Abstract
The full likelihood approach in statistical analysis is regarded as the most efficient means for estimation and inference. For complex length-biased failure time data, computational algorithms and theoretical properties are not readily available, especially when a likelihood function involves infinite-dimensional parameters. Relying on the invariance property of length-biased failure time data under the semiparametric density ratio model, we present two likelihood approaches for the estimation and assessment of the difference between two survival distributions. The most efficient maximum likelihood estimators are obtained by the em algorithm and profile likelihood. We also provide a simple numerical method for estimation and inference based on conditional likelihood, which can be generalized to k-arm settings. Unlike conventional survival data, the mean of the population failure times can be consistently estimated given right-censored length-biased data under mild regularity conditions. To check the semiparametric density ratio model assumption, we use a test statistic based on the area between two survival distributions. Simulation studies confirm that the full likelihood estimators are more efficient than the conditional likelihood estimators. We analyse an epidemiological study to illustrate the proposed methods.
Keywords: Conditional likelihood, Density ratio model, em algorithm, Length-biased sampling, Maximum likelihood approach
1. Introduction
Length-biased sampling has been recognized in the statistics and scientific literature for decades (Zelen, 2004; Scheike & Keiding, 2006). An epidemiological study serves as an illustration: in a prevalent cohort study, individuals age 65 and older were recruited and screened for dementia (Wolfson et al., 2001). Individuals who were confirmed to have dementia were prospectively followed to assess their time to death or censoring. Individuals who survived until the time of recruitment were sampled into the study, whereas those who died before recruitment were not included. This resulted in selection bias, because the observed time intervals from the onset of dementia to death were longer for individuals in the study cohort compared with those dementia patients in the general population. With three different dementia diagnoses, it is of interest to compare the survival distributions from different groups of patients.
For conventional survival data, the partial likelihood is rank invariant with a binary covariate under the proportional hazards model (Peto & Peto, 1972). However, when right-censored survival data are subject to length-biased sampling, the invariance is lost because the size of the failure time also plays a key role in the inference. The proportional hazards assumption does not hold in the hazard functions of the observed biased failure time data, even though the unbiased failure time satisfies the proportional hazards model. For uncensored length-biased data, Chen (2010) and Mandel & Ritov (2010) proposed estimation approaches that rely on the invariance of the covariate effects under the accelerated failure time model. For censored length-biased data, the accelerated failure time model does not enjoy invariance due to the presence of dependent right censoring.
The density ratio model, which relates the distributions of two arms under a flexible semi-parametric structure, has been explored for uncensored data and traditional right-censored data (Anderson, 1979; Qin et al., 2002; Shen et al., 2007), but to our knowledge has never been investigated for length-biased survival data. In fact, the density ratio model has the invariance property for length-biased data; this naturally facilitates the construction of likelihood functions.
Assume T̃ to be the time interval measured from an initiating event to failure within the population of interest. Let fu(t) and f (t) define the probability density function for unbiased time, T̃ and the length-biased time, T, respectively, in the first group. Then
| (1) |
Let Fu be the cumulative distribution function of T̃. For the second group, let gu and Gu denote the unbiased density and the cumulative distribution functions. The corresponding f and g represent the density functions for length-biased samples, and F and G denote the distribution functions of T in each group. Under the density ratio model, the ratio of two density functions of T̃ in the target population is assumed to have the form
| (2) |
where γ (t) is a known monotone function for transforming the observed time t. Different forms of γ (t) correspond to some conventionally used densities; the common forms of γ in applications include γ (t) = t or γ (t) = log(t) (Anderson, 1979; Kay & Little, 1987; Qin et al., 2002). When γ (t) = t, β is the log odds ratio for density of T̃ = t + 1 against density of T̃ = t between the two groups. In this case, model (2) is also equivalent to a semiparametric model in which the log ratio of two density functions is linear in t, where β is the slope in the linear function. When γ (t) = log(t) and 0 < β ⩽ 1, gu(t) is proportional to its length of t with the power of β. When both density functions are exponential, they belong to the intersection of the density ratio models and proportional hazards models. For any form of γ (t), β = 0 leads to the equality of the two density functions, because αu = αu(β). A unique property of (2) is its invariance for length-biased T. The density ratio of biased samples can be associated with the same model structure, except for a different intercept, g(t) = exp{α + βγ (t)} f (t), where α = αu + log(μf) − log(μg) and μg = ∫ tgu(t) dt.
In the present paper, we propose a conditional estimating equation approach to estimating the nonparametric baseline density function fu(t) and parameters (αu, β) for length-biased data under this model. We also develop a full likelihood approach under the density ratio model. We obtain the maximum likelihood estimators by extending Vardi’s em algorithm for nonparametric estimation under the renewal process together with the profile likelihood method. We assess the adequacy of the density ratio model using a test based on the area between two survival curves.
