Abstract
Human ATRX mutations are associated with cognitive deficits, developmental abnormalities, and cancer. We show that the Atrx-null embryonic mouse brain accumulates replicative damage at telomeres and pericentromeric heterochromatin, which is exacerbated by loss of p53 and linked to ATM activation. ATRX-deficient neuroprogenitors exhibited higher incidence of telomere fusions and increased sensitivity to replication stress–inducing drugs. Treatment of Atrx-null neuroprogenitors with the G-quadruplex (G4) ligand telomestatin increased DNA damage, indicating that ATRX likely aids in the replication of telomeric G4-DNA structures. Unexpectedly, mutant mice displayed reduced growth, shortened life span, lordokyphosis, cataracts, heart enlargement, and hypoglycemia, as well as reduction of mineral bone density, trabecular bone content, and subcutaneous fat. We show that a subset of these defects can be attributed to loss of ATRX in the embryonic anterior pituitary that resulted in low circulating levels of thyroxine and IGF-1. Our findings suggest that loss of ATRX increases DNA damage locally in the forebrain and anterior pituitary and causes tissue attrition and other systemic defects similar to those seen in aging.
Introduction
Faithful replication of DNA maintains genomic stability, limits the accumulation of cancer-promoting mutations, and can extend life span. Replication stress can occur spontaneously from stalling of the replication machinery as it encounters obstacles on the DNA template, such as DNA lesions, intricate secondary structures, or DNA-bound proteins (1). Impaired progression of the replication machinery can also occur when cells are exposed to exogenous damage or drugs that limit the nucleotide pool, or that cause interstrand crosslinking. The stabilization of paused replication forks to allow restart involves activation of checkpoint signaling to provide the cell extra time to respond (2). Failure to stabilize stalled forks causes fork collapse, a process involving dissociation of the polymerases, exposure of extended single-stranded DNA (ssDNA) regions, improper processing, and DNA breakage. Stalled replication can have serious consequences, such as genomic instability, chromosomal rearrangements from illegitimate recombination events, and reduced cell viability (1). Constitutive heterochromatin, telomeric DNA, and other DNA structures can pose particular challenges to the replication machinery. To overcome such hindrances, specialized factors are required to resolve difficult DNA structures or to promote access of restart pathway proteins to stalled forks (3).
ATRX is a Rad54-like ATP-driven DNA translocase belonging to the Swi/Snf family of chromatin remodelers (4). The biological impact of ATRX mutations in humans appears to vary according to the extent and timing of disruption. In humans, ATRX somatic frameshift and nonsense mutations that completely abolish protein function have been identified in pancreatic neuroendocrine tumors (PanNETs) (5–7), pediatric and adult glioblastomas, and other cancers of the CNS (8–12), underscoring the anti-tumorigenic roles of ATRX. Tumors with ATRX mutations often harbor long telomeres, which are characteristic of the telomere maintenance mechanism known as alternative lengthening of telomeres (ALT) (6, 7, 11, 13, 14). Conversely, hypomorphic inherited mutations of the ATRX gene cause a rare developmental disorder, ATR-X syndrome (OMIM 301040), with diagnostic features of severe cognitive deficits, microcephaly, seizures, short stature, developmental delay, and α-thalassemia, without increased neoplasia incidence (15–17). Thus, inactivating mutations of the ATRX gene during development are likely embryonic lethal, while diminished ATRX activity leads to severe developmental and cognitive abnormalities. ATRX somatic inactivating mutations that arise later and consequently bypass embryonic development appear to drive cancer progression, particularly in the CNS.
The cellular functions of ATRX are not completely clear; however, several lines of evidence point to a role in the maintenance of genomic integrity. Decreasing levels of ATRX by RNA interference induces spindle defects, as well as chromosomal alignment and segregation problems during mitosis and meiosis, and complete loss of ATRX in neuroprogenitor cells (NPCs) can also induce mitotic abnormalities (18–21). Furthermore, ATRX-depleted mouse embryonic stem cells have unstable telomeres (22). The ATRX protein can bind to specific genomic sites, including imprinting control regions and the Rhox5 promoter (23, 24), but is also highly enriched at repetitive regions, including ribosomal DNA repeats, pericentromeric heterochromatin, and telomeric chromatin, where it was proposed to cooperate with DAXX to incorporate the histone variant H3.3 into chromatin (22, 25–29).
In the present report, we demonstrate that ATRX deletion in NPCs causes excessive replication–related DNA damage, which is exacerbated by loss of p53, replication stress–inducing drugs, and telomestatin (TMS), a G-quadruplex (G4) ligand. Unexpectedly, mutant mice lacking ATRX in the forebrain and pituitary had low circulating IGF-1, thyroxine (T4), and glucose levels and displayed degenerative phenotypes previously described in mouse models of progeria. Collectively, our data reveal that ATRX protects cells from replicative stress–induced DNA damage and telomeric fusions and that loss of ATRX in the embryonic CNS and anterior pituitary elicits systemic endocrine and metabolic abnormalities.
Results
Endogenous DNA damage accumulates in the Atrx-null NPCs.
AtrxLoxP female mice (30) were mated to Foxg1Cre male mice (31) to generate male progeny lacking ATRX in the embryonic forebrain starting at E8.5 (referred to as cKO mice). Since ATRX depletion can induce telomeric instability in embryonic stem cells (22), we stained brain cryosections from E13.5 embryos with antibodies for the phosphorylated histone variant H2AX (γH2AX), a canonical marker for double-stranded breaks (DSBs) (32). This analysis showed a high level of DNA damage throughout the cortex, hippocampal hem (future hippocampus), and basal ganglia in the cKO embryonic forebrain that was significantly increased compared with that in controls (Figure 1A). The level of DNA damage was appreciably lower in the neonatal brain; nevertheless, comparatively higher levels of γH2AX were observed in the neonatal hippocampus, a region that is still proliferative at this stage of development, as well as in the cortex of mutants (Figure 1B). Thus, the pattern of DNA damage was generally confined to proliferative areas of the developing brain. To determine whether DNA damage incurred during the embryonic period persists in postmitotic cells, we examined γH2AX and ATRX staining patterns in P7 control and cKO forebrain cryosections. This analysis demonstrated that ATRX is indeed not expressed at the protein level and revealed that no damage was present or remained unrepaired in the absence of ATRX in the postnatal juvenile brain (Supplemental Figure 1, A and B; supplemental material available online with this article; doi: 10.1172/JCI65634DS1). Given the post-replicative state of the brain at this time, these data suggest that endogenous DNA damage due to loss of ATRX occurs primarily in proliferating cells in the embryonic and perinatal period.
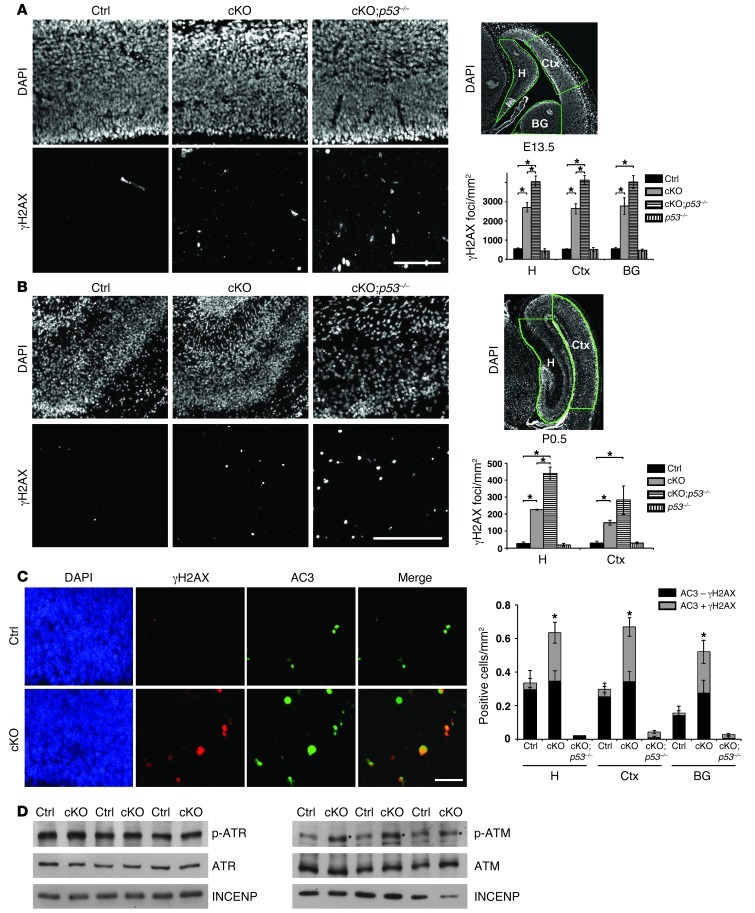
Figure 1. Increased DNA damage leads to ATM activation and p53-dependent apoptosis in the Atrx-null embryonic brain.
(A) Immunostaining for γH2AX in E13.5 control (Ctrl), cKO, and cKO;p53–/– compound mutant cortical cryosections. Scale bar: 100 μm. DAPI staining of E13.5 forebrain highlights in green the hippocampal hem (H), cortex (Ctx), and basal ganglia (BG) regions where γH2AX foci per unit area were scored. Control, cKO, and cKO;p53–/– (n = 3); p53–/– (n = 2). (B) γH2AX staining in P0.5 control, cKO, and cKO;p53–/– cortical cryosections. Scale bar: 200 μm. DAPI staining of P0.5 forebrain highlights in green the hippocampus (H) and cortex (Ctx) regions where γH2AX foci per unit area were scored. Control and cKO (n = 3); cKO;p53–/– and p53–/– (n = 2). (C) Co-immunofluorescence detection of γH2AX (red) and activated caspase-3 (AC3; green) in E13.5 cortical cryosections. Scale bar: 30 μm. AC3+ cells were scored for the presence (AC3 + γH2AX) or absence (AC3 – γH2AX) of DNA damage (n = 3). (D) Western blot analysis of nuclear protein extracts obtained from E13.5 telencephalon (n = 3). While levels of ATR and phospho-ATR were not increased (left panels), phospho-ATM was noticeably increased in the cKO extracts compared with controls (indicated by an asterisk). Original magnification, ×100 (A and B); ×200 (C). *P < 0.05.
Accumulation of DNA damage in Atrx-null NPCs induces ATM activation and is exacerbated by p53 deletion.
