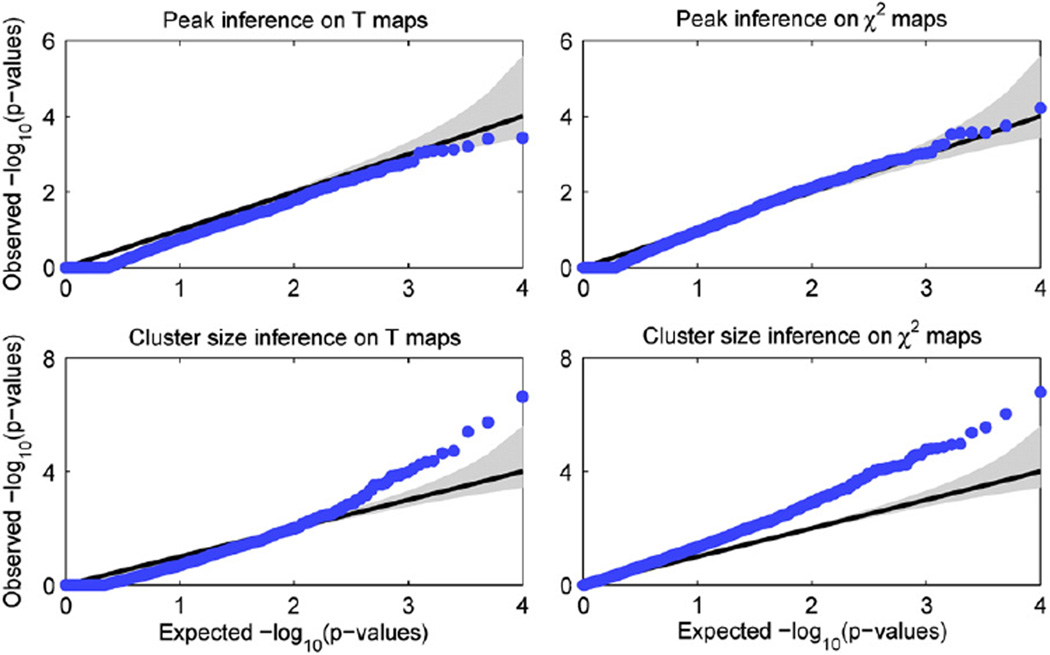
Fig. 3 shows the Q–Q plots for the four combinations of voxel vs. cluster and single-locus T vs. multi-locus χ2 tests on real TBM images, with the confidence intervals based on the Beta distribution shown by the gray region. It can be seen that for voxel-wise inferences, small p-values are well behaved for values more extreme than 0.1 if slightly conservative for the single-locus model, while for cluster size inferences, the results show dramatic anti-conservativeness and are invalid. Note that random field theory provides a good approximation only when the peak is high and, in particular, when the global maximum is small the algorithm might return p-values greater than 1, reflected in the truncated values in the lower left of the Q–Q plot.
Fig. 3.
Real data null simulations to evaluate validity of brain-wise FWE-corrected p-values. Q–Q plots of for the four combinations of voxel vs. cluster and single-locus T vs. multi-locus χ2 tests, showing that voxel-wise inferences appear to be valid, while cluster size inferences are dramatically anti-conservative and invalid. For each case 10,000 simulations were carried out, with SNPs randomly selected and their subject IDs shuffled; cluster forming threshold was set at 0.001. Gray region shows 95% confidence bands (see text for details).
Validation of the “small effect” assumption
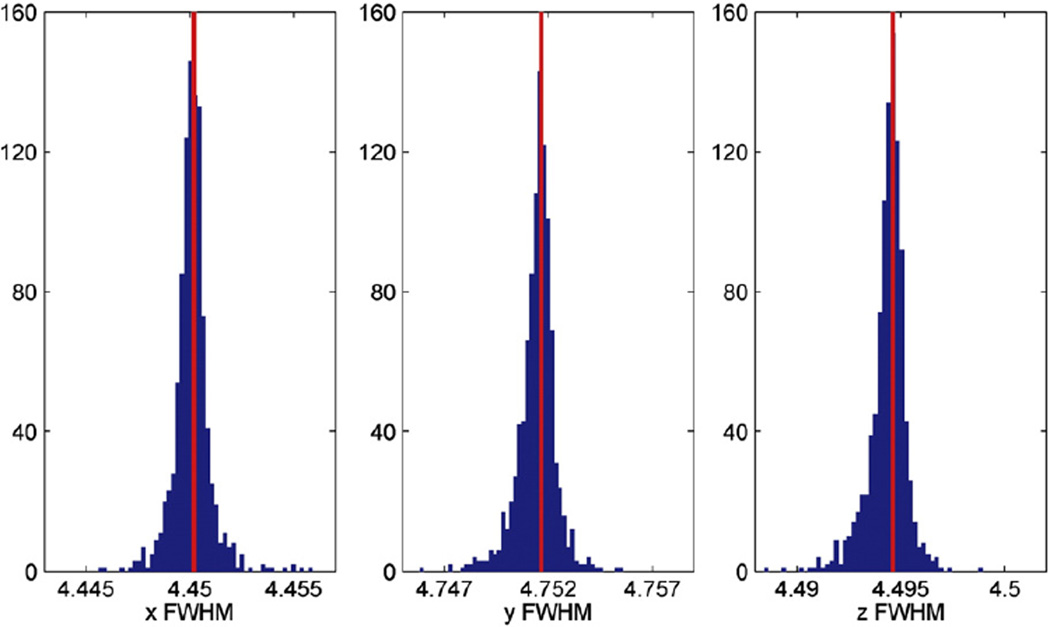
Fig. 4 shows the histograms of the FWHM in voxel units estimated in three directions, for 1000 randomly selected SNPs (blue) and the FWHM from the no-SNP model marked in red. It can be seen that accounting for SNP information has only a very small effect on the estimates, leading to a difference of FWHM within 0.005 voxel in any direction.
Fig. 4.
Validation of the “small effect” assumption for FWHM estimation. Histograms of the FWHM in voxels estimated in three directions, for 1000 randomly selected SNPs (blue) and the FWHM from the no-SNP model marked in red. The FWHM from the no-SNP model appears to be an excellent surrogate for individual SNP models, rarely differing by more than 0.005 in any direction.
Details of the nonstationarity cluster size “small effect” validations are in Appendix B. In short, we found that using the no-SNP's voxel-wise smoothness estimates (in place of each SNP's estimates) resulted in a tiny, at most conservative perturbation on nonstationary cluster size significance.
Validation of fast permutation and parametric tail estimation
Fig. 5 shows the “gold standard” distribution from 1 million permutations (blue), and the GPD fit based on one independent sample of 100,000 permutations (black line), as well as shaded region showing the 95% confidence band (gray) based on 2.5 and 97.5 percentiles of the 1000 repeated GPD fits (each based on 100,000 permutations). The Kolmogorov–Smirnov test found no difference between the tails of the individual and pooled permutation distributions (all p-values were greater than 0.2). The Anderson–Darling 5-sample test also found no evidence that the samples were drawn from different distributions (p-value greater than 0.12).
Fig. 5.
Validation of parametric tail approximation to nonparametric permutation distribution. The “gold standard” distribution from 1 million permutations (blue), and the GPD fit based on one independent sample of 100,000 permutations (black line) shows excellent agreement. The shaded region shows the 95% confidence band (gray) based on 2.5 and 97.5 percentiles of the 1000 repeated GPD fits (each based on 100,000 permutations). The tails were truncated and 250 of the most extreme permutation values were used in all parametric fittings.
Brain-wide genome-wide association studies
We first review the real-data results using single-locus model, then with the LSKM multi-locus method.
Single-locus results
Table 2 shows the 20 top associated SNPs, as ranked by their minimum voxel-wise FWE-corrected as found by random field theory. No brain-wide genome-wide significant association was identified.
Table 2.
