Abstract
Giant axonal neuropathy (GAN) is an early-onset neurological disorder caused by mutations in the GAN gene (encoding for gigaxonin), which is predicted to be an E3 ligase adaptor. In GAN, aggregates of intermediate filaments (IFs) represent the main pathological feature detected in neurons and other cell types, including patients’ dermal fibroblasts. The molecular mechanism by which these mutations cause IFs to aggregate is unknown. Using fibroblasts from patients and normal individuals, as well as Gan–/– mice, we demonstrated that gigaxonin was responsible for the degradation of vimentin IFs. Gigaxonin was similarly involved in the degradation of peripherin and neurofilament IF proteins in neurons. Furthermore, proteasome inhibition by MG-132 reversed the clearance of IF proteins in cells overexpressing gigaxonin, demonstrating the involvement of the proteasomal degradation pathway. Together, these findings identify gigaxonin as a major factor in the degradation of cytoskeletal IFs and provide an explanation for IF aggregate accumulation, the subcellular hallmark of this devastating human disease.
Introduction
Abnormal aggregates of intermediate filaments (IFs) are pathological hallmarks of tissue-specific diseases, including keratin aggregates in Mallory bodies in diseased hepatocytes (1); Rosenthal fibers in the glial cells of Alexander disease patients, which contain glial fibrillary acidic protein (GFAP) (2); and neuronal IF aggregates in the neurons of patients with amyotrophic lateral sclerosis (ALS), Alzheimer’s disease (3), Parkinson’s disease, dementia with Lewy bodies, neuronal IF inclusion disease (NIFID), giant axonal neuropathy (GAN) (4–6), some forms of Charcot-Marie-Tooth disease (CMT), diabetic neuropathy, and spinal muscular atrophy (7, 8). Although numerous mutant genes have been identified that cause these diseases, few have provided insights into the mechanisms responsible for IF aggregate formation. GAN is unique with respect to these diseases, as it affects numerous classes of cytoskeletal IFs (4–6, 9–15). We therefore reasoned that a detailed analysis of this disease might provide insights into common mechanisms responsible for the formation of IF aggregates in different tissues of GAN patients and into the regulation of the turnover of this major class of cytoskeletal proteins.
GAN is a recessive disease (4, 5, 16, 17) caused by mutations in the GAN gene (encoding for gigaxonin), which is located on chromosome 16q24. Over 30 distinct mutations distributed throughout the GAN gene have been identified (9, 11, 16). GAN patients are born apparently normal, but typically by 3–5 years of age, they display progressive muscle weakness, diminished tendon reflexes, and pronounced gait disturbances. Subsequently, they become wheelchair bound, experience dysmetria and seizures, and ultimately require feeding and breathing tubes. Patients typically die during the second or third decade of life. Histopathology reveals the presence of large axonal swellings (“giant axons”) filled with type IV neuronal IFs — the neurofilament (NF) triplet proteins NF light, medium, and heavy chain (NF-L, NF-M, and NF-H, respectively) — in the CNS and peripheral nervous system (PNS) (4, 5, 18). In addition to the NF triplet proteins, the type III IF protein peripherin is also a component of the endogenous IF networks in neurons of the PNS (19). Thus, it is likely that the aggregates detected in the giant axons of PNS neurons in GAN patients contain both the triplet proteins and peripherin. Most patients also have characteristic kinky hair, which suggests that type I and II keratin IFs are affected (12, 13). Moreover, as first shown more than 30 years ago by Pena (6, 15) and subsequently verified by others (10, 20–22), cultured skin fibroblasts from patients contain large IF aggregates of type III vimentin (encoded by VIM) (6, 10, 15, 20–22), and similar IF aggregates have been detected in Schwann cells, melanocytes, endothelial cells, muscle cells, and Langerhans cells (14, 23). Taken together, these observations suggest that gigaxonin mutations affect many members of the cytoskeletal IF protein family.
Based on sequence homology, gigaxonin appears to be an E3 ubiquitin ligase adaptor and thus is likely involved in degrading protein substrates via the ubiquitin-proteasome system (UPS). Gigaxonin belongs to the BTB-KELCH domain family of adaptor proteins. The KELCH domain interacts with the protein targeted for degradation, whereas the BTB domain interacts with the Cullin-3 component of the ubiquitin ligase complex (24–27). In support of gigaxonin’s role in regulating the turnover of IF proteins, histological examination of Gan–/– mice has revealed pathological features similar to those found in GAN patients. These include accumulation of IF proteins, formation of aggregates in the nervous system, loss of peripheral axons, and muscle atrophy (28, 29). Further evidence supporting a role for gigaxonin in regulating IF protein levels is based on quantitative measurements in Gan–/– mice, which have shown up to a 7-fold increase in IF protein levels in the brain, cerebellum, and spinal cord (28, 29). Importantly, the increase in the levels of IF proteins is not associated with a corresponding increase of IF mRNAs (28). Together, these results suggest that complete elimination of gigaxonin leads to increased IF protein levels with subsequent aggregation.
Even though the major alterations that take place in the cells of GAN patients appear to involve IFs (10, 15, 20), it has been reported that gigaxonin is involved in degradation of the microtubule-associated proteins MAP1B (30), MAP8 (31), and tubulin folding cofactor B (TBCB) (32). However, other studies have shown that microtubules (MTs) appear normal (15) and TBCB levels are not altered in GAN cells (20). Similarly, the relative amounts of MAP1B, MAP8, and TBCB do not increase in GAN mouse models that display a dramatic alteration in NF organization (29). Furthermore, Pena previously provided evidence that actin networks appear normal in GAN cells (15).
Here, we focused our attention on the mechanisms responsible for the alterations in IF organization using fibroblasts obtained from GAN patients. These cells provide a useful experimental model since they can be cultured easily and they contain large vimentin IF (VIF) aggregates that resemble the NF aggregates seen in nerve cell axons. Our present findings showed that the most likely mechanism responsible for the aggregates of IFs in GAN is attributable to a critically important role for gigaxonin in regulating the levels of IF proteins via a proteasomal-dependent degradation pathway.
Results
VIFs are aggregated in GAN patients’ cells.