Persistent DNA damage can result in the phosphorylation and upregulation of p53 by the ATR/Chk1 or ATM/Chk2 pathways, leading to cell cycle arrest or apoptosis (33, 34). We previously demonstrated that loss of ATRX results in a p53-dependent apoptotic response in cultured primary mouse NPCs and in vivo in the forebrain at E13.5 (18). To determine whether accumulated DNA damage could be the underlying cause of apoptosis, we performed double immunofluorescence of γH2AX and cleaved caspase-3, a marker of apoptotic cells. The majority of cells undergoing apoptosis (i.e., cleaved caspase-3+ cells) in the cKO telencephalon also stained positive for γH2AX, indicating that cell death is likely a downstream consequence of DNA damage (Figure 1C). To substantiate this conclusion and examine the role of p53, we generated ATRX/p53 compound mutant mice and assessed levels of DNA DSBs in the brain. γH2AX signal in cKO;p53–/– embryos was increased compared with that in cKO mice, suggesting that the accumulation of DNA damage in the ATRX-deficient forebrain directly triggers a p53-dependent apoptotic response and that the absence of p53 prevents the removal of these cells by apoptosis (Figure 1, A and B, right panels and graphs). We next explored the possible involvement of ATR and ATM signaling in the response to increased DNA damage. Western blot analysis of control and cKO E13.5 telencephalon protein extracts demonstrated similar activation of ATR in control and cKO samples. On the other hand, a clear increase in ATM activation was observed in the cKO embryonic telencephalon (Figure 1D). Taken together, these findings show that loss of ATRX in the embryonic brain results in the accumulation of DNA damage, which triggers activation of ATM and p53, culminating in neuroprogenitor cell death.
DNA damage response at telomeres and telomeric DNA end fusions.
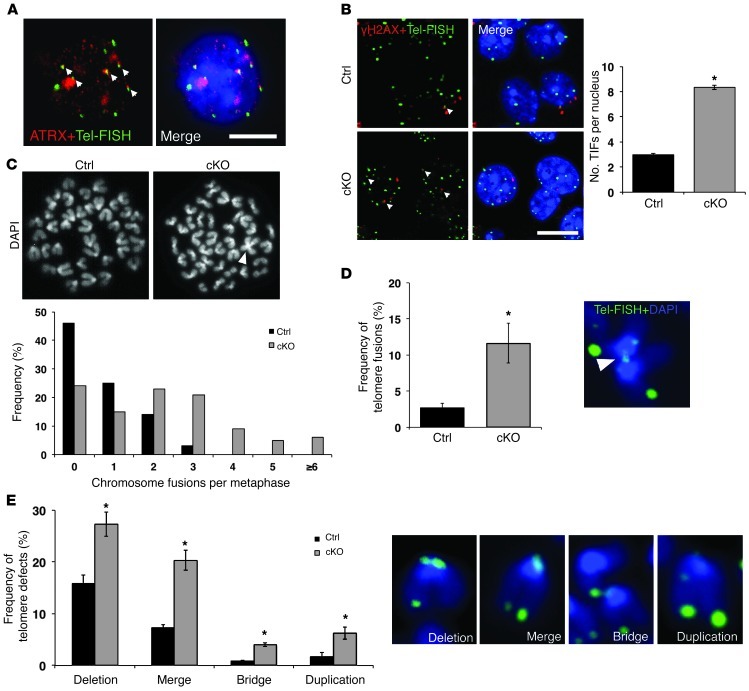
To determine the specific genomic sites of DNA damage response incurred by loss of ATRX, we examined telomeres, since ATRX enrichment has been previously reported at these genomic regions. However, since it has been proposed that ATRX associates with telomeres only in pluripotent embryonic stem cells (22), we first assessed whether ATRX protein is present at telomeres in NPCs. ATRX immunofluorescence performed in conjunction with telomere FISH (Tel-FISH) in control NPCs demonstrated colocalization of ATRX and a subset of telomeres (Figure 2A). Next, we used Tel-FISH in combination with γH2AX immunostaining to determine whether the DNA repair machinery is activated at telomeric chromatin in cultured cKO NPCs. Confocal imaging showed that the number of telomere dysfunction–induced foci (TIFs) per nucleus was increased in the cKO compared with control NPCs (Figure 2B). In fact, 66.1% of all γH2AX foci were observed at telomeres in the cKO cells (Supplemental Figure 2B). ATRX protein also binds pericentromeric heterochromatin, and we repeated this experiment using major satellite-specific labeled DNA probes (MSat-FISH). Again, we found that γH2AX+/mSAT+ foci were more frequent in cKO NPCs and that a large proportion of DNA damage foci (69.51% ± 1.27%) corresponded to pericentromeric heterochromatin in cKO NPCs (Supplemental Figure 2, A and B). These numbers reflect the acrocentric nature of mouse chromosomes, with some γH2AX foci overlapping with the p arm telomere and the adjacent pericentromeric region. Our results demonstrate that heterochromatic regions of the genome are prone to DNA damage in the absence of ATRX. To further explore the function of ATRX in genome stability, we examined metaphase spreads from control and cKO NPC cultures. ATRX immunostaining of control metaphase spreads revealed strong enrichment of the protein at pericentromeric heterochromatin and at some telomeres (Supplemental Figure 2C). Analysis of DAPI-stained spreads revealed an increased frequency of chromosome fusions in cKO metaphase spreads compared with controls, as evidenced by overlapping chromosomes (Figure 2C). Centromere-to-centromere, centromere–to–q arm, and q arm–to–q arm fusions were observed. The number of chromosomes per metaphase spread was not affected in cKO NPCs (Supplemental Figure 2D). To further explore the nature of the fusions, we performed TelFISH on control and cKO metaphase spreads and detected an increased incidence of telomeric DNA end fusions (Figure 2D). Other common defects included telomere deletions or duplications, as well as merged or bridged telomeres (Figure 2E). Overall, the frequency of all telomere defects was more than doubled in the cKO NPCs compared with controls, and defects involved the p and q arm telomeres at an equal frequency (Supplemental Figure 2E).
Figure 2. Increased DNA damage and telomere defects in cKO NPCs.
(A) Confocal immuno-FISH images of ATRX (red) and telomeres (Tel-FISH; green) in NPCs demonstrates colocalization of the ATRX protein with a subset of telomeres. Scale bar: 5 μm. (B) Confocal immuno-FISH images of γH2AX (red) and telomeres (Tel-FISH; green) shows increased incidence of TIF (γH2AX/Tel-FISH colocalization) in cKO compared with control NPCs (300 nuclei counted, n = 3 control/cKO littermate-matched pairs). Scale bar: 10 μm. (C) DAPI staining of control and cKO metaphase spreads shows representative chromosome fusion in cKO NPC metaphase (arrowhead). Frequency of fusions per metaphase was increased in cKO metaphases compared with control (control: 88 metaphases, cKO: 108 metaphases counted, n = 3). (D) Tel-FISH (green) demonstrates increased telomeric fusions in cKO metaphase chromosomes compared with control (1,475 chromosomes counted; n = 3). (E) Telomere defects (deletion, merge, bridge, and duplication) were scored in control and cKO metaphase chromosomes. Representative images of defects appear to the right of quantification. In all cases, cKO chromosomes showed an increase in telomeric defects compared with control (1,475 chromosomes counted; n = 3). Original magnification, ×1,000 (A and B); ×630 (C–E). *P < 0.05.
ATRX is not required for the repair of DSBs.
Accumulation of DNA DSBs could occur if ATRX plays a direct role in DSB repair mechanisms. Two non–mutually exclusive pathways carry out repair of DNA DSBs: homologous recombination (HR) in proliferative cells; and non-homologous end joining (NHEJ) in postmitotic cells (35). Since γH2AX foci were primarily concentrated in proliferative zones of the developing brain, we reasoned that ATRX may be specifically involved in the repair of DSBs via HR. To address this possibility, we cultured primary NPCs for 2 days in N2 neurobasal medium and exposed proliferative NPCs to γ-irradiation to induce DSBs and activate the HR pathway. The DNA damage response (i.e., γH2AX signal accumulation) was measured between 0 and 6 hours after irradiation, when damage induced by low doses of γ-irradiation should be actively resolved in control cells (Supplemental Figure 3). Both control and cKO NPC colonies showed a dose-dependent increase in γH2AX signal 3 hours after irradiation, which was subsequently resolved (i.e., decreased γH2AX staining) by 6 hours after treatment (Supplemental Figure 3). Therefore, we concluded that ATRX is not required for repair of exogenous irradiation–induced DSBs.
ATRX protects cells from replication stress.
Since ATRX does not appear to function in the repair of DSBs, we reasoned that it might help prevent DNA damage to preserve genomic integrity. Given that damage is restricted to proliferating regions of the cKO embryonic brain, we tested the possibility that cKO NPCs are particularly sensitive to DNA damage during S phase. We inactivated ATRX in AtrxloxP mouse embryonic fibroblasts (MEFs) with adenoviral delivery of Cre recombinase and treated them with hydroxyurea (HU), a drug that depletes deoxyribonucleotide pools, inhibits DNA synthesis, and causes replication fork stalling (36). MEFs were treated with increasing doses of HU for 24 hours, and cell viability was assessed 2 days later. MEFs lacking ATRX (Ad-CreGFP) were more sensitive to HU than untreated cells or cells treated with Ad-GFP. Ad-CreGFP treated MEFs were not more sensitive to γ-irradiation than Ad-GFP–transduced or untransduced cells (Figure 3A). We also treated control and cKO NPCs with HU and mitomycin C (MMC), a DNA crosslinker that also causes replication fork stalling. Both HU and MMC treatment caused decreased viability of cKO NPCs compared with control cells. As in MEFs, exposure to various doses of γ-irradiation did not alter cKO NPC viability compared with control NPCs, demonstrating a specific sensitivity to drugs that induce replication fork stalling (Figure 3B).
