Abstract
Atrial fibrillation (AF) is a highly prevalent arrhythmia with pronounced morbidity and mortality. Inward-rectifier K+ current (IK1) is believed to be an important regulator of reentrant-spiral dynamics and a major component of AF-related electrical remodeling. MicroRNA-26 (miR-26) is predicted to target the gene encoding KIR2.1, KCNJ2. We found that miR-26 was downregulated in atrial samples from AF animals and patients and this downregulation was accompanied by upregulation of IK1/KIR2.1 protein. miR-26 overexpression suppressed expression of KCNJ2/KIR2.1. In contrast, miR-26 knockdown, inhibition, or binding-site mutation enhanced KCNJ2/KIR2.1 expression, establishing KCNJ2 as a miR-26 target. Knockdown of endogenous miR-26 promoted AF in mice, whereas adenovirus-mediated expression of miR-26 reduced AF vulnerability. Kcnj2-specific miR-masks eliminated miR-26–mediated reductions in Kcnj2, abolishing miR-26’s protective effects, while coinjection of a Kcnj2-specific miR-mimic prevented miR-26 knockdown-associated AF in mice. Nuclear factor of activated T cells (NFAT), a known actor in AF-associated remodeling, was found to negatively regulate miR-26 transcription. Our results demonstrate that miR-26 controls the expression of KCNJ2 and suggest that this downregulation may promote AF.
Introduction
Recent studies have uncovered an important role of microRNAs (miRNAs) in regulating cardiac excitability and arrhythmogenesis in various cardiac diseases, including myocardial infarction (1), cardiac hypertrophy (2), diabetic cardiomyopathy (3), atrial fibrillation (AF) (4, 5), and other cardiac conditions (6–8). These studies have primarily focused on the muscle-specific miRNAs miR-1 and miR-133, with the exception of miR-328, which also contributes to shaping cardiac electrophysiology (5).
AF is a highly prevalent condition associated with pronounced morbidity and mortality that can cause or exacerbate heart failure and is an important risk factor for stroke (9). AF is characterized by atrial electrical remodeling (mediated predominantly by ion-channel alterations), which favors arrhythmia recurrence and maintenance (9, 10). A prominent feature of the electrical remodeling associated with AF is abbreviation of the effective refractory period (ERP) favoring reentry, primarily due to shortening of atrial action potential duration (APD) (9, 10). Numerous studies have demonstrated that increased inward-rectifier K+ current (IK1), along with increased expression of the principal underlying subunit KCNJ2 mRNA and its encoded Kir2.1 protein, is a prominent feature of AF-related atrial electrical remodeling (10–17). IK1 is the key K+ current responsible for setting the resting membrane potential, controlling cardiac excitability, and modulating late-phase repolarization and APD in cardiac cells. Augmentation of inward-rectifier K+ currents such as IK1, which shortens atrial APD and stabilizes rotor dynamics, is an important factor favoring AF maintenance (18, 19). Furthermore, IK1 is a central regulator of cardiac excitability and arrhythmogenesis and a promising target for new antiarrhythmic approaches (20).
The mechanism of IK1 dysregulation in AF is poorly understood. We performed a computational analysis of miRNAs altered in AF to identify miRNA candidates for IK1 dysregulation (See Supplemental Methods; supplemental material available online with this article; doi: 10.1172/JCI62185DS1), which suggested that miR-26 has the potential to repress KCNJ2/Kir2.1/IK1. miR-26 belongs to a cardiac-enriched (21), but noncardiac-specific, miRNA family composed of 3 members, miR-26a-1, miR-26a-2 and miR-26b, sharing identical seed sequences (2 to 8 nucleotides at the 5′ end determining gene targeting) (22) and with only 2 nucleotide differences between miR-26a and miR-26b (Supplemental Figure 1), suggesting that these miRNAs likely have the same set of target genes. miR-26 sequences are highly conserved across species (Supplemental Figure 1). The 3′ UTR of KCNJ2 mRNA contains 2 binding sites for miR-26a and 2 for miR-26b (Supplemental Figure 2). We therefore hypothesized that miR-26 family miRNAs participate in AF pathophysiology by regulating the expression of KCNJ2/Kir2.1/IK1. Both miR-26a and miR-26b are downregulated in hypertrophied hearts (23) and aortic-valve disease (24). However, the potential roles of miR-26 in cardiomyocyte function and cardiac pathophysiology have not been studied.
Results
Dysregulation of miR-26 family miRNAs and KCNJ2/Kir2.1 in AF.
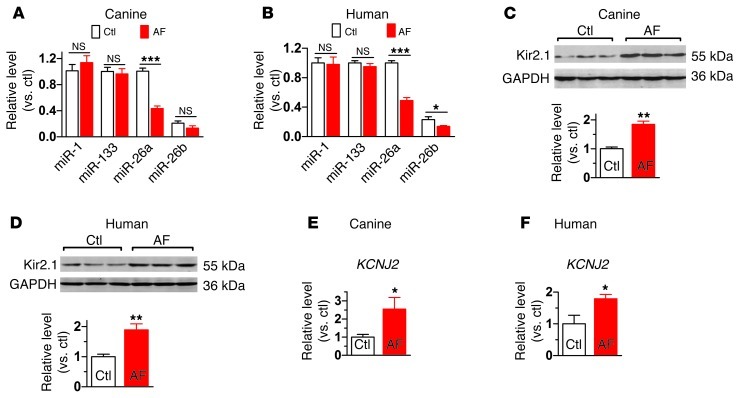
In an effort to explore the roles of miRNAs in AF, we found that miR-26 family members were significantly downregulated (>50%) in a canine AF model (miR-26a; Figure 1A) and in AF patients (miR-26a and miR-26b; Figure 1B and see Supplemental Table 1 for patient information), indicating possible involvement in AF pathophysiology. The level of miR-26a was approximately 4 times higher than that of miR-26b in our canine and human preparations (Supplemental Figure 1). In contrast, miR-1 and miR-133 levels were unaltered in AF samples (Figure 1, A and B). Moreover, in line with previous studies (10–17), we found Kir2.1 protein as well as KCNJ2 mRNA to be upregulated in both AF dogs and patients (Figure 1, C–F). These results are consistent with the notion that miR-26 might contribute to KCNJ2 dysregulation in AF, a possibility that we proceeded to test directly.
Figure 1. Downregulation of miR-26 and upregulation of KCNJ2/Kir2.1 in AF.
(A and B) qPCR for various miRNAs showing downregulation of miR-26 in atrial samples from a canine model (n = 10 for control [Ctl] and n = 9 for AF) and from AF and sinus rhythm patients (n = 5 per group), respectively. For description of patient population, see Supplemental Table 1. (C and D) Western blot analysis showing significant upregulation of Kir2.1 in atrial tissues from AF dogs (n = 9/group) and AF patients (n = 6 for control and n = 5 for AF). (E and F) qPCR showing significant upregulation of KCNJ2 transcripts in atrial tissues from AF dogs (n = 9/group) and AF patients (n = 6 for control and n = 5 for AF). *P < 0.05, **P < 0.01, ***P < 0.001 vs. control. Values are expressed as mean ± SEM.
Validation of KCNJ2 as a target for miR-26.
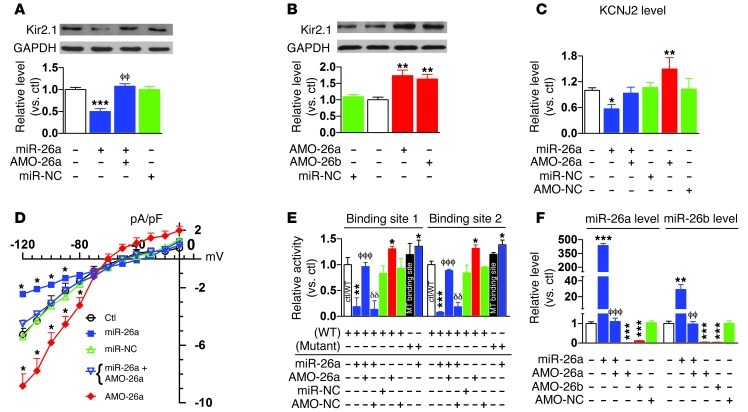
Based on computational analyses revealing 2 miR-26–binding sites in the 3′ UTR of KCNJ2 (Supplemental Figure 2), we conducted Western blot analysis and luciferase reporter gene assays to experimentally clarify whether KCNJ2 is in fact a target for miR-26. As shown in Figure 2A, transfection of miR-26a (100 nM) into H9c2 rat ventricular cells produced marked downregulation of Kir2.1 protein compared with sham-treated control cells. This repression was efficiently rescued by suppressing expression of miR-26 with its antisense inhibitor AMO-26a (10 nM). More strikingly, upregulation of Kir2.1 protein was consistently seen with AMO-26a or AMO-26b alone to knock down endogenous miR-26 and mimic the effect of AF (Figure 2B), indicating the relief of tonic repression of Kir2.1 by miR-26. KCNJ2 mRNA expression was also decreased by miR-26a, reflecting mRNA destabilization (Figure 2C). We further went on to verify the effects of miR-26 at the functional level. IK1 was recorded in neonatal rat ventricular cells using whole-cell patch-clamp techniques. As shown in Figure 2D, the cells transfected with miR-26 had smaller IK1 density than control cells, and the difference was eliminated by cotransfection with AMO-26a. Application of AMO-26a alone increased IK1 density. The regulation of the KCNJ2 gene by miR-26 was confirmed by luciferase assay, which indicated that both of the binding sites for miR-26 responded similarly to miR-26a expression changes (Figure 2E). Mutation of the binding sites (Supplemental Figure 2A) abolished the suppressant effect of miR-26 on the KCNJ2 target gene in luciferase assay (Figure 2E), exposing a small stimulatory effect. The specificity of miR-26 action was indicated by an absence of change with the negative control miRNA (miR-NC) and AMO (AMO-NC) (Supplemental Figure 2B). The efficacy of miR-26a, AMO-26a, or AMO-26b transfer in altering miR-26a and miR-26b expression was verified by real-time RT-PCR experiments (Figure 2F).
Figure 2. Regulation of Kir2.1 expression by miR-26.