2. Conditional likelihood method
Suppose that T = A + V is the observed failure time and U = A + C is the censoring time, where V is the time from examination to failure, and C is the time from examination to censoring. Here, A is called the truncation variable and V is the residual survival time. Assume that residual censoring time C and (A, V) are independent. The joint density distributions of (A, V) and (A, T) have the same formula when there is no censoring,
| (3) |
Given (3), the probability of observing a biased failure time is
The conditional density function of observing uncensored T can be expressed as
| (4) |
where Z = 0 or 1 is the group indicator, , Sc(· | Z) is the survival distribution of C given Z, λf = pr(T < U | Z = 0) and
| (5) |
where and λg = pr(T < U | Z = 1). Under the density ratio model assumption in (2), the ratio of the two conditional density functions retains a model framework similar to that of the marginal density function for the unbiased observation T̃,
where α1 = αu + log(λf μf) − log(λgμg) and w(t) = log{wg(t)} − log{wf (t)}. When the censoring distribution is the same for two arms, there is a further simplification with w(t) = 0.
Let Yi = min(Ti, Ui), Ai, δi = I (Ti ⩽ Ui), Zi (i = 1, . . . , n) be the observed data, equivalently denoted by (Yi, Ai, δi, Zi) for the ith subject. Conditional on the observed failure times: {s1, . . . , sm1, . . . , sm}, m = m1 + m2 where m1 is the number of failures among n1 subjects in arm 1, and m2 is the number of failures among n2 subjects in arm 2, and the likelihood for F1 and (α1, β) is proportional to
where p1i = d F1(si) and F1(t) = ∑si⩽t p1i. The corresponding loglikelihood function is
| (6) |
subject to the constraints p1i ⩾ 0(i = 1, . . . , m), and
where the second constraint comes from the fact that ∫ g1(t) dt = 1. After profiling p1i, we have the logistic type of loglikelihood (Qin & Zhang, 1997),
where ρ = m2/m1. If w(t) is known, the score equations for (α1, β) are
| (7) |
| (8) |
The expectations for the right side of equations (7) and (8) are zero under model (2), so the estimating equation from the score function is unbiased. If f1 and g1 have finite mean, finite and nonzero variance, 0 < Ef1 {γ (t)}2 < ∞ and 0 < Eg1 {γ (t)}2 < ∞, we can derive the existence of Ef1 {m(t)}2 and Eg1 {m(t)}2, where m(t) = γ (t) exp{α1 + βγ (t) + w(t)}/[1 + ρ exp{α1 + βγ (t)w(t)}]. Under the above regularity conditions, the derivatives of (7) and (8) with respect to (α1, β) yield a positive definite matrix, so the solution is unique and leads to consistent estimators of (α1, β).
When w(t) is unknown, we replace it by its consistent estimator w̃(t) = log{w̃g(t)} − log{w̃f (t)}, where w̃f and w̃g are the area under the Kaplan–Meier estimators of the residual censoring time of each arm. We obtain the estimators, (α̃1, β̃) by solving equations (7) and (8). Subsequently, we can estimate F1(t) by F̃1(t) = ∑i:si⩽t p̃1i, from
and estimate G1(t) = by G̃1(t) = ∑i:si⩽t q̃1i, where
The quantities, λf and λg can be consistently estimated by and . Because fu and gu are proper density functions with ∫ fu(t) dt = 1 and ∫ gu(t) dt = 1, consistent estimators of μf and μg can be derived from (4) and (5) as
Provided there is a positive follow-up time t, wf (t) and wg(t) can be consistently estimated by w̃f (t) and w̃g(t), which are bounded away from zero. Given these consistent estimators, the consistent estimators of Fu(t) and Gu(t) can subsequently be obtained through (4) and (5),
Our motivating example involves three dementia diagnoses in the study cohort; a comparison of the survival distributions among the three types is of scientific interest. We generalize the proposed two-arm estimation and inference procedures to those for k-arm studies where k > 2. Details for the global k-arm estimation and inference methods can be found in the Supplementary Material.
3. Full likelihood approach
3.1. Maximum likelihood estimate
Conditioning on the observed failure times, the proposed estimation procedure is straightforward and can be implemented easily, as shown in § 2. However, conditional inference is often less efficient than inference based on the full likelihood (Wang, 1991; Zeng & Lin, 2007). We formulate the full likelihood function under the density ratio model by generalizing the nonparametric maximum likelihood estimation method of Vardi (1989).
From (3), the full likelihood for the observed data can be formally expressed as
where F̄u and Ḡu are the survivor functions of T̃ for the two arms. Because of the relationship between the probability density function of the unbiased time and that of the biased time given in (1),
Under the density ratio model, dG(t) = exp{α + βγ (t)} d F(t) and removing the constant Yi, the full likelihood of the observed data is proportional to
| (9) |
To find the nonparametric maximum likelihood estimator for F and the unknown parameters (α, β) from (9), it is sufficient to maximize the discrete version of F on the point mass support of 0 < t1 < ⋯ < th, where {t1, . . . , th} denote the uniquely observed failure and censoring times from both arms in increasing order (Vardi, 1989). Let pi = d F(ti) (i = 1, . . . , h) and qi = dG(ti) = pi exp{α + βγ (ti)}.
The discrete version of the full likelihood can be expressed as
| (10) |
where
are the multiplicities of the failure times and censoring times at tj in each arm. Thus, maximizing (9) reduces to maximizing (10) with respect to pj (j = 1, . . . , h), β and α subject to the constraints
| (11) |
When α = β = 0, the likelihood (10) is equivalent to the nonparametric likelihood for length-biased data proposed by Vardi (1989).