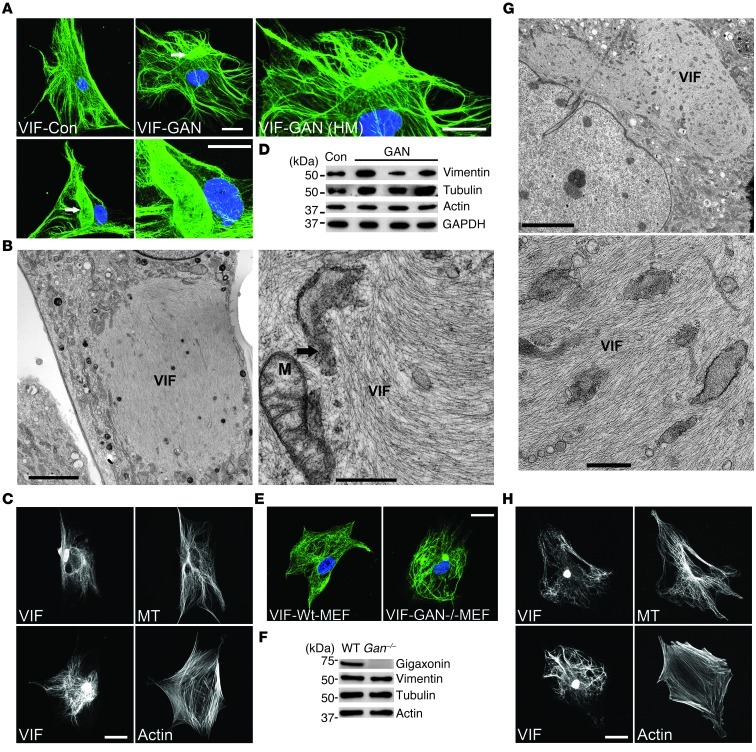
Fibroblasts derived from skin biopsies taken from 3 GAN patients (referred to herein as GAN cells) were studied by immunofluorescence to determine the organization of VIF. The majority of GAN cells contained unusually thick bundles of VIF not seen in control cells. In addition to these bundles, GAN cells frequently contained large perinuclear aggregates (Figure 1A). The number of cells containing aggregates increased as a function of passage number in culture. For example, in GAN cell line 08F699 (having mutations IVS6_1g_a and L510X), approximately 20.89% ± 2.05% of cells (n = 300) contained VIF aggregates at passage 3, whereas at passage 9, approximately 50.9% ± 4.25% had aggregates (P = 0.0069; n = 300), as determined by immunofluorescence. Transmission EM (TEM) confirmed the presence of large accumulations of 10-nm-diameter VIFs in juxtanuclear aggregates and unusually thick IF bundles dispersed throughout the cytoplasm (Figure 1B and data not shown). Further confirmation of the polymerized state of vimentin in GAN cells was obtained from biochemical analyses of IF-enriched cytoskeletal preparations (33). Virtually all of the vimentin was in low-speed pellet fractions, indistinguishable from controls and demonstrating its insoluble polymerized form, with almost no detectable vimentin in the supernatant (data not shown).
Figure 1. VIFs are aggregated in both GAN cells and Gan–/– MEFs.
(A) VIF networks in a control (Con) cell and in 2 different GAN cells, the latter of which were aggregated into bundles and large bodies (arrows). HM, higher magnification. Representative images, 5 preparations of 3 GAN patient lines. (B) TEM showing aggregated VIF in GAN cells. Mitochondrion (M) and elements of the endoplasmic reticulum (arrow) are indicated in the right panel. (C) MTs (top) and actin (bottom) in double-stained GAN cells. Representative images, 5 preparations. (D) Western blot analyses of control fibroblasts (AG08470) and 3 GAN cell lines. Fold changes (± SD) in GAN line vimentin levels relative to control were as follows: F07476 (left), 1.623 ± 0.161; 08F699 (middle), 0.687 ± 0.05; F09133 (right), 1.183 ± 0.064 (P = 0.0039). Representative blots, 3 preparations. (E) VIFs were only aggregated in Gan–/– MEFs, not WT MEFs. Representative images, 5 preparations. (F) Western blotting of WT and Gan–/– MEF lysates. Representative blots, 3 preparations. (G) TEM showing VIF aggregates in Gan–/– MEFs. (H) MT (top) and actin (bottom) organization in Gan–/– MEFs. Representative images, 5 preparations. Scale bars: 10 μm (A, C, E, and H); 5.0 μm (B, left, and G, left); 0.5 μm (B, right, and G, right).
Frequently, mitochondria were found either embedded in or surrounding the VIF aggregates (Figure 1B). Many of these mitochondria were abnormally swollen with reduced numbers of cristae (data not shown). Elements of the smooth and rough endoplasmic reticulum were also seen surrounding or within the VIF aggregates (Figure 1B). In contrast to the dramatic changes seen in VIF in GAN cells, actin/microfilament (actin/MF) organization and MTs were indistinguishable from controls, as determined by fluorescence microscopy (Figure 1C). Quantitative immunoblotting of the GAN cell lines showed that the levels of vimentin compared with controls varied from 0.6- to 1.6-fold (Figure 1D). Of the 3 lines, 2 contained increased levels of vimentin; however, it is not presently clear whether these differences are functionally relevant.
VIF are also aggregated in Gan–/– mouse embryonic fibroblasts.
Because defective gigaxonin has a major effect on VIF organization in GAN cells, we hypothesized that the complete elimination of gigaxonin should have similar effects on cells derived from mouse models. Indeed, Gan–/– mouse embryonic fibroblasts (MEFs) (28) contained VIF aggregates that also increased as a function of passage number (passage 3, ∼21.4% ± 1.1%, n = 315; passage 6, ∼47.5% ± 0.30%, n = 475; P = 0.00047), as assessed by both light microscopy and EM (Figure 1, E and G). EM revealed that IF aggregates in Gan–/– MEFs appeared similar to those observed in GAN cells (Figure 1G). In addition, MTs and MFs appeared normal in these MEFs, and their vimentin levels were similar to those of controls (Figure 1, F and H). Thus, either disease-causing mutations in gigaxonin or its complete knockout caused abnormal aggregates of VIF.
Expression of WT gigaxonin causes loss of VIF networks and clearance of vimentin from cells.
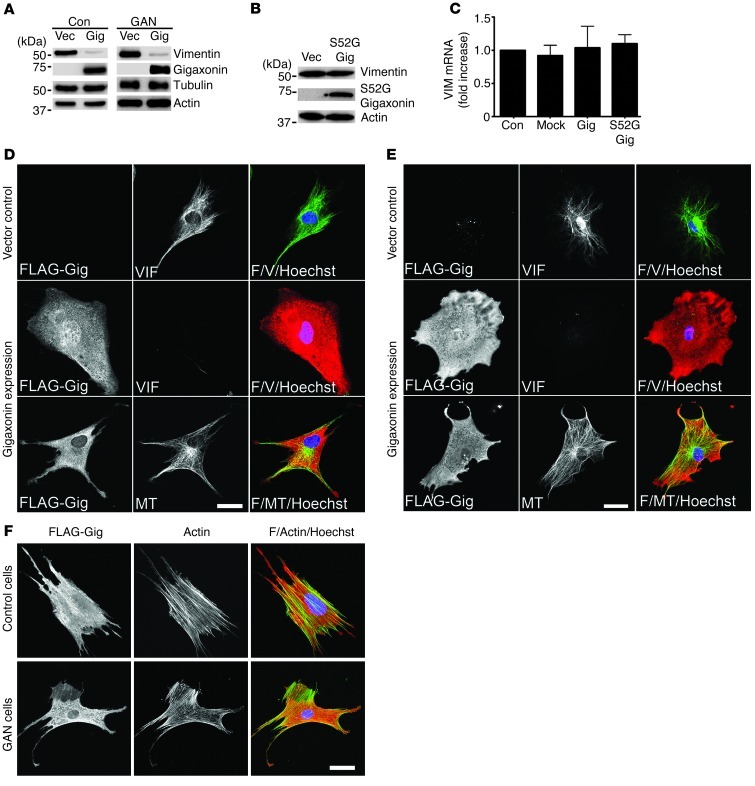
Given our observations that mutations in or deletions of gigaxonin caused IF aggregation, we sought to test the possibility that restoration of normal VIF networks might be achieved by introducing WT gigaxonin into GAN cells or Gan–/– MEFs. However, we found that expressing WT gigaxonin did not just clear the VIF aggregates, but in fact had the more dramatic effect of causing almost complete loss of vimentin in both control and GAN cells (Figure 2, A, D, and E). Although VIF disappeared by 72 hours, MTs and MFs appeared normal (Figure 2, D–F, and refs. 15, 20), and no changes in the levels of tubulin and actin were detected by immunoblotting (Figure 2A). We also tested whether expression of WT gigaxonin resulted in a loss of TBCB, as previously reported (32). Immunoblotting revealed no changes in TBCB levels in control or GAN cells under the same experimental conditions (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI66387DS1). This latter finding confirmed the results of other studies (20, 29). To further confirm the role of gigaxonin in regulating IF protein levels, a mutant form, S52G gigaxonin (9), was expressed in BJ5ta cells. Under these conditions, no change in vimentin levels were detected by immunoblotting (Figure 2B and Supplemental Figure 2). Thus, this mutant form of gigaxonin, unlike WT gigaxonin, could not clear vimentin. Overall, these results demonstrated that gigaxonin has a substantial effect on VIFs, but not on the other major cytoskeletal proteins, tubulin and actin.