(A and B) Immunoblots showing effects of miR-26a (n = 5) and antisense (AMO-26a; n = 6) on Kir2.1 protein expression in H9c2 rat ventricular cells. miR-26a (100 nM) and AMO-26a (10 nM) were transfected with lipofectamine. Control, mock-treated with lipofectamine; miR-NC, negative-control miRNA; AMO-NC, negative-control AMO. **P < 0.01, ***P < 0.001 vs. control; ϕϕP < 0.01 vs. miR-26a alone. (C) qPCR showing effects on KCNJ2 transcript levels in H9c2 cells. *P < 0.05, **P < 0.01 vs. control; n = 4 per group. (D) IK1 density in cultured neonatal rat ventricular cardiomyocytes. IK1 was elicited by 100-ms pulses to voltages indicated. *P < 0.05 vs. control; n = 11/group. (E) Luciferase reporter activities from H9c2 cells cotransfected with miR-26a (10 nM) or AMO-26a (10 nM) and chimeric vectors carrying luciferase gene and a fragment containing one of the binding motifs. Control/WT, control with WT binding sites; MT, mutated miR-26–binding sites in the 3′ UTR of KCNJ2. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control; ϕϕϕP < 0.001 vs. miR-26a alone; δδP < 0.01 vs. miR-26a + AMO26a; n = 3/group for mutated constructs and 4/group for other groups. (F) Verification of changes in miR-26 expression by qPCR. AMO-26a and AMO-26b target both miR-26a and miR-26b. **P < 0.01, ***P < 0.001 vs. control; ϕϕP < 0.01, ϕϕϕP < 0.001 vs. miR-26a alone; n = 4/group. Values are mean ± SEM.
In subsequent control experiments, we found no effects of miR-26 on the protein levels of other K+ subunits, such as HERG (encoded by KCNH2) for rapid delayed rectifier K+ current, KvLQT1 (encoded by KCNQ1) for slow delayed rectifier K+ current, and Kv4.3 (encoded by KCND3) for transient outward K+ current (Supplemental Figure 3). These results are consistent with computational predictions excluding these genes as targets for miR-26 and indicate that the effect of miR-26 on KCNJ2/Kir2.1/IK1 is target specific.
Regulation of AF vulnerability by miR-26.
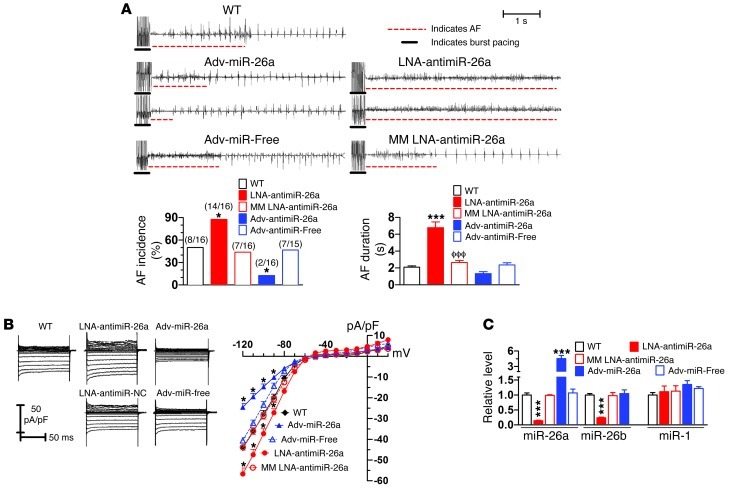
Upregulation of Kir2.1 due to downregulation of miR-26 is expected to promote AF. To test this notion, we injected locked nucleic acid–modified AMO-26 (LNA–anti–miR-26; Supplemental Figure 4) into mice through tail veins to knock down endogenous miR-26 (mimicking the AF effect), with an approach that is documented to be highly efficient in knocking down targeted miRNAs with long-lasting efficacy under in vivo conditions (25–27). This type of LNA-modified molecule has been shown by numerous studies to have improved cellular uptake and stability as well as target affinity and specificity. Attempts to induce AF by intracardiac pacing were then made. AF vulnerability was substantially enhanced in the mice treated with LNA–anti–miR-26, as indicated by an increased proportion of animals with successful induction of AF by electrical stimulation and prolonged AF duration once induced (Figure 3A) as compared with sham-operated, age-matched WT mice. In contrast, with miR-26 overexpression by tail-vein injection of miR-26a–expressing adenovirus (adv–miR-26a; Supplemental Figure 5), AF incidence was significantly reduced (Figure 3A). Mean AF duration also decreased, albeit without a statistically significant difference from WT, likely because of the very small number of miR-26–overexpressing mice that showed quantifiable AF (n = 2; Figure 3A) (please note that for clarity of display, data for AF incidence and duration for relevant corresponding groups are shown as separate sets in Figures 3 and 4; however, all statistical comparisons were performed simultaneously for all groups, as shown in Supplemental Figure 6). Effective cellular uptake of adv–miR-26a was verified (Supplemental Figure 7). Whole-cell patch-clamp recordings confirmed increased IK1 density in atrial cells after LNA–anti–miR-26a treatment, and the opposite effect was seen after adv–miR-26a treatment (Figure 3B). Corresponding effects of LNA–anti–miR-26a and adv–miR-26a on Kir2.1 protein levels were confirmed by Western blot analysis (Supplemental Figure 8, A and B). MM–LNA–anti–miR-26 (where MM indicates mismatched) (Supplemental Figure 4) failed to produce the changes described above, as did the adenovirus carrying the miR-26a–free vector (Supplemental Figure 5). Quantitative real-time RT-PCR (qPCR) confirmed the downregulation of both miR-26a and miR-26b in atrial samples from the mice injected with LNA–anti–miR-26a as well as overexpression of miR-26a in the animals infected by adv–miR-26a (Figure 3C). Neither miR-26a upregulation with adv–miR-26a nor downregulation with LNA–anti–miR-26a altered cardiac structure or function as determined echocardiographically (Supplemental Table 2). Of the ECG intervals, miR-26a manipulation affected only the QT interval, with forced miR-26a upregulation (which downregulated KCNJ2) increasing QT interval and miR-26a downregulation decreasing QT interval (Supplemental Table 3).
Figure 3. Regulation of AF vulnerability by miR-26.
(A) Effects of miR-26 and its antisense on AF in mice. Upper panels: representative atrial electrogram recordings. WT, control mice receiving vehicle injections; adv–miR-free, adenovirus vector without miR-26; MM LNA–anti–miR-26a, mismatched LNA–anti–miR-26a as negative control constructs. Burst pacing is highlighted by solid underlines, whereas dashed underlines indicate AF. Lower panels: percentage of animals with successful AF induction (left: results are shown as n/N, where n = number inducible into AF/total of N mice) and AF duration in animals with successful AF induction (right). *P < 0.05, ***P < 0.001 vs. WT; ϕϕϕP < 0.001 vs. LNA–anti–miR-26a. Note: here and in Figure 4, related data sets for AF incidence and AF duration are shown separately for clarity in display. However, statistical comparisons were performed between all animals with interventions simultaneously (see Supplemental Figure 6 for all comparisons), with the statistical comparisons here reflecting the results of simultaneous comparisons of all data in Supplemental Figure 6. (B) IK1 in atrial myocytes isolated from mice treated with various constructs. Left panels: IK1 recordings. Right panel: IK1 density-voltage relationships. Results for adv–miR-26a and LNA–anti–miR-26a are shown with solid lines and the symbols defined on the figure; results for their controls (adv-miR free and MM LNA–anti–miR-26a) are shown with dashed lines. *P < 0.05 vs. WT; n = 12 cells/group. (C) qPCR verification of atrial miR-26 expression changes resulting from various constructs (note: anti–miR-26a is complementary to both miR-26a and miR-26b). ***P < 0.001 vs. WT; n = 8 mice/group. Values are mean ± SEM.
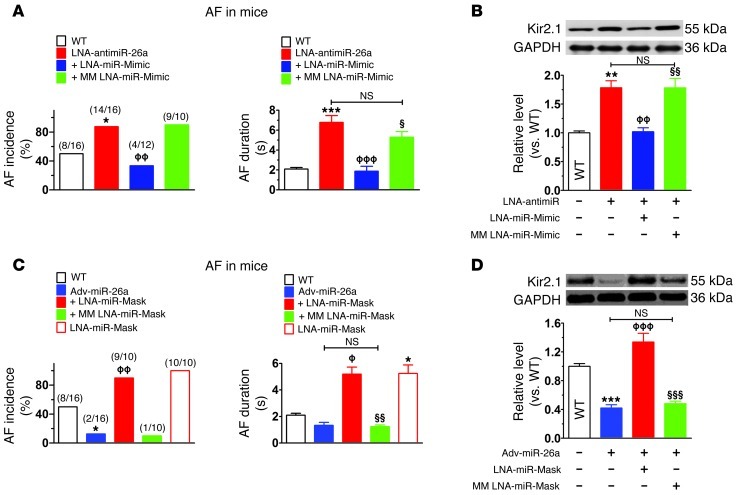
Figure 4. Verification of the specificity of miR-26/KCNJ2/Kir2.1 in the control of AF.