3.2. Implementation of the EM algorithm
Vardi (1989) showed a natural connection between the nonparametric likelihood of length-biased data and the likelihood under multiplicative censorship. For a right-censored observation, U can be considered as incomplete failure time data in an em algorithm and equivalently expressed as U = ϕT, ϕ ∼ Un(0, 1), T ∼ d F(t), where T and ψ are independent and T is the biased failure time for a subject with Z = 0. The joint density of (U, T) and the marginal density of U are, respectively,
The conditional density function of T given right-censored time U can thus be expressed as
| (12) |
For subjects in the treatment group with Z = 1, similar equations can be derived given T ∼ dG(t). Applying the em algorithm in Vardi (1989), we can estimate the nonparametric density function F and parameters (α, β) from the likelihood function (10).
Denote the observed data by 𝒪 = {Yi, δi, Zi : i = 1, . . . , n}. A key step in the em algorithm is to replace the current estimated probability mass at tj by its expected and normalized conditional multiplicity of the complete data observation at tj using (12). At the e-step, we start with arbitrary (i = 1, . . . , h), α0 and β0 satisfying (11). Since not all Ti are observable, we thus replace those unobserved I (Ti = tj) by their expectations. The expectation of in each arm given the observed data and current parameter estimators is
where . The conditional expectation of the complete data loglikelihood is
| (13) |
where . Define the sum of the wj as .
At the m-step, we maximize the loglikelihood (13) under the constraints specified in (11) by including the Lagrange multipliers in the weighted likelihood (Owen, 1988) as follows:
Taking the derivative of ℓL with respect to pj, we have
together with the constraints (11), leading to λ1 = w+. Consequently,
| (14) |
After profiling over the pj s in (13) and ignoring the constant terms, we have
| (15) |
Taking the derivative of ℓL with respect to α,
and together with (14), we obtain . After inserting the Lagrange multipliers into (15), the loglikelihood function with respect to α and β under the constraints is
| (16) |
where . We can estimate α, β from (16) and then derive pj from (14). Given the estimated α, β and pj, and are updated by repeating the e-step. The iteration is repeated until a prespecified criterion is satisfied. The maximum likelihood estimate is denoted by θ̂n = (α̂, β̂, F̂), where F̂ (t) = ∑tj⩽tp̂j. The corresponding unbiased distribution function is estimated by
3.3. Large sample properties
Let θ0 ≡ (α0, β0, F0) denote the true values of θ ≡ (α, β, F). Under the regularity conditions specified in the Appendix, we can establish the asymptotic properties of the estimated maximum likelihood estimator θ̂n for t ∈ [0, τ], where τ is a finite upper bound of the support for the population survival times.
Theorem 1. Under Conditions A1–A4, the maximum likelihood estimator θ̂n is consistent, defined by
converging to 0 almost surely and uniformly as n → ∞.
Theorem 2. Under Conditions A1–A4, n1/2(θ̂n − θ0) converges weakly to a tight zero-mean Gaussian process −ϕ′ψ0 { (𝕎)} as n → ∞.
Definitions of ϕ′ψ0 { (𝕎)} and 𝕎 are provided in the Appendix. The consistency of θ̂n is proved by the Kullback–Leibler information approach (Murphy, 1994; Parner, 1998). The weak convergence of θ̂n is established by applying the Z-theorem for the infinite-dimensional estimating equations (van der Vaart & Wellner, 1996). We present the proof of Theorem 1 and an outline proof for Theorem 2 in the Appendix. Although the variance matrix is derived from the weak convergence of θ̂n, its computation is complex. Alternatively, we use the simple boot-strap resampling method to compute the variances for the estimated parameters (Bilker & Wang, 1996; Gross & Lai, 1996; Asgharian et al., 2002). Given the established weak convergence for the proposed estimators in Theorem 2 under the regularity conditions, this bootstrap is valid for estimating the variances of the estimators (van der Vaart & Wellner, 1996, Ch. 3.6).
4. Inference
4.1. Hypothesis testing
To test the difference in the survival distributions, we describe likelihood ratio tests under the conditional likelihood framework, when the censoring distributions in the two arms are the same, w(t) = 0. For the conditional probability density function, f1(t) = g1(t) is equivalent to fu(t) = gu(t), when w(t) = 0. If f1(t) = g1(t) and wf (t) = wg(t), then λf = λg because both f (t) and g(t) are proper density functions. Thus, f (t) = g(t), and fu(t) = gu(t). We therefore use the conditional likelihood ratio statistic to test the equality f1(t) = g1(t), i.e., H0 : α1 = β = 0. Let ℓc denote the loglikelihood of (6). Under the null hypothesis H0, ℓc reduces to ℓc(0, 0, F̂1), where F̂1 is the nonparametric estimator of F1 under α1 = β = 0. The asymptotic distribution of Rc = 2{ℓc(α̃1, β̃, F̃1) − ℓc(0, 0, F1)} is because β = 0 implies α1 = 0. When the censoring distributions of the two arms are not equal, w(t) ≠ 0, the asymptotic distribution of Rc for testing β = 0 will not be chi-squared in general, but we can obtain a critical value for the test statistic using the bootstrap. Intuitively, the full likelihood ratio statistic Rf, which is defined similarly to Rc, may be used to test H0. Our simulation results showed that this test statistic can be well approximated by a distribution when w(t) = 0, though we cannot prove this property theoretically.