Voxel-wise inference with the single-locus model: The top 20 most associated SNPs ranked by voxel-wise from RFT.
| Chr | SNP (gene) | Min. puc | Min. voxel | Talairach coordinates (mm) |
|---|---|---|---|---|
| 6 | rs2132683 | 2.419 × 10−10 | 6.388 × 10−6 | [−36, −68, 4] |
| 6 | rs713155 | 2.945 × 10−10 | 7.649 × 10−6 | [4, −52, 0] |
| 1 | rs476463 (CSMD2) | 3.010 × 10−10 | 7.817 × 10−6 | [12, −36, 64] |
| 7 | rs2429582 (AK025672) | 1.002 × 10−10 | 1.016 × 10−5 | [68, −4, −8] |
| 3 | rs9990343 | 5.061 × 10−10 | 1.256 × 10−5 | [28, 0, 56] |
| 11 | rs490592 | 1.322 × 10−9 | 3.061 × 10−5 | [20, 24, 32] |
| 20 | rs11696501 | 1.339 × 10−9 | 3.101 × 10−5 | [−28, −44, −4] |
| 3 | rs10511089 | 1.704 × 10−9 | 3.849 × 10−5 | [8, −92, 12] |
| 8 | rs4534106 | 2.161 × 10−9 | 4.814 × 10−5 | [−60, −28, −4] |
| 6 | rs11970254 | 2.175 × 10−9 | 4.846 × 10−5 | [64, −8, 4] |
| 9 | rs7025303 (X75342) | 2.192 × 10−9 | 4.872 × 10−5 | [−12, −68, 28] |
| 9 | rs7873102 (X75342) | 2.251 × 10−9 | 4.982 × 10−5 | [−12, −68, 28] |
| 1 | rs710865 (KIAA0090) | 2.519 × 10−9 | 5.560 × 10−5 | [−28, −44, −40] |
| 6 | rs9362770 | 2.831 × 10−9 | 6.179 × 10−5 | [4, −52, 0] |
| 20 | rs2073233 | 3.023 × 10−9 | 6.528 × 10−5 | [36, −104, 16] |
| 9 | rs776019 (X75342) | 3.500 × 10−9 | 7.527 × 10−5 | [−12, −68, 28] |
| 2 | rs12479254 (BOK) | 3.689 × 10−9 | 7.902 × 10−5 | [32, −44, −8] |
| 16 | rs11643520 | 4.186 × 10−9 | 8.827 × 10−5 | [−60, −76, −12] |
| 5 | rs4296809 | 4.200 × 10−9 | 8.857 × 10−5 | [36, −40, 4] |
| 9 | rs776015 | 4.362 × 10−9 | 9.195 × 10−5 | [−12, −68, 28] |
Table 3 shows the 20 top associated SNPs ranked by their minimum cluster size FWE-corrected as found with our nonparametric permutation described above. The volume of the largest cluster is shown in both voxel and RESEL count. Again no brain-wide genome-wide significant association was identified. Note that a larger volume in voxels does not necessarily lead to a larger size in RESELs since the cluster might be located in smooth regions where the RESEL count is small. Also observe that large clusters do not necessarily have high voxel-wise peaks.
Table 3.
Cluster size inference with the single-locus model: The top 20 most associated SNPs ranked by cluster from permutation.
| Chr | SNP (gene) | # of clusters | Largest cluster in RESELs (voxels) | Min. puc in the largest cluster | Min. cluster |
|---|---|---|---|---|---|
| 16 | rs7196505 | 5 | 12.8028 (1451) | 8.271 × 10−7 | 3.535 × 10−6 |
| 16 | rs2160315 | 9 | 12.1533 (1392) | 1.310 × 10−6 | 4.330 × 10−6 |
| 5 | rs4866878 | 4 | 10.7658 (1495) | 1.163 × 10−8 | 6.933 × 10−6 |
| 11 | rs490592 | 8 | 10.7446 (1406) | 2.434 × 10−7 | 6.986 × 10−6 |
| 8 | rs2279970 | 18 | 9.6840 (1049) | 3.429 × 10−8 | 1.044 × 10−5 |
| 11 | rs555097 | 24 | 9.6079 (1363) | 8.762 × 10−7 | 1.076 × 10−5 |
| 8 | rs10504140 (ST18) | 19 | 8.8353 (1604) | 1.179 × 10−7 | 1.485 × 10−5 |
| 5 | rs4296809 | 4 | 8.5050 (1155) | 4.200 × 10−9 | 1.719 × 10−5 |
| 2 | rs2300477 (MEIS1) | 23 | 8.4740 (926) | 7.728 × 10−8 | 1.743 × 10−5 |
| 3 | rs2929358 (AK097644) | 18 | 8.1639 (1099) | 2.117 × 10−7 | 2.010 × 10−5 |
| 12 | rs1108923 (ACCN2) | 4 | 7.8121 (993) | 1.789 × 10−6 | 2.377 × 10−5 |
| 8 | rs4639533 (ST18) | 14 | 7.6760 (1628) | 2.046 × 10−7 | 2.542 × 10−5 |
| 2 | rs1347044 (NAP5) | 4 | 7.4795 (685) | 1.546 × 10−5 | 2.805 × 10−5 |
| 3 | rs303860 (C3orf31) | 1 | 7.2479 (1566) | 5.894 × 10−7 | 3.160 × 10−5 |
| 11 | rs10501511 | 2 | 7.1783 (1376) | 5.453 × 10−7 | 3.278 × 10−5 |
| 18 | rs17525599 (AL833817) | 7 | 7.1288 (1192) | 5.036 × 10−7 | 3.365 × 10−5 |
| 8 | rs7001339 | 7 | 7.1250 (1356) | 9.435 × 10−7 | 3.372 × 10−5 |
| 19 | rs10402592 (SPC24) | 10 | 7.1123 (1421) | 3.132 × 10−7 | 3.394 × 10−5 |
| 1 | rs4839223 (SLC6A17) | 9 | 6.9098 (1273) | 1.082 × 10−6 | 3.786 × 10−5 |
| 5 | rs13154903 | 5 | 6.8379 (1311) | 4.169 × 10−8 | 3.939 × 10−5 |
Multi-locus results
Table 4 shows the top 20 most associated genes from voxel-wise inferences using LSKM by combining all the SNPs located in each gene. The top 12 genes are significant after brain-genome-wise correction.
Table 4.
Voxel-wise inference with the multi-locus model: The 20 most associated genes ranked by voxel-wise from RFT.