(A) Inhibitory effect of LNA–miR-mimic on induction (left) and maintenance in inducible animals (right) of AF induced in mice also treated with LNA–anti–miR-26a. MM LNA–miR-mimic, mismatched miR-mimic (negative control). Number inducible/total used is indicated by n/N values within brackets. *P < 0.05, ***P < 0.001 vs. WT; ϕϕP < 0.01, ϕϕϕP < 0.001 vs. LNA–anti–miR-26a alone; §P < 0.05 vs. LNA–anti–miR-26a + LNA–miR-mimic. (B) Western blot verifying ability of miR-mimic to knock down Kir2.1. **P < 0.01 vs. WT; ϕϕP < 0.01 vs. LNA–anti–miR-26a alone; §§P < 0.01 vs. LNA–anti–miR-26a + LNA–miR-mimic; n = 6 for each group. (C) LNA–miR-mask abolishes the protective effect of adv–miR-26a against AF induction (left) and maintenance (right). +LNA–miR-mask,mice injected with LNA–miR-mask (5 mg/kg/d daily for 3 days before injection of adv–miR-26a); LNA–miR-mask, mice injected with LNA–miR-mask alone; MM miR-mask, mismatched miR-mask (negative control). *P < 0.05 vs. WT; ϕP < 0.05, ϕϕP < 0.01 vs. adv–miR-26a alone; §§P < 0.01 vs. adv–miR-26a + LNA–miR-mask. (D) Western blot verifying ability of miR-mask to protect against Kir2.1 knockdown by adv–miR-26a. ***P < 0.001 vs. WT; ϕϕϕP < 0.001 vs. adv–miR-26a alone; §§§P < 0.001 vs. adv–miR-26a + LNA–miR-mask; n = 6/group. Values are mean ± SEM. Group definitions as in Figure 3. Note: the experiments shown in Figure 3A and Figure 4, A and C, were done contemporaneously. Thus, the same WT group data serve as controls in each case, and the same LNA–anti–miR-26a and adv–miR-26a data are shown in Figure 3 and Figure 4. Related data sets for AF incidence and AF duration are shown separately for clarity in display. However, statistical comparisons were performed between all animals with interventions simultaneously (see Supplemental Figure 6).
While the above results suggested a substantial role for miR-26 in controlling AF initiation and maintenance in mice, whether the effects are attributable specifically to targeting of KCNJ2/Kir2.1/IK1 remained to be determined. To shed light on this issue, we conducted the following 2 types of experiments. First, we wanted to see whether knockdown of miR-26 is still able to promote AF if Kir2.1 upregulation resulting from miR-26 downregulation is prevented by using the miRNA mimic (miR-mimic) techniques developed in previous work (28) to specifically retain miR-26 downregulation of Kir2.1. We designed a miR-mimic to specifically target the 3′ UTR of KCNJ2 without interacting with potential miR-26 target sites of any other transcripts of known function (Supplemental Figure 9), thus mimicking the effect of miR-26 on KCNJ2 without altering other targets. We began by coinjecting LNA–anti–miR-26 and LNA–miR-mimic into mice and assessed changes in vulnerability to AF. Whereas LNA–anti–miR-26 enhanced AF incidence and duration relative to WT control animals, this effect was eliminated by coinjection with LNA–miR-mimic (Figure 4A). The ability of the miR-mimic to prevent Kir2.1 expression enhancement caused by LNA–anti–miR-26 was confirmed by Western blot analysis showing reductions of atrial Kir2.1 protein levels to WT levels (Figure 4B). The mismatched LNA–miR-mimic (LNA–MM–miR-mimic) failed to alter the arrhythmia-promoting or Kir2.1 protein–upregulating effects of LNA–anti–miR-26. Second, we tested directly whether overexpression of miR-26 is still able to suppress AF if its repressive effect on KCNJ2 is specifically prevented,while its inhibitory effects on other target genes are maintained. To achieve this goal, we used the miRNA-masking oligodeoxynucleotide (ODN) (miR-mask) strategy, or target-protection approach, that has been developed to produce target gene–specific protection against a miRNA (28). Two miR-masks (Supplemental Figure 9) were designed to be fully complementary to the 2 binding sites for miR-26 at the 3′ UTR of KCNJ2 without binding to any other functionally identified targets and were LNA modified for greater in vivo efficacy. These miR-masks were composed of single-stranded DNA, which is unable to incorporate into the RNA-induced silencing complex (RISC) that is essential for miRNA action. Thus, the miR-masks bind specifically and tightly to the miR-26–binding sequence of KCNJ2 (preventing miR-26 binding), but do not bind to other miR-26 targets, and do not mimic miR-26 effects on KCNJ2 translation or mRNA stability. Consequently, the miR-masks prevent the effect of miR-26 on KCNJ2 without preventing effects on other miR-26 targets. As shown in Figure 4C, in mice pretreated with the LNA–miR-masks via tail-vein injection, adv–miR-26 failed to protect against AF induction and maintenance, in contrast with the clear protection produced by adv–miR-26 in nonpretreated mice. Moreover, administration of the LNA–miR-masks alone increased AF inducibility and maintenance (Figure 4C), indicating that endogenous background miR-26 expression offers significant protection against AF. The ability of the LNA–miR-masks to prevent the repressive effect of miR-26 on Kir2.1 was verified in mice treated with the construct together with adv–miR-26 (Figure 4D).
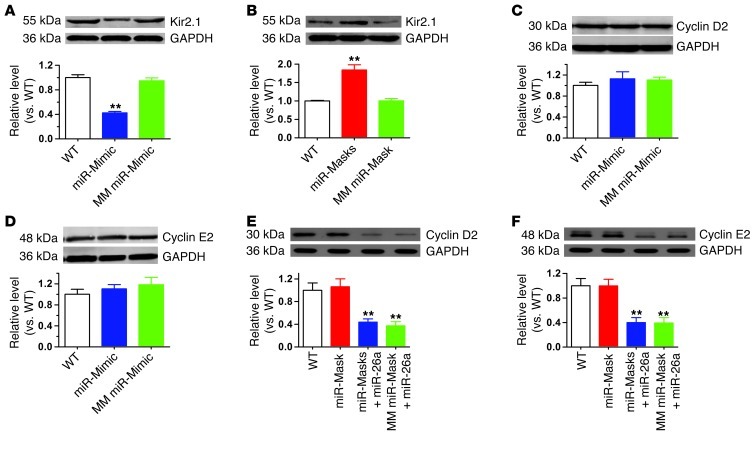
Figure 4 shows the ability of miR-mimic and miR-mask to counteract the effects of anti–miR-26a and adv–miR-26a, respectively. The ability of the LNA–miR-mimic alone (Figure 5A) to downregulate and the LNA–miR-masks alone (Figure 5B) to upregulate atrial Kir2.1 protein levels in mice were also verified. Further control experiments were conducted to confirm biologically the gene specificity of LNA–miR-mimic and LNA–miR-masks on KCNJ2 by verifying the lack of action on other miR-26 targets predicted bioinformatically not to interact (as described in Supplemental Methods and Supplemental Figure 9). First, administration of LNA–miR-mimic (Figure 5, C and D) showed no effects on the expression of protein products of 2 previously validated target genes of miR-26, Cyclin D2 and Cyclin E2 (29). Second, in contrast to Kir2.1, the repressive effects of adv–miR-26a on Cyclin D2 and Cyclin E2 protein expression (Supplemental Figure 8, C and D) were not affected by coapplication of the LNA–miR-masks (Figure 5, E and F) in agreement with the absence of recognition sites for the LNA–miR-masks on these 2 genes.
Figure 5. Verification of the specificity of LNA–miR-mimic and LNA–miR-masks on KCNJ2/Kir2.1.
(A and B) Verification of the ability of the LNA–miR-mimic alone to reduce (A) and the LNA–miR-mask alone to increase (B) atrial Kir2.1 protein levels by Western blot on protein samples from atrial tissues. The respective negative control constructs were also examined. **P < 0.01 vs. WT; n = 6 /group. (C and D) Verification of the specificity of the LNA–miR-mimic to repress Kir2.1 without affecting the other untargeted genes, Cyclin D2 and Cyclin E2. (E and F) Verification of the specificity of the LNA–miR-masks to block action of miR-26 on Kir2.1 without altering its effects on 2 other proven target genes, Cyclin D2 and Cyclin E2 (27). miR-26a, adv–miR-26a; miR-masks, LNA–miR-masks; MM miR-mask, mismatched LNA–miR-mask. **P < 0.01 vs. WT; n = 6/group. Values are expressed as mean ± SEM.
Potential mechanism of miR-26 downregulation in AF.
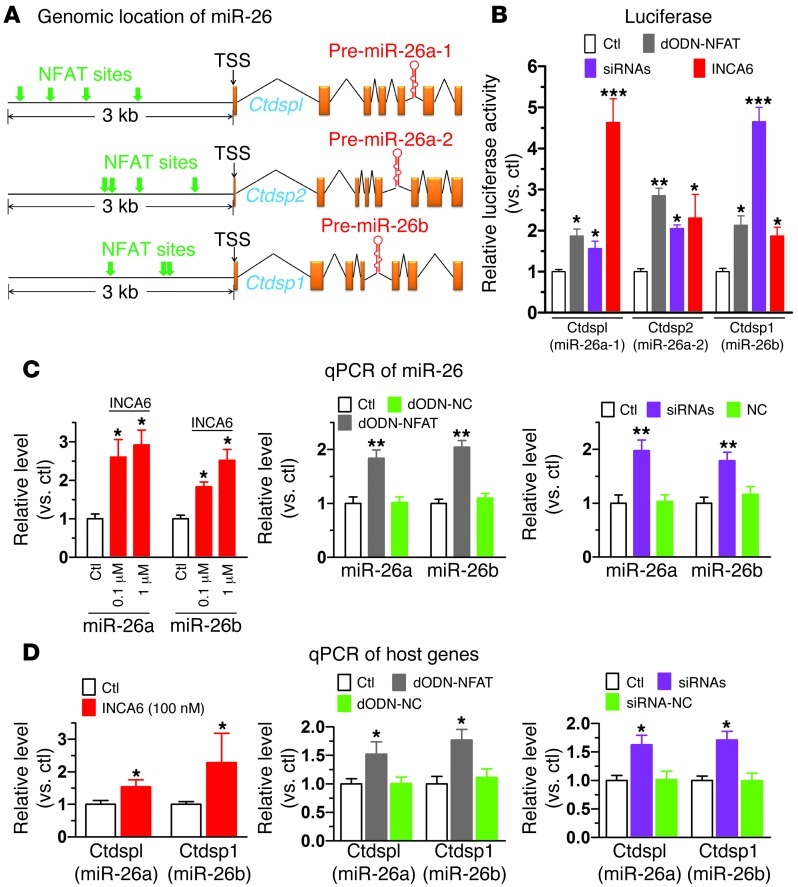
Having established the role of miR-26 in controlling AF through targeting KCNJ2, we went on to elucidate potential molecular mechanisms underlying the dysregulation of miR-26 in AF. We suspected that the 3 members of the miR-26 family, miR-26a-1, miR-26a-2, and miR-26b, might be transcriptionally regulated in humans by a common transcription factor, since both miR-26a and miR-26b were significantly downregulated in AF (Figure 1B). Computational analysis using MatInspector V2.2 revealed a common feature of the 5′ flanking regions of the human host genes for the miR-26 family members: they all contain a cluster of putative cis-acting elements for nuclear factor of activated T cells (NFAT) within 5 kb upstream of the transcriptional start sites across different species (Figure 6A and Supplemental Table 4). Intriguingly, enhanced NFAT activity has been frequently implicated in AF promotion and is believed to couple rapid atrial activity to atrial remodeling via Ca2+-sensitive signaling through calcineurin (30–32). We therefore explored the role of NFAT in regulating expression of miR-26a and miR-26b.