4.2. Model checking
To assess the parametric component for the link function γ (t) imposed on the ratio of the two density functions in model (2), we use a general model checking method similar to that in Shen et al. (2007). The test statistic is based on a quadratic form of the cumulative difference between the nonparametric estimate of the distribution function and its estimate under the density ratio model assumption. The goodness-of-fit test is
| (17) |
where F̂u is the estimated distribution function of T̃ using the full likelihood under the density ratio model assumption, and F̂υ is Vardi’s nonparametric estimate using data from arm 1 only. Similarly, we can replace F̂u(t) by F̃u(t) in (17) to assess the goodness-of-fit for the conditional likelihood estimator. Because the derivation of the asymptotic distribution of An is tedious and not the focus of this paper, we use the bootstrap to find its critical values.
5. Numerical studies and applications
5.1. Simulations
We compare the performance of the estimation and inference of the full likelihood and the conditional likelihood, under various scenarios. We use a Weibull distribution for fu, so that f is also Weibull. The density gu is exp{αu + βγ (t)} fu(t) for Z = 1, where γ (t) = t.
For Z = 0 or 1, we generate independent pairs (Ã, T̃), then keep those with à < T̃, where à is generated independently from a uniform distribution on (0, τ0) and T̃ is generated from the density fu or gu depending on Z, where τ0 is the upper bound of T̃. The censoring times C are generated independently from uniform distributions corresponding to censoring percentages ranging from 15 to 43%. The censoring indicator is δ = I (T ⩽ C + A). For each scenario, we use sample sizes n1 = n2 =50 or 100, and repeat the simulation 2000 times.
In Table 1, we summarize the standard deviation of the estimated parameters for both methods. We also provide the size and power of the conditional likelihood ratio statistics for testing the null hypothesis H0 : αu = β = 0. The empirical bias slightly increases with increasing censoring for the conditional likelihood estimator. For both methods, the point estimators are fairly unbiased, and the biases are not significantly different from zero, so we do not present them. The standard error of each estimate is found using 500 bootstrap replicates. Compared with the conditional likelihood approach, the estimators from the full likelihood are more efficient; the standard deviations for the former approach are up to 1.65 times greater. The efficiency gain is more significant when censoring increases. The significance levels of the conditional likelihood ratio tests are maintained around the nominal levels. We also compare the estimated mean and the baseline cumulative distributions, F̂u(t) and F̃u(t) at the 50 and 75% quantiles. The standard deviations of the full likelihood estimator are smaller than those of the conditional likelihood estimators when data have heavy censoring, but the difference in efficiency is otherwise small. For both methods, the estimation efficiency for (β, F) increases with the percentage of uncensored data. For the conditional likelihood, although wf (t) and wg(t) may not be estimated with great accuracy when most of data are uncensored, ŵf (t) and ŵg(t) are always robust and have relatively small impact on the estimation efficiency for (β, F).
Table 1.
Estimated standard deviations of the parameter estimators under the density ratio model, size and power of the likelihood ratio tests under the nominal level of 1 and 5%
| Full likelihood | Conditional likelihood | |||||||
|---|---|---|---|---|---|---|---|---|
| Standard deviation | Rf : χ2-test | Standard deviation | Rc : χ2-test | |||||
| Cens. | α̂u | β̂ | 1% | 5% | α̃u | β̃ | 1% | 5% |
| αu = β = 0, n0 = n 1 = 50 | ||||||||
| 15% | 0.17 | 0.17 | 1.4 | 6.5 | 0.20 | 0.19 | 1.1 | 6.3 |
| 25% | 0.18 | 0.17 | 1.2 | 5.8 | 0.22 | 0.21 | 0.8 | 5.6 |
| 43% | 0.19 | 0.18 | 1.4 | 5.6 | 0.31 | 0.30 | 1.2 | 6.8 |
| αu = β = 0, n0 = n 1 = 100 | ||||||||
| 15% | 0.11 | 0.11 | 1.5 | 6.4 | 0.12 | 0.12 | 0.8 | 5.6 |
| 25% | 0.11 | 0.11 | 1.3 | 6.0 | 0.14 | 0.14 | 1.0 | 4.8 |
| 43% | 0.12 | 0.12 | 1.5 | 6.2 | 0.19 | 0.19 | 1.0 | 6.0 |
| αu = −0.405, β = 0.333, n0 = n1 = 50 | ||||||||
| 15% | 0.21 | 0.15 | 58.6 | 79.6 | 0.23 | 0.17 | 41.1 | 66.1 |
| 25% | 0.21 | 0.15 | 54.1 | 75.4 | 0.25 | 0.18 | 35.4 | 60.1 |
| 43% | 0.23 | 0.16 | 38.6 | 59.6 | 0.33 | 0.25 | 20.0 | 41.8 |
| αu = −0.405, β = 0.333, n0 = n1 = 100 | ||||||||
| 15% | 0.13 | 0.10 | 91.0 | 97.5 | 0.15 | 0.11 | 81.8 | 93.5 |
| 25% | 0.15 | 0.11 | 87.2 | 94.8 | 0.16 | 0.12 | 68.0 | 87.6 |
| 43% | 0.15 | 0.11 | 62.8 | 73.8 | 0.21 | 0.16 | 41.5 | 67.3 |
5.2. Example
In a multicentre prospective observational study conducted throughout Canada during 1991–1996, more than 14 000 individuals aged 65 years or older participated in a health survey (Wolfson et al., 2001). Among them, 1132 participants who were identified to have dementia were followed until their death or censoring in 1996. We have complete data on a total of 818 individuals, consisting of the date of dementia onset, date of screening for dementia, date of death or censoring and the dementia diagnosis. Three categories of dementia diagnosis were identified among these patients: probable Alzheimer’s disease, with n = 393, possible Alzheimer’s disease, with n = 252 and vascular dementia, with n = 173. Several studies have confirmed the stationarity assumption, indicating that the dataset collected from this cohort of individuals represents a collection of length-biased samples (Asgharian et al., 2006). The distributions of the censoring time measured from the date of screening for dementia to censoring are essentially the same for the three diagnosis groups.