Figure 2. Gigaxonin expression causes vimentin clearance.
(A) Immunoblotting of control and GAN cells 72 hours after initiating expression of vector control (Vec) or FLAG-tagged WT gigaxonin (Gig). Representative blots, 4 experiments. (B) Immunoblotting of cells expressing vector or FLAG-tagged S52G gigaxonin (S52G Gig). Representative blots, 3 experiments. (C) RT-PCR analysis of VIM mRNA in BJ5ta cells. Average data ± SD, 3 experiments. Fold increases relative to control (normalized to 1) were as follows: mock, 0.92 ± 0.268; WT gigaxonin (P = 0.6459 vs. mock), 1.04 ± 0.321; S52G gigaxonin, 1.1 ± 0.135 (P = 0.3588 vs. mock). (D and E) Immunofluorescence of (D) BJ5ta cells and (E) GAN cells, 72 hours after initiating expression of vector or WT gigaxonin, with anti-FLAG and anti-vimentin or anti-tubulin (MT). Overlays are shown at right (F, FLAG; V, vimentin). Representative images, 5 preparations. (F) BJ5ta (top) and GAN (bottom) cells stained for gigaxonin and actin after 72-hour expression of WT gigaxonin. Overlays are shown at right. Representative images, 4 preparations. Scale bars: 10 μm (D–F).
Because gigaxonin is a putative E3 ligase adaptor, its expression is likely to cause IF clearance via a degradative pathway. However, in principle, it is also possible that the decrease in vimentin protein is caused by a reduction in transcription of VIM mRNA. To test this, VIM mRNA levels were determined by RT-PCR after expression of WT or S52G gigaxonin in BJ5ta cells. Neither WT nor mutant gigaxonin had any detectable effect on VIM mRNA levels (Figure 2C). This provides further evidence in support of a role for gigaxonin in mediating the degradation of vimentin.
Time course of vimentin clearance due to gigaxonin expression.
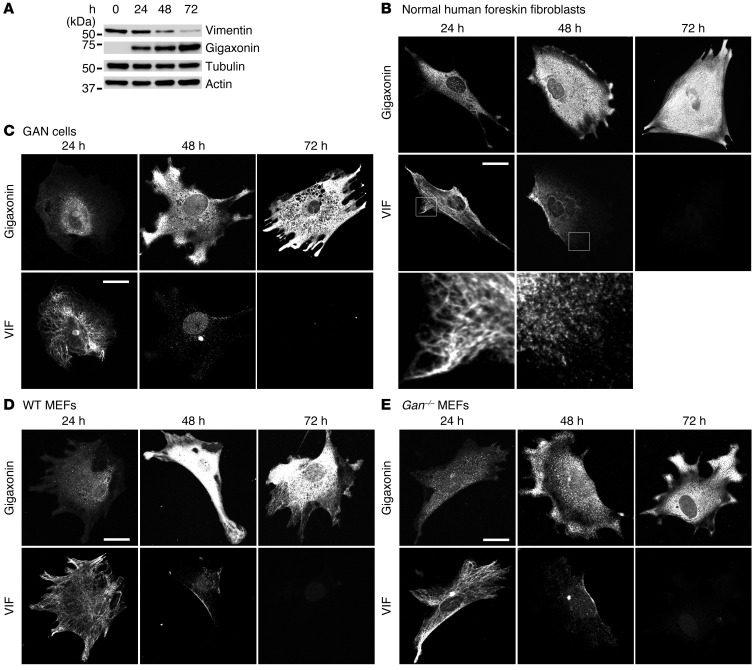
To define the time course of the loss of the VIF network and vimentin protein clearance, GAN and control cells were analyzed at different time points after initiation of gigaxonin expression. Immunoblotting revealed a time-dependent loss of vimentin over 24–72 hours, resulting in levels only approximately 8% of those seen in controls (P = 0.00097; compare Figure 3A and Figure 2A). This disappearance of vimentin was inversely related to the time course and levels of WT gigaxonin expression. Parallel immunofluorescence studies showed that at 24 hours after induction of gigaxonin expression, VIFs were still observed, but by 48 hours, mature IFs disappeared, and only short IFs and nonfilamentous vimentin particles remained (Figure 3B and ref. 34). By 72 hours, there was no detectable vimentin (Figure 3B). These observations suggest that VIF first disassembled into short IFs and nonfilamentous particles that were subsequently further degraded. This series of VIF degradation steps was similar for GAN cells and WT and Gan–/– MEFs (Figure 3, C–E). These observations also demonstrated that in GAN cells and Gan–/– MEFs, the aggregates were the last vimentin structures to disappear after gigaxonin expression (Figure 3, C and E), most likely attributable to the high concentration of VIF in the aggregates relative to the rest of the cell. In contrast, time course analysis after S52G gigaxonin expression in BJ5ta cells revealed no change in vimentin protein levels and no obvious structural changes in VIF (Figure 2B and Supplemental Figure 2, A and B). These results confirmed that disease-causing mutations in gigaxonin result in the loss of its function, insofar as its ability to clear vimentin is concerned.
Figure 3. Time course of VIF changes after WT gigaxonin expression.
(A) Quantitative immunoblot analyses of lysates from BJ5ta cells prepared 0, 24, 48, and 72 hours after initiation of WT gigaxonin expression. Vimentin, tubulin, and actin were detected with specific antibodies, and gigaxonin was detected with anti-FLAG. Vimentin levels decreased to 88% ± 7.7%, 37.5% ± 3.7%, and 7.7% ± 4.5% of controls after 24, 48, and 72 hours, respectively (P = 0.00097). Representative blots, 3 experiments. (B) BJ5ta cells were fixed 24, 48, and 72 hours after initiation of WT gigaxonin expression and were double labeled with anti-FLAG and anti-vimentin. Vimentin staining patterns of boxed regions are shown at higher magnification below (enlarged approximately ×5-fold): VIFs were still intact at 24 hours, whereas at 48 hours, vimentin was mainly present as short filaments and particles. (C–E) Time course of vimentin clearance, assessed by immunofluorescence, in (C) GAN cells, (D) WT MEFs, and (E) Gan–/– MEFs. Cells were fixed 24, 48, and 72 hours after initiation of WT gigaxonin expression and were double labeled with anti-FLAG and anti-vimentin. Representative images, 4 experiments. Scale bars: 10 μm (B–E).
Interactions between gigaxonin and vimentin.