Figure 6. Transcriptional regulation of miR-26 by NFAT.
(A) Schematic genomic maps of the 3 miR-26 family members showing human host genes, intronic locations of pre–miR-26a/b, and putative NFAT binding sites (indicated by arrows) in the 5′ flanking regions. (B) Effects of NFAT inhibition on promoter activities of the host genes of human miR-26 family members: Ctdspl/miR-26a-1, Ctdsp2/miR-26a-2, and Ctdsp1/miR-26b, determined by luciferase activity assay using pGL3 vector carrying the promoter regions containing NFAT-binding sites. NFAT was inhibited by INCA6 (100 nM), sequestered by dODN (dODN-NFAT; 10 nM), or silenced by siRNAs to NFATc3 and NFATc4 (10 nM). Negative controls failed to affect luciferase activity (data not shown). *P < 0.05, **P < 0.01, ***P < 0.001 vs. control; n = 4/group. (C) Effects of NFAT inhibition by INCA6 (left panel; n = 5/group), dODN-NFAT (10 nM; middle panel; n = 6/group), and siRNA (10 nM; right panel; n = 6/group) on miR-26a and miR-26b levels, determined by qPCR in H9c2 cells. *P < 0.05, **P < 0.01 vs. control. (D) Effects of NFAT inhibition on expression of miR-26 host genes, determined by qPCR in H9c2 cells. Note that NFAT inhibition does not affect miR-1 levels (Supplemental Figure 13). *P < 0.05 vs. control; n = 6/group. Values are mean ± SEM.
We first identified the transcription start sites (TSSs) of the host genes using 5′ RACE techniques: CtdspI for miR-26a-1, Ctdsp2 for miR-26a-2, and Cdtsp1 for miR-26b (Supplemental Figure 10, A–D, and Supplemental Table 6). We then subcloned the promoter fragments spanning the putative NFAT-binding sites upstream of the TSSs (Supplemental Figure 10, B–D) into pGL3 luciferase vector and conducted luciferase assays to assess the effects of a decoy ODN that contains the ideal binding site for NFAT (dODN-NFAT; for sequence, see Supplemental Figure 11) to sequester NFAT and block its function as well as to study the effects of siRNAs targeting NFATc3 and NFATc4, to knock down NFAT expression (Supplemental Figure 12), and to analyze the actions of the cell-permeable NFAT inhibitor INCA6. As depicted in Figure 6B, dODN-NFAT, NFAT-siRNAs, or INCA6 robustly increased luciferase activity of the miR-26 host-gene promoter constructs. Next, we evaluated the effects of INCA6 on the expression levels of miR-26a and miR-26b in H9c2 cells. As shown in Figure 6C, 24-hour incubation of cells with INCA6 produced concentration-dependent increases in both miR-26a and miR-26b expression. Similar to INCA6, transfection of H9c2 cells with dODN-NFAT or NFAT-siRNAs caused around 2-fold upregulation of both miR-26a and miR-26b (Figure 6C). As expected, the host genes demonstrated parallel changes in their expression (Figure 6D). In contrast to miR-26, miR-1 expression, as a negative control, was not significantly affected by dODN-NFAT, siRNAs, and INCA6 (Supplemental Figure 13). These results indicate that NFAT negatively regulates transcription of the miR-26 family members. Negative control–scrambled dODN and siRNA did not affect miR-26 promoter activity or the expression level of miR-26 (Figure 6, B and C).
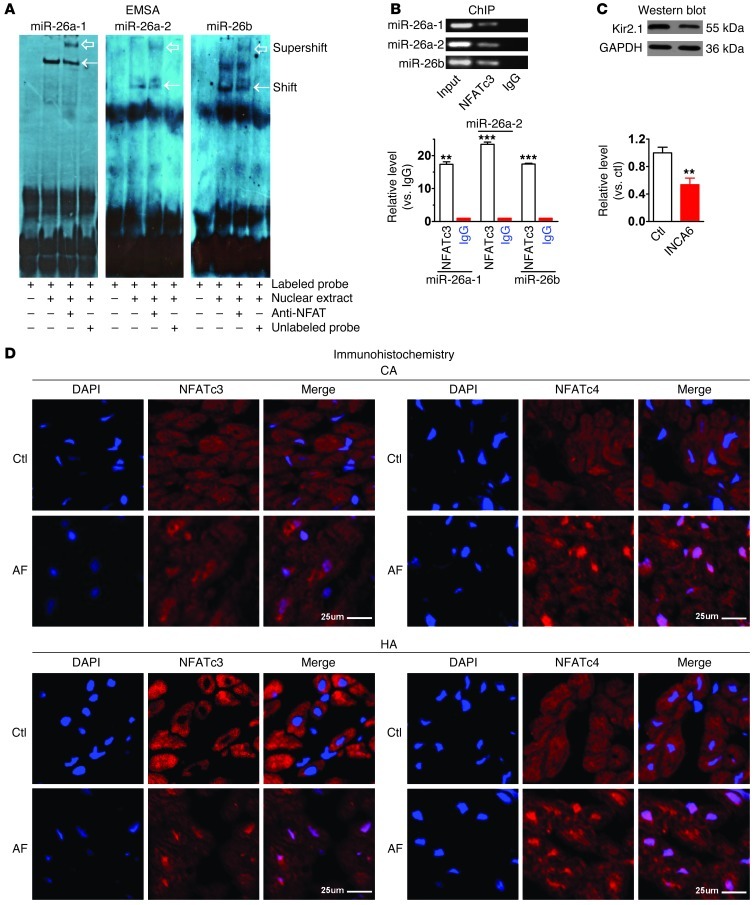
To verify the physical interaction between NFAT and the cis-acting elements of miR-26, we first carried out EMSA in conjunction with supershift to assess the ability of NFAT to bind the putative cis-acting elements in the 5′ flanking regions of the miR-26a and miR-26b host genes. We incubated each probe, a synthesized, digoxigenin-labeled (DIG-labeled) putative NFAT cis-acting element (double-stranded ODN fragment), with nuclear extract from HeLa cells. As depicted in Figure 7A, a shifted band representing protein-DNA binding was identified, which was eliminated upon addition of excessive nonlabeled probes, but was supershifted by the antibody to NFAT. We then subsequently conducted ChIP to further verify the protein-DNA interactions that occur inside the nucleus of living cells. Our results confirmed the binding of NFAT to each of the 3 cis-acting elements in the 5′ flanking regions of the 3 miR-26 members (Figure 7B).
Figure 7. NFAT-dependent regulation of miR-26 in AF.
(A) EMSA to test in vitro binding of NFATc3 to its cis-acting element in the promoter region of human host genes for miR-26a-1, miR-26a-2, and miR-26b. Note shifted band representing protein-DNA binding (arrows), abolished with excess nonlabeled probe. Supershifted band (hollow arrows) represents NFATc3-cis-acting element binding in presence of anti-NFATc3 antibody. (B) ChIP testing in vivo binding of NFAT to human host genes of miR-26a-1, miR-26a-2, and miR-26b. Top: PCR products of 5′ flanking region encompassing NFAT-binding sites following immunoprecipitation with anti-NFATc3 antibody. Bottom: host gene binding measured by qPCR following ChIP, expressed as fold-changes over IgG-negative control. Input: genomic DNA without immunoprecipitation. **P < 0.01, ***P < 0.001 vs. IgG; n = 3/group. (C) Western blot showing Kir2.1 downregulation by INCA6 (100 nM). **P < 0.01 vs. control; n = 5/group. Values in B and C are mean ± SEM. (D) Immunohistochemical images showing nuclear translocation of NFATc3 and NFATc4 in atrial samples from AF dogs (CA) and patients (HA). Blue, nuclear staining (DAPI); red, NFAT staining; violet, NFAT nuclear localization in merged images. Findings similar to those in D were obtained in 3 subjects per group. Scale bars: 25 μm.
We reasoned that if NFAT is indeed a negative regulator of miR-26, inhibition of NFAT activity should downregulate Kir2.1 protein by relieving the inhibitory effect on miR-26, provided that other regulatory factors are kept unchanged. This hypothesis was supported by the data shown in Figure 7C: application of INCA6 significantly decreased Kir2.1 protein expression. Finally, we confirmed NFAT modulation in AF, as evidenced by enhanced translocation of NFATc3 and NFATc4 from cytoplasm to nucleus in atrial samples of AF dogs and patients on immunohistochemical examination (Figure 7D). The experiments shown in Figures 6 and 7 indicate that NFAT is a negative regulator of miR-26 transcription and that, since NFAT is translocated to cardiomyocyte nuclei in AF, it is a strong candidate to underlie AF-induced miR-26 dysregulation.