Applying the methods proposed for an overall analysis, we use the group diagnosed with possible Alzheimer’s disease as the baseline cohort with a link function of γ (t) = t or log(t). The corresponding parameter estimates and their standard errors are given in Table 2. We test the hypothesis that all three diagnoses of dementia are equidistributed. Under the log-transformation γ (t) = log(t), the conditional likelihood ratio test indicates a marginal significant difference in the survival distributions, p-value = 0.05. The model-based survival distribution estimates for the three groups with γ (t) = log(t) are provided in the Supplementary Material.
Table 2.
Parameter estimators, standard deviations and p-values from the conditional likelihood ratio test under the density ratio model for the dementia study
| γ(t) | α̃u1 (sd) | β̃1 (sd) | α̃u2 (sd) | β̃2 (sd) | p-value, Rc |
|---|---|---|---|---|---|
| t | 0.16 (0.11) | −0.03 (0.02) | 0.28 (0.16) | −0.05 (0.03) | 0.11 |
| log(t) | 0.22 (0.21) | −0.15 (0.15) | 0.54 (0.24) | −0.42 (0.18) | 0.05 |
Baseline group is ‘Possible Alzheimer’s’; αu1 and β1, the coefficients for ‘Probable Alzheimer’s’ vs. ‘Possible Alzheimer’s’; αu2 and β2, the coefficients for ‘Vascular’ vs. ‘Possible Alzheimer’s’; p-value, the critical value of Rc is determined by χ2 statistic with one degree of freedom; sd, standard deviation.
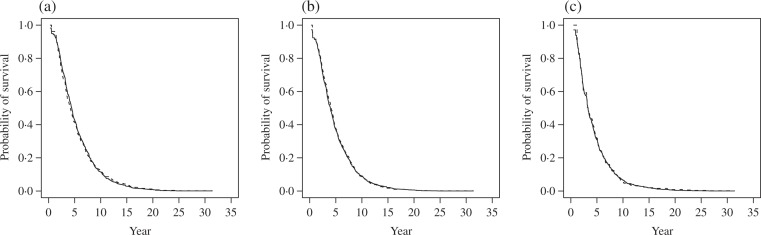
Using the test statistic described in § 4.2, we check the fit of the density ratio model with γ (t) = t and γ (t) = log(t). The tests indicate that both link functions fit the data well; the p-values from the goodness-of-fit test range from 0.59 to 0.92. Comparing the density ratio model-based survival curve for each arm against Vardi’s nonparametric survival estimate, we find that the model with the link function γ (t) = log(t) leads to a slightly better fit in all three pairwise comparisons. As shown in Fig. 1, the survival distribution estimate using the full likelihood approach under the density ratio model with γ (t) = log(t) is almost identical to Vardi’s nonparametric estimate of the survival distribution for each disease diagnosis.
Fig. 1.
Estimated survival curves using Vardi’s method (dotted line) and by the full likelihood approach under the density ratio model with γ (t) = log(t) (solid line) for (a) possible Alzheimer’s disease, (b) probable Alzheimer’s disease and (c) vascular dementia.
Under the density ratio model assumption, the estimated parameters for the three pairwise comparisons among the diagnosis groups are listed in Table 3. The estimates using the full likelihood and conditional likelihood methods are similar, whereas the standard deviations of the full likelihood estimators are smaller than those obtained from the conditional likelihood.
Table 3.
Pairwise models: parameter estimators, standard deviations and p-values from the likelihood ratio test statistics for full and conditional likelihood methods for the dementia study
| Vascular vs. probable | Vascular vs. possible | Probable vs. possible | ||||
|---|---|---|---|---|---|---|
| Full | Conditional | Full | Conditional | Full | Conditional | |
| γ(t) = t | ||||||
| α̂u(sd) | 0.09 (0.11) | 0.11 (0.15) | 0.24 (0.10) | 0.25 (0.14) | 0.19 (0.09) | 0.18 (0.13) |
| β̂(sd) | −0.02 (0.02) | −0.02 (0.03) | −0.05 (0.02) | −0.05 (0.03) | −0.04 (0.02) | −0.03 (0.02) |
| p-value | 0.32 | 0.37 | 0.01 | 0.06 | 0.03 | 0.13 |
| γ(t) = log(t) | ||||||
| α̂u(sd) | 0.29 (0.18) | 0.34 (0.21) | 0.50 (0.20) | 0.52 (0.24) | 0.26 (0.17) | 0.21 (0.20) |
| β̂(sd) | −0.24 (0.14) | −0.27 (0.17) | −0.39 (0.15) | −0.39 (0.17) | −0.19 (0.12) | −0.15 (0.14) |
| p-value | 0.08 | 0.09 | 0.01 | 0.02 | 0.13 | 0.29 |
Possible, possible Alzheimer’s disease; Probable, probable Alzheimer’s disease; p-value, conditional likelihood ratio test Rc or full likelihood ratio test Rf for assessing the equality of two survival distributions; sd, standard deviation.