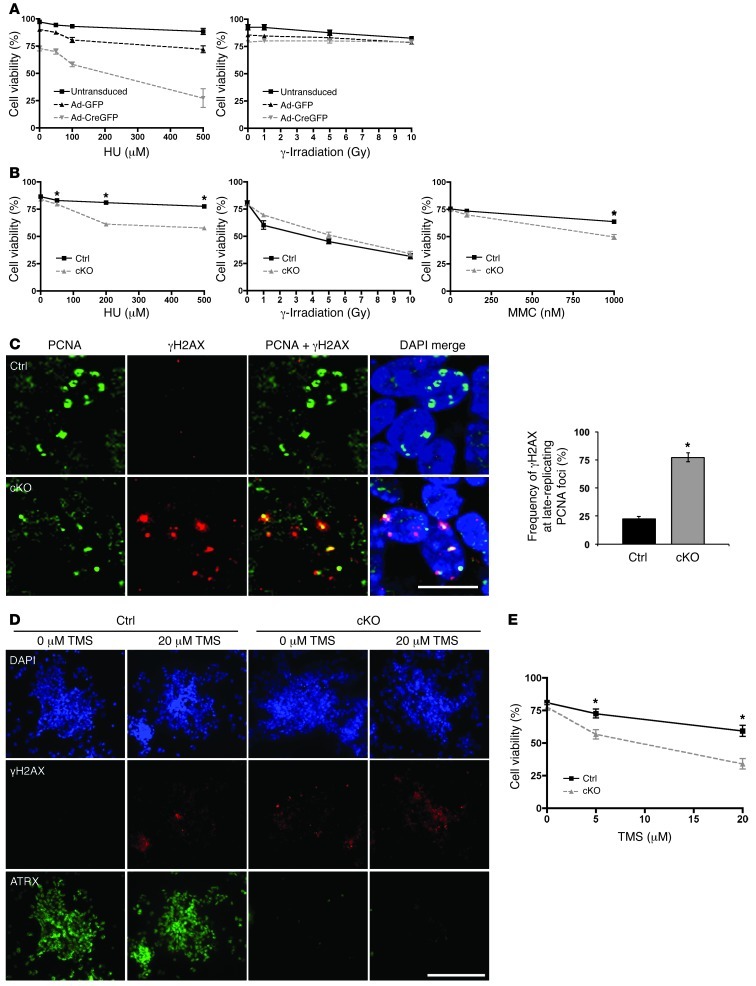
Figure 3. ATRX-deficient cells are hypersensitive to replication stress–inducing agents and the G4-DNA ligand TMS.
(A) AtrxloxP/Y MEFs were untransduced or transduced with adenovirus expressing Cre recombinase fused to GFP (Ad-CreGFP) or Ad-GFP and subsequently treated with HU for 24 hours or γ-irradiated at the indicated doses. Cell viability was measured at 24 hours after HU treatment (n = 4) and at 6 hours after irradiation (n = 3) via trypan blue dye exclusion. (B) Control and cKO NPCs were treated with HU or MMC for 24 hours or γ-irradiated at the indicated doses. Cell viability was measured at 24 hours after HU and MMC treatment and at 6 hours after irradiation (n = 3). (C) Co-immunofluorescence detection of PCNA, a marker of replication foci, and γH2AX in control and cKO E13.5 cortical cryosections. Results were quantified by measuring the ratio of γH2AX staining that localized to late-replicating PCNA foci to total γH2AX staining per cell, to account for overall lower levels of γH2AX signal in control cells (300 nuclei counted, n = 3). Scale bar: 12 μm. (D) Control and cKO NPCs were treated with 20 μm TMS for 2 hours, and γH2AX signal was imaged 6 hours after treatment. Scale bar: 70 μm. (E) Control and cKO NPCs were treated with TMS for 24 hours, and cell viability was measured 24 hours after treatment (n = 3). Original magnification, ×600 (C); ×100 (D). *P < 0.05.
Blocked replication forks could be a source of genomic instability in the cKO cells, since they can lead to collapse of the replisome and the formation of DSBs (37). Replicating cortical NPCs lacking ATRX protein were assessed for DNA damage (γH2AX) at replication foci using an anti-PCNA antibody and imaged using confocal microscopy (Figure 3C). PCNA is a component of the replication machinery and exhibits a distinct staining pattern during S phase. In late S phase, the replication foci are observed in regions of the nucleus containing heterochromatin and as such colocalize with DAPI-bright heterochromatin bundles (38). Atrx-null NPCs accumulated more DNA damage at these late replication foci compared with control cells, confirming an increase in DSBs during the replication process (Figure 3C). We also performed FACs analysis to determine whether S phase is extended in these cells. We detected a small but significant decrease in the G1 population and a modest increase in S phase population in the cKO NPCs at 4 days in vitro. This suggests that difficulties in replicating heterochromatin result in a slight delay in S phase progression of cKO cells (Supplemental Figure 3B). We conclude from these findings that loss of ATRX results in an accumulation of endogenous replicative damage in cultured NPCs, specifically at late-replicating chromatin.
G4-DNA stabilization exacerbates DNA damage and decreases viability of Atrx-null NPCs.
A potential function of ATRX is to stabilize stalled forks or facilitate the replication of complex secondary DNA structures, such as G4-DNA, which are found at telomeres, among other genomic sites (39). To test whether ATRX facilitates the replication of G4-DNA, we treated control and Atrx-null NPCs with TMS, a natural product isolated from Streptomyces anulatus that binds with high affinity to G4 structures (40). Neuroprogenitors dissected from control and cKO embryonic forebrain were cultured and treated with 20 μM TMS for 24 hours. γH2AX staining was increased (Figure 3D), and cell viability was decreased (Figure 3E) in the TMS-treated cKO NPCs compared with TMS-treated control cells. These findings suggest that ATRX deficiency in NPCs synergizes with G4-DNA stabilization to cause DNA damage and cell death.
Loss of ATRX shortens life span and induces progeroid-like phenotypes.
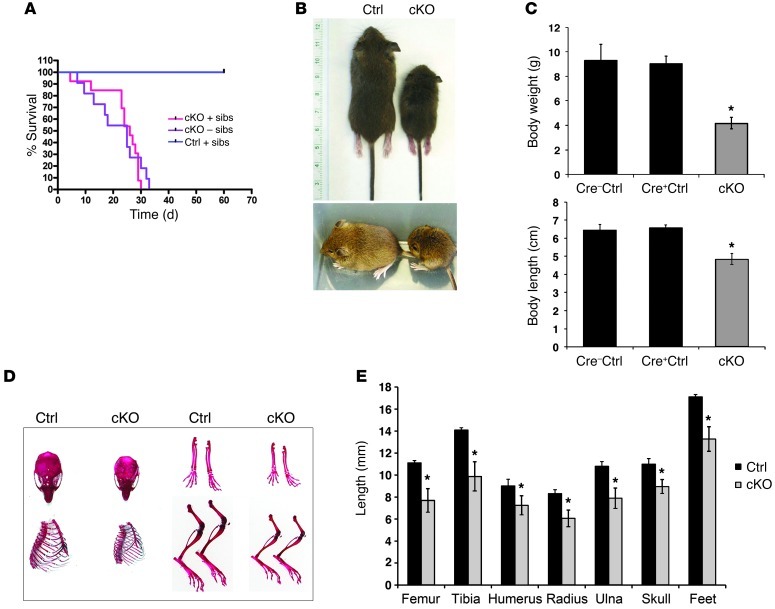
While a portion of mutant pups die in the neonatal period (30), many cKO male mice survived, but rarely longer than 30 days after birth (P30), with an average life span of 22.7 ± 1.7 days (Figure 4A). Moistening of food pellets to help with intake of solids did not improve life span, nor did the removal of siblings from the litter to reduce competition for breast milk (Figure 4A). Mutant mice at P20 had milk in the stomach, suggesting that starvation was not the cause of death or growth retardation. P17 cKO mice had severely stunted growth compared with control littermates, as determined by measurements of body weight and length, as well as length of various skeletal elements (Figure 4, B–E). Reduced body size was apparent at birth (30), but a more dramatic effect was seen from P5 onward. The tibia, femur, humerus, radius, ulna, and foot were significantly shorter in the cKO mice at P17 (Figure 4E). We confirmed specific deletion of ATRX in the forebrain of cKO mice at several postnatal time points, while ATRX remained expressed in other tissues, such as the liver, heart, thymus, spleen, testes, and skeletal growth plate of cKO mice (Supplemental Figure 4, A–D).
Figure 4. Reduced growth and life span in mice lacking ATRX in the forebrain.
(A) Kaplan-Meier survival curve of Cre+ control (n = 12) and cKO mice raised with (+sibs, n = 13) or without (–sibs, n = 11) siblings. Survival of cKO mice was significantly decreased compared with that of control mice (P = 0.0001). The survival of cKO mice was not significantly different whether they were raised with or without siblings (P = 0.4974). (B) Representative pictures of P17 control and cKO littermates, illustrating size difference of the mice. (C) Body weight (g) and length (cm) measurements of control (Cre–, n = 25; Cre+, n = 8) and cKO (n = 24) mice. No significant difference was observed between Cre– and Cre+ control mice. (D) Skeletal elements of control and cKO mice were stained with alizarin red and alcian blue. (E) Length measurements of P17 control and cKO skeletal elements. (n = 5). *P < 0.05.
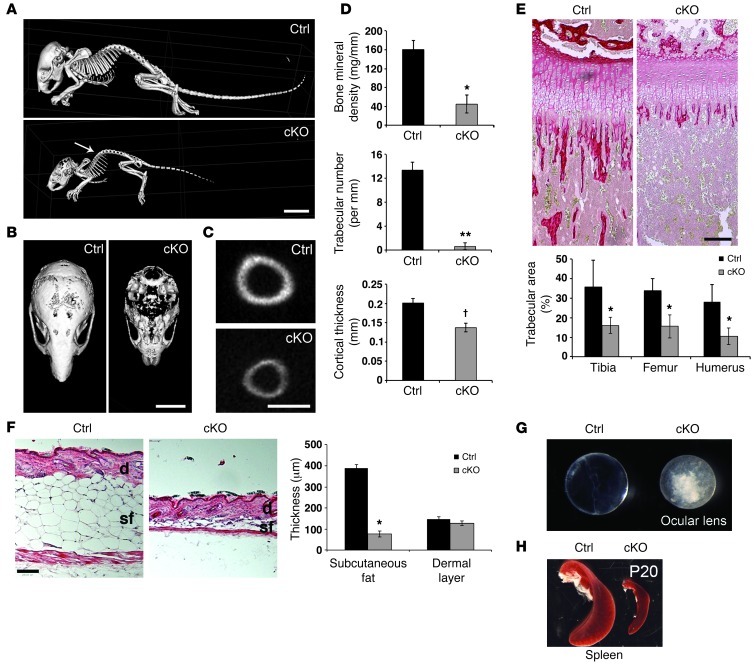
Microcomputed tomography (microCT) analyses showed that cKO mutant mice displayed kyphosis, an abnormal curvature of the spinal column (Figure 5A), as well as decreased bone mineral density (BMD), lower trabecular number, and reduced bone cortical thickness (Figure 5, A–D). Loss of trabecular bone was confirmed by picrosirius red staining of tibia, femur, and humerus of control and cKO mice at P17 (Figure 5E). To determine whether this effect stemmed from a developmental problem or was due to tissue degeneration, we also assessed trabecular bone at earlier ages. We found no difference in neonatal and P7 mice and detected an intermediate phenotype at P10, suggesting that bone development occurs initially normally, with evidence of deterioration from P10 onward (Supplemental Figure 5A).
Figure 5. Postnatal phenotypes in Atrx-cKO mice.