Discussion
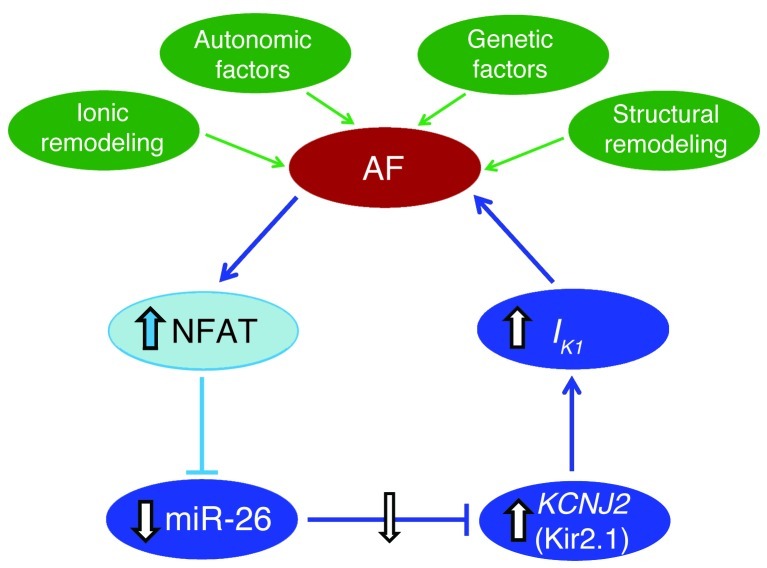
Taken together, our results identify miR-26 as a potentially important regulator of KCNJ2 gene expression and, via IK1, a determinant of AF susceptibility. In addition, our findings identify miR-26 as a potential mediator of the electrophysiological effects of Ca2+-dependent NFAT signaling, believed to be an important player in AF perpetuation (30–32). Our study therefore reveals what we believe to be novel molecular control mechanisms for AF at the miRNA level, as summarized schematically in Figure 8.
Figure 8. Molecular mechanism underlying the AF-promoting effect of miR-26 downregulation (shown in blue).
AF activates NFAT, enhancing NFAT translocation into the nucleus, where it transcriptionally represses the expression of miR-26 genes. Reduced miR-26 expression then derepresses its target gene KCNJ2 at both mRNA and protein levels, causing an augmentation of IK1. The augmented IK1 favors the maintenance of AF. Note: the roles of components illustrated in darker blue were shown by manipulation both in vivo and in vitro; those in lighter blue were demonstrated in vitro only. In green are underlying pathophysiological factors leading to initial AF occurrence, which also contribute to the properties and probability of AF in the presence of enhanced IK1.
AF can be induced by a variety of factors, including atrial dilation, oxidative stress, atrial ischemia or hypoxia, and intracellular Ca2+ overload; all of these ultimately act by altering, directly or indirectly, atrial electrical activity, principally by modulating ion channel function and/or expression (9, 10, 33). Many ion currents are altered by atrial electrical remodeling during AF (10). IK1 is a particularly important mechanistic determinant of AF-supporting reentry because, in addition to accelerating repolarization, enhanced IK1 increases Na+ current availability, accelerating and stabilizing reentrant rotors (18). IK1 enhancement is not only a potentially important contributor to AF associated with atrial remodeling (18, 33), but is also the causative factor in familial AF, resulting from gain-of-function mutations associated with Kir2.1 channelopathies (34–36).
More generally, IK1 plays a central role in cardiac excitability and arrhythmogenesis (20, 37). Although IK1 alterations are important in the failing heart, they are not explained by changes in candidate-subunit transcript levels (38), suggesting posttranscriptional regulation. Similarly, KCNJ2 protein expression changes in patients with AF may exceed mRNA alterations (16), pointing to posttranscriptional mechanisms such as miRNA-induced inhibition of translation. Our findings indicate that miR-26 regulates this important K+ channel under specific disease conditions and provide insights into the transcriptional control of miR-26. Further studies of miR-26 dysregulation of IK1 in other pathological contexts are clearly of potential interest.
Downregulation of L-type Ca2+ current is well established as an important component of the ionic remodeling associated with AF (9, 33). The Ca2+-calcineurin-NFAT system plays a central role in detecting the rapid repetitive atrial activation that occurs during AF and in coupling AF to L-type Ca2+ current downregulation (30–32). The contribution of IK1 upregulation has been established more recently (10), but is recognized to be of substantial importance to AF pathophysiology (18). Our findings here suggest that the calcineurin-NFAT system engaged by AF might also induce IK1 changes via transcriptional regulation of miR-26, indicating that this Ca2+-dependent system may be a common pathway for at least 2 of the important changes in ion-channel function associated with AF.
We found that adenoviral gene transfer of miR-26 via tail-vein injection was able to upregulate cardiac miR-26 expression and suppress AF inducibility and maintenance. AF therapy is presently quite inadequate, with conventional antiarrhythmic drugs having limited efficacy and potentially significant toxicity (9, 39). There is therefore great interest in novel therapeutic approaches to arrhythmia, one of which in early development is gene therapy (39, 40). Our findings suggest that miR-26 could conceivably be a candidate for AF-targeting gene therapy in the future.
The expression of miR-1, which is upregulated in myocardial infarction and established by a previous study to target KCNJ2/Kir2.1/IK1 (1), was unaltered in our samples from dogs and patients with AF (Figure 1A). Girmatsion et al. (4), however, reported that the expression of miR-1 was reduced by approximately 86% in left atrial samples of AF patients undergoing mitral valve repair or bypass grafting. There was a corresponding increase in Kir2.1 protein expression. Based on their data, the authors proposed that downregulation of miR-1 is responsible for upregulation of Kir2.1/IK1 in AF patients. However, their study leaves a number of important unresolved issues. First, no direct evidence for an AF-promoting role of miR-1 downregulation was presented. Second, Kir2.1 was found to be upregulated by only 1.5-fold at the protein level but by 3-fold at the mRNA level in the Girmatsion study (4). These results are inconsistent with the expected effect of miR-1 because miR-1 has been documented not to affect KCNJ2 transcript levels (1). Third, the authors showed that the expression of connexin-43, another validated target of miR-1 (1), was unaltered. Finally, whether the observed upregulation of Kir2.1 expression in their preparations was caused by miR-1 downregulation was not tested. The lack of miR-1 expression change in our patient and dog samples indicates that miR-1 changes are unlikely to have been a major contributor to AF in our subjects.
Although we identified miR-26 downregulation in AF as an important potential contributor to IK1 upregulation and AF promotion, we are in no way claiming that miR-26/IK1 changes are the only factor governing AF development. AF is clearly a complex condition that results from multiple potential etiological contributors that can interact with each other (36). In addition to IK1 changes, the electrical disturbances in AF-related remodeling can stem from changes in ICaL, constitutive IKACh, and alterations in connexin function (36). Indeed, a comparison of the time course of changes in AF duration, miR-26 expression, and Kir2.1 protein expression in our canine AF model (Supplemental Figure 14) shows that AF vulnerability is significantly increased at 1 week of atrial tachypacing, before significant changes in miR-26 and Kir2.1 appear, indicating that other factors are certainly involved. Nevertheless, further statistically significant changes occur in AF vulnerability between weeks 1 and 6, concomitant with miR-26/Kir2.1 changes, consistent with the profibrillatory properties of miR-26 downregulation/Kir2.1 upregulation seen in our mouse model (P < 0.001). Furthermore, structural alterations and changes in intracellular Ca2+ handling can also play a substantial role, and the mechanistic contributors to AF vary, depending on underlying diseases and genetic factors (33, 36). Nor is IK1 the only potential target for miR-26 in AF. For example, we recently demonstrated that miR-26 also targets the nonselective cation-channel subunit TRPC3, through which it can alter fibroblast function and thereby contribute to AF promotion (41). However, the miR-26 interaction with the TRPC3 gene is not predicted to be mimicked by the KCNJ2-binding miR-mimic, nor is it blocked by the miR-masks that we used in the present study. Thus, the protective effects against KCNJ2 upregulation and AF promotion that we observed in our mouse model with the miR-mimic and the opposite phenomena with the miR-masks are not mediated by TRPC3 changes. Similarly, our miR-mimic did not bind to any other functional transcripts in GenBank, and the miR-mask interaction with the miR-26–binding sites on KCNJ2 did not apply to any other functional transcripts in GenBank. Therefore, our results strongly suggest that the effects of miR-26 alterations that we observed on AF in our in vivo mouse model were mediated through changes in KCNJ2/IK1. Our observations of the AF-promoting effect of miR-26 downregulation/IK1 upregulation are limited to changes in AF vulnerability and AF sustainability once induced. We never saw spontaneously initiated AF in this study, and thus our observations provide no information about the mechanisms of spontaneous AF onset in humans. They rather relate to the factors determining the substrate for AF initiation and persistence upon induction by spontaneous atrial ectopy/tachyarrhythmia.
We demonstrated in vitro that NFAT is a negative regulator of miR-26: NFAT inhibition upregulated miR-26 transcription and reciprocally downregulated KCNJ2 expression. However, we were unable to create an in vivo model to test whether we could mimic the effects on miR-26/KCNJ2/AF vulnerability seen with AF by enhancing NFAT signaling. Until this is accomplished, the role of NFAT in AF-related miR-26 regulation and associated AF promotion remains to be fully established. Another potential limitation to our findings is that the underlying heart diseases of patients from whom we obtained atrial-tissue samples (Supplemental Table 1) could have influenced the results.
In conclusion, we have discovered that miR-26 regulates the KCNJ2 gene and the IK1 channel that it encodes. The application of innovative miR-mimic and miR-mask methods allowed us to confirm that KCNJ2 changes are necessary and sufficient to explain AF vulnerability increases that result from miR-26 upregulation and decreases caused by miR-26 downregulation in an in vivo mouse model. miR-26 is consistently decreased in AF patients and an animal model of AF, and AF-related miR-26 dysregulation is likely due to suppression of miR-26 transcription by the Ca2+-dependent transcription factor NFAT. These findings provide what we believe are new insights into the molecular mechanisms underlying a common and important cardiac arrhythmia and may have implications for other arrhythmia conditions in which IK1 is dysregulated.
Methods
AF induction in mice.
C57BL/6 mice were anesthetized with 1% sodium pentobarbital through tail-vein injection (30 mg/kg). Intracardiac pacing was performed in these animals by inserting an 8-electrode catheter (1.1 F, octapolar EP catheter; SciSense) through the jugular vein and advancing it into the right atrium and ventricle with protocols similar to those reported in previous studies (5, 42). Inducibility of atrial arrhythmias was tested by applying 6-second tachypacing bursts through the catheter electrodes using an automated stimulator interfaced with the data acquisition system (GY6000; HeNan HuaNan Medical Science & Technology Ltd.). The first 6-second burst had a cycle length of 40 ms, decreasing in each successive burst with a 2-ms decrement down to a cycle length of 20 ms. Successful induction of AF was defined as the creation of rapid irregular atrial rhythm lasting for 1 second or more, calculated from the direct atrial activation recordings.