| Chr | Gene | # of SNP in the gene | Min. puc | Min. voxel | Min. voxel | Talairach coordinates (mm) |
|---|---|---|---|---|---|---|
| 12 | GRIN2B | 131 | 1.167 × 10−13 | 3.213 × 10−9 | 5.798 × 10−5 | [20, −84, 44] |
| 9 | X75342 | 25 | 1.963 × 10−13 | 6.414 × 10−9 | 1.157 × 10−4 | [−12, −72, 32] |
| 7 | AK025672 | 42 | 2.214 × 10−12 | 1.072 × 10−7 | 0.0019 | [68, −4, −12] |
| 1 | PGM1 | 13 | 8.274 × 10−12 | 2.290 × 10−7 | 0.0041 | [−48, −76, −16] |
| 1 | BC022483 | 4 | 5.062 × 10−12 | 3.222 × 10−7 | 0.0058 | [12, 40, 4] |
| 1 | AJ249210 | 22 | 8.488 × 10−12 | 3.441 × 10−7 | 0.0062 | [18, 13, 21] |
| 13 | FARP1 | 101 | 1.601 × 10−11 | 6.010 × 10−7 | 0.0108 | [−40, −84, −12] |
| 18 | C18orf58 | 28 | 1.910 × 10−11 | 8.953 × 10−7 | 0.0162 | [−40, −44, 20] |
| 5 | AK092765 | 7 | 8.528 × 10−11 | 1.505 × 10−6 | 0.0272 | [0, 24, 8] |
| 6 | FUT9 | 38 | 7.529 × 10−11 | 1.770 × 10−6 | 0.0319 | [−44, −60, −12] |
| 10 | U69546 | 52 | 9.544 × 10−11 | 2.271 × 10−6 | 0.0410 | [32, −92, −4] |
| 10 | AK131357 | 22 | 9.058 × 10−11 | 2.422 × 10−6 | 0.0437 | [−20, −56, −40] |
| 1 | BSND | 3 | 5.155 × 10−11 | 3.091 × 10−6 | 0.0558 | [28, 32, −12] |
| 1 | KIAA0090 | 7 | 6.186 × 10−11 | 3.736 × 10−6 | 0.0674 | [−28, −44, −40] |
| 2 | GALNT14 | 47 | 1.465 × 10−10 | 5.591 × 10−6 | 0.1009 | [8, −52, 72] |
| 6 | AK126371 | 7 | 2.521 × 10−10 | 6.326 × 10−6 | 0.1141 | [40, −4, −36] |
| 2 | EPHA4 | 41 | 1.705 × 10−10 | 7.940 × 10−6 | 0.1433 | [12, −52, 72] |
| 12 | THRAP2 | 24 | 3.657 × 10−10 | 8.322 × 10−6 | 0.1502 | [−64, −20, 36] |
| 8 | COL14A1 | 44 | 1.900 × 10−10 | 8.901 × 10−6 | 0.1606 | [−28, −20, 28] |
| 14 | ACIN1 | 6 | 2.847 × 10−10 | 9.994 × 10−6 | 0.1803 | [76, −44, −4] |
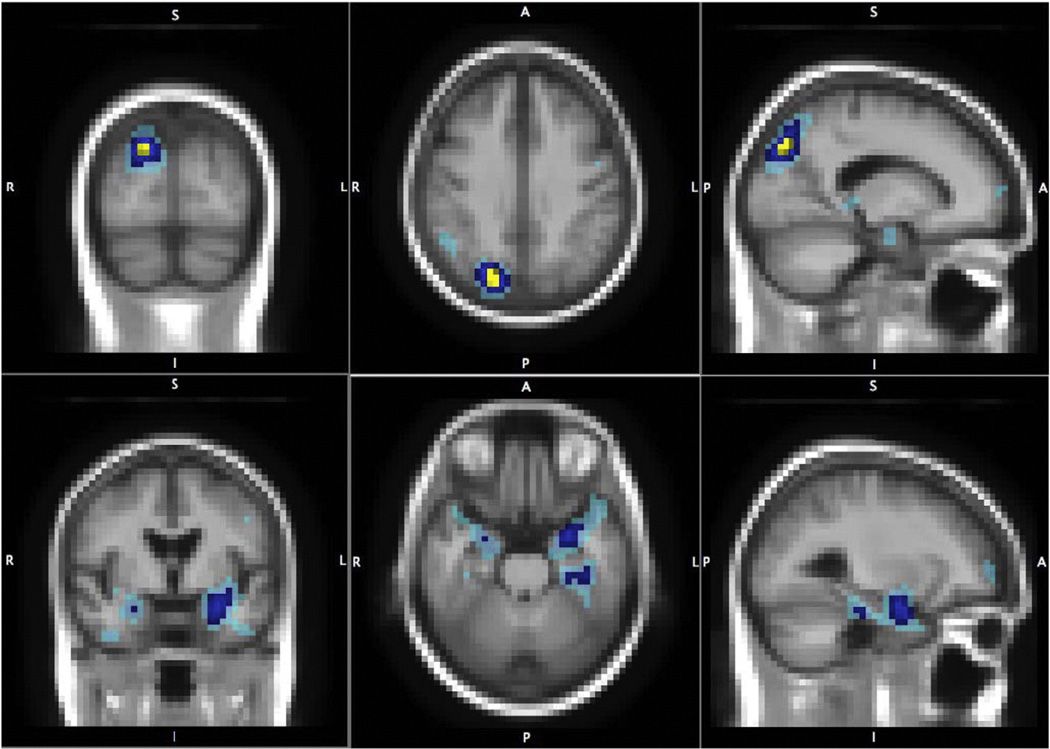
To further understand the top hit from the voxel-wise analysis, Table 5 shows the top 10 most associated SNPs located on the gene GRIN2B with their minor allele frequency, the number of subjects in each genotypic group, the minimum uncorrected LSKM p-value puc (i.e. LSKM model applied with 1 SNP), the image-wise corrected p-value and the location of the peak in the brain. Note that all the SNPs are common variants and the smallest phenotypic group has more than 60 subjects, suggesting that the GRIN2B effect is not simply driven by rare SNPs or a small number of subjects in the population. This also provides a post-hoc approach to locate the driving markers in the gene and validate the findings. Two SNPs, rs10845840 and rs11055612, were identified by a former study to have association with the temporal lobe atrophy using the same ADNI dataset (Stein et al., 2010b). In the present study, the effect of each single SNP does not have genome-wide significance. However, reviewing the single-locus statistic images for these top SNPs (not shown), most show an effect in the parietal and temporal lobes (see Table 5). When the LSKM test-statistic combines all the GRIN2B SNPs together, however, significant effects are found in these two regions (Fig. 6).
Table 5.
Voxel-wise inference with the multi-locus model in GRIN2B: The top 10 most associated SNPs in GRIN2B ranked by voxel-wise .
| SNP | MAF | Number of subjects in each genotypic group1 |
Min. LSKM puc | Corrected voxel | Talairach coordinates (mm) | ||
|---|---|---|---|---|---|---|---|
| Maj | Het | Min | |||||
| rs10845840 | 0.4412 | 226 | 375 | 139 | 5.388 × 10−9 | 1.694 × 10−4 | [−40, 16, −24] |
| rs7301754 | 0.3101 | 356 | 299 | 80 | 7.422 × 10−9 | 2.817 × 10−4 | [24, −84, 48] |
| rs2216344 | 0.4534 | 218 | 369 | 151 | 1.976 × 10−8 | 5.087 × 10−4 | [16, −88, 40] |
| rs220573 | 0.3480 | 316 | 333 | 91 | 2.211 × 10−8 | 8.079 × 10−4 | [20, −84, 44] |
| rs220575 | 0.3480 | 316 | 333 | 91 | 2.211 × 10−8 | 8.079 × 10−4 | [20, −84, 44] |
| rs11055612 | 0.4953 | 179 | 381 | 176 | 6.147 × 10−8 | 1.300 × 10−3 | [−40, 16, −24] |
| rs918168 | 0.2959 | 363 | 316 | 61 | 7.654 × 10−8 | 4.722 × 10−4 | [−40, 48, 4] |
| rs2300267 | 0.4041 | 271 | 336 | 131 | 1.730 × 10−7 | 4.100 × 10−3 | [20, −84, 44] |
| rs2160730 | 0.3730 | 288 | 346 | 103 | 2.038 × 10−7 | 4.900 × 10−3 | [4, 48, 36] |
| rs11055651 | 0.3378 | 295 | 328 | 86 | 2.521 × 10−7 | 2.600 × 10−3 | [48, −80, −28] |
Homozygous major allele, Maj; Heterozygous, Het; Homozygous minor allele, Min. Note that the number of subjects in each genotype group may not add up to the total number of subjects if data is missing for that SNP in some subjects.
Fig. 6.
Multi-locus LSKM results for gene GRIN2B. The parietal (upper panels) and temporal (lower panels) foci that the most associated gene GRIN2B affects in the brain. Brain-wide genome-wide significant voxels are in yellow; brain-wide (post hoc gene-wise) significant voxels are in dark blue; and 0.001 uncorrected significant voxels are in light blue. Both parietal and temporal lobes are known to be affected in Alzheimer's disease.
In addition to GRIN2B, several other genes were detected to have significant association after correction for the whole genome. Many of them have been widely studied, closely related to AD or identified by other genome-wide association studies. See the Discussion part for details on their biological significance.
Finally, we attempted to generalize our nonparametric permutation approach to the multi-locus LSKM model, but found that different combinations of SNPs show substantially different null distributions with the result that the pooled distribution is not representative of individual null distributions. For example, we constructed the null maximum cluster size distributions for 10 adjacent SNPs on the genome with small average MAFs and another 10 adjacent SNPs with large MAFs based on 100,000 permutation samples, and found significantly different tail distributions (Kolmogorov–Smirnov test p<0.05).
Discussion
In this paper, we presented a fast implementation of random field theory and permutation inference for imaging genetics applications. Both voxel- and cluster-wise inferences were introduced to detect localized high-intensity effects or spatial distributed signals. This approach was combined with a multi-locus model based on LSKMs to model the interaction of genetic variants in each gene. The method was illustrated on both T and χ2 maps, generated from single-locus and multi-locus model respectively and validated on both generated Gaussian images and TBM data. When random field theory fails to work on real data, a fast permutation procedure was also proposed and a parametric tail approximation was used to estimate small p-values with a moderate number of permutation samples. By applying the methods to testing the association between 448,294 SNPs and 18,043 genes in 31,662 voxels of the entire brain across 740 elderly subjects from the Alzheimer's Disease Neuroimaging Initiative (ADNI), several significantly associated genes were identified. Some of them have been widely studied and closely related to AD, suggesting the validity of the approach.