(A) Whole skeletal isosurface images of P17 control and cKO mice were generated using microCT. Arrow points to kyphosis. Scale bar: 10 mm. (B) Horizontal view of the cKO skull illustrates decreased bone mineralization. Scale bar: 5 mm. (C) Tibial cross section shows decreased cortical thickness. Scale bar: 1 mm. (D) Decreased BMD (*P = 0.002), trabecular number (**P = 0.002), and cortical thickness (†P = 0.0001) in the cKO mice compared with controls. Data were obtained from hind legs using MicroView 3D software (n = 3). (E) Picrosirius red staining of P17 control and cKO tibia (representative image), femur, and humerus reveals a drastic loss of trabecular bone area in cKO compared with control mice (n = 4; *P < 0.05). Scale bar: 200 mm. (F) H&E staining of P20 skin cryosections shows loss of subcutaneous fat in cKO compared with control mice (n = 3; *P = 0.0002). Dermal thickness was not significantly different (n = 3; *P = 0.3545). Scale bar: 300 mm. sf, subcutaneous fat; d, dermis. (G) Dark field image of P20 control and cKO ocular lenses demonstrates appearance of cataracts (loss of lens transparency) in cKO compared with control. (H) Dark field image of P20 control and cKO spleen demonstrates the disproportionally smaller size of cKO spleen. Original magnification, ×50 (E and F). See also Supplemental Figure 5.
Extensive loss of subcutaneous fat (<25% of controls) was a constant characteristic of the mutant mice, while thickness of the dermis was not significantly affected (Figure 5F). Again, by measuring subcutaneous fat at various time points, we observed that this phenotype was exacerbated at P20 compared with P10 (Supplemental Figure 5, B and C). Mutant mice rarely opened their eyes, and analysis of the lens showed the formation of cataracts (Figure 5G). Accounting for decreased total body weight of the cKO mice, the spleen and liver were consistently smaller and the heart was significantly enlarged at P20 (Figure 5H and Supplemental Figure 5D). These effects were also seen at P10 but were not as dramatic, as evidenced by a lack of significant difference between control and cKO organ size (Supplemental Figure 5D). The phenotypes described above strongly resemble those reported in several mutant mice displaying accelerated aging, but as the deterioration and death of the mice occurred rather quickly, it is difficult to completely distinguish between developmental and progeria-like phenotypes.
Endocrine defects in Atrx-cKO mice.
To investigate the mechanism(s) by which ATRX disruption in the CNS could cause such drastic postnatal phenotypes, we first wanted to confirm that the skeletal growth phenotype was caused by non-cell-autonomous mechanisms. Indeed, tibiae from cKO and control mice grew at similar rates in ex vivo organ cultures and had similar growth plate length or organization (Figure 6A), substantiating that bone growth defects are caused by systemic alterations.
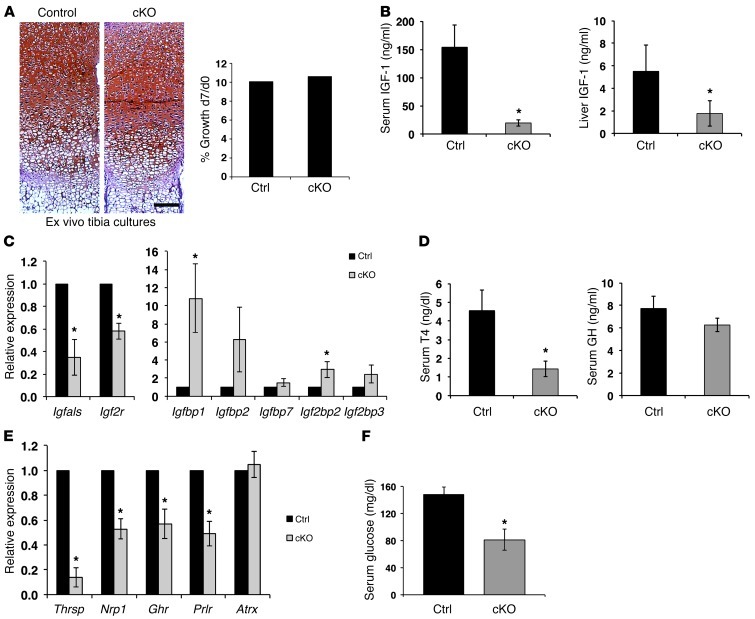
Figure 6. Endocrine defects and hypoglycemia in Atrx-cKO mice.
(A) Longitudinal growth of control and cKO tibia was measured after 7 days (d7) of ex vivo culture. Results are expressed as the ratio of length at d7 to that at d0. No difference in growth was detected between control and cKO mice (n = 3). Scale bar: 100 μm. (B) Serum and liver IGF-1 levels are decreased in cKO mice (n = 3). (C) Expression of several IGF-1 pathway genes is altered in cKO liver compared with controls (n = 3). Real-time data were normalized to Gapdh expression. (D) Circulating T4 levels are significantly decreased in P20 cKO mice compared with controls, while GH levels are only mildly affected (n = 3). (E) Thyroid hormone target genes exhibit decreased expression in the liver of P20 cKO mice compared with controls (n = 3). Real-time data were normalized to Gapdh expression. (F) Glucose levels are reduced in P20 cKO serum compared with controls (n = 5). Original magnification, ×50 (A). *P < 0.05.
Given the small size of the cKO mice, we suspected that the endocrine system might be defective. The insulin-like growth factor 1/growth hormone (IGF-1/GH) somatotroph axis has previously been linked to growth, aging, and life span (41). We observed impairment in IGF-1 levels and signaling in the cKO mice. ELISA assays at P17 showed a dramatic reduction of serum and liver IGF-1 levels compared with those in control samples (Figure 6B). In addition, quantitative RT-PCR (qRT-PCR) analyses revealed marked transcriptional alterations of key genes for somatotroph signaling in the liver (Figure 6C).
Thyroid hormone is an important regulator of Igf1 expression and skeletal growth during the prepubertal growth period, while GH effects are limited at that time (42). We observed that circulating thyroxine (T4) levels were dramatically decreased in the cKO mice, while the GH level was only slightly diminished, and the difference did not reach statistical significance (Figure 6D). In addition, expression of several thyroid hormone–responsive genes such as Thrsp, Nrp1, Ghr, Prlr was decreased in the liver (Figure 6E). It is important to note that Atrx transcript and protein levels were normal in livers of cKO mice (Supplemental Figure 4C and Figure 6E), indicating that these transcriptional effects are not due to spurious deletion of Atrx in the liver itself. Given that hypoglycemia is a feature of several mutant mice displaying segmental progeria (43–45) and that thyroid hormone is essential for the maintenance of glucose homeostasis, we measured blood glucose and found that it was significantly lower in P20 cKO compared with control mice (Figure 6F).
ATRX is deleted and causes DNA damage in the embryonic anterior pituitary.
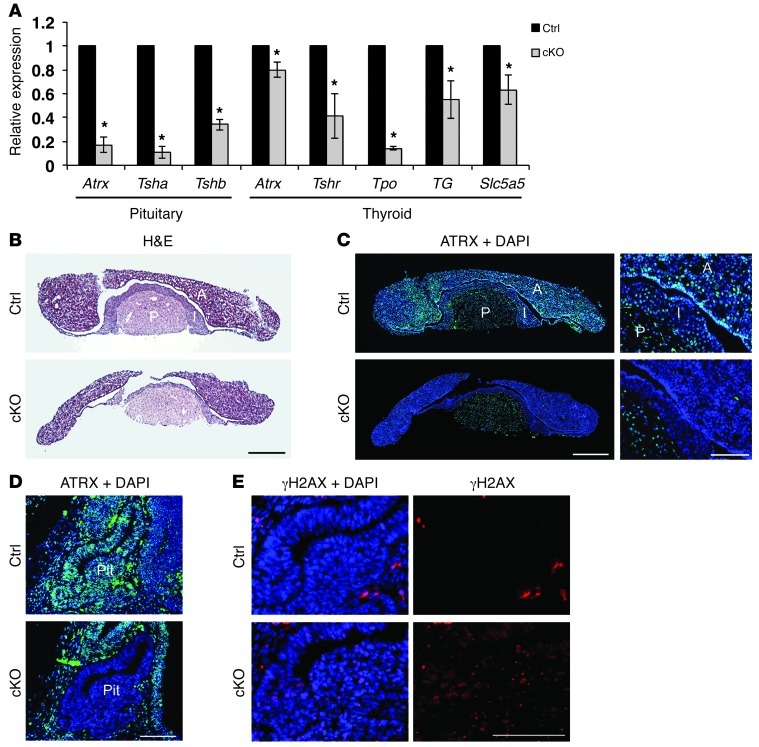
The control of thyroid hormone levels and actions is complex and relies on the production of thyroid-releasing hormone (TRH) by the hypothalamus, leading to thyroid-secreting hormone (TSH) production by the pituitary, which in turn induces T3 and T4 synthesis by the thyroid gland (46). To determine the cause of low circulating T4 levels, we first investigated Atrx and Tsh expression levels in the pituitary of control and cKO mice. Atrx expression was decreased, as were the levels of Tsh transcripts (Figure 7A). In addition, several genes responsible for T4 production (Tshr, Tpo, Tg, and Slc5a5) were expressed at lower levels in the thyroid of cKO mice compared with control mice (Figure 7A). These findings suggest that Atrx deletion in the pituitary impairs TSH production, leading to decreased signaling to the thyroid and low T4 production. ATRX immunofluorescence of P20 pituitary cryosections confirmed ATRX expression in the pituitary of control mice and that it was absent in the anterior and intermediate pituitary of cKO mice (Figure 7, B and C). We also observed deletion of ATRX in the embryonic anterior pituitary at E13.5 (Figure 7D), which corresponded to a dramatic increase in γH2AX staining (Figure 7E). These results suggest that ATRX is deleted early during embryonic development of the anterior pituitary of cKO mice, where it causes increased DNA damage and results in abnormal function of the thyrotrophs in postnatal cKO mice.
Figure 7. Loss of ATRX in the developing anterior pituitary causes DNA damage, reduced Tsh expression, and altered thyroid function.