To study AF changes with miR-26 manipulation, adv–miR-26a was injected into mice through a tail vein (100 μl; titer: 1010 pfu/ml) once daily for 3 consecutive days. Manipulation of miR-26a was similarly achieved by tail-vein injection of LNA–anti–miR-26a and/or LNA–miR-mimic or adv–miR-26a and/or LNA–miR-mask at 5 mg/kg/d once a day for 3 days. AF was measured and/or atrial-tissue samples for biochemical analysis were obtained 3 days after the last dose of the constructs.
Human atrial samples from patients with AF.
Human tissues (right atrium appendage) were provided by the Second Affiliated Hospital of the Harbin Medical University using procedures approved by the Ethics Committee of Harbin Medical University. The tissues were obtained from 22 individuals undergoing heart surgery, 10 in sinus rhythm and 12 with AF (Supplemental Table 1). These preparations were used to isolate total RNA for real-time RT-PCR quantification of miRNAs and to extract cytosolic proteins for measurement of Kir2.1.
Dog AF model.
Mongrel dogs of either sex, weighing between 15 and 25 kg, were implanted with right atrial pacemakers (Shanghai Fudan University, Shanghai, China) under sodium pentobarbital (30 mg/kg i.v.) anesthesia. AF was induced by up to 6 weeks of atrial tachypacing at 400 bpm. Dogs with spontaneously persisting AF, defined as continuous spontaneous AF maintenance for at least 2 hours, were used in this study (5).
Cell culture.
The H9c2 (rat embryonic ventricular) cell line used in this study was purchased from ATCC and cultured in DMEM.
Cardiomyocyte isolation and primary cell culture.
The enzymatic dispersion techniques used to isolate single atrial myocytes from neonatal rats and adult mice have been previously described in detail (5). Briefly, 1- to 3-day-old rats were decapitated and their hearts were aseptically removed. The atria were dissected, minced, and trypsinized at 37°C for 10 minutes. Dissociated cells were plated in 24-well plates in DMEM (Invitrogen) containing 10% FBS and 0.1 mM bromodeoxyuridine (Sigma-Aldrich) and the nonadherent cardiomyocytes were removed. The cells (1 × 105/well) were seeded in a 24-well plate for further experiments. This procedure yielded cultures with 90% ± 5% myocytes, as assessed by microscopic observation of cell beating. The cardiomyocytes were also verified by positive staining with an anti–α-actin monoclonal antibody through immunocytochemistry.
For the isolation of adult mouse heart cells, mice that had been treated with varying constructs were anesthetized with sodium pentobarbital (30 mg/kg i.p.). The hearts were rapidly removed and retrogradely perfused through the aorta using a modified Langendorff apparatus. The preparation was perfused with standard Tyrode’s solution (126 mM NaCl, 5.4 mM KCl, 10 mM HEPES, 0.33 mM NaH2PO4•2H2O, 1.0 mM MgCl2•6H2O, 1.8 mM CaCl2, and 10 mM glucose; pH adjusted to 7.4 with NaOH) for 5 minutes, then switched to Ca2+-free Tyrode’s solution until beating stopped, followed by perfusion with the same solution containing collagenase II (7 mg/50 ml) and BSA. The freshly isolated atrial myocytes were gently centrifuged and resuspended in Kraftbruhe (KB) storage solution (70 mM glutamic acid, 15 mM taurine, 30 mM KCl, 10 mM KH2PO4, 0.5 mM MgCl2, 0.5 mM EGTA, 10 mM HEPES, and 10 mM glucose; pH adjusted to 7.35 with KOH). All solutions were aerated with 100% oxygen and warmed to 37 ± 0.5°C. Only single rod-shaped, Ca2+-tolerant, and quiescent cells with clear cross-striations were selected for patch-clamp recording.
Computational prediction of miRNA target genes and cis-acting elements for transcription factors.
We used 7 established miRNA target prediction algorithms: DIANA-microT3.1, miRanada, MirTarget2, PicTar, PITA, microcosm, and TargetScan5.1. Only the miRNAs that are predicted to target a given gene by at least 4 of the 7 algorithms were considered as candidates for further analysis (43).
The binding sites for various transcription factors in the promoter regions of the host genes of miR-26a-1, miR-26a-2, and miR-26b from different species (human, canine, rat, and mouse) were analyzed with MatInspector V2.2 (Genomatix) (44). Analyses were made to the 5′ flanking regions 5 kb upstream of the transcriptional start sites of the host genes (Supplemental Table 4).
Synthesis of miRNAs and anti-miRNA antisense inhibitors.
miR-26a and miR-26b were synthesized by Integrated DNA Technologies Inc., and their respective antisense oligonucleotides, AMO-26a and AMO-26b, were synthesized by Exiqon. Five nucleotides or deoxynucleotides at both ends of the antisense molecules were locked (LNA; the ribose ring is constrained by a methylene bridge between the 2′-O- and the 4′-C atoms). Additionally, a scrambled RNA was used as a negative control. For in vivo experiments, LNA-modified constructs, LNA–anti–miR-26a, LNA–miR-mimic, and LNA–miR-masks, were synthesized by Exiqon. The LNA–anti–miR-26a and LNA–miR-masks were single-stranded DNA analogs complementary to mature miR-26 (5′-UUCAAGUAAUCCAGGAUAGGCU-3′) and the miR-26–binding sites on KCNJ2, respectively, which were chemically modified with 2′-OMe phosphoramidites and locked nucleic acids (the ribose ring is constrained by a methylene bridge between the 2′-O- and the 4′-C atoms) (28). For negative control experiments, a mismatched miR-26 LNA-antimiR, MM-LNA–anti–miR-26 (5′-TCCTGGATACTATGT-3′) (with underlines indicating nucleotides that were locked), was also synthesized. LNA–miR-mimic is a double-stranded RNA fragment with locked nucleotides 1 at the 5′ end and 2 at the 3′ end. The LNA–anti–miR-26, LNA–miR-mimic, or LNA–miR-masks were injected into mice through the tail vein at a dosage of 5 mg/kg/d in 0.2 ml saline once a day for 3 consecutive days.
qPCR analysis.
The TaqMan MicroRNA RT Kit and TaqMan MicroRNA Assay Kit (Applied Biosystems) were used in conjunction with real-time PCR with TaqMan for quantification of miRNAs in our study, as previously described in detail (1). Total RNA samples were isolated with Ambion’s mirVana miRNA Isolation Kit, from human right atrial appendages, canine left atrial preparations, cultured neonatal rat atrial myocytes, H9c2 cells, and mouse left atrium. Reactions contained TaqMan MicroRNA Assay qRT-PCR Primer Sets specific for human, canine, rat, and mouse miRNAs and a scrambled miRNA as a negative control. qRT-PCR was performed on a thermocycler (Mx3005P Realtime PCR System; Stratagene) for 40 cycles. Fold variations in miRNA expression between RNA samples were calculated after normalizing to the internal control, snU6. The Ct is defined as the fractional cycle number at which the fluorescence passes the fixed threshold.
For quantification of transcripts of the host genes of the miR-26 family members, qPCR was carried out with total RNA samples treated with DNase I. TaqMan quantitative assay of transcripts was performed with real-time 2-step RT-PCR, involving an initial reverse transcription with random primers and subsequent PCR amplification of the targets. An expression level of b2m was used as an internal control.
Western blot analysis.
Protein samples (membrane and cytosolic samples separately) were extracted from canine left atria, human right atrial appendages, cultured neonatal rat atrial myocytes, and murine atrial tissues of WT and transgenic mice for immunoblotting analysis, with procedures essentially the same as described in detail elsewhere (1). The protein content was determined by BCA Protein Assay Kit using bovine serum albumin as an internal standard. Protein samples (∼50 μg) were fractionated by SDS-PAGE (6%–12% polyacrylamide gels) and transferred to PVDF membranes (Millipore). The samples were then incubated overnight at 4°C with the primary antibodies diluted 1:200. Affinity purified mouse monoclonal anti-Kir2.1 antibody was purchased from the UC Davis/NIH NeuroMab Facility. Rabbit polyclonal anti-HERG1 was purchased from Alomone Labs. Goat polyclonal anti-KvLQT1 and anti-Kv4.3, mouse monoclonal anti-NFATc3, and rabbit polyclonal anti-NFATc4 were purchased from Santa Cruz Biotechnology Inc., and rabbit polyclonal anti–cyclin D2 and anti–cyclin E2 were purchased from Cell Signaling Technology. The day after incubation with the primary antibody, the membrane was incubated with secondary antibody (Santa Cruz Biotechnology Inc.) diluted in PBS for 2 hours at room temperature. Finally, the membrane was rinsed with PBS before scanning with an Infrared Imaging System (LI-COR Biosciences). GAPDH was used as an internal control for protein input, using anti-GAPDH monoclonal antibody (Fitzgerald Industries International Inc.). Western blot bands were quantified using QuantityOne software by measuring the band intensity (area × OD) for each group and normalizing to GAPDH. The final results are expressed as fold changes by normalizing the data to the control values.
Construction of luciferase-miRNA–target site fusion plasmids.
To construct reporter vectors bearing miRNA-target sites, we synthesized fragments containing the exact target sites for miR-26a and miR-26b in the 3′ UTR of KCNJ2 and the fragments carrying nucleotide replacement mutations at the seed sites (Supplemental Figure 2) synthesized by Invitrogen. Each construct contained 1 of the 2 predicted binding sites for miR-26a and miR-26b (Supplemental Figure 2). These inserts were ligated into HindIII and SpeI sites in the pMIR-REPORT luciferase miRNA expression reporter vector (Ambion) (1).
Construction of promoter-luciferase fusion plasmids.