A head-to-head comparison of our single-locus results based on voxel-wise inferences with those of the vGWAS study (Stein et al., 2010a) shows a large overlap on the top SNPs identified. No brain-wide genome-wide significant association was found in both studies but RFT-corrected p-values are slightly more significant than a brain-wise Bonferroni correction, indicating a power gain by using the spatial information in 3D images.
A comparison of our single-locus results based on cluster size inferences with the findings of the vGWAS study (Stein et al., 2010a) identifies only two SNPs in common, rs4296809 and rs490592, ranking in the top 20 in both studies. These two SNPs had the largest “winning” volumes in the vGWAS study although they did not have the highest intensity (Stein et al., 2010a), and similarly we found these two SNPs brain-wide significant (although not brain-wide genome-wide significant) by cluster extent. This sheds light on the difference between cluster size inferences and voxel-wise intensity studies. Cluster size inferences favor those SNPs that have distributed effects on brain structure. There may be relevant SNPs that influence brain structure but are never the top SNP at any voxel, and thus missed by voxel-wise studies.
A comparison of our multi-locus results based on voxel-wise inferences with the vGeneWAS study (Hibar et al., 2011b) finds no overlap at all. An important difference is that the vGeneWAS work used principal components regression to extract orthogonal components that explain a desired proportion of variance of a set of predictors, and then built a multiple partial-F regression model. The different findings, and our work's greater statistical significance, may be attributed to LSKM being able to capture the interaction of multiple SNPs.
Finally, note that while we have used Bonferroni with NSNP or Ngene, previous works used a reduced “effective N” over SNPs or genes (Hibar et al., 2011b; Stein et al., 2010a), and hence our brain- and genome-wise FWE p-values could be improved slightly.
Methodology assessment
While the single-locus model did not identify any significant SNPs with voxel or nonstationary cluster size inferences, several genes we identified in our multi-locus analysis have been shown to be associated to AD after full image-wise, genome-wise correction, while previous work has not found any (Hibar et al., 2011b). The increased sensitivity should be attributed to both the more sensitive LSKM methods and the random field theory. The LSKM provides a flexible approach to model high-dimensional interactions, and the kernel based on identical by state across subjects is likely a more biologically plausible way to account for the interaction and correlation between SNPs in each gene than simply explaining the variance of data empirically (Hibar et al., 2011b). The random field theory, on the other hand, fully takes advantage of the spatial information in images and tackles the multiple testing problem by accounting for searching over space. This implicitly takes into account the high correlation among voxels and thus provides more statistical power than correcting on voxel-wise results. Furthermore, the fast implementation of random field theory reduces the computational efforts dramatically. It took less than 2 days to search the whole genome using LSKMs on one desktop. SNP-by-SNP association tests can also be completed within 3 to 4 days if the tasks are distributed to 10 CPUs. Although we only illustrated the fast implementation of random field theory by T and χ2 fields based on a single-locus and a multi-locus model, it is applicable to any statistic produced by a general linear model, e.g., F test. However, the results from random field theory are based on a series of assumptions and approximations. When dealing with a specific dataset, null simulations should be carried out before making any statistical inferences to ensure that the false positive rate is not inflated and small p-values are well-behaved. In the present study, we found that the nonstationary cluster size test does not work on the TBM dataset, with false positives exceeding the nominal rate. But when simulated on generated Gaussian data, nonstationary cluster size inferences were valid and only slightly conservative. This indicates that cluster size inferences might be sensitive to the RPV structure in nonstationary datasets, but that it may work well in fMRI data which has more homogeneous smoothness. This needs to be tested in the future.
The fast implementation of the random field theory is based on an assumption that each SNP has only a subtle effect on the smoothness of standard residual images and will hardly change the smoothness estimates. Our simulations have shown that a “No-SNP” model's residuals have nearly identical smoothness to models with a SNP regressor. However, there is still a subtle dependence between cluster size and local smoothness when comparing different models with two different SNPs. For large clusters, there is a systematic deflation of cluster size when an alternative, independent RPV image is used to measure its size. On balance, these results show that the “small effect” assumption is appropriate and at worst a bit conservative for estimating cluster size.
Biological significance
Several genes were identified to have significant associations. The most significantly associated gene in the study is GRIN2B, which encodes the N-methyl-d-aspartate (NMDA) glutamate receptor NR2B subunit and affects both the parietal and temporal lobes in human brains (Fig. 6). This protein is well known to be involved in learning and memory (Tang et al., 1999), structural plasticity of the brain (Lamprecht and LeDoux, 2004) and excitotoxic cell death (Parsons et al., 2007), and has age-dependent prevalence in the synapse (Yashiro and Philpot, 2008). Therefore, the relationship between NR2B subunit gene (GRIN2B) variants and AD has attracted a large amount of attention and interests. Many studies have confirmed that NR2B subunit is down-regulated significantly in susceptible regions of AD brains (Bi and Sze, 2002; Farber et al., 1998; Hynd et al., 2004). Actually GRIN2B is already a therapeutic target in Alzheimer's disease and has also been spotted by several studies in the literature (Jiang and Jia, 2009; Stein et al., 2010b).
The gene PGM1 encodes the protein Phosphoglucomutase-1. The level of this enzyme was found to be significantly altered in the hippocampus of the patients who suffered from AD compared with control hippocampus using two-dimensional gel electrophoresis and mass spectrometry techniques (Sultana et al., 2007). Down regulation of this gene might have an effect on the memory and cognitive functions in human brains.
The gene EPHA4 encodes the EPH receptor A4 (Ephrin type-A receptor 4) in humans. This gene belongs to the ephrin receptor subfamily of the protein-tyrosine kinase family (Fox et al., 1995). EPH and EPH-related receptors have been implicated in mediating developmental events in the nervous system and regulating excitatory neuro-transmission. Early reduction of hippocampal EPH receptors was found to underlie cognitive impairment in AD by a modeling study (Simón et al., 2009). EPHA4 is also involved in intracellular signaling in the morphogenesis of dendritic spines and the processing of EPHA4 by gamma-secretase might affect the pathogenesis of AD (Inoue et al., 2009). A whole genome association study of brain-wide imaging phenotypes based on the ADNI cohort also identified SNPs that are proximal to the EPHA4 gene (Shen et al., 2010).
Many other genes identified, including FARP1, FUT9, BSND, GALNT14, THRAP2, COL14A1 and ACIN1 etc., have well understood gene functions but yet to be directly related to AD, which deserves further investigations.
Acknowledgments
Data used in preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (http://adni.loni.ucla.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.ucla.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott; Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Amorfix Life Sciences Ltd.; AstraZeneca; Bayer HealthCare; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd. and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Rev March 26, 2012 Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129 and K01 AG030514.
T Ge is supported by the China Scholarship Council (CSC). J Feng is a Royal Society Wolfson Research Merit Award holder, partially supported by an EU grant BION, a UK EPSRC grant and National Centre for Mathematics and Interdisciplinary Sciences (NCMIS) in the Chinese Academy of Sciences. DP Hibar is supported by an NSF Graduate Fellowship. PM Thompson is supported by NIH grants HD050735, EB008281 and AG040060. TE Nichols is supported by an MRC grant G0900908.
Appendix A. The score statistic and its Satterthwaite approximation
As shown by Liu et al. (2007), we do not actually need to estimate the function h(·) in Eq. (5). By using a connection to Linear Mixed Models, we can create a score test for the covariance induced by h in the ordinary least square (OLS) model fit ignoring h. Specifically, the analysis of the LSKM model Eq. (5) for a given set of SNPs can be transformed into the following linear mixed model:
| (12) |
where Y, X0 and βK inherit the same meaning above, h is an N × 1 random vector following multivariate normal distribution with mean 0 and covariance matrix τK. εK is also an N × 1 random vector following multivariate normal distribution with mean 0 and covariance matrix . A comparison of Eq. (5) and Eq. (12) indicates that they have exactly the same form except that h is now treated as a random effect. Therefore, the statistical test ℋ0 : h(·) = 0 concerning whether the effect of multiple SNPs is associated with the imaging traits is equivalent to testing the variance component τ as ℋ0 : τ = 0 versus its alternative ℋ1 : τ > 0. Liu et al. (2007) proposed a score statistic which takes the form:
| (13) |
where β̂0 and are the maximum-likelihood estimates of β0 and under the null hypothesis. In other words, they are obtained from the linear regression model Y = X0β0 + ε0.