(A) Quantitative RT-PCR analysis of P23 control and cKO shows loss of Atrx expression in the pituitary (5.9-fold decrease) and thyroid (1.25-fold decrease). Tsha and Tshb subunits showed decreased expression in the pituitary, and a number of downstream targets of TSH showed decreased expression in the thyroid of cKO mice compared with controls (n = 3). (B) H&E staining of P23 control and cKO pituitary. Scale bar: 500 μm. (C) Immunofluorescence detection of ATRX expression in P23 control and cKO pituitary demonstrates specific loss of ATRX in the anterior and intermediate pituitary. Scale bars: 500 μm (left panels) and 50 μm (right panels). A, anterior; I, intermediate; P, posterior pituitary. (D) Immunofluorescence detection of ATRX in E13.5 control and cKO sagittal embryonic pituitary cryosections shows loss of ATRX expression. Scale bar: 100 μm. Pit, pituitary. (E) Immunofluorescence detection of γH2AX in E13.5 sagittal embryonic pituitary cryosections shows increased DNA damage in cKO embryonic pituitary compared with control. Scale bar: 100 μm. Original magnification, ×25 (C, left panels); ×100 (C, right panels, and E); ×50 (D). *P < 0.05.
Discussion
We have shown that ATRX deficiency causes replicative DNA damage at telomeres and pericentromeric heterochromatin and increases the incidence of telomeric fusions. Moreover, deletion of the Atrx gene in the embryonic nervous system and anterior pituitary attenuates postnatal endocrine and metabolic signaling with concurrent growth reduction and progressive progeria-like tissue deterioration. Control experiments confirmed that loss of ATRX is restricted to the forebrain and anterior pituitary. Furthermore, our previous studies have shown that specific and efficient inactivation of the Atrx gene in cartilage does not cause any of the skeletal and growth phenotypes described here (47), providing further evidence that these defects are non-cell-autonomous.
We do not yet fully comprehend the mechanisms responsible for the accumulation of DNA damage observed in the Atrx-null embryonic forebrain and pituitary. We established that loss of ATRX does not prevent DNA repair, since γH2AX foci did no persist in the postnatal brain at P7 and Atrx-null cultured NPCs could resolve DSBs induced by γ-irradiation. However, we provide evidence that the accumulation of damage is partly due to replication stress linked to G4-DNA stability. Telomeres are nucleoprotein complexes that protect chromosome ends from degradation and end fusions. They consist of repetitive DNA high in guanine and cytosine nucleotide residues and have the ability to form G4s when in the single-stranded form (39). The presence of these bulky DNA adducts can potentially impede DNA replication. We demonstrate that Atrx-null NPCs are hypersensitive to the potent G4 ligand TMS, suggesting that in the absence of ATRX, cells have difficulty in resolving G4-DNA. This conclusion is supported by the demonstration that the ATRX protein can bind G4-DNA structures in vitro (48). Since G4-DNA structures form at telomeres, the γH2AX signals we observed at telomeres in Atrx-null NPCs may result from a failure to properly resolve G4 structures, leading to telomere uncapping and initiation of the DNA damage response. This in turn may explain the increase in telomere fusions observed in Atrx-null NPCs due to repeating break-fusion-break cycles. In contrast, heterochromatic regions flanking centromeres are AT rich, and a role for ATRX in facilitating replication of G4-DNA structures cannot explain the increased genetic damage at these sites. ATRX potentially plays a more general role in heterochromatin remodeling/replication during late S phase or may be required to facilitate replication of other secondary structures. ATRX is associated with the DAXX histone chaperone and was proposed to participate in the incorporation of histone variant H3.3 at highly repetitive regions, including pericentric heterochromatin and telomeric chromatin (28, 49). There is evidence that promyelocytic nuclear bodies (PML-NBs) are involved in facilitating heterochromatin remodeling/replication in late S/G2 phase (50). ATR-X syndrome patient mutations have been shown to alter ATRX targeting to PML-NBs, suggesting an important requirement for ATRX at these subnuclear domains (51). ATRX and DAXX localize to PML-NBs during S phase and may be required for remodeling of heterochromatin during replication, a process likely to involve nucleosome remodeling and histone variant deposition or ejection (52).
An important and unexpected finding from our studies is that specific loss of ATRX early during embryonic brain and pituitary development can influence postnatal health and life span. Systemic loss or mutation of several factors required for telomeric stability, response to replication stress, or nucleotide excision repair (NER) cause phenotypes that resemble premature aging in mice (44, 53–60) and segmental progeria syndromes in humans, including XPF/ERCC1 in xeroderma pigmentosum, ERCC6/8 in Cockayne syndrome, and ATR in Seckel syndrome (61).
The link between excessive DNA damage and a suppressed IGF-1 response, although reported in many progeria models, has not yet been satisfactorily explained. Here, we provide evidence that DNA damage in the embryonic pituitary leads to decreased expression of Tsh, with the predictable outcome of low thyroxine production by the thyroid. Recent reports have demonstrated that thyroid hormone is more critical than growth hormone in the regulation of IGF-1 levels at prepubertal stages of development (62). Genetically modified mice that are deficient in thyroid hormone show greater than 50% reduction in Igf1 expression in liver and bone, and T4 treatment can reverse this effect (62). In addition, thyroid hormone and its receptor can bind intron 1 of the Igf1 gene and stimulate its expression (62). In line with this model, circulating thyroid hormone levels were decreased in the Atrx-cKO mice, and several thyroid hormone–responsive genes were decreased in the liver. We also detected altered expression of several genes involved in IGF-1 signaling in the liver, including reduced Igfals and increased Igfbp1 expression. Notably, Ercc1–/– mice exhibit a quite rapid postnatal degeneration phenotype similar to that of the Atrx-cKO mice, and Thrsp is the most downregulated gene in the Ercc1-null liver, exhibiting a 15-fold decrease (63). This suggests that diminished thyroid hormone action might be implicated in both models, and it will be important in the future to examine thyroxine levels in other models of progeria. Taken together, our findings suggest that DNA damage incurred in the embryonic anterior pituitary leads to defective expression of Tsh postnatally, causing hypothyroidism, decreased IGF-1 signaling, and hypoglycemia. Decreased IGF-1 in the serum can negatively impact skeletal growth development, trabecular content, and subsequent mineralization, as seen in our cKO mice (64). It should be noted that thyroid hormone can also act directly on target tissues such as growth plate chondrocytes, independent of liver-derived IGF-1. Reduced bone growth, and possibly other phenotypes observed in our mutant mice, might therefore be caused by a combination of reduced circulating IGF-1 and reduced thyroid hormone receptor activation in the target tissue (65, 66).
Hyperactivity of the p53 tumor suppressor gene shortens life span and accelerates aging in mice (67). We had previously reported that p53 is activated in the absence of ATRX in the embryonic brain and that loss of p53 rescued cell death in the ATRX-deficient mouse brain (18). Thus, p53 is required for the removal of cells with excessive DNA damage in the Atrx-null brain, which explains the enhanced accumulation of DNA damage we observed upon simultaneous loss of ATRX and p53. Decreased telomere stability and length are also key determinants of life span and have been reported in cancer as well as several types of segmental progeria syndromes, such as Werner syndrome, Cockayne syndrome, dyskeratosis congenita, and Hutchinson-Gilford progeria (68–75). If telomeric damage is the key driver of life span, our findings would then suggest that telomeric abnormalities confined to specific brain and/or pituitary cells are in themselves sufficient to induce several aging-like phenotypes prematurely, a provocative idea that would have to be confirmed or dismissed in future work.
Problems in replication have been shown to lead to chromosome segregation failures in ensuing mitoses (76). Similarly, several G4-DNA ligands have been demonstrated to cause telomeric fusions and telomere aggregate formation that eventually lead to chromosomal instability, anaphase bridges, and mitotic catastrophe (77–81). It is conceivable that replicative damage is the underlying cause of mitotic defects that we previously reported in ATRX-deficient human cells, such as chromosome missegregation, anaphase bridges, and micronucleus formation (19). Functions for ATRX in restricting replicative stress, telomere fusions, and mitotic defects may also help explain the tumor-suppressive roles recently ascribed to the ATRX protein. Mutations in the ATRX gene were identified in several types of cancers, including pediatric brain tumors (5–7, 9, 10). A common denominator in the majority of Atrx-null tumors was the frequent appearance of large telomeric foci, a hallmark of alternative lengthening of telomeres (ALT), providing further evidence that ATRX function is intimately linked to telomere biology, possibly as a suppressor of illegitimate recombination events. The hypersensitivity of Atrx-null neuroprogenitors to TMS is an important finding in that regard, as it indicates that Atrx-null tumors may be susceptible to treatment with G4-DNA binding ligands.
Methods
Mouse husbandry, genotyping, and tissue preparation.
Mice were exposed to 12-hour light/12-hour dark cycles and fed tap water and regular chow ad libitum. The AtrxloxP mice have been described previously (30, 82). AtrxloxP mice, when mated to mice expressing Cre recombinase under the control of the Foxg1 promoter (Foxg1KiCre/+) (31), produce male progeny (AtrxloxP/YFoxg1KiCre/+) with Atrx deficiency in the forebrain (AtrxFoxg1Cre). To account for heterozygosity of Foxg1 due to knock-in of the Cre recombinase gene, Cre+ males were used as controls. Trp53tm1Tyj/J mice were obtained from The Jackson Laboratory, and the mutant allele was introduced into the AtrxloxP and Foxg1KiCre/+ mice. Subsequent mating of this progeny yielded mice that are p53-null in all tissues and Atrx/p53 double-null in the forebrain. For timed matings, midday of discovery of the vaginal plug was considered to be E0.5. Yolk sac or tail DNA was used for genotyping. Atrx, Cre, and p53 genotyping was performed by PCR as previously described (18, 30).
Immunofluorescence and antibodies.
Embryos were fixed overnight in 4% paraformaldehyde, washed 3 times in 1× PBS for 5 minutes each, and dehydrated in 30% sucrose PBS. Postnatal mice were perfused with 4% paraformaldehyde before PBS washes and dehydration. Brains were flash-frozen in liquid nitrogen using Cryomatrix (Thermo Scientific) cryoprotectant and sectioned as described previously (83). For immunofluorescence staining of cryosections, antigen retrieval was performed on slides by warming 10 mM sodium citrate pH 6 solution to approximately 95°C and microwaving slides in solution for 10 minutes on low. After cooling, slides were washed and incubated with primary antibody overnight at 4°C. For detection of PCNA, primary antibody was incubated in 10 U/ml DNase I (Sigma-Aldrich) as previously described (18). Slides were washed in PBS and incubated with secondary antibody for 1 hour. Sections were counterstained with DAPI (Sigma-Aldrich) and mounted with Vectashield (Vector Laboratories) or SlowFade Gold (Invitrogen). For immunofluorescence staining of primary cortical culture, cells were incubated in blocking solution (PBS with 0.3% Triton-X and 5% normal goat serum) for 30 minutes and subsequently incubated with primary antibody overnight at 4°C. Cells were washed in PBS and incubated with secondary antibody for 1 hour. Cells were counterstained with DAPI and mounted with Vectashield.