6. Discussion
Other than applications in standard case-control studies, the two-sample density ratio model has been used for the estimation of malaria attributable fractions (Qin & Leung, 2005) and the analysis of genetic linkage studies (Qin & Zhang, 2005; Anderson, 1979). In this paper, by taking advantage of the invariance property, we have proposed two likelihood approaches for analysing length-biased failure data. We generalized Vardi’s em algorithm together with the profile likelihood method, so that the unknown parameters and baseline nonparametric distribution of T can be jointly estimated under the density ratio model. Inference based on the full likelihood method is more efficient than that based on the conditional likelihood, because the censored data are fully utilized in the em algorithm. Moreover, the full maximum likelihood estimator of F is on the support of both failure and censoring times, whereas the conditional maximum likelihood estimator of F is on the failure times only. Finally, the full likelihood method is more robust to various censoring patterns, because it does not require the estimation of w(t), which is a function of the residual censoring distribution. Although the full likelihood estimator has efficiency and robustness advantages over the conditional likelihood estimate, the conditional approach is appealing due to its straightforward computation under the density ratio model. Kalbfleisch (1978) recommended the conditional approach in a partially specified generalized linear model, of which the two-sample density ratio model is a special case.
There is a unique feature for length-biased data different from conventional survival data. The normalizing factor in the length-biased density function, μf, can be estimated consistently if there is a positive follow-up time, regardless of how short this is. Because of symmetry between the truncation time A and the residual survival time V under length-biased sampling, information of V can be recovered from A, which is treated as free follow-up time. Such a data structure facilitates the estimability of the mean of the population failure times.
As suggested by one reviewer, it would be interesting to generalize the density ratio model to semiparametric regression models by taking covariates other than group indicators into consideration. One possibility is to model covariates in a parametric form, which may be incorporated into a general density ratio structure.
Acknowledgments
This work was supported in part by grants from the National Institutes of Health. We are grateful to three referees, an associate editor and the editor for helpful comments and suggestions. We thank Professors Asgharian and Wolfson and the investigators from the Canadian Study of Health and Aging for providing us with the dementia data.
Appendix.
We assume the following regularity conditions.
Condition A1. The true parameters (ξ0, F0) belong to ℬ × ℱ = {(ξ, F) : ξ ∈ ℬ and F is a distribution function with continuous and differentiable density function}, where ξ = (α, β)T and ℬ is a compact set in ℝ2.
Condition A2. The known monotone function γ (t) is differentiable and bounded for t ∈ [0, τ].
Condition A3. The residual censoring time C has a continuous survival function and satisfies pr(C > V) > 0 and E(C | Z) > 0.
Condition A4. The Fisher information −E{∂2ℓf(ξ, F̂)/∂ξ2} evaluated at ξ0 is positive, where ℓf(ξ, F) is defined in expression (A2).
Condition A1 ensures the smoothness of the underlying distribution functions. Condition A3 states that at least some subjects are uncensored at the end of the study. The condition on E(C | Z) > 0 ensures a positive follow-up time in each arm, so that the upper bound for the finite support of failure time denoted by τ can be consistently estimated by τ̂ = max1,...,n{Yi}. Condition A4 is a classical condition for semi-parametric models (Andersen et al., 1992, Condition VII2.1.(e), p. 497) and implies that the information matrix for ξ is positive definite when the cumulative distribution function is known.
Proof of Theorem 1. Denote the maximum likelihood estimator based on n observed samples by θ̂n = α̂, β̂, F̂). Estimation of the cumulative distribution function F is via a discretized version F̂ (t) = ∑ti⩽tp̂i. To obtain the maximum likelihood estimator of F subject to the constraint and , we consider the unconstrained loglikelihood function ℓf (α, β, F, λ3, λ4) by incorporating the Lagrange multipliers λ3 and λ4,
| (A1) |
Taking the derivative of ℓf (α, β, F, λ3, λ4) with respect to pj, setting it equal to zero, multiplying pj on both sides, summing over j and taking the constraints into account, we obtain λ3 = n. Taking the derivative of ℓf (α, β, F, λ3, λ4) with respect to α, together with two constraints, we obtain λ4 = n2. After inserting the Lagrange multipliers into (A1), the loglikelihood function ℓf (α, β, F) under the constraints is
| (A2) |
Let P ≡ (p1, . . . , ph). The maximizer of ℓf (α, β, F) for given (α, β) is denoted by P̂ (α, β) and the corresponding estimator of F is F̂ (α, β). The loglikelihood function ℓf (α, β, F) is strictly concave in P, implying a unique maximizer P̂ (α, β) for each (α, β) in the compact set ℬ. The compactness of ℬ and the continuity of the profile likelihood ℓf {α, β, F(α, β)} implies the existence of the maximum likelihood estimator for (α, β, F). Condition A4 ensures the uniqueness of the maximum likelihood estimator (Rothenberg, 1971).