The Satterthwaite method is then used to approximate the distribution of Qτ by a scaled chi-squared distribution , where the scale parameter κ and the degrees of freedom v are calculated by matching the mean and variance of Qτ and . Liu et al. (2007) showed that κ = Ĩττ/2ẽ and ν = 2ẽ2/Ĩττ, where Ĩττ = Iττ − Iτσ2Iσ2σ2−1Iτσ2T, Iττ = tr(P0K)2/2, Iτσ2 = tr(P0KP0)/2, , ẽ = tr(P0K)/2, and is the residual forming matrix under the null hypothesis. The significance of the test can then be assessed by comparing the scaled score statistic to the chi-squared distribution with εK,i degrees of freedom.
Appendix B. Understanding the “small-effect” assumption
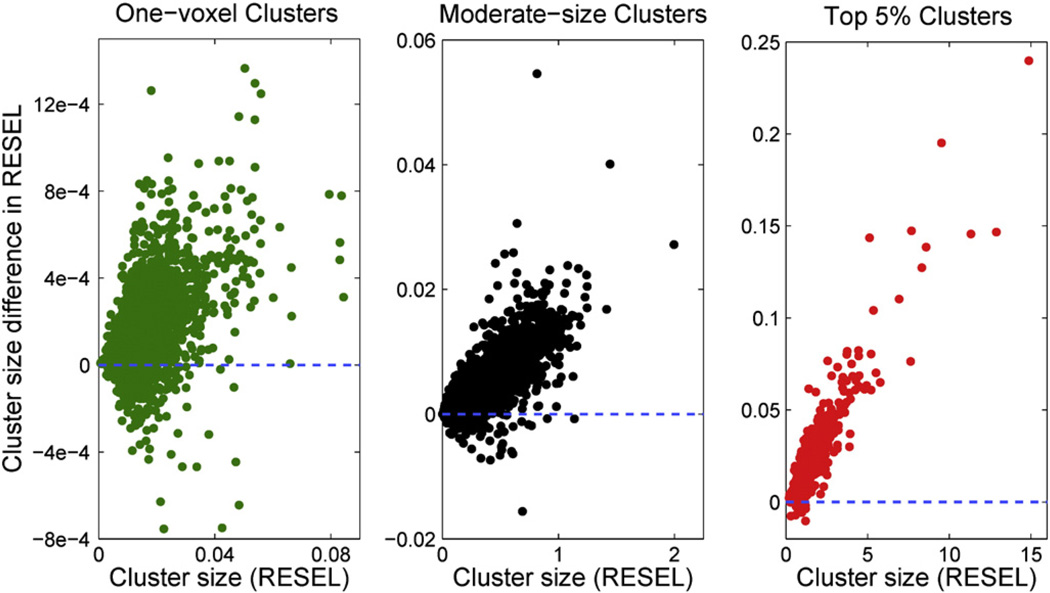
To understand the effect of estimating smoothness with a “No-SNP” model, some further simulations were carried out. For each iteration of the simulation, two SNPs, denoted as SNP-A and SNP-B, were randomly selected and two models, Model-A and Model-B, were set up to test the association between the SNP and the imaging traits. Model-A plays the role of the SNP of interest that we want to make inference on, and Model-B plays the role of the “No-SNP” model used only to create a RPV image. We included a SNP in this “No-SNP” model to match the degrees-of-freedom between the two models and rule-out any systematic difference in the residual standard deviation between the two models. The corresponding RPV images, RPV-A and RPV-B, were estimated after regressing out the SNP information along with demographic covariates. Then all the clusters observed in Model-A were measured using both RPV-A and RPV-B. This procedure was repeated for 1000 times. We found that 86.2% of the clusters in Model-A have a larger cluster size in RESEL units when measured with their own RPV image than with Model-B's RPV. Given that there is very little variation in smoothness between SNP models (see Fig. 4), this suggests a dependence between estimated cluster size and RPV variation.
To understand this dependence, we further categorized all clusters into three classes according to their size in voxel units. The first category contains all one-voxel clusters. The third category comprises clusters whose sizes are in top 5% of all the clusters observed. All the remaining moderate size clusters belong to the second category. Fig. 7 shows the difference in cluster size measured in RESELs, RPV-A–RPV-B, plotted against the true cluster size measured by RPV-A. 76.0% of the one-voxel clusters have larger cluster size when measured with their own RPV images, 90.0% of moderate-sized clusters have a larger size, and 98.7% of the top clusters have a larger size. This is unexpected, since there is negligible difference in smoothness between SNPs (see Fig. 4). Given that a cluster in Model-A has exactly the same voxels when measured with RPV-A or RPV-B, the difference in cluster size in RESELs is only explained by RPV-A being larger then RPV-B in that region; that is, Model-A's residual errors are rougher than Model-B's precisely where Model-A has large clusters. One explanation for this apparent positive correlation between cluster size (in RESELs) and local roughness is the role of unmodelled variation of SNP-A in Model-B. A large cluster in Model-A means there is a contiguous region where SNP-A explains variation in the data, but since Model-B lacks SNP-A, Model-B must have some spatially structured lack-of-fit; this lack-of-fit will add spatial structure to the residuals of Model-B in this region and can inflate the local smoothness estimate (i.e. decrease roughness).
Fig. 7.
Intrinsic correlation between the cluster size and the corresponding RPV image. Clusters observed in Model-A were measured by RPV-A and an alternative RPV image, RPV-B, estimated from a different model. Three scatter plots are presented where the difference in cluster size measured in RESELs, RPV-A–RPV-B, is plotted against the true cluster size measured by RPV-A. The left panel is for all one-voxel clusters. The right panel is for the top 5% largest clusters. All moderate size clusters are shown in the middle panel.
In short, it appears that the impact of the “small effect” assumption is to at most underestimate cluster size in RESELs, and thus produce a slightly conservative p-value.
Appendix C. Random field theory
Random field theory is a powerful tool to approximate the probability of voxels exceeding some threshold or the probability of clusters exceeding a certain size. These tools depend on a number of assumptions including:
Lattice approximation — Images are realizations of a smooth random field sampled at regular points on a lattice; sampling is assumed to be fine enough to capture the local features of the field;
Large search region — Search volume is large compared to the smoothness of the images;
Image smoothness — Images are smooth at the voxel scale, that is, their smoothness in terms of FWHM is relatively large compared to the voxel size;
High threshold — Threshold for both voxel and cluster size inferences should be sufficiently high since many theoretical approximations are derived asymptotically (Adler, 1981; Cao, 1999);
Stationarity — Uniform smoothness for stationary cluster size inferences. This ensures that the null distribution of cluster size is homogeneous.
These assumptions usually present practical difficulties for the use of random field theory in low smoothness or with low degrees of freedom (Hayasaka and Nichols, 2003). Careful evaluations are advised when applying the method on new settings (Meyer-Lindenberg et al., 2008; Silver et al., 2011).