The following primary antibodies were used: anti-ATRX, rabbit polyclonal (1:200; Santa Cruz Biotechnology Inc.); anti–cleaved caspase-3 (Asp175), rabbit monoclonal (1:400; Cell Signaling Technology); anti-γH2AX (Ser139), rabbit (1:100; Cell Signaling Technology); anti-γH2AX (Ser139), mouse monoclonal (1:400; Millipore); and anti-PCNA, mouse monoclonal (1:100; Santa Cruz Biotechnology Inc.). The secondary antibodies used were goat anti-rabbit–Alexa Fluor 594 (1:800 dilution; Molecular Probes) and goat anti-mouse–Alexa Fluor 488 (1:800 dilution; Molecular Probes).
Microscopy.
Experiments demonstrating colocalization of γH2AX and PCNA, γH2AX and major satellite repeats, and γH2AX and telomeres were captured using a laser scanning confocal microscope (FV1000, Olympus). z-Stacks were obtained at 0.25-μm z intervals generally spanning 10–20 μm. Overlapping signal was scored as a colocalization event. For colocalization of γH2AX and PCNA, only cells in which PCNA staining was characteristic of late S phase were counted. Each cell (n = 300) was scored for the number of γH2AX foci that overlapped with PCNA foci divided by the total number of γH2AX foci per cell, to obtain a measure of the cellular levels of DNA damage (γH2AX) at late-replicating chromatin. All other images were captured using an inverted microscope (DMI 6000b, Leica). Digital microscopy images were captured with a digital camera (ORCA-ER, Hamamatsu). Openlab imaging software was used for manual and automated image capture, and processing was performed using Volocity software (PerkinElmer). For quantification of γH2AX foci per area, at least 6 serial cortical cryosections were assessed for γH2AX foci within the indicated regions. To obtain a relative measure of DNA damage, the ratio of γH2AX foci to area (mm2) was calculated. The same method was used to quantify the ratio of cleaved capsase-3+ and γH2AX+ cells, and for this experiment a Student’s t test was used to compare only the proportion of caspase-3+γH2AX+ cells between control and Atrx-null mice.
Western blot analysis.
Nuclear protein was extracted from E13.5 telencephalon using a standard extraction kit (Thermo Scientific) and quantified using a Bradford assay. Protein (10 μg) was resolved on 6% SDS-PAGE and transferred onto nitrocellulose membranes (Bio-Rad). The membranes were probed with anti-ATM, rabbit monoclonal (1:500, Millipore); anti–phospho-ATM (S1981), mouse monoclonal (1:250, Rockland); anti-ATR, goat polyclonal (1:250, Santa Cruz Biotechnology Inc.); anti–phospho-ATR (S428), rabbit polyclonal (1:250, Santa Cruz Biotechnology Inc.); and anti-INCENP, rabbit polyclonal (1:10,000, Sigma-Aldrich) antibodies, followed by the appropriate horseradish peroxidase–conjugated secondary antibodies (1:4,000, GE Healthcare). The membrane was incubated in ECL before exposure to X-ray film. See complete, unedited blots in the supplemental material.
Primary NPC cultures and manipulation.
Cortical progenitor cultures were prepared as described previously (84, 85) using cortices dissected from E13.5 embryos. Cells were seeded on polyornithine-coated (Sigma-Aldrich) plastic plates or glass coverslips. Primary cortical cultures were treated with acute γ-irradiation (0, 1, 5, and 10 Gy) at 2 days after plating using a Cobalt-60 irradiator (Theratron 60, Atomic Energy of Canada Limited) located in the London Regional Cancer Center, London, Ontario, Canada. Cells were fixed in 4% paraformaldehyde for 10 minutes, washed in PBS, and stored in PBS at 4°C at 0, 3, and 6 hours after treatment and processed for immunofluorescence. HU (Sigma-Aldrich), MMC (Sigma-Aldrich), γ-irradiation, and TMS (synthesized in the Biomedicinal Information Research Center, National Institute of Advanced Industrial Science and Technology, Tokyo, Japan) sensitivity assays in cultured NPCs were conducted 48 hours after plating cells. NPC cultures were treated with appropriate doses of HU, γ-irradiation, or TMS for the indicated length of time, and cell viability was measured using the trypan blue dye exclusion method. Cell counts were determined with a hemacytometer.
FISH.
For DNA-FISH experiments, primary cortical cultures were permeabilized in CSK buffer with 0.5% Triton-X (Sigma-Aldrich) prior to fixation. For TelFISH, a fluorophore-labeled DNA oligonucleotide probe, TAACCC7-Alexa 488-3′ (Integrated DNA Technologies), was dissolved at 1 pmol/μl in hybridization buffer. Hybridization was performed as previously described (86). For major satellite FISH, immunofluorescence was performed first, followed by fixation with 4% paraformaldehyde for 10 minutes and subsequent hybridization with a major satellite repeat probe as previously described (87).
MEF cultures and manipulations.
E13.5 embryos were minced, digested in trypsin for 45 minutes at 37°C, and resuspended in DMEM (Sigma-Aldrich) containing 10% FBS, 50 U/ml penicillin, 50 μg/ml streptomycin, and 2 mM l-glutamine. Primary AtrxloxP/Y and wild-type MEFs were seeded at 1.5 × 106 cells per 6-well plate and transduced with adenovirus expressing Cre recombinase and a GFP reporter gene (Ad-CreGFP) or GFP only (Ad-GFP control) approximately 12 hours later. Three days after transduction, MEFs were counted and reseeded at 1.5 × 106 cells per 6-well plate (P5/P0). HU sensitivity in MEFs was determined by transducing 1.5 × 106 cells with adenovirus, culturing for 72 hours, and treating cells with appropriate doses of HU for 24 hours, replacing medium, and measuring viability via trypan blue dye exclusion 2 days later. Sensitivity to γ-irradiation in MEFs was assessed by transducing 1.5 × 106 cells with adenovirus, waiting 72 hours, and treating cells with appropriate doses of γ-irradiation and measuring viability via trypan blue dye exclusion 48 hours later.
qRT-PCR.
Total RNA was obtained from control and cKO liver with the RNeasy Mini Kit (QIAGEN) and reverse transcribed into cDNA as described previously (8). Control reactions without reverse transcriptase were prepared in parallel. cDNA was amplified with gene-specific primers under the following conditions: 25–35 cycles of 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 1 minute. For qRT-PCR, cDNA was amplified with iQ SYBR Green Master Mix (Bio-Rad) by using the standard curve Ct method of quantification. Experiments were performed on a Chromo-4 thermocycler (MJ Research) and analyzed with Opticon Monitor 3 and GeneX (Bio-Rad) software. Gene expression analysis was repeated in triplicate for each sample. Conditions for amplification were as follows: 35 cycles of 95°C for 10 seconds, 55°C for 20 seconds, 72°C for 30 seconds, and a final melting curve generated in increments of 0.5°C per plate read. Standard curves were generated for each primer pair with 3-fold serial dilutions of control cDNA. Primer efficiency was calculated as: E = (10[–1/slope] – 1) * 100%, where a desirable slope is –3.32 and r2 > 0.990. All data were corrected against Gapdh as an internal control. Error bars represent SEM.
Alcian blue and alizarin red staining.
Skinned and eviscerated mouse carcasses at P17 were fixed overnight in 95% ethanol and transferred to acetone. Fixed skeletons were stained in alizarin red and alcian blue, as described previously (88). Images were captured using an Olympus SP-57OUZ digital camera. Skeletal elements from at least 4 different littermate pairs were imaged using a Zeiss Stereo Zoom Microscope Stemi SV6, and measurements of individual bones were undertaken with a ruler accurate to 0.1 mm.
microCT.
Mice were euthanized on P17, skinned, eviscerated, and stored in formalin. Whole skeletons were scanned on a GE Locus Ultra scanner at 120 kV and 20 mA, with a 0.154-mm3 voxel resolution with a total of 900 slices per scan. To assess bone density, hind limbs were scanned on a GE Locus scanner at 80 kV and 0.450 mA with a 0.020-mm3 voxel resolution with 900 slices per scan. BMD, cortical thickness, and trabecular numbers were calculated using MicroView 3D visualization and analysis software (version 2.1.2, GE Healthcare Biosciences) (89). BMD was determined from proximal tibias using the bone analysis module within MicroView by defining the region of interest (ROI) as the area between 0.3 mm and 0.6 mm proximal to the growth plate to include the trabecular spongiosa. The ROI was a cylinder fitting within the central region of the trabecular bone, excluding the cortical bone. Cortical bone–mimicking epoxy (SB3, Gammex RMI) was used for calibration. Trabecular number was determined using the stereology function within MicroView, using the same ROI as used for BMD. Cortical thickness was determined at the tibial mid-diaphysis, defined as the distance halfway between the two ends of the tibia. Thickness of the bone was measured radially in each quadrant and averaged.
Picrosirius red staining of bone sections.
Fore- and hind limbs were dissected at P0, P7, P10, and/or P17 upon sacrifice and processed for histological analyses by 0.1% picrosirius red (Sigma-Aldrich, catalog P6744-16A) as described previously (90). Trabecular area (μm2) within an area of interest (AOI) was measured by observers blinded to the experimental protocol using Openlab 4.0.4 software (Improvision), with the AOI set from just below the hypertrophic chondrocytes to 200 μm below the growth plate in the trabecular bone area of the mineralized zone. The AOI measurements were adapted from refs. 91, 92. Trabecular area was normalized to total AOI and expressed as a percentage. All images were taken with a Retiga EX camera (Leeds Precision Instruments Inc.) connected to a DMRA2 microscope (Leica).
Ex vivo tibia cultures.
Tibiae were isolated from P0 mutant and control mice using a Zeiss Stereo Zoom Microscope Stemi SV6 and dissection tools, as described previously (93). Isolated tibiae were plated in 12-well culture dishes and cultured with 1 ml per well of sterile organ culture α-MEM medium (Gibco, Life Technologies, catalog 1257063) containing ascorbic acid (0.005%, Sigma-Aldrich, A4034), β-glycerophosphate (0.02%, Sigma-Aldrich, 819830), BSA (0.2%, Fisher Scientific, 9048486), 200 nM glutamine (0.25 ml, Gibco, Life Technologies, 25030081), and penicillin streptomycin (0.4 ml, Gibco, Life Technologies, 15140122) for 7 days at 37°C, 5% CO2. Organ culture medium was refreshed on days 2, 4, and 6. Tibia samples were fixed in 70% ethanol overnight at 4°C on day 7. Samples were processed, embedded in paraffin, and sectioned at the Molecular Pathology Core Facility, Robarts Research Institute, London, Ontario, Canada. Tibia lengths were measured on days 0 and 7 using the Zeiss Stereo Zoom Microscope Stemi SV6 with a ruler accurate to 0.1 mm.