We prove the consistency of θ̂n by the Kullback–Leibler information approach (Murphy, 1994; Parner, 1998). Since the maximum likelihood estimator θ̂n is bounded, by Helly’s selection theorem, there exists a convergent subsequence of θ̂n, denoted by θ̂nk ≡ (α̂nk, β̂nk, F̂nk), such that (α̂nk, β̂nk) converges to some (a*, β*) in ℬ and F̂nk converges to a distribution function F* in ℱ. The second step is to show that any such convergent subsequence of θ̂n must converge to θ0, i.e., θ* ≡ (a*, β*, F*) = θ0.
The empirical Kullback–Leibler distance ℓf (α̂nk, β̂nk, F̂nk) − ℓf (ᾱ, β̄, F̄) is always nonnegative for any (ᾱ, β̄, F̄) in the parameter space, since (α̂nk, β̂nk, F̂nk) maximizes the loglikelihood function ℓf (α, β, F). We choose (ᾱ, β̄) = (α0, β0) and let
and E0 is the expectation under the true parameter values. If θ0 was used as the initial value in the em algorithm in § 3.2, then F̄ (t) is the one-step estimator of the cumulative distribution function. It can be shown that F̄ (t) converges to F0 almost surely and uniformly in t. By the properties of the maximum likelihood estimator,
| (A3) |
By taking limits on both sides, we can show that (nk)−1{ℓf (θnk) − ℓf (α0, β0, F̄)} must converge to the negative Kullback–Leibler distance between Pθ* and Pθ0 almost surely by the strong law of large numbers, where Pθ is the probability measure under the parameter θ. Expression (A3) implies that the Kullback–Leibler distance between Pθ* and Pθ0 is nonpositive. However, the limiting version of ℓf is maximized at θ0, implying that Pθ* = Pθ0 almost surely. Thus, the model identifiability yields θ* = θ0. Since every convergent subsequence of θ̂n converges to θ0, the entire sequence θ̂n must converge to θ0 for any t ∈ [0, τ]. The convergence is almost sure, since we only use the strong law of large numbers at most countably many times. The continuity and monotonicity of F0 ensure the uniform convergence of F̂ (t) in t.
Proof of Theorem 2. The technical details are provided in the Supplementary Material. We only list the main steps here. For theoretical developments, it is helpful to express the likelihood in terms of the hazard function. Let Λ denote the cumulative hazard function for the length-biased time T. Using counting process notation, Ni (t) = I (Yi ⩽ t)δi, for i = 1, . . . , n, the loglikelihood for ψ ≡ (α, β, Λ) is
where ξ = (α, β)T, X (u) = (1, γ (u))T and Ki(u) = (1 − δi) I (u ⩾ Yi) exp{Zi XT(u) ξ − Λ(u)} dΛ(u). Denote the maximum likelihood estimator based on n observed samples by ψ̂n = (α̂, β̂, Λ̂). We prove the weak convergence of ψ̂n by using the Z-theorem (van der Vaart & Wellner, 1996, Theorem 3.3.1) for infinite-dimensional estimating equations.
Denote the score equation vector of the loglikelihood ℓf (ψ) by U(ξ, Λ) ≡ {U1(ξ, Λ), U2(t; ξ, Λ)}, where U1(ξ, Λ) is the score function of ξ,
and U2(t; ξ, Λ) is the score function for the infinite-dimensional parameter Λ. To derive U2(t; ξ, Λ), we consider a one-dimensional submodel given as dΛη,h = (1 + ηh) dΛ, where h is a bounded and integrable function. When setting h(·) = I (· ⩽ t) and evaluating the derivative of the loglikelihood with respect to η at η = 0,
where Yi (u) = I (Yi ⩽ u) is the at risk function for the ith subject. Denote the expectation of score function under the true parameter values (α0, β0, Λ0) by U0(ξ, Λ) ≡ {U10(ξ, Λ), U20(t; ξ, Λ)}. We prove Theorem 2 by verifying the required conditions in Theorem 3.3.1 of van der Vaart & Wellner (1996). We first verify that U0(ξ, Λ) is Fréchet differentiable and its Fréchet derivative can be calculated through the one-parameter submodel given by ξη = ξ0 + ηξ and Λη = Λ0 + ηΛ:
where
In the Supplementary Material, we prove that the inverse operator exists and takes the following form:
where Φ = d22(Λ) − d21 d12.
We then show the weak convergence of the score equation vector evaluated at the true value, U(ξ0, Λ0). Last we prove the following stochastic approximation for the score equation vector
We conclude that under the Conditions A1–A4, n1/2(ψ̂n − ψ0) converges weakly to a tight mean zero Gaussian process (𝕎). By the functional delta method, n1/2(θ̂n − θ0) converges weakly to a tight mean zero Gaussian process −ϕ′ψ0{ (𝕎)}, where ϕ is the transformation from ℬ × 𝒜 → ℬ × ℱ, and ϕ′ψ0 is its Hadamard derivative at ψ0.