C.1. Voxel-wise inferences
Voxel-wise inference refers to assessing the probability of observing at least one activation/significant association for a particular random field which could be Gaussian, T, F, χ2 etc. Denote the corresponding test statistic as Z, this is equivalent to estimate the probability P(max Z > uc), where uc is a pre-defined threshold. No exact result for this probability exists. However, at high threshold, it has been shown that each connected component of the excursion set is convex, i.e., the holes and hollows disappear (Adler, 1981; Cao, 1999). Hence, the number of local maxima and the number of connected excursion components, denoted as m, converge. Furthermore, for even higher threshold, m counts one if the global maximum exceeds uc and zero otherwise. Therefore, P(max Z > uc) can be approximated by E{m}, the expectation of m. The Euler characteristic (EC) counts the number of connected components of the excursion set, minus the number of “holes” plus the number of “hollows” (Worsley, 1996; Worsley et al., 1992), and thus coincides with m at high threshold. More importantly, EC is amiable to probabilistic analysis under the assumption that the excursion set does not intersect with the boundary (Adler, 1981). When the search region is sufficiently large with respect to the smoothness of the images, the violation of this assumption results in minor error. To summarize, at high threshold uc, we have (Worsley et al., 1996)
| (14) |
where D is the dimension of search area, Rd is the unitless resel count, which is a d-dimensional feature of the search area V, closely related to the smoothness of images. ρd is the EC density which only depends on the threshold uc and the type of statistical field. For detailed calculations of Rd, d = 0, 1, 2, 3 and a list of ρd, d = 0, 1, 2, 3 for different fields, see Worsley et al. (1996).
C.2. Stationary cluster size inferences
For cluster size inference, the particular problem to address is the probability of observing at least one cluster above a certain size s (Friston et al., 1994; Friston et al., 1996). This is equivalent to the probability that the largest cluster has a greater size than s, i.e., P(max S > s). Again no exact result exists. To estimate this probability, three variables are of interest: The number of voxels above a threshold uc denoted as N, the number of connected component of excursion set m as above, and the number of voxels in a randomly selected cluster, denoted S. The expectations of the three random variables are linked by
| (15) |
E{N} can simply be obtained by
| (16) |
where V is the search volume and F(·) is the cumulative distribution function depending on the type of the underlying random field. E{m} is estimated by the expectation of Euler characteristic as in Eq. (14). It is known that at high threshold, the number of components of excursion set m follows a Poisson distribution (Adler, 1981; Cao, 1999):
| (17) |
Therefore, P(max S > s) can be calculated as
| (18) |
This is known as the Poisson clumping heuristic (Aldous, 1989). Finally, one needs to estimate the probability P(S > s) in Eq. (18). It has been shown that at high levels near local maxima, the behavior of the random fields can be approximated by a particular quadratic form or its inverse, depending on the particular type of the field (Adler, 1981; Cao, 1999). Hence, the size distribution of the connected components of the excursion set S can be calculated using this approximation. This is directly linked to the distribution of the height of the maxima above the threshold where asymptotical results for high level excursion have been obtained for different fields. For a full result of the distribution of S of a variety of random fields, see Adler (1981) and Cao (1999).
C.3. Nonstationary cluster size inferences
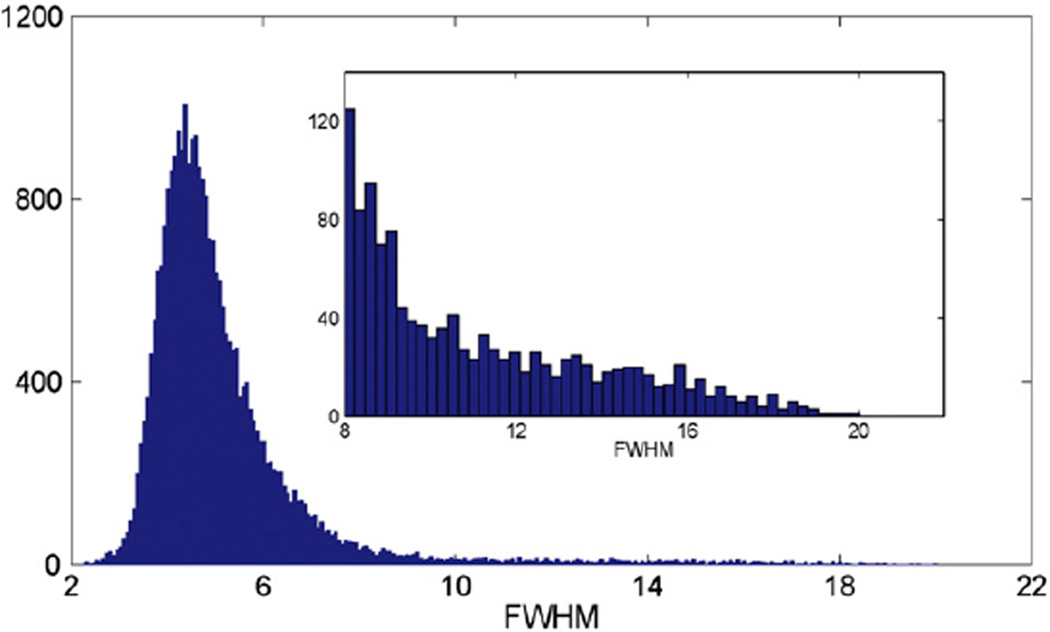
Fig. 8 shows a histogram of the full width at half maximum (FWHM) for each voxel, which is a measure of the local smoothness of the volumetric tissue difference images. It can be seen that the smoothness of the images is not uniform which violates the assumption for stationary cluster size inferences. Nonstationarity will not affect the results of voxel-wise inferences, but it will bias the results for cluster size inferences since clusters tend to be larger in smoother regions and small in rough regions (Hayasaka et al., 2004; Worsley et al., 1999). To overcome this, a concept of resolution elements (RESELs) was introduced and defined by
| (19) |
where V is the search volume in voxel units and D is the dimension of the search area (Worsley et al., 1999). By reducing the search volume to a single voxel, the local smoothness of the images can be expressed as RESEL per voxel (RPV). The cluster size in term of RESELs is then defined as
| (20) |
where C is a cluster. The cluster size S in voxel unit in Eq. (18) is then replaced by the cluster size in RESEL R. Once the distribution of R is approximated, nonstationary cluster size inferences can be carried out. It is clear to see that voxels in smoother regions tend to have smaller RPV while voxels in rougher regions tend to have larger RPV. Therefore, measuring cluster size in RESEL is equivalent to distorting a nonstationary image to stationary by adjusting the distance between voxels and measuring the cluster volume in the resulting image (Taylor and Adler, 2003; Worsley et al., 1999). This stretches rough areas and shrinks smooth areas so that on average stationarity can be achieved.
Fig. 8.
Nonstationarity of the volumetric tissue difference image. A histogram of the FWHM in voxels. The embedded panel is a zoom-in of the right tail behavior.
C.4. Estimation of smoothness
Both voxel and cluster size inferences require the estimation of image smoothness, in terms of FWHM or an RPV image. The smoothness of images is estimated from the covariance matrix of partial derivatives of residual fields, i.e., after removing structures from images by regressing out covariates (Kiebel et al., 1999). Specifically, the spatial derivatives of a random field G are defined as
| (21) |
In real data, the covariance matrix Λ is estimated based on standardized residual images, which is defined at each voxel v as
| (22) |
where η is the error degrees of freedom, σ̂(v) is the estimated standard deviation of e(v) at voxel v (Worsley et al., 1999). In 3D images, partial derivatives are approximated by taking differences between u(v) and adjacent voxels in x, y and z directions and dividing it by the voxel dimension:
| (23) |
Then if the field is stationary, an estimate of |Λ| is given by pooling voxels in the search volume and averaging the determinants over space:
| (24) |
where V is the number of voxels. If the field is nonstationary, a voxel-wise estimate of |Λ(v)| is given by
| (25) |
Finally, FWHM or RPV is estimated in terms of |Λ̂| by
| (26) |
and
| (27) |
(Worsley, 2002). Note that while is unbiased, is actually a biased estimator of FWHM(v) and needs to be corrected by a factor which is basically a small degrees of freedom correction (Worsley, 2002).
Footnotes
Note that in the LSKM setting, nonparametric refers to a highly flexible approach to fitting a curve through data, and not to making weak assumptions on the statistical distribution of the errors.
This is an application of Bonferroni over two tests; as the opposite extremes of a large image are nearly independent (See e.g., (Adler and Taylor, 2007), Page 146–147, and the references therein), this should be an accurate approximation.