Measurements of IGF-1, GH, T4, and glucose.
Plasma and liver samples were collected from P17 mice. Blood was collected from the inferior vena cava. EDTA pH 7.0 was added to the blood sample and centrifuged at 21,000 g for 10 minutes at 4°C. Plasma supernatant was collected and kept frozen at –80°C. Liver samples were collected and homogenized by 2 freeze-thaw cycles. Liver homogenate supernatant was collected and assayed immediately. Plasma and liver IGF-1 content were measured using a mouse IGF-1 ELISA kit (R&D Systems, catalog MG100). Plasma GH and T4 were assayed using Millipore (EZRMGH-45K) and Calbiotech (T4044T) ELISA kits, respectively, according to the manufacturers’ instructions. Blood glucose levels were measured immediately prior to sacrifice using the OneTouch FastTake Meter according to the manufacturer’s instructions.
Statistics.
Statistical analysis was performed using GraphPad Prism software (4.02; GraphPad Software Inc.), and all results are expressed as the mean ± SEM unless indicated otherwise. Two independent data sets were compared with the Student’s t test (unpaired, 2-tailed). Statistical analyses of Kaplan-Meier survival curves were performed using the log-rank test and the Gehan-Breslow-Wilcoxon test. P values of 0.05 or less were considered to indicate significance.
Study approval.
All procedures involving animals were conducted in accordance with the regulations of the Animals for Research Act of the Province of Ontario and approved by the University of Western Ontario Animal Care and Use Committee.
Supplementary Material
Acknowledgments
This work was supported by a Paediatrics Graduate Studentship at the University of Western Ontario, a Curtis Cadman Studentship, and a Queen Elizabeth II Ontario Graduate Scholarship in Science and Technology (QEIIOGSST) to L.A. Watson; a CIHR Masters Scholarship to J.R. Li; and an Ontario Graduate Scholarship to L.A. Solomon. Operating funds were from the Canadian Institutes for Health Research (CIHR-MOP93697 to N.G. Bérubé and CIHR-MOP102539 to N.G. Bérubé and F. Beier).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(5):2049–2063. doi:10.1172/JCI65634.
References
- 1.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11(3):208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 2.Allen C, Ashley AK, Hromas R, Nickoloff JA. More forks on the road to replication stress recovery. J Mol Cell Biol. 2011;3(1):4–12. doi: 10.1093/jmcb/mjq049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buonomo SB. Heterochromatin DNA replication and Rif1. Exp Cell Res. 2010;316(12):1907–1913. doi: 10.1016/j.yexcr.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 4.Xue Y, et al. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc Natl Acad Sci U S A. 2003;100(19):10635–10640. doi: 10.1073/pnas.1937626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yachida S, et al. Small cell and large cell neuroendocrine carcinomas of the pancreas are genetically similar and distinct from well-differentiated pancreatic neuroendocrine tumors. Am J Surg Pathol. 2012;36(2):173–184. doi: 10.1097/PAS.0b013e3182417d36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heaphy CM, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333(6041):425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiao Y, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331(6021):1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiao Y, et al. Frequent ATRX, CIC, and FUBP1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3(7):709–722. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 10.Cheung NK, et al. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA. 2012;307(10):1062–1071. doi: 10.1001/jama.2012.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kannan K, et al. Whole-exome sequencing identifies ATRX mutation as a key molecular determinant in lower-grade glioma. Oncotarget. 2012;3(10):1194–1203. doi: 10.18632/oncotarget.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molenaar JJ, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483(7391):589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 13.Lovejoy CA, et al. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 2012;8(7):e1002772. doi: 10.1371/journal.pgen.1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen DN, et al. Brain Pathol. Molecular and morphologic correlates of the alternative lengthening of telomeres phenotype in high-grade astrocytomas [published online ahead of print August 29, 2012]. doi: 10.1111/j.1750-3639.2012.00630.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbons RJ, Picketts DJ, Villard L, Higgs DR. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with alpha-thalassemia (ATR-X syndrome). Cell. 1995;80(6):837–845. doi: 10.1016/0092-8674(95)90287-2. [DOI] [PubMed] [Google Scholar]
- 16.Gibbons RJ, Picketts DJ, Higgs DR. Syndromal mental retardation due to mutations in a regulator of gene expression. Hum Mol Genet. 1995;4 spec no:1705–1709. doi: 10.1093/hmg/4.suppl_1.1705. [DOI] [PubMed] [Google Scholar]
- 17.Gibbons RJ, Higgs DR. Molecular-clinical spectrum of the ATR-X syndrome. Am J Med Genet. 2000;97(3):204–212. doi: 10.1002/1096-8628(200023)97:3<204::AID-AJMG1038>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 18.Seah C, et al. Neuronal death resulting from targeted disruption of the Snf2 protein ATRX is mediated by p53. J Neurosci. 2008;28(47):12570–12580. doi: 10.1523/JNEUROSCI.4048-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ritchie K, Seah C, Moulin J, Isaac C, Dick F, Berube NG. Loss of ATRX leads to chromosome cohesion and congression defects. J Cell Biol. 2008;180(2):315–324. doi: 10.1083/jcb.200706083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baumann C, Viveiros MM, De La Fuente R. Loss of maternal ATRX results in centromere instability and aneuploidy in the mammalian oocyte and pre-implantation embryo. PLoS Genet. 2010;6(9):e1001137. doi: 10.1371/journal.pgen.1001137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De La Fuente R, Viveiros MM, Wigglesworth K, Eppig JJ. ATRX, a member of the SNF2 family of helicase/ATPases, is required for chromosome alignment and meiotic spindle organization in metaphase II stage mouse oocytes. Dev Biol. 2004;272(1):1–14. doi: 10.1016/j.ydbio.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 22.Wong LH, et al. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010;20(3):351–360. doi: 10.1101/gr.101477.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kernohan KD, et al. ATRX partners with cohesin and MeCP2 and contributes to developmental silencing of imprinted genes in the brain. Dev Cell. 2010;18(2):191–202. doi: 10.1016/j.devcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 24.Bagheri-Fam S, et al. Defective survival of proliferating Sertoli cells and androgen receptor function in a mouse model of the ATR-X syndrome. Hum Mol Genet. 2011;20(11):2213–2224. doi: 10.1093/hmg/ddr109. [DOI] [PubMed] [Google Scholar]
- 25.Berube NG, Smeenk CA, Picketts DJ. Cell cycle-dependent phosphorylation of the ATRX protein correlates with changes in nuclear matrix and chromatin association. Hum Mol Genet. 2000;9(4):539–547. doi: 10.1093/hmg/9.4.539. [DOI] [PubMed] [Google Scholar]
- 26.McDowell TL, et al. Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc Natl Acad Sci U S A. 1999;96(24):13983–13988. doi: 10.1073/pnas.96.24.13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eustermann S, et al. Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin. Nat Struct Mol Biol. 2011;18(7):777–782. doi: 10.1038/nsmb.2070. [DOI] [PubMed] [Google Scholar]
- 28.Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci U S A. 2010;107(32):14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baumann C, Schmidtmann A, Muegge K, De La Fuente R. Association of ATRX with pericentric heterochromatin and the Y chromosome of neonatal mouse spermatogonia. BMC Mol Biol. 2008;9:29. doi: 10.1186/1471-2199-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berube NG, et al. The chromatin-remodeling protein ATRX is critical for neuronal survival during corticogenesis. J Clin Invest. 2005;115(2):258–267. doi: 10.1172/JCI22329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hebert JM, McConnell SK. Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev Biol. 2000;222(2):296–306. doi: 10.1006/dbio.2000.9732. [DOI] [PubMed] [Google Scholar]
- 32.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273(10):5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 33.Shiloh Y. ATM and ATR: networking cellular responses to DNA damage. Curr Opin Genet Dev. 2001;11(1):71–77. doi: 10.1016/S0959-437X(00)00159-3. [DOI] [PubMed] [Google Scholar]
- 34.Roos WP, Kaina B. DNA damage-induced cell death by apoptosis. Trends Mol Med. 2006;12(9):440–450. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18(1):134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 36.Koc A, Wheeler LJ, Mathews CK, Merrill GF. Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. J Biol Chem. 2004;279(1):223–230. doi: 10.1074/jbc.M303952200. [DOI] [PubMed] [Google Scholar]
- 37.Cahill D, Connor B, Carney JP. Mechanisms of eukaryotic DNA double strand break repair. Front Biosci. 2006;11:1958–1976. doi: 10.2741/1938. [DOI] [PubMed] [Google Scholar]
- 38.Takanari H, Yamanaka H, Mitani H, Izutsu K. Replication sites as revealed by double label immunofluorescence against proliferating cell nuclear antigen (PCNA) and bromodeoxyuridine (BrdU) in synchronized CHO cells and vincristine-induced multinucleate cells. Biol Cell. 1994;82(1):23–31. doi: 10.1016/0248-4900(94)90062-0. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Patel DJ. Guanine residues in d(T2AG3) and d(T2G4) form parallel-stranded potassium cation stabilized G-quadruplexes with anti glycosidic torsion angles in solution. Biochemistry. 1992;31(35):8112–8119. doi: 10.1021/bi00150a002. [DOI] [PubMed] [Google Scholar]
- 40.Shin-ya K, et al. Telomestatin, a novel telomerase inhibitor from Streptomyces anulatus. J Am Chem Soc. 2001;123(6):1262–1263. doi: 10.1021/ja005780q. [DOI] [PubMed] [Google Scholar]
- 41.Niedernhofer LJ, et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444(7122):1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- 42.Xing W, et al. Genetic evidence that thyroid hormone is indispensable for prepubertal insulin-like growth factor-I expression and bone acquisition in mice. J Bone Miner Res. 2012;27(5):1067–1079. doi: 10.1002/jbmr.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Pluijm I, et al. Impaired genome maintenance suppresses the growth hormone — insulin-like growth factor 1 axis in mice with Cockayne syndrome. PLoS Biol. 2007;5(1):e2. doi: 10.1371/journal.pbio.0050002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van de Ven M, et al. Adaptive stress response in segmental progeria resembles long-lived dwarfism and calorie restriction in mice. PLoS Genet. 2006;2(12):e192. doi: 10.1371/journal.pgen.0020192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mostoslavsky R, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124(2):315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 46.Gaitonde DY, Rowley KD, Sweeney LB. Hypothyroidism: an update. Am Fam Physician. 2012;86(3):244–251. [PubMed] [Google Scholar]