Supplementary material
Supplementary material available at Biometrika online includes a proof of the estimability of the mean of T̃, the inference method for k-arms, additional simulation results and a detailed proof of Theorem 2.
References
- Anderson JA. Multivariate logistic compounds. Biometrika. 1979;66:17–26. [Google Scholar]
- Andersen PK, Borgan Ø, Gill RD, Keiding N. Statistical Models Based on Counting Processes. New York: Springer; 1992. [Google Scholar]
- Asgharian M, M’Lan CE, Wolfson DB. Length-biased sampling with right censoring: An unconditional approach. J Am Statist Assoc. 2002;97:201–9. [Google Scholar]
- Asgharian M, Wolfson DB, Zhang X. Checking stationarity of the incidence rate using prevalent cohort survival data. Statist Med. 2006;25:1751–67. doi: 10.1002/sim.2326. [DOI] [PubMed] [Google Scholar]
- Bilker WB, Wang MC. A semiparametric extension of the Mann–Whitney test for randomly truncated data. Biometrics. 1996;50:10–20. [PubMed] [Google Scholar]
- Chen YQ. Semiparametric regression in size-biased sampling. Biometrics. 2010;66:149–58. doi: 10.1111/j.1541-0420.2009.01260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross ST, Lai TL. Bootstrap methods for truncated and censored data. Statist. Sinica. 1996;6:509–30. [Google Scholar]
- Kalbfleisch JD. Likelihood methods and nonparametric tests. J Am Statist Assoc. 1978;73:167–70. [Google Scholar]
- Kay R, Little S. Transformations of the explanatory variables in the logistic regression model for binary data. Biometrika. 1987;74:495–501. [Google Scholar]
- Mandel M, Ritov Y. The accelerated failure time model under biased sampling. Biometrics. 2010;66:149–58. doi: 10.1111/j.1541-0420.2009.01366_1.x. [DOI] [PubMed] [Google Scholar]
- Murphy SA. Consistency in a proportional hazards model incorporating a random effect. Ann Statist. 1994;22:712–31. [Google Scholar]
- Owen AB. Empirical likelihood ratio confidence intervals for a single functional. Biometrika. 1988;75:237–49. [Google Scholar]
- Parner E. Asymptotic theory for the correlated gamma-frailty model. Ann Statist. 1998;26:183–214. [Google Scholar]
- Peto R, Peto J. Asymptotically efficient rank invariant test procedures. J. R. Statist. Soc. A. 1972;135:185–207. [Google Scholar]
- Qin J, Leung D. A semiparametric two-component ‘compound’ mixture model and its application to estimating malaria attributable fractions. Biometrics. 2005;61:456–64. doi: 10.1111/j.1541-0420.2005.00330.x. [DOI] [PubMed] [Google Scholar]
- Qin J, Zhang B. A goodness-of-fit test for logistic regression models based on case-control data. Biometrika. 1997;84:609–18. [Google Scholar]
- Qin J, Zhang B. Marginal likelihood, conditional likelihood and empirical likelihood: connections and applications. Biometrika. 2005;92:251–70. [Google Scholar]
- Qin J, Berwick M, Ashbolt R, Dwyer T. Quantifying the change of melanoma incidence by Breslow thickness. Biometrics. 2002;58:665–70. doi: 10.1111/j.0006-341x.2002.00665.x. [DOI] [PubMed] [Google Scholar]
- Rothenberg TJ. Identification in parametric models. Econometrica. 1971;39:577–91. [Google Scholar]
- Scheike TH, Keiding N. Design and analysis of time-to-pregnancy. Statist Meth: Med Res. 2006;15:127–40. doi: 10.1191/0962280206sm435oa. [DOI] [PubMed] [Google Scholar]
- Shen Y, Qin J, Costantino J. Inference of tamoxifen’s effects on prevention of breast cancer from a randomized controlled trial. J Am Statist Assoc. 2007;102:1235–44. doi: 10.1198/016214506000001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart AW, Wellner JA. Weak Convergence and Empirical Processes with Applications to Statistics. New York: Springer; 1996. [Google Scholar]
- Vardi Y. Multiplicative censoring, renewal processes, deconvolution and decreasing density: nonparametric estimation. Biometrika. 1989;76:751–61. [Google Scholar]
- Wang MC. Nonparametric estimation from cross-sectional survival data. J Am Statist Assoc. 1991;86:130–43. [Google Scholar]
- Wolfson C, Wolfson DB, Asgharian M, M’Lan C, Ostbye T, Rockwood K, Hogan D, The Clinical Progression of Dementia Study Group A reevaluation of the duration of survival after the onset of dementia. New Engl J Med. 2001;344:1111–6. doi: 10.1056/NEJM200104123441501. [DOI] [PubMed] [Google Scholar]
- Zelen M. Forward and backward recurrence times and length biased sampling: age specific models. Lifetime Data Anal. 2004;10:325–34. doi: 10.1007/s10985-004-4770-1. [DOI] [PubMed] [Google Scholar]
- Zeng D, Lin DY. Maximum likelihood estimation in semiparametric regression models with censored data. J. R. Statist. Soc. B. 2007;69:507–64. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material available at Biometrika online includes a proof of the estimability of the mean of T̃, the inference method for k-arms, additional simulation results and a detailed proof of Theorem 2.