References
- Adler RJ. The Geometry of Random Fields. New York: Wiley; 1981. [Google Scholar]
- Adler RJ, Taylor JE. Random Fields and Geometry. Springer Verlag; 2007. [Google Scholar]
- Akil H, Brenner S, Kandel E, Kendler KS, King MC, Scolnick E, Watson JD, Zoghbi HY. The future of psychiatric research: genomes and neural circuits. Science. 2010;3270(5973):1580. doi: 10.1126/science.1188654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldous D. Probability approximations via the Poisson clumping heuristic. Appl. Math. Sci. 1989;77 [Google Scholar]
- Anderson MJ, Robinson J. Permutation tests for linear models. Aust. N. Z. J. Stat. 2001;430(1):75–88. [Google Scholar]
- Bi H, Sze CI. N-methyl-aspartate receptor subunit NR2A and NR2B messenger RNA levels are altered in the hippocampus and entorhinal cortex in Alzheimer's disease. J. Neurol. Sci. 2002;2000(1–2):11–18. doi: 10.1016/s0022-510x(02)00087-4. [DOI] [PubMed] [Google Scholar]
- Braskie MN, Jahanshad N, Stein JL, Barysheva M, McMahon KL, de Zubicaray GI, Martin NG, Wright MJ, Ringman JM, Toga AW, et al. Common Alzheimer's disease risk variant within the CLU gene affects white matter microstructure in young adults. J. Neurosci. 2011;310(18):6764. doi: 10.1523/JNEUROSCI.5794-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J. The size of the connected components of excursion sets of χ2, t and F fields. Adv. Appl. Probab. 1999;31:579–595. [Google Scholar]
- Cao J, Worsley KJ. Applications of random fields in human brain mapping. Spat. Stat. Methodol. Asp. Appl. 2001;159:170–182. [Google Scholar]
- Cardon LR, Craddock N, Deloukas P, Duncanson A, Kwiatkowski DP, McCarthy MI, Ouwehand WH, Samani NJ, Todd JA, Donnelly P, et al. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;4470(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David HA, Nagaraja HN. Order Statistics. Wiley Online, Library; 1970. [Google Scholar]
- Fan J, LV J. Sure independence screening for ultrahigh dimensional feature space. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 2008;700(5):849–911. doi: 10.1111/j.1467-9868.2008.00674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber NB, Newcomer JW, Olney JW. The glutamate synapse in neuropsychiatric disorders: focus on schizophrenia and Alzheimer's disease. Prog. Brain Res. 1998;116:421–437. doi: 10.1016/s0079-6123(08)60453-7. [DOI] [PubMed] [Google Scholar]
- Filippini N, Rao A, Wetten S, Gibson RA, Borrie M, Guzman D, Kertesz A, Loy-English I, Williams J, Nichols T, et al. Anatomically-distinct genetic associations of APOE 4 allele load with regional cortical atrophy in Alzheimer's disease. Neuroimage. 2009;440(3):724–728. doi: 10.1016/j.neuroimage.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Fox GM, Holst PL, Chute HT, Lindberg RA, Janssen AM, Basu R, Welcher AA. cDNA cloning and tissue distribution of five human eph-like receptor protein-tyrosine kinases. Oncogene. 1995;100(5):897. [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JRB, Elliott KS, Lango H, Rayner NW, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;3160(5826):889. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;4490(7164):851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, Worsley KJ, Frackowiak RSJ, Mazziotta JC, Evans AC. Assessing the significance of focal activations using their spatial extent. Hum. Brain Mapp. 1994;1:210–220. doi: 10.1002/hbm.460010306. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Holmes A, Poline JB, Price CJ, Frith CD. Detecting activations in pet and FMRI: levels of inference and power. Neuroimage. 1996;40(3):223–235. doi: 10.1006/nimg.1996.0074. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Ashburner JT, Kiebel SJ, Nichols TE, Penny WD. Statistical Parametric Mapping: the Analysis of Functional Brain Images. Elsevier/Academic Press; 2007. [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am. J. Psychiatry. 2003;1600(4):636. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Gumbel EJ. Statistics of Extremes. Dover Pubns; 2004. [Google Scholar]
- Hayasaka S, Nichols TE. Validating cluster size inference: random field and permutation methods. Neuroimage. 2003;200(4):2343–2356. doi: 10.1016/j.neuroimage.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Hayasaka S, Phan KL, Liberzon I, Worsley KJ, Nichols TE. Nonstationary cluster-size inference with random field and permutation methods. Neuroimage. 2004;220(2):676–687. doi: 10.1016/j.neuroimage.2004.01.041. [DOI] [PubMed] [Google Scholar]
- Hibar DP, Kohannim O, Stein JL, Chiang MC, Thompson PM. Multilocus genetic analysis of brain images. Front. Genet. 2011a;20(73) doi: 10.3389/fgene.2011.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar DP, Stein JL, Kohannim O, Jahanshad N, Saykin AJ, Shen L, Kim S, Pankratz N, Foroud T, Huentelman MJ, et al. Voxelwise gene-wide association study (vGeneWAS): multivariate gene-based association testing in 731 elderly subjects. Neuroimage. 2011b;56:1875–1891. doi: 10.1016/j.neuroimage.2011.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerl RW. Ridge analysis 25 years later. Am. Stat. 1985;39:186–192. [Google Scholar]
- Holmes AP. PhD thesis. 1994. Statistical issues in functional brain mapping. [Google Scholar]
- Holmes AP, Blair RC, Watson G, Ford I. Nonparametric analysis of statistic images from functional mapping experiments. J. Cereb. Blood Flow Metab. 1996;160(1):7–22. doi: 10.1097/00004647-199601000-00002. [DOI] [PubMed] [Google Scholar]
- Hynd MR, Scott HL, Dodd PR. Differential expression of N-methyl-d-aspartate receptor NR2 isoforms in Alzheimer's disease. J. Neurochem. 2004;900(4):913–919. doi: 10.1111/j.1471-4159.2004.02548.x. [DOI] [PubMed] [Google Scholar]
- Inoue E, Deguchi-Tawarada M, Togawa A, Matsui C, Arita K, Katahira-Tayama S, Sato T, Yamauchi E, Oda Y, Takai Y. Synaptic activity prompts γ-secretase-mediated cleavage of epha4 and dendritic spine formation. J. Cell Biol. 2009;1850(3):551. doi: 10.1083/jcb.200809151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Jack CR, Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, et al. The Alzheimer's disease neuroimaging initiative (ADNI): MRI methods. J. Magn. Reson. Imaging. 2008;270(4):685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Jia J. Association between NR2B subunit gene (GRIN2B) promoter polymorphisms and sporadic Alzheimer's disease in the North Chinese population. Neurosci. Lett. 2009;4500(3):356–360. doi: 10.1016/j.neulet.2008.10.075. [DOI] [PubMed] [Google Scholar]
- Joyner AH, et al. A common MECP2 haplotype associates with reduced cortical surface area in humans in two independent populations. Proc. Natl. Acad. Sci. 2009;1060(36):15483. doi: 10.1073/pnas.0901866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy PE, Cade BS. Randomization tests for multiple regression. Commun. Stat. Simul. Comput. 1996;250(4):923–936. [Google Scholar]
- Kiebel SJ, Poline JB, Friston KJ, Holmes AP, Worsley KJ. Robust smoothness estimation in statistical parametric maps using standardized residuals from the general linear model. Neuroimage. 1999;100(6):756–766. doi: 10.1006/nimg.1999.0508. [DOI] [PubMed] [Google Scholar]
- Knijnenburg TA, Wessels LFA, Reinders MJT, Shmulevich I. Fewer permutations, more accurate p-values. Bioinformatics. 2009;250(12):i161. doi: 10.1093/bioinformatics/btp211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohannim O, Hibar DP, Stein JL, Jahanshad N, Jack CR, Weiner MW, Toga AW, Thompson PM. Boosting power to detect genetic associations in imaging using multi-locus, genome-wide scans and ridge regression. Biomedical Imaging: From Nano to Macro, 2011 IEEE International Symposium on. IEEE. 2011:1855–1859. [Google Scholar]