Fragments corresponding to the promoter regions spanning the putative NFAT-binding sites upstream of the TSSs of Ctdspl (for miR-26a-1), Ctdsp2 (for miR-26a-2) and Ctdsp1 (for miR-26b) were synthesized by Invitrogen. The fragments were subcloned into luciferase-containing pGL3-promoter vector (Promega) for study of the role of NFAT in regulating transcription of the miR-26 family members and their host genes, according to procedures described elsewhere (44, 45). The sequences of the promoter fragments used were as follows (boldface letters indicate the NFAT-binding sites): Ctdspl (for miR-26a-1), 5′-ATTATTCAGTCTATCTTGAATGTGCTGTAAGGACTGGGATAAAGATATATTTTTTCCTCATGGATAGGTACTTGTCCCAACAATTTTTGA; Ctdsp2 (for miR-26a-2), 5′-TGTTTCCAAATTGCCTCTTACCAACCATGCAGTCAGAGAGGCCAGAGGAAAGGGGATACAGCAGGTAGGGAACCAAGTGAGAGTCAGTGG; Ctdsp1 (for miR-26b), 5′-AAGACCCATTTTACAGATGAGGTAGTGCTATCTCCAAGTCCTCAACGAGGAAACCGAGAAGCCTCTAGTCCCGGGTCTTCAGAAAACGCA.
Construction of adenovirus and infection.
The procedures were similar to those in the study reported by van Rooij et al. (46) and our previous study (5). Mouse miR-26a-1 precursor DNA (5′-GGATCCGTTCCGGCACCGGAGCAAGTTCATTAGGTCCTATCCGACACGTCCAGGGTTCCCCGGATAAGAACCAATGAACGTGCCCCTGCGCCCGGACTTTTTTAAGCTT-3′) was synthesized by GenScript. The fragment was first inserted into adenovirus shuttle plasmid pDC316-EGFP-U6 (Microbix Biosystems Inc.). pDC316-EGFP-U6 was then cotransfected with the infectious adenovirus genomic plasmid pBHGloxΔE1,3Cre into 293 cells by liposome reagent. Following cotransfection of these 2 DNAs, homologous recombination occurred to generate a recombinant adenovirus in which the transgene (pre–miR-26) was incorporated into the viral genome, replacing the ΔE1 region (Supplemental Figure 5).
Preparation of dODNs.
Single-stranded phosphorothioate dODNs were synthesized by Integrated DNA Technologies Inc. (Supplemental Figure 11). The ODN was dried and dissolved in sterilized Tris-EDTA buffer (10 mM Tris + 1 mM EDTA) (44, 47). The supernatant was purified using Micro Bio-spin30 columns (Bio-Rad) and quantified by spectrophotometry. The double-stranded dODN was then prepared by annealing complementary single-stranded ODN by heating to 95°C for 10 minutes followed by cooling to room temperature slowly over 2 hours. The dODN was prepared at a concentration of 50 μM in saline. For negative control, a scrambled dODN was also used.
siRNAs to NFATc3 and NFATc4.
siRNAs (Supplemental Figure 11 for sequences) and the scrambled negative control siRNA (Cat no. 12935-300 Stealth RNAi Negative Control Duplexes, Medium GC Duplex) were synthesized by Invitrogen.
Transfection procedures.
Neonatal rat atrial myocytes and H9c2 cells were transfected with 100 nM miRNA and/or 10 nM AMOs, siRNAs, or negative control constructs with Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions, as in previous studies (1, 48). Cells were used for luciferase assay 24 hours after the transfection or were collected for total RNA or protein purification 48 hours after the transfection.
For dODN studies, cells were washed with serum-free medium once and then incubated with 500 μl fresh FBS-free medium (39, 42). dODNs and Lipofectamine 2000 (0.25 μl; Invitrogen) were separately mixed with 25 μl of Opti-MEM I Reduced Serum Medium (Gibco; Invitrogen) for 5 minutes. Then the 2 mixtures were combined and incubated for 20 minutes at room temperature. The Lipofectamine/ODN mixture was added dropwise to the cells and incubated at 37°C for 5 hours. Subsequently, 25 μl fresh medium containing 30% FBS was added to the well and the cells were maintained in culture until use.
Luciferase activity assay.
For luciferase assay involving miRNA function, H9c2 cells were transfected with the pMIR-REPORT luciferase miRNA expression reporter vector carrying the 3′ UTR of KCNJ2 or promoter-luciferase fusion plasmid pGL3, as previously described in detail (1, 44, 45).
ChIP.
ChIP assays were conducted with the Millipore Magna ChIP Kit according to the manufacturer’s instructions (Millipore) as previously described (48). Briefly, H9c2 cells were grown to subconfluency, washed, and fixed in 1% formaldehyde for 10 minutes to crosslink nucleoprotein complexes, and scraped in PBS containing protease inhibitor cocktail. Pelleted cells were then lysed and sonicated in detergent lysis buffer. Sheared DNA-protein complexes were immunoprecipitated by incubating the lysates with 2 μg antibodies against NFATc3 (Santa Cruz Biotechnology Inc.) or IgG (as a control) overnight. Magnetic Protein G beads provided with the Millipore Magna ChIP Kit (Millipore) were used, and after extensive washing, crosslinks were removed at 62°C for 2 hours while agitating in 100 μl ChIP elution buffer and 1 μl Proteinase K. The DNA was isolated using the QIAquick PCR Purification Kit (QIAGEN), and the presence of the miR-26 promoter was analyzed by PCR amplification using 10% of purified DNA. The primers used for miR-26 promoter sequences containing NFAT cis-acting elements are listed in Supplemental Table 5. The PCR products were analyzed by gel electrophoreses on a 1% agarose gel with GelRed Nucleic Acid Gel Stain.
EMSA.
EMSA was performed with the DIG Gel Shift Kit (Roche), as described previously (44, 48). Varying amounts of nuclear protein extracts from HeLa cells were incubated with DIG-labeled double-stranded oligonucleotides containing the putative NFAT cis-acting element. For competition experiments, 100-fold excess of unlabeled double-stranded NFAT consensus oligonucleotides and, for super-shift experiments, 1 μg of NFATc3 antibody (Santa Cruz Biotechnology Inc.) were added to the reaction. The generated chemiluminescent signals were recorded on x-ray film.
Whole-cell patch-clamp recording.
Patch-clamp techniques were applied to isolated atrial myocytes from mice subjected to tail-vein injection of LNA-construct probes and/or adenovirus-transferred miRNA probes. We used procedures that have been described previously in detail (1, 5, 38). Briefly, the pipettes of patch electrodes had tip resistances of 2 to 3 MΩ when filled with pipette solution. Isolated cells were placed in a 1-ml chamber mounted on an inverted microscope (IX-70; Olympus) and perfused with Tyrode’s solution. Whole-cell recording was performed using an Axopatch 200B amplifier (Axon Instruments). Signals were filtered at 1 kHz, and data were acquired by A/D conversion (Digidata 1320; Axon Instruments). Ion currents were recorded in the whole-cell voltage-clamp mode. For recordings of IK1, the pipette solution contained 20 mM KCl, 110 mM K-aspartate, 1 mM MgCl2, 5 mM MgATP, 0.1 mM GTP, 5 mM Na2 phosphocreatine, 10 mM EGTA, and 10 mM HEPES (pH 7.2 with KOH); the external Tyrode’s solution contained 126 mM NaCl, 5.4 mM KCl, 2 mM CaCl2, 0.8 mM MgCl2, 10 mM HEPES, and 10 mM dextrose (pH 7.4 with NaOH). CdCl2 (250 μM) was included to inhibit ICaL. Experiments were conducted at 36 ± 1°C. Junction potentials were zeroed before formation of the membrane-pipette seal and were not corrected for data analyses. Series resistance and capacitance were compensated, and leak currents were subtracted. Cells with considerable leak currents were rejected for analysis. The data were collected in an IBM-compatible computer and analyzed with the use of pCLAMP 9.2.
IK1 was recorded with 100-ms square-wave pulses to voltages ranging from –120 mV to +10 mV with a holding potential of –20 mV at a frequency of 0.1 Hz (1). For all recordings, sodium current was inactivated with the use of a holding potential of –20 mV. Since our study was designed for group comparisons of the experimental results, the currents were all recorded immediately after membrane rupture and series resistance compensation in order to minimize time-dependent rundown of currents. Individual currents were normalized to the membrane capacity to control for differences in cell size and are expressed as current densities (pA/pF).
Statistics.
Group data are expressed as mean ± SEM. Two-group–only comparisons were performed by unpaired Student’s t test. Multiple-group comparisons for real-time RT-PCR and Western blot experiments were analyzed with 1-way ANOVA followed by Bonferroni’s post hoc tests. The statistical comparisons for AF incidence were performed with the χ2 test. To account for multiple testing, we selected for comparison only results of primary biological significance and applied a correction using the Holm-Bonferroni method (Supplemental Figure 6A). The statistical significances for multiple-group comparisons of AF duration with all the groups shown in Figure 3A and Figure 4, A and C, were evaluated in a single 1-way ANOVA with post hoc Tukey’s tests (Supplemental Figure 6B). A P value of less than 0.05 was considered significant.
Study approval.
All animal handling and human tissue sample procurement procedures were approved prior to study onset by the Animal Care and Use and Human Research Ethics Committees at the Harbin Medical University and the Montreal Heart Institute.
Supplementary Material
Acknowledgments
We thank Xiaofan Yang and Jianchun Zhang for technical support, and Marie-Claude Guertin, Director of the Biostatistics Unit of the Montreal Heart Institute Coordinating Center, for expert biostatistical advice. This work was supported in part by the Canadian Institute of Health Research (MOP 44365), the European-North American Atrial Fibrillation Research Alliance (ENAFRA; no. 07CVD03) of Fondation Leducq, the Quebec Heart and Stroke Foundation (to S. Nattel), Creative Research Groups of the National Natural Science Foundation of China (81121003 to B. Yang), and the National Natural Science Foundation of China (81130088 to B. Yang; 30971252 to Y. Lu).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(5):1939–1951. doi:10.1172/JCI62185.