- 47.Solomon LA, Li JR, Berube NG, Beier F. Loss of ATRX in chondrocytes has minimal effects on skeletal development. PLoS One. 2009;4(9):e7106. doi: 10.1371/journal.pone.0007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Law MJ, et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell. 2010;143(3):367–378. doi: 10.1016/j.cell.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 49.Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24(12):1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luciani JJ, et al. PML nuclear bodies are highly organised DNA-protein structures with a function in heterochromatin remodelling at the G2 phase. J Cell Sci. 2006;119(pt 12):2518–2531. doi: 10.1242/jcs.02965. [DOI] [PubMed] [Google Scholar]
- 51.Berube NG, et al. Patient mutations alter ATRX targeting to PML nuclear bodies. Eur J Hum Genet. 2008;16(2):192–201. doi: 10.1038/sj.ejhg.5201943. [DOI] [PubMed] [Google Scholar]
- 52.Ishov AM, Vladimirova OV, Maul GG. Heterochromatin and ND10 are cell-cycle regulated and phosphorylation-dependent alternate nuclear sites of the transcription repressor Daxx and SWI/SNF protein ATRX. J Cell Sci. 2004;117(pt 17):3807–3820. doi: 10.1242/jcs.01230. [DOI] [PubMed] [Google Scholar]
- 53.de Boer J, et al. Premature aging in mice deficient in DNA repair and transcription. Science. 2002;296(5571):1276–1279. doi: 10.1126/science.1070174. [DOI] [PubMed] [Google Scholar]
- 54.Niedernhofer LJ. Nucleotide excision repair deficient mouse models and neurological disease. DNA Repair (Amst). 2008;7(7):1180–1189. doi: 10.1016/j.dnarep.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Niedernhofer LJ. Tissue-specific accelerated aging in nucleotide excision repair deficiency. Mech Ageing Dev. 2008;129(7–8):408–415. doi: 10.1016/j.mad.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harada YN, et al. Postnatal growth failure, short life span, and early onset of cellular senescence and subsequent immortalization in mice lacking the xeroderma pigmentosum group G gene. Mol Cell Biol. 1999;19(3):2366–2372. doi: 10.1128/mcb.19.3.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weeda G, et al. Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr Biol. 1997;7(6):427–439. doi: 10.1016/S0960-9822(06)00190-4. [DOI] [PubMed] [Google Scholar]
- 58.Murga M, et al. A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nat Genet. 2009;41(8):891–898. doi: 10.1038/ng.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O’Driscoll M. Mouse models for ATR deficiency. DNA Repair (Amst). 2009;8(11):1333–1337. doi: 10.1016/j.dnarep.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 60.Jaarsma D, et al. Age-related neuronal degeneration: complementary roles of nucleotide excision repair and transcription-coupled repair in preventing neuropathology. PLoS Genet. 2011;7(12):e1002405. doi: 10.1371/journal.pgen.1002405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neveling K, Bechtold A, Hoehn H. Genetic instability syndromes with progeroid features. Z Gerontol Geriatr. 2007;40(5):339–348. doi: 10.1007/s00391-007-0483-x. [DOI] [PubMed] [Google Scholar]
- 62.Wang L, Shao YY, Ballock RT. Thyroid hormone-mediated growth and differentiation of growth plate chondrocytes involves IGF-1 modulation of beta-catenin signaling. J Bone Miner Res. 2012;25(5):1138–1146. doi: 10.1002/jbmr.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kamileri I, et al. Defective transcription initiation causes postnatal growth failure in a mouse model of nucleotide excision repair (NER) progeria. Proc Natl Acad Sci U S A. 2012;109(8):2995–3000. doi: 10.1073/pnas.1114941109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mohan S, Kesavan C. Role of insulin-like growth factor-1 in the regulation of skeletal growth. Curr Osteoporos Rep. 2012;10(2):178–186. doi: 10.1007/s11914-012-0100-9. [DOI] [PubMed] [Google Scholar]
- 65.Wojcicka A, Bassett JH, Williams GR. Biochim Biophys Acta. Mechanisms of action of thyroid hormones in the skeleton [published online ahead of print May 25, 2012]. doi: 10.1016/j.bbagen.2012.05.005 . [DOI] [PubMed] [Google Scholar]
- 66.Wit JM, Camacho-Hubner C. Endocrine regulation of longitudinal bone growth. Endocr Dev. 2011;21:30–41. doi: 10.1159/000328119. [DOI] [PubMed] [Google Scholar]
- 67.Tyner SD, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415(6867):45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 68.Chang S, et al. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nat Genet. 2004;36(8):877–882. doi: 10.1038/ng1389. [DOI] [PubMed] [Google Scholar]
- 69.Crabbe L, Jauch A, Naeger CM, Holtgreve-Grez H, Karlseder J. Telomere dysfunction as a cause of genomic instability in Werner syndrome. Proc Natl Acad Sci U S A. 2007;104(7):2205–2210. doi: 10.1073/pnas.0609410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Damerla RR, Knickelbein KE, Strutt S, Liu FJ, Wang H, Opresko PL. Werner syndrome protein suppresses the formation of large deletions during the replication of human telomeric sequences. Cell Cycle. 2012;11(16):3036–3044. doi: 10.4161/cc.21399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Opresko PL, et al. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol Cell. 2004;14(6):763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 72.Benson EK, Lee SW, Aaronson SA. Role of progerin-induced telomere dysfunction in HGPS premature cellular senescence. J Cell Sci. 2011;123(pt 15):2605–2612. doi: 10.1242/jcs.067306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Batenburg NL, Mitchell TR, Leach DM, Rainbow AJ, Zhu XD. Cockayne Syndrome group B protein interacts with TRF2 and regulates telomere length and stability. Nucleic Acids Res. 2012;40(19):9661–9674. doi: 10.1093/nar/gks745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shay JW, Wright WE. Mutant dyskerin ends relationship with telomerase. Science. 1999;286(5448):2284–2285. doi: 10.1126/science.286.5448.2284. [DOI] [PubMed] [Google Scholar]
- 75.Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402(6761):551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 76.Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol. 2009;11(6):753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- 77.Kim MY, Gleason-Guzman M, Izbicka E, Nishioka D, Hurley LH. The different biological effects of telomestatin and TMPyP4 can be attributed to their selectivity for interaction with intramolecular or intermolecular G-quadruplex structures. Cancer Res. 2003;63(12):3247–3256. [PubMed] [Google Scholar]
- 78.Incles CM, Schultes CM, Kempski H, Koehler H, Kelland LR, Neidle S. A G-quadruplex telomere targeting agent produces p16-associated senescence and chromosomal fusions in human prostate cancer cells. Mol Cancer Ther. 2004;3(10):1201–1206. [PubMed] [Google Scholar]
- 79.Tahara H, Shin-Ya K, Seimiya H, Yamada H, Tsuruo T, Ide T. G-Quadruplex stabilization by telomestatin induces TRF2 protein dissociation from telomeres and anaphase bridge formation accompanied by loss of the 3′ telomeric overhang in cancer cells. Oncogene. 2006;25(13):1955–1966. doi: 10.1038/sj.onc.1209217. [DOI] [PubMed] [Google Scholar]
- 80.Gauthier LR, et al. Rad51 and DNA-PKcs are involved in the generation of specific telomere aberrations induced by the quadruplex ligand 360A that impair mitotic cell progression and lead to cell death. Cell Mol Life Sci. 2011;69(4):629–640. doi: 10.1007/s00018-011-0767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hampel SM, et al. Mechanism of the antiproliferative activity of some naphthalene diimide g-quadruplex ligands. Mol Pharmacol. 2013;83(2):470–480. doi: 10.1124/mol.112.081075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Garrick D, et al. Loss of Atrx affects trophoblast development and the pattern of X-inactivation in extraembryonic tissues. PLoS Genet. 2006;2(4):e58. doi: 10.1371/journal.pgen.0020058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Berube NG, Jagla M, Smeenk C, De Repentigny Y, Kothary R, Picketts DJ. Neurodevelopmental defects resulting from ATRX overexpression in transgenic mice. Hum Mol Genet. 2002;11(3):253–261. doi: 10.1093/hmg/11.3.253. [DOI] [PubMed] [Google Scholar]
- 84.Gloster A, El-Bizri H, Bamji SX, Rogers D, Miller FD. Early induction of Talpha1 alpha-tubulin transcription in neurons of the developing nervous system. J Comp Neurol. 1999;405(1):45–60. doi: 10.1002/(SICI)1096-9861(19990301)405:1<45::AID-CNE4>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 85.Slack RS, El-Bizri H, Wong J, Belliveau DJ, Miller FD. A critical temporal requirement for the retinoblastoma protein family during neuronal determination. J Cell Biol. 1998;140(6):1497–1509. doi: 10.1083/jcb.140.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang LF, Ogawa Y, Ahn JY, Namekawa SH, Silva SS, Lee JT. Telomeric RNAs mark sex chromosomes in stem cells. Genetics. 2009;182(3):685–698. doi: 10.1534/genetics.109.103093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Isaac CE, et al. The retinoblastoma protein regulates pericentric heterochromatin. Mol Cell Biol. 2006;26(9):3659–3671. doi: 10.1128/MCB.26.9.3659-3671.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang G, Woods A, Agoston H, Ulici V, Glogauer M, Beier F. Genetic ablation of Rac1 in cartilage results in chondrodysplasia. Dev Biol. 2007;306(2):612–623. doi: 10.1016/j.ydbio.2007.03.520. [DOI] [PubMed] [Google Scholar]
- 89.Ulici V, Hoenselaar KD, Gillespie JR, Beier F. The PI3K pathway regulates endochondral bone growth through control of hypertrophic chondrocyte differentiation. BMC Dev Biol. 2008;8:40. doi: 10.1186/1471-213X-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yan Q, Feng Q, Beier F. Reduced chondrocyte proliferation, earlier cell cycle exit and increased apoptosis in neuronal nitric oxide synthase-deficient mice. Osteoarthritis Cartilage. 2012;20(2):144–151. doi: 10.1016/j.joca.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 91.Sawyer A, Lott P, Titrud J, McDonald J. Quantification of tartrate resistant acid phosphatase distribution in mouse tibiae using image analysis. Biotech Histochem. 2003;78(5):271–278. doi: 10.1080/10520290310001646668. [DOI] [PubMed] [Google Scholar]
- 92.O’Connor RD, Zayzafoon M, Farach-Carson MC, Schanen NC. Mecp2 deficiency decreases bone formation and reduces bone volume in a rodent model of Rett syndrome. Bone. 2009;45(2):346–356. doi: 10.1016/j.bone.2009.04.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Agoston H, et al. C-type natriuretic peptide regulates endochondral bone growth through p38 MAP kinase-dependent and -independent pathways. BMC Dev Biol. 2007;7:18. doi: 10.1186/1471-213X-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.