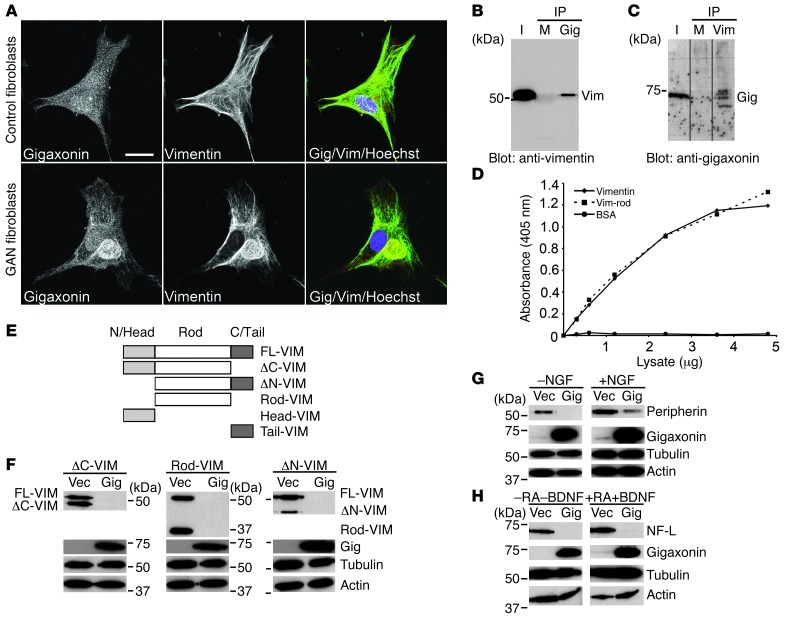
With respect to the localization of gigaxonin, immunofluorescence revealed that it was evenly distributed throughout the cytoplasm in control cells. However, in GAN cells, gigaxonin was concentrated in the VIF aggregates (Figure 4A). Therefore, we sought to determine whether gigaxonin interacts with vimentin in co-IP experiments. WT gigaxonin was expressed in BJ5ta cells for 36 hours, a time point at which cells still contained vimentin (Figure 3A). Cells were lysed at this time, and IP using anti-gigaxonin antibodies revealed the presence of vimentin by immunoblotting, and the reciprocal experiment (IP using anti-vimentin) showed the presence of gigaxonin (Figure 4, B and C). Additional evidence for an interaction between gigaxonin and vimentin in cell lysates was obtained using ELISA (Figure 4D). Taken together, these results revealed an interaction between vimentin and gigaxonin. However, further studies are required to determine whether this interaction is direct or mediated by other proteins. In addition, we do not know the assembly state of vimentin involved in the interaction with gigaxonin under normal in vivo physiological conditions. In IP experiments, vimentin was completely solubilized in the RIPA buffer used to optimize conditions for vimentin/gigaxonin interactions.
Figure 4. Gigaxonin interacts with vimentin, peripherin, and NF-L.
(A) Control and GAN cells double labeled with anti-gigaxonin and anti-vimentin. Association between gigaxonin and vimentin was only observed in the GAN cell aggregates. Representative images, 4 preparations for each of 3 GAN cell lines. (B) IPs using anti-gigaxonin were blotted with anti-vimentin. I, input (cell lysate). Rabbit IgG was used as control (M). Representative blot, 3 experiments. (C) IPs using anti-vimentin were blotted with anti-gigaxonin. Mouse IgG was used as control (M). Representative blot, 3 experiments. Lanes were run on the same gel but were noncontiguous (black lines). (D) ELISA demonstrated that gigaxonin bound to purified vimentin and the vimentin rod domain. Average data, 3 experiments. (E) Schematic of the vimentin deletion constructs FLAG-FL-VIM, FLAG-ΔC-VIM, FLAG-ΔN-VIM, FLAG-Rod-VIM, FLAG-Head-VIM, and FLAG-Tail-VIM. (F) Immunoblotting of lysates from 3T3 cell lines with FLAG-ΔC-VIM, FLAG-Rod-VIM, and FLAG-ΔN-VIM after WT gigaxonin expression for 72 hours, using antibodies to vimentin, FLAG (gigaxonin), tubulin, and actin. Representative blots, 3 experiments. (G) Immunoblotting of PC12 cell lysates prepared 72 hours after initiating WT gigaxonin expression using anti-peripherin, tubulin, actin, and gigaxonin. Similar results were obtained in the absence and presence of NGF. Representative blots, 3 experiments. (H) Immunoblotting of SH-SY-5Y cell lysates prepared 72 hours after WT gigaxonin expression using antibodies to NF-L, tubulin, actin, and gigaxonin. NF-L clearance was the same in undifferentiated (-RA-BDNF) and differentiated (+RA+BDNF) cells. Representative blots, 3 experiments. Scale bar: 10 μm (A).
The central rod domain of vimentin is required for its clearance by gigaxonin.
In order to identify which region of vimentin is important for its clearance by gigaxonin, FLAG-tagged deletion constructs of its 3 major domains (Figure 4E) were prepared: the non–α-helical N-terminal domain (referred to herein as the FLAG-ΔN-VIM construct), the non–α-helical C-terminal domain (FLAG-ΔC-VIM), and the central rod domain, with both the N- and C-termini deleted (FLAG-Rod-VIM). These constructs were used to establish lines of mouse 3T3 fibroblasts. After expression of WT gigaxonin for 72 hours, all 3 deletion constructs of vimentin were cleared, as determined by immunoblotting. As expected, the endogenous full-length vimentin was also cleared within these same cells (Figure 4F). These results suggest that, as the common domain in all 3 deletion constructs, the central rod domain of vimentin is required for its clearance by gigaxonin (Figure 4, E and F).
Additionally, we expressed FLAG-tagged non–α-helical N and C termini alone (referred to herein as Head and Tail, respectively). We found that the FLAG-Head-VIM and FLAG-Tail-VIM constructs could only be expressed at extremely low levels in 3T3 cells, undetectable by immunoblotting; by immunofluorescence, they were mainly associated with the endogenous VIF network (data not shown). Therefore, we prepared FLAG-Head-VIM and FLAG-Tail-VIM deletion construct–expressing lines using Vim–/– MEFs in order to visualize them by immunofluorescence in the absence of an extensive endogenous VIF network (see Methods). As a further control, we also prepared individual lines of Vim–/– MEFs expressing FLAG-tagged full-length vimentin (FLAG-FL-VIM), FLAG-ΔN-VIM, FLAG-ΔC-VIM, and FLAG-Rod-VIM. As expected, FLAG-ΔC-VIM formed filaments (35, 36), whereas FLAG-ΔN-VIM and FLAG-Rod-VIM appeared particulate (nonfilamentous) throughout the cell (Supplemental Figure 3A). Immunofluorescence confirmed that these latter 3 constructs were cleared by gigaxonin expression (Figure 4F and Supplemental Figure 3B). The observation that all 3 constructs were cleared indicates that being incorporated into a filament is not essential for degradation, since the nonfilamentous forms of vimentin that contained the rod domain were efficiently degraded. In contrast, neither the nonfilamentous N terminus (FLAG-Head-VIM) nor the C terminus (FLAG-Tail-VIM) was cleared (Supplemental Figure 3B). These latter constructs appeared to be concentrated in the nucleus, most likely a result of diffusion reflecting their small size. Together, these results provide further evidence that the central rod domain of vimentin is required for clearance by gigaxonin. Additional support for gigaxonin’s interaction with the central rod domain came from our ELISA results (Figure 4D). Since the central rod domain is highly conserved among all types of cytoskeletal IFs, its interaction with gigaxonin may help to explain both the accumulation and the clearance of different IF types in the affected cell types in GAN patients and Gan–/– mice (see below).