References
- 1.Yang B, et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. . Nat Med. 2007;13(4):486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 2.Carè A, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13(5):613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 3.Feng B, et al. miR133a regulates cardiomyocyte hypertrophy in diabetes. Diabetes Metab Res Rev. 2010;26(1):40–49. doi: 10.1002/dmrr.1054. [DOI] [PubMed] [Google Scholar]
- 4.Girmatsion Z, et al. Changes in microRNA-1 expression and IK1 up-regulation in human atrial fibrillation. . Heart Rhythm. 2009;6(12):1802–1809. doi: 10.1016/j.hrthm.2009.08.035. [DOI] [PubMed] [Google Scholar]
- 5.Lu Y, et al. MicroRNA-328 contributes to adverse electrical remodeling in atrial fibrillation. Circulation. 2010;122(23):2378–2387. doi: 10.1161/CIRCULATIONAHA.110.958967. [DOI] [PubMed] [Google Scholar]
- 6.Terentyev D, et al. miR-1 overexpression enhances Ca2+ release and promotes cardiac arrhythmogenesis by targeting PP2A regulatory subunit B56alpha and causing CaMKII-dependent hyperphosphorylation of RyR2. . Circ Res. 2009;104(4):514–521. doi: 10.1161/CIRCRESAHA.108.181651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Y, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129(2):303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 8.Matkovich SJ, et al. MicroRNA-133a protects against myocardial fibrosis and modulates electrical repolarization without affecting hypertrophy in pressure-overloaded adult hearts. Circ Res. 2010;106(1):166–175. doi: 10.1161/CIRCRESAHA.109.202176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nattel S. New ideas about atrial fibrillation 50 years on. Nature. 2002;415(6868):219–226. doi: 10.1038/415219a. [DOI] [PubMed] [Google Scholar]
- 10.Nattel S, Maguy A, Le Bouter S, Yeh YH. Arrhythmogenic ion-channel remodeling in the heart: heart failure, myocardial infarction, and atrial fibrillation. Physiol Rev. 2007;87(2):425–456. doi: 10.1152/physrev.00014.2006. [DOI] [PubMed] [Google Scholar]
- 11.Zhang H, Garratt CJ, Zhu J, Holden AV. Role of up-regulation of IK1 in action potential shortening associated with atrial fibrillation in humans. . Cardiovasc Res. 2005;66(3):493–502. doi: 10.1016/j.cardiores.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 12.Bosch RF, Zeng X, Grammer JB, Popovic CM, Kuhlkamp V. Ionic mechanisms of electrical remodelling in human atrial fibrillation. Cardiovasc Res. 1999;44(1):121–131. doi: 10.1016/S0008-6363(99)00178-9. [DOI] [PubMed] [Google Scholar]
- 13.Workman AJ, Kane KA, Rankin AC. The contribution of ionic currents to changes in refractoriness of human atrial myocytes associated with chronic atrial fibrillation. Cardiovasc Res. 2001;52(2):226–235. doi: 10.1016/s0008-6363(01)00380-7. [DOI] [PubMed] [Google Scholar]
- 14.Cha TJ, Ehrlich JR, Zhang L, Nattel S. Atrial ionic remodelling induced by atrial tachycardia in the presence of congestive heart failure. Circulation. 2004;110(12):1520–1526. doi: 10.1161/01.CIR.0000142052.03565.87. [DOI] [PubMed] [Google Scholar]
- 15.Dobrev D, et al. Human inward rectifier potassium channels in chronic and postoperative atrial fibrillation. Cardiovasc Res. 2002;54(2):397–404. doi: 10.1016/S0008-6363(01)00555-7. [DOI] [PubMed] [Google Scholar]
- 16.Gaborit N, et al. Human atrial ion channel and transporter subunit gene-expression remodeling associated with valvular heart disease and atrial fibrillation. Circulation. 2005;112(4):471–481. doi: 10.1161/CIRCULATIONAHA.104.506857. [DOI] [PubMed] [Google Scholar]
- 17.Atienza F, et al. Activation of inward rectifier potassium channels accelerates atrial fibrillation in humans: evidence for a reentrant mechanism. Circulation. 2006;114(23):2434–2442. doi: 10.1161/CIRCULATIONAHA.106.633735. [DOI] [PubMed] [Google Scholar]
- 18.Pandit SV, et al. Ionic determinants of functional reentry in a 2-D model of human atrial cells during simulated chronic atrial fibrillation. Biophys J. 2005;88(6):3806–3821. doi: 10.1529/biophysj.105.060459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katsouras G, et al. Differences in atrial fibrillation properties under vagal nerve stimulation versus atrial tachycardia remodeling. Heart Rhythm. 2009;6(10):1465–1472. doi: 10.1016/j.hrthm.2009.07.034. [DOI] [PubMed] [Google Scholar]
- 20.Noujaim SF, et al. Specific residues of the cytoplasmic domains of cardiac inward rectifier potassium channels are effective antifibrillatory targets. FASEB J. 2010;24(11):4302–4312. doi: 10.1096/fj.10-163246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 23.Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100(3):416–424. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 24.Nigam V, et al. Altered microRNAs in bicuspid aortic valve: a comparison between stenotic and insufficient valves. J Heart Valve Dis. 2010;19(4):459–465. [PMC free article] [PubMed] [Google Scholar]
- 25.Lanford RE, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327(5962):198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elmén J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452(7189):896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 27.Grünweller A, Hartmann RK. Locked nucleic acid oligonucleotides: the next generation of antisense agents? Bio Drugs. 2007;21(4):235–243. doi: 10.2165/00063030-200721040-00004. [DOI] [PubMed] [Google Scholar]
- 28.Wang Z. The principles of miRNA-masking antisense oligonucleotides technology. Methods Mol Biol. 2011;676:43–49. doi: 10.1007/978-1-60761-863-8_3. [DOI] [PubMed] [Google Scholar]
- 29.Kota J, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137(6):1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin CC, et al. Activation of the calcineurin-nuclear factor of activated T-cell signal transduction pathway in atrial fibrillation. Chest. 2004;126(6):1926–1932. doi: 10.1378/chest.126.6.1926. [DOI] [PubMed] [Google Scholar]
- 31.Qi XY, et al. Cellular signaling underlying atrial tachycardia remodeling of L-type calcium current. Circ Res. 2008;103(8):845–854. doi: 10.1161/CIRCRESAHA.108.175463. [DOI] [PubMed] [Google Scholar]
- 32.Tavi P, et al. Pacing-induced calcineurin activation controls cardiac Ca2+ signaling and gene expression. . J Physiol. 2004;554(pt 2):309–320. doi: 10.1113/jphysiol.2003.053579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nattel S, Burstein B, Dobrev D. Atrial remodelling and atrial fibrillation: Mechanisms and implications. Circ Arrhythm Electrophysiol. 2008;1(1):62–73. doi: 10.1161/CIRCEP.107.754564. [DOI] [PubMed] [Google Scholar]
- 34.Priori SG, et al. A novel form of short QT syndrome (SQT3) is caused by a mutation in the KCNJ2 gene. Circ Res. 2005;96(7):800–804. doi: 10.1161/01.RES.0000162101.76263.8c. [DOI] [PubMed] [Google Scholar]
- 35.Xia M, et al. A Kir2.1 gain-of-function mutation underlies familial atrial fibrillation. Biochem Biophys Res Commun. 2005;332(4):1012–1019. doi: 10.1016/j.bbrc.2005.05.054. [DOI] [PubMed] [Google Scholar]
- 36.Wakili R, Voigt N, Kääb S, Dobrev D, Nattel S. Recent advances in the molecular pathophysiology of atrial fibrillation. J Clin Invest. 2011;121(8):2955–2968. doi: 10.1172/JCI46315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dhamoon AS, Jalife J. The inward rectifier current (IK1) controls cardiac excitability and is involved in arrhythmogenesis. . Heart Rhythm. 2005;2(3):316–324. doi: 10.1016/j.hrthm.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z, Yue L, White M, Pelletier G, Nattel S. Differential expression of inward rectifier potassium channel mRNA in human atrium versus ventricle and in normal versus failing hearts. Circulation. 1998;98(22):2422–2428. doi: 10.1161/01.CIR.98.22.2422. [DOI] [PubMed] [Google Scholar]
- 39.Dobrev D, Carlsson L, Nattel S. Novel molecular targets for atrial fibrillation therapy. Nat Rev Drug Discov. 2012;11(4):275–291. doi: 10.1038/nrd3682. [DOI] [PubMed] [Google Scholar]
- 40.Amit G, Qin H, Donahue JK. Biological therapies for atrial fibrillation. J Cardiovasc Pharmacol. 2008;52(3):222–227. doi: 10.1097/FJC.0b013e318182c8e0. [DOI] [PubMed] [Google Scholar]
- 41.Harada M, et al. Transient receptor potential canonical-3 channel-dependent fibroblast regulation in atrial fibrillation. Circulation. 2012;126(17):2051–2064. doi: 10.1161/CIRCULATIONAHA.112.121830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schrickel JW, et al. Induction of atrial fibrillation in mice by rapid transesophageal atrial pacing. Basic Res Cardiol. 2002;97(6):452–460. doi: 10.1007/s003950200052. [DOI] [PubMed] [Google Scholar]
- 43.Luo X, Zhang H, Xiao J, Wang Z. Regulation of human cardiac ion channel genes by microRNAs: Theoretical perspective and pathophysiological implications. Cell Physiol Biochem. 2010;25(6):571–586. doi: 10.1159/000315076. [DOI] [PubMed] [Google Scholar]
- 44.Lin H, Xiao J, Luo X, Pan Z, Yang B, Wang Z. Transcriptional control of pacemaker channel genes HCN2 and HCN4 by Sp1 and implications in re-expression of these genes in hypertrophic heart. . Cell Physiol Biochem. 2009;23(4–6):317–326. doi: 10.1159/000218178. [DOI] [PubMed] [Google Scholar]
- 45.Luo X, Xiao J, Lin H, Lu Y, Yang B, Wang Z. Genomic structure, transcriptional control and tissue distribution of human ERG1 and KCNQ1 genes. . Am J Physiol. 2007;294(3):H1371–H1380. doi: 10.1152/ajpcell.00398.2007. [DOI] [PubMed] [Google Scholar]
- 46.van Rooij E, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA. 2006;103(48):18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao H, et al. A single decoy oligodeoxynucleotides targeting multiple oncoproteins produces strong anti-cancer effects. Mol Pharmacol. 2006;70(5):1621–1629. doi: 10.1124/mol.106.024273. [DOI] [PubMed] [Google Scholar]
- 48.Xiao J, Lin H, Luo X, Luo X-Y, Wang Z. miR-605 joins p53 network to form a p53:miR-605:Mdm2 positive feedback loop in response to stress. . EMBO J. 2011;30(3):524–532. doi: 10.1038/emboj.2010.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.