- Kohannim O, Hibar DP, Jahanshad N, Stein JL, Hua X, Toga AW, Jack CR, Jr, Weiner MW, Thompson PM, et al. ISBI 2012. Barcelona, Spain: 2012. Predicting temporal lobe volume on MRI from genotypes using L1−L2 regularized regression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwee LC, Liu D, Lin X, Ghosh D, Epstein MP. A powerful and flexible multilocus association test for quantitative traits. Am. J. Hum. Genet. 2008;820(2):386–397. doi: 10.1016/j.ajhg.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamprecht R, LeDoux J. Structural plasticity and memory. Nat. Rev. Neurosci. 2004;50(1):45–54. doi: 10.1038/nrn1301. [DOI] [PubMed] [Google Scholar]
- Leow A, Huang SC, Geng A, Becker J, Davis S, Toga A, Thompson P. Information Processing in Medical Imaging. Springer; 2005. Inverse consistent mapping in 3D deformable image registration: its construction and statistical properties; pp. 493–503. [DOI] [PubMed] [Google Scholar]
- Leow AD, Yanovsky I, Chiang MC, Lee AD, Klunder AD, Lu A, Becker JT, Davis SW, Toga AW, Thompson PM. Statistical properties of jacobian maps and the realization of unbiased large-deformation nonlinear image registration. Med. Imaging, IEEE Trans. 2007;260(6):822–832. doi: 10.1109/TMI.2007.892646. [DOI] [PubMed] [Google Scholar]
- Liu D, Lin X, Ghosh D. Semiparametric regression of multidimensional genetic pathway data: least-squares kernel machines and linear mixed models. Biometrics. 2007;630(4):1079–1088. doi: 10.1111/j.1541-0420.2007.00799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, et al. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM) Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001;3560(1412):1293. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Nicodemus KK, Egan MF, Callicott JH, Mattay V, Weinberger DR. False positives in imaging genetics. Neuroimage. 2008;400(2):655–661. doi: 10.1016/j.neuroimage.2007.11.058. [DOI] [PubMed] [Google Scholar]
- Nichols T, Hayasaka S. Controlling the familywise error rate in functional neuroimaging: a comparative review. Stat. Methods Med. Res. 2003;120(5):419–446. doi: 10.1191/0962280203sm341ra. [DOI] [PubMed] [Google Scholar]
- Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Hum. Brain Mapp. 2002;150(1):1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons CG, Stöffler A, Danysz W. Memantine: a NMDA receptor antagonist that improves memory by restoration of homeostasis in the glutamatergic system— too little activation is bad, too much is even worse. Neuropharmacology. 2007;530(6):699–723. doi: 10.1016/j.neuropharm.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Pickands J., III Statistical inference using extreme order statistics. Ann. Stat. 1975:119–131. [Google Scholar]
- Poline JB, Mazoyer BM. Analysis of individual positron emission tomography activation maps by detection of high signal-to-noise-ratio pixel clusters. J. Cereb. Blood Flow Metab. 1993;130(3):425–437. doi: 10.1038/jcbfm.1993.57. [DOI] [PubMed] [Google Scholar]
- Poline JB, Worsley KJ, Evans AC, Friston KJ. Combining spatial extent and peak intensity to test for activations in functional imaging. Neuroimage. 1997;50(2):83–96. doi: 10.1006/nimg.1996.0248. [DOI] [PubMed] [Google Scholar]
- Potkin SG, Guffanti G, Lakatos A, Turner JA, Kruggel F, Fallon JH, Saykin AJ, Orro A, Lupoli S, Salvi E, et al. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for alzheimer's disease. PLoS One. 2009;40(8):e6501. doi: 10.1371/journal.pone.0006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roland PE, Levin B, Kawashima R, Åkerman S. Three-dimensional analysis of clustered voxels in 15O-butanol brain activation images. Hum. Brain Mapp. 1993;10(1):3–19. [Google Scholar]
- Schaid D. Genomic similarity and kernel methods ii: methods for genomic information. Hum. Hered. 2010a;70:132–140. doi: 10.1159/000312643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid DJ. Genomic similarity and kernel methods i: advancements by building on mathematical and statistical foundations. Hum. Hered. 2010b;70:109–131. doi: 10.1159/000312641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Kim S, Risacher SL, Nho K, Swaminathan S, West JD, Foroud T, Pankratz N, Moore JH, Sloan CD, et al. Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: a study of the ADNI cohort. NeuroImage. 2010;53(3):1051–1063. doi: 10.1016/j.neuroimage.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver M, Montana G, Nichols TE. False positives in neuroimaging genetics using voxel-based morphometry data. Neuroimage. 2011;540(2):992–1000. doi: 10.1016/j.neuroimage.2010.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simón AM, de Maturana RL, Ricobaraza A, Escribano L, Schiapparelli L, Cuadrado-Tejedor M, Pérez-Mediavilla A, Avila J, Del Río J, Frechilla D. Early changes in hippocampal eph receptors precede the onset of memory decline in mouse models of alzheimer's disease. J. Alzheimers Dis. 2009;170(4):773–786. doi: 10.3233/JAD-2009-1096. [DOI] [PubMed] [Google Scholar]
- Smith R. Thresholdmethods for sample extremes. In: De Oliveira JT, editor. Statistical Extremes and Applications. Dordrecht, The Netherlands: D. Reidel; 1984. pp. 6211–6638. [Google Scholar]
- Stein JL, Hua X, Lee S, Ho AJ, Leow AD, Toga AW, Saykin AJ, Shen L, Foroud T, Pankratz N, et al. Voxelwise genome-wide association study (vGWAS) Neuroimage. 2010a;530(3):1160–1174. doi: 10.1016/j.neuroimage.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JL, Hua X, Morra JH, Lee S, Hibar DP, Ho AJ, Leow AD, Toga AW, Sul JH, Kang HM, et al. Genome-wide analysis reveals novel genes influencing temporal lobe structure with relevance to neurodegeneration in alzheimer's disease. Neuroimage. 2010b;510(2):542–554. doi: 10.1016/j.neuroimage.2010.02.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana R, Boyd-Kimball D, Cai J, Pierce WM, Klein JB, Merchant M, Butterfield DA. Proteomics analysis of the alzheimer's disease hippocampal proteome. J. Alzheimers Dis. 2007;110(2):153–164. doi: 10.3233/jad-2007-11203. [DOI] [PubMed] [Google Scholar]
- Tang YP, Shimizu E, Dube GR, Rampon C, Kerchner GA, Zhuo M, Liu G, Tsien JZ. Genetic enhancement of learning and memory in mice. Nature. 1999;4010(6748):63–69. doi: 10.1038/43432. [DOI] [PubMed] [Google Scholar]
- Taylor JE, Adler RJ. Euler characteristics for Gaussian fields on manifolds. Ann. Probab. 2003;31:533–563. [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B (Methodol.) 1996;58:267–288. [Google Scholar]
- Vounou M, Nichols TE, Montana G. Discovering genetic associations with high-dimensional neuroimaging phenotypes: a sparse reduced-rank regression approach. Neuroimage. 2010;530(3):1147–1159. doi: 10.1016/j.neuroimage.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsley KJ. Local maxima and the expected Euler characteristic of excursion sets of χ2 , F and T fields. Adv. Appl. Probab. 1994:13–42. [Google Scholar]
- Worsley KJ. The geometry of random images. Chance. 1996;9:27–40. [Google Scholar]
- Worsley KJ. Non-stationary FWHM and its effect on statistical inference of fMRI data. Neuroimage. 2002;15:S346. [Google Scholar]
- Worsley KJ, Evans AC, Marrett S, Neelin P. A three-dimensional statistical analysis for CBF activation studies in human brain. J. Cereb. Blood Flow Metab. 1992;12(6):900–918. doi: 10.1038/jcbfm.1992.127. [DOI] [PubMed] [Google Scholar]
- Worsley KJ, Marrett S, Neelin P, Vandal AC, Friston KJ, Evans AC, et al. A unified statistical approach for determining significant signals in images of cerebral activation. Hum. Brain Mapp. 1996;40(1):58–73. doi: 10.1002/(SICI)1097-0193(1996)4:1<58::AID-HBM4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Worsley KJ, Andermann M, Koulis T, MacDonald D, Evans AC. Detecting changes in nonisotropic images. Hum. Brain Mapp. 1999;80(2–3):98–101. doi: 10.1002/(SICI)1097-0193(1999)8:2/3<98::AID-HBM5>3.0.CO;2-F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashiro K, Philpot BD. Regulation of NMDA receptor subunit expression and its implications for LTD, LTP, and metaplasticity. Neuropharmacology. 2008;550(7):1081–1094. doi: 10.1016/j.neuropharm.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]