Gigaxonin expression causes clearance of neuron-specific IFs.
Because the neurological consequences of GAN are associated with IF aggregates in the axons of PNS and CNS neurons in patients as well as in Gan–/– mice (Supplemental Figure 5 and refs. 28, 29), we sought to determine whether gigaxonin expression can cause the clearance of IFs composed of peripherin and the NF triplet proteins. We established a PC12 cell line stably expressing FLAG-tagged WT gigaxonin. Immunoblotting revealed that expression of gigaxonin caused the loss of peripherin in these cells, which was not affected by their differentiation status (Figure 4G). These results were also confirmed by immunofluorescence (Supplemental Figure 4, A and B). Similarly, analysis of a neuroblastoma cell line, SH-SY-5Y, that stably expressed WT gigaxonin revealed clearance of NF-L protein by both immunoblotting and immunofluorescence (Figure 4H and Supplemental Figure 4, C and D).
Mechanisms involved in the clearance of vimentin by gigaxonin.
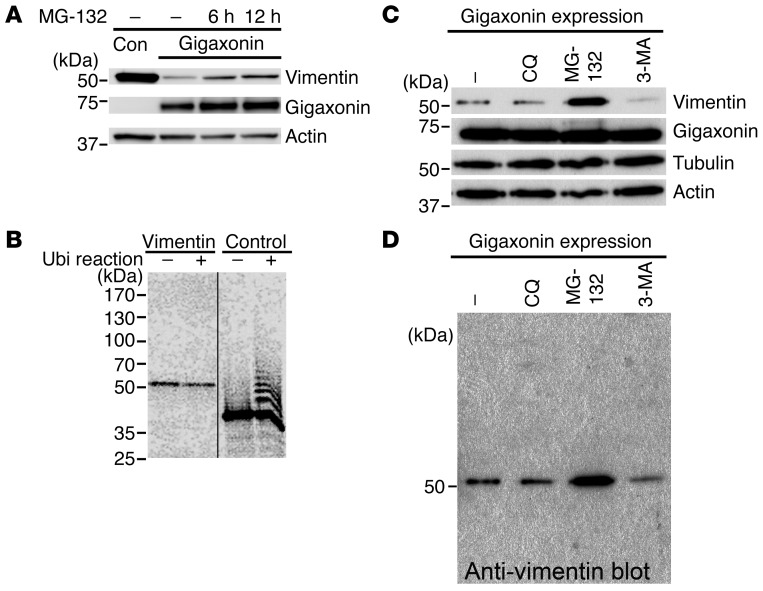
Based on the primary structure of gigaxonin, which predicts that it is an E3 ubiquitin ligase adaptor, and the finding that different types of IF proteins were degraded when gigaxonin was expressed in cells, we performed additional experiments to determine whether gigaxonin-mediated clearance of IF proteins involves the UPS. BJ5ta cells that had been cleared of vimentin by expression of WT gigaxonin for 72 hours were treated with MG-132 to inhibit proteasomes for 6–12 hours. Prior to the addition of MG-132, vimentin levels were barely detectable. Immunoblotting showed the appearance of increasing amounts of vimentin concomitant with 6–12 hours of proteasome inhibition (Figure 5A). These experiments were not carried out for the longer time periods required to fully restore vimentin levels, due to the significant deleterious effects of MG-132 on cell viability (37). As a further control, Gan–/– MEFs were also treated with MG-132 for 12 hours. Immunoblot analysis showed no detectable increase in vimentin (data not shown), which further suggests that gigaxonin is essential for vimentin degradation by proteasomes. These findings suggest that the degradation of vimentin involves the typical UPS pathway, and it is therefore expected that vimentin should be ubiquitinated. Our attempts to show vimentin ubiquitination used IP with anti-vimentin or anti-ubiquitin 36 hours after gigaxonin expression in BJ5ta cells. In addition, we carried out experiments using His-tagged ubiquitin in vivo followed by IP. These assays provided no evidence for vimentin ubiquitination in vivo (data not shown; see Methods). Therefore, we attempted to use an in vitro ubiquitination assay using reticulocyte lysates. Similarly, we could not detect ubiquitination of vimentin (Figure 5B).
Figure 5. Mechanism responsible for VIF clearance by gigaxonin.
(A) Immunoblotting of control and WT gigaxonin expressing BJ5ta cells treated with MG-132 using antibodies to vimentin, actin, or FLAG (gigaxonin). MG-132 treatment caused substantial recovery of vimentin in gigaxonin-expressing cells. Representative blots, 4 experiments. (B) In vitro ubiquitination assay showing ubiquitination ladder for the positive control sic 60–barnase–DHFR, but not for vimentin in the same preparation. Representative autoradiogram, 3 experiments. Lanes were run on the same gel but were noncontiguous (black line). (C) Immunoblotting of lysates of BJ5ta cells that had expressed gigaxonin for 72 hours and subsequently were treated with chloroquine (CQ; lysosome inhibitor), MG-132 (proteasome inhibitor), or 3-MA (autophagy inhibitor) for 12 hours. Blots were probed with antibodies to vimentin, actin, tubulin, or gigaxonin. The untreated cell lysate (–) contained very small amounts of vimentin. Vimentin recovery was seen only upon inhibition of proteasomes, not by inhibition of lysosomes or autophagy. Representative immunoblots, 4 experiments. (D) Longer exposure of the anti-vimentin full-length immunoblot in C, showing absence of a typical ubiquitinated protein ladder for vimentin. Representative blots from 4 experiments.
To determine whether other degradative pathways play a role in vimentin clearance, BJ5ta cells were cleared of vimentin by expression of WT gigaxonin for 72 hours. Parallel cultures of these cells were then separately treated for 12 hours with inhibitors for autophagy (3-methyl adenine; 3-MA) or lysosomal degradation (chloroquine) or with MG-132 as a control. Immunoblotting revealed the reappearance of vimentin only with MG-132 treatment (Figure 5C). It should also be noted that there was no evidence for the typical ladder of vimentin — which would indicate ubiquitination — when these same samples were immunoblotted with anti-vimentin (Figure 5D).
Discussion
Our results showed that the normal function of gigaxonin, a predicted E3 ligase adaptor protein, was to target certain IF proteins (e.g., vimentin, peripherin, and NF-L) for degradation. They also provided a framework for understanding the molecular basis of the formation of the large IF aggregates seen in different cell types of GAN patients. In both PNS and CNS, large axonal aggregates of IFs are the pathological hallmark of GAN. Although the exact mechanism behind the formation of these aggregates is not known, our results suggest that under normal conditions, gigaxonin regulates the degradation of IF proteins within axons. Therefore, when gigaxonin is defective, as in GAN, it leads to local accumulation of IFs along axons. Support for this possibility stems from the finding that the UPS is present along axons (e.g., ref. 38). Since GAN is a progressive disorder beginning at an early age with no detectable phenotype, it is likely that giant axons form slowly over prolonged periods beginning in early childhood and into the teenage years. The normal slow turnover of IF proteins (39–44) could therefore lead to the gradual accumulation of polymerized IFs into aggregates in the absence of normally functioning gigaxonin. Support for this possibility comes from our observation of GAN patient fibroblasts, which showed a passage-dependent increase in the number of cells containing VIF aggregates. The formation of large IF aggregates within axons could lead to altered axonal transport and progressive neuronal dysfunction, accounting for the hallmark features of the disease, particularly the early-onset neuropathy. In support of defective transport mechanisms, we found accumulation of mitochondria in close proximity to and within the IF aggregates. This most likely reflects the previously reported interactions, binding, and anchorage of mitochondria to both VIFs (45) and NF IFs (46). It is also important to note that the nervous system contains both neuronal and glial cells. Therefore the glial-specific IF protein GFAP, as well as VIFs in astrocytes (47), could also contribute to the GAN phenotype.
The finding that GAN was caused by mutations in a single gene suggests that gene replacement may be a successful therapeutic approach. Indeed, overexpression of WT gigaxonin did lead to the disappearance of VIF aggregates, as well as those containing peripherin and NF IFs. Although we did not test whether GFAP IFs are cleared in glial cells, it is also conceivable that the similar aggregates of this type III IF protein might be cleared by gigaxonin in patients with Alexander disease (2). However, as in any gene rescue therapy, one must carefully consider dosage, since high levels of gigaxonin expression can lead to a complete loss of the normal IF system in numerous cell types. Although the overexpression of gigaxonin has provided us with the most convincing evidence in support of its role in IF protein degradation, it is possible that excess expression of gigaxonin may have side effects that we have been unable to detect. We feel that this is unlikely, as the overexpression of gigaxonin for prolonged periods (up to a month) had no obvious effects on cell growth (data not shown). We also showed that overexpression of the S52G mutant of gigaxonin, at the same level as WT gigaxonin overexpression, had no effect on the organization of VIF networks, and there was no degradation of vimentin. These findings emphasize that the specific targets for gigaxonin are members of the cytoskeletal IF protein family. In further support of the specificity for IF cytoskeletal systems, we could detect no decreases in the level of either tubulin or actin when gigaxonin was expressed in cells. However, we did detect an increase in the amount of tubulin in GAN cells expressing defective gigaxonin, even though the overall organization of MTs appeared normal. Based on the well-established interactions between IFs and MTs, it is conceivable that alterations in IF protein turnover caused by gigaxonin mutations could stimulate tubulin expression by unknown feedback mechanisms (34, 48–50).
The finding that gigaxonin expression degraded IF proteins was consistent with their higher expression levels and IF aggregate formation previously seen in the differentiated tissues of adult Gan–/– mice (28, 29). However, the Gan–/– MEFs analyzed in the present study showed no difference in vimentin levels compared with WT MEFs. This discrepancy may be related to the early developmental stage at which the MEFs were obtained. For example, there could be less accumulation of IFs in embryonic fibroblasts relative to that seen in adult differentiated cells. The fact that human patients appear normal during their first few years of life also suggests that it takes a significant amount of time to accumulate IF aggregates within neurons that can cause clinically detectable neurological phenotypes. Further studies are required to understand the role and regulation of gigaxonin during early development and, in GAN patients, as a function of disease progression.
IFs have been shown to be composed of stable proteins with a half-life ranging from 18 hours to 8 days (39–42). However, very little is known about the pathways involved in their degradation in normal cells. Although our present results demonstrated that gigaxonin targets IF proteins for degradation via the UPS, the precise mechanism for proteasome targeting remains unknown. An insight regarding this mechanism comes from the observation that numerous nonfilamentous vimentin particles and short IFs appeared as the VIF network was being cleared by expressing gigaxonin. This finding suggests that long mature IFs are first disassembled into smaller structures, which have previously been identified as assembly/disassembly intermediates (51), before being degraded. However it is not known whether short IFs, unit length filaments (ULFs), tetramers, dimers, monomers, or other intermediates are targeted for degradation. The mechanism by which gigaxonin expression leads to VIF disassembly is also unknown.
It is conceivable that interactions between gigaxonin and vimentin participate in VIF disassembly, especially since our evidence suggests that this E3 ligase adaptor interacts with the highly conserved central rod domain, a region critically involved in regulating IF assembly into higher-order structures through the formation of coil-coiled interactions (52, 53). Subsequently, in its role as an E3 ligase adaptor, gigaxonin could target the smallest disassembly units for degradation. It is also possible that phosphorylation of vimentin may be involved in the process of its degradation by proteasomes, since the major known regulator of IF disassembly is phosphorylation (54–56). However, the state of vimentin phosphorylation in the presence of elevated gigaxonin remains unknown. Recent studies involving IF protein degradation in muscle atrophy provide additional insights into a possible relationship between phosphorylation and degradation of IF proteins (57). Specifically, desmin, the protein comprising the IF of skeletal muscle, was found to be phosphorylated, disassembled, and ubiquitinated by the E3 ligase TRIM32 and subsequently degraded (57). In contrast, our inability to detect ubiquitination of vimentin suggests that alternate mechanisms may be involved in targeting it to the proteasome. For example, gigaxonin may be involved in the ubiquitination of other proteins that bind to and target vimentin to the proteasome. In support of this possibility, it has previously been shown that direct target protein ubiquitination is not required for proteasomal degradation (58–61). Rather, an intermediate protein that is ubiquitinated binds to the target protein and escorts it to the proteasome for degradation. One of the best-studied examples of this type of pathway involves the human papilloma virus oncoprotein E7 and the degradation of its binding partner, the tumor-suppressor protein retinoblastoma (Rb) (62, 63). Therefore, Rb is not directly ubiquitinated, but is targeted to the proteasome by binding to ubiquitinated E7. A comparable mechanism may exist for the degradation of IF proteins mediated by gigaxonin. Nonetheless, the fact that gigaxonin is predicted to be an E3 ligase adaptor suggests that vimentin should become ubiquitinated. However, it is well known that ubiquitination of a protein is often difficult to detect for numerous technical reasons, including specific deubiquitinase activity (64). Therefore, further studies are required to determine whether vimentin is ubiquitinated.
Although it is not known how gigaxonin is regulated in normal cells, it has previously been shown that it is expressed at extremely low levels (∼7,500 molecules/cell; ref. 20). In contrast, IF proteins such as vimentin can represent 2%–3% or more of total cell protein (65), yet only a small fraction appears to be in a soluble form (66). These limiting amounts of gigaxonin may therefore only be capable of targeting this small “soluble” pool of vimentin, which can then be degraded by the proteasome. In addition, the levels of gigaxonin in cells may be regulated at the transcriptional level and/or by its turnover. Better understanding of how gigaxonin is regulated is an essential element of understanding its role in the degradation of VIFs and of peripherin and NF IF proteins.
In summary, we identified gigaxonin as the first E3 ligase adaptor that targets cytoskeletal IF proteins for degradation. Our findings provided insight into the mechanisms responsible for the degradation of several major types of IF proteins and provided a framework for understanding the mechanisms responsible for the progressive neurological disorder GAN. Based on these findings, it is possible that the abnormal regulation of gigaxonin may also be involved in the numerous other diseases known to involve accumulations of IF and/or their constituent proteins, such as Alzheimer’s disease and Parkinson’s disease (see Introduction and refs. 3, 7).
Methods
Cell culture.
BJ5ta human foreskin fibroblasts and NIH-3T3 mouse fibroblasts (ATCC) were maintained in DMEM (Invitrogen) supplemented with 10% calf serum (HyClone). MEFs from control or Gan–/– mice (28) were cultured in DMEM containing 10% FCS. GAN patient fibroblast cell lines included F07476 (containing a 57- to 131-kb microdeletion, encompassing at least exons 3–11, as well as p.Glu486Lys in exon 9), 08F699 (containing the mutations IVS6_1g_a and L510X), and F09133 (containing the mutations S52G and C393X) were obtained from R. Van Coster (Ghent University Hospital, Ghent, Belgium). These GAN cells were maintained in Medium 199 (Invitrogen) supplemented with 10% FCS. Control skin fibroblasts from a 10-year-old female subject (AG08470; Coriell Institute for Medical Research) were grown in DMEM containing 10% FCS. PC12 cells were grown as previously described (67), and SH-SY-5Y cells (ATCC) were grown in DMEM (Invitrogen) supplemented with 10% FCS. Cell lines expressing recombinant proteins were maintained in their respective culture medium supplemented with 2 μg/ml puromycin (Sigma-Aldrich). Vim–/– MEFs (gift of J.E. Eriksson and E. Torvaldson, Åbo Akademi University, Turku, Finland) were grown in DMEM containing 10% FCS. PC12 cells were differentiated with nerve growth factor (NGF; Invitrogen) as described previously (67–69). SH-SY-5Y cells were differentiated by culturing them in medium supplemented with 10 μM retinoic acid (RA; Sigma-Aldrich) for 1 week and then 10 μM RA plus 50 ng/ml brain-derived neurotrophic factor (BDNF; Sigma-Aldrich) for an additional week. Dorsal root ganglia (DRG) cultures were prepared from E13.5 embryos. DRGs were dissected and dissociated for 15 minutes at 37°C in 0.05% trypsin plus 0.53 mM EDTA (Invitrogen). After trypsinization, DRGs were rinsed with L-15 containing trypsin inhibitor (1 mg/ml; Sigma-Aldrich) and then triturated. Neurons were plated onto 4-well plates (LabTek) precoated with poly-d-lysine and extracellular matrix (Sigma-Aldrich) and maintained in neurobasal medium supplemented with B-27 supplement plus 0.5 mM glutamine and 15 ng/ml NGF. Culture media were supplemented with penicillin/streptomycin (Sigma-Aldrich), and all cell cultures were maintained at 37°C in a humidified CO2 incubator.
Antibodies and reagents.
Mouse anti-vimentin, rat and mouse anti–α-tubulin, mouse anti–β-tubulin, rabbit anti-gigaxonin, rabbit anti-FLAG, mouse anti–NF-L, rabbit anti-TBCB, and mouse anti-actin were obtained from Sigma-Aldrich. Mouse anti-GAPDH (Biolegend), chicken anti-vimentin (Covance), rabbit anti–vimentin 314, and rabbit anti–peripherin 199 have been described previously (54, 70). Secondary antibodies included Alexa Fluor 488–, Alexa Fluor 568–, FITC-, and rhodamine-conjugated goat anti-mouse, anti-chicken, and anti-rabbit IgG (Invitrogen). Secondary antibodies for immunoblotting included peroxidase-conjugated goat anti-mouse and anti-rabbit (Jackson ImmunoResearch). TRITC-conjugated phalloidin and Hoechst 33258 (Invitrogen) were used to stain actin and nuclei, respectively.
In some experiments, treatment with MG-132 (12 μM; Calbiochem; 10 or 20 mg/ml stock in DMSO) was used to inhibit proteasomes, chloroquine (150 μM; Sigma-Aldrich) was used to inhibit lysosome function, and 3-MA (5 mM; Sigma-Aldrich) was used to inhibit autophagy. Cells were exposed to each of these inhibitors for 6–12 hours. In the MG-132 experiments, control cells were treated with a comparable concentration of DMSO.
DNA constructs.
Plasmid vectors for eukaryotic expression of WT gigaxonin (pTL3.1-gigaxonin) or mutant gigaxonin (pTL3.1–293X gigaxonin and pTL3.1–R138H gigaxonin) were previously described (10). The lentiviral vector pLEX-MCS–FLAG-tagged WT gigaxonin was generated as follows. FLAG-tagged gigaxonin was amplified by PCR using primers 5′-CCCCCCACTAGTATGGACTACAAAGACGATGAC-3′ (forward) and 5′-CCCCCCGCGGCCGCTCAAGGGGAATGAACACGAA-3′ (reverse). The PCR product was then digested with SpeI and NotI and cloned into the pLEX-MCS backbone that was previously digested with the same restriction enzymes. The lentiviral vector expressing mutant FLAG-tagged S52G gigaxonin was prepared by introducing the 154A→G point mutation in the WT sequence of gigaxonin using the QuickChange Lightning Site-Directed Mutagenesis Kit (Stratagene). Lentiviral vectors coding for nontagged WT gigaxonin (pLEX-MCS gigaxonin) were prepared by digesting pTL3.1 with BamHI and NotI to release WT gigaxonin, then cloning it into the pLEX-MCS vector that had been double digested with BamHI and NotI.
FLAG-ΔC-VIM (vimentin with its non–α-helical C-terminal domain [aa 1–411] deleted), FLAG-ΔN-VIM (vimentin with its non–α-helical N-terminal domain [aa 103–466] deleted), and FLAG-Rod-VIM (vimentin rod domain; aa 103–411) were cloned into the lentiviral pLEX-MCS vector between BamHI and NotI sites using the InFusion HD Cloning System (Clontech). Each fragment was amplified using Phusion High Fidelity Polymerase, and the FLAG tag was included in the 5′ primer for each deletion fragment. FLAG-FL-VIM (full-length vimentin), FLAG-Head-VIM (vimentin head domain only [aa 1–102]), and FLAG-Tail-VIM (vimentin tail domain only [aa 411–466]) were amplified by PCR and cloned into pLEX-MCS vectors between BamHI and NotI sites. Individual clones were confirmed by sequencing.
Lentiviruses were produced according to the manufacturer’s instructions by cotransfecting plasmids pLEX-MCS (Thermo Scientific), along with the helper plasmids pVSVG and pAX2, into 293FT cells (Invitrogen) using Xfect Transfection Reagent (Clontech). Culture supernatant was collected 2 days later, and target cells were incubated with the viral supernatant supplemented with 8 μg/ml polybrene (Sigma-Aldrich) for 4–8 hours, after which the virus containing medium was replaced with the growth medium. This technique was used to establish 3T3 cell lines expressing FLAG-ΔN-VIM, FLAG-ΔC-VIM, and FLAG-rod-VIM constructs and Vim–/– cell lines expressing FLAG-ΔN-VIM, FLAG-ΔC-VIM, FLAG-rod-VIM, FLAG-Head-VIM, FLAG-Tail-VIM, and FLAG-FL-VIM constructs. In some cases, these lines were also infected with lentivirus expressing WT gigaxonin. Other lines used included BJ5ta, GAN 08F699, GAN F07476, 3T3, PC12, and SH-SY-5Y cells expressing WT gigaxonin and BJ5ta cells expressing S52G gigaxonin.
Immunofluorescence, image acquisition, and electron microscopy.
Cells were processed for indirect single and double immunofluorescence after fixation in methanol or 3.7% formaldehyde as previously described (34, 50). The fixed, immunostained cells were imaged using a Zeiss confocal LSM510 META (Carl Zeiss) microscope with oil immersion objective lenses (Plan-Apochromat, ×63 and ×100, 1.40 NA; Carl Zeiss) (54). Electron microscopy was performed as previously described (71).
SDS-PAGE and immunoblotting.
Cells grown in 60- or 100-mm culture dishes were lysed in Laemmli sample buffer, and proteins were separated by SDS-PAGE and immunoblotted as previously described (54, 72).
Image processing.
Image J (NIH) and Photoshop CS3 (Adobe Systems) were used for figure preparation. Immunoblots were quantified using Kodak molecular imaging network software (Kodak).
IP experiments.
Cell extracts for IP were prepared from BJ5ta cells expressing gigaxonin for 36 hours. 100-mm culture dishes were washed twice with ice-cold PBS and then lysed on ice in 600 μl RIPA buffer (1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate, 150 mM NaCl, 50 mM Tris-HCl, pH 8.0) containing protease (EDTA-free mini tablets; Roche) and phosphatase inhibitors (Cocktail sets I & II; Calbiochem). The lysates were centrifuged at 20,000 g for 30 minutes, and the supernatants were removed in order to determine protein concentration using the BCA protein assay kit (Pierce; Thermo Scientific). The clarified supernatant was subjected to IP with protein A sepharose 4 fast flow resin (Amersham Biosciences) according to the manufacturer’s instructions. Briefly, 20 μg rabbit anti-gigaxonin IgG or anti-vimentin IgG was allowed to bind to approximately 100 μg of cell extract proteins in 1.5 ml of 100 mM sodium phosphate buffer (pH 7.0) for 3 hours at 4°C and then mixed with protein A sepharose resin for another 16 hours at 4°C. For controls, 20 μg normal rabbit/mouse IgG was used. The resin was washed thoroughly, and samples were boiled in SDS-PAGE sample buffer. Mouse monoclonal anti-vimentin IgG or rabbit anti-gigaxonin IgG were used for immunoblotting after SDS-PAGE.
mRNA analysis.
Total RNA was prepared from cells using TRIzol (Invitrogen) and further purified to remove any contaminating DNA (RNeasy kit; Invitrogen). For quantitative RT-PCR, 1 μg total RNA was used to generate cDNA with the SuperScript III First-Strand kit (Invitrogen), and 1 μl of the total reaction volume was used for quantitative RT-PCR. All amplifications were performed on a Mastercyler EP Realplex (Eppendorf) using a master-mix containing SYBR Green and Rox (Applied Biosystem). RNA expression levels were analyzed using the following primers: VIM forward, 5′-AAGAGAACTTTGCCGTTGAA-3′; VIM reverse, 5′-GTGATGCTGAGAAGTTTCGT-3′; GAPDH forward, 5′-CTGCACCACCAACTGCTTAG-3′; GAPDH reverse, 5′-AGGTCCACCACTGACACGTT-3′; GAN forward, 5′-GGGTAGCGAGATGGTAACTTG-3′; GAN reverse, 5′-CGGATGGAAGGAGTGGTTTAG-3′.
The ΔΔCt method was used for relative quantification, with GAPDH expression levels serving as reference (73). Each sample was run in triplicate, and any Ct values that diverged 4 or more cycles were excluded for subsequent analyses. Data represent average ± SD from 3 independent experiments.
ELISA.
ELISA was carried out in 96-well Micro-test III assay plates (Falcon 3910; BD Labware) at room temperature on a Titer Plate shaker. Full-length vimentin and the vimentin rod domain (aa 103–411) were expressed in bacteria and purified as described previously (74). The wells of the microtiter plates were coated with 1 μg full-length vimentin or the rod domain in 100 μl PBS for 5 hours. The wells were then blocked 3 times with 0.05% Tween-20 in PBS (PBS-T; 300 μl/well) for 1 hour. To prepare cell extracts, FLAG-tagged gigaxonin expressing BJ5ta cells grown on a 100-mm dish were washed twice with 12 ml ice-cold Tris-buffered saline (TBS) and extracted with 1 ml PBS containing 1% Triton X-100, 0.5 M NaCl, 10 mM MgCl2, and protease inhibitors as above. The cell extracts (0–5 μg) in 100 μl of 0.5% BSA in PBS-T were overlaid and allowed to bind for 16 hours at room temperature. Wells were washed 3 times with PBS-T (300 μl/well) for 30 minutes, and then rabbit anti-FLAG IgG (125 μl of 0.5 μg/ml) in PBS-T was added and allowed to bind for 2 hours at room temperature. The wells were washed 3 times with PBS-T (300 μl/well) and incubated with 125 μl alkaline phosphatase–conjugated anti-rabbit IgG (1:5,000 dilution in PBS-T containing 0.5% BSA) for 1 hour. The plates were then washed 3 times with alkaline phosphatase buffer (100 mM Tris-HCl, 100 mM NaCl, 5 mM MgCl2, pH 9.5; 300 μl/well) for 30 minutes, and 100 μl of 1 mg/ml p–nitrophenylphosphate disodium in the alkaline phosphatase buffer was added to each well. The developed color was read in a MRX Revelation microplate reader at 405 nm (Dynex Corp.). For controls, cell extracts were overlaid on BSA-coated wells (75).
In vitro ubiquitination assay.
For substrate expression, we used the following constructs: pCDNA3-myc-VIM; pCDNA3–myc–Cullin-3 (Addgene catalog no. 19893; ref. 76); pET30A–WT gigaxonin. As a control, we used a construct encoding sic 60–barnase–DHFR fusion — consisting of 60 aas from the bacterial Sic1 protein (containing a PPXY Rsp5 ubiquitination motif); followed by a catalytically inactive (H102A) Bacillus amyloliquefaciens barnase, with all lysine replaced by arginine, methionine, or alanine residues; followed by E. coli dihydrofolate reductase (DHFR) — that has previously been used for ubiquitination and unfolding studies (77). Radioactive vimentin and control substrates (in pGEM3zf[+]) were expressed from a T7 promoter by in vitro transcription and translation using a TNT Coupled Reticulocyte Lysate system (Promega) or an E. coli T7 S30 Extract system (Promega) containing [35S]-methionine. The ubiquitination reaction mixture contained 100 nM UbE1, 200 ng UbcH5a (E2), 1 mg/ml ubiquitin, and 2 μM ubiquitin aldehyde (all from Boston Biochem); 20 mM creatine phosphate and 0.2 mg/ml creatine phosphokinase (both from Roche); 4 mM ATP (Sigma-Aldrich); and 1 μM DTT in ubiquitination buffer (25 mM Tris-Cl, pH 7.5; 50 mM NaCl; and 4 mM MgCl2), as described previously (78). In addition, in vitro–transcribed and –translated gigaxonin and Cullin-3 (supplemented with nonradioactive methionine) or Rsp5 (E3) were added to assay for the ubiquitination of vimentin or the control substrates (77, 78). The reaction was incubated at 30°C for 90 minutes, and then samples were placed in Laemmli sample buffer and analyzed by SDS-PAGE. Radioactive proteins were detected by autoradiography (Instant Imager; Packard).
In vivo ubiquitination assay.
In vivo ubiquitination was performed as described previously (79). In brief, HEK293T cells (100 mm) were cotransected with pCI-6xHis-Ub and FLAG-tagged gigaxonin or with FLAG-tagged gigaxonin and pCDNA3.1 (empty vector). 24 hours after transfection, cells were incubated with 10 μM MG-132 for 4 hours, then washed twice with PBS. Cell were extracted in 2 ml lysis buffer containing 50 mM Tris-HCl (pH 7.4), 0.25 M NaCl, 0.1% Triton X-100, 1 mM EDTA, 50 mM NaF, 1 mM DTT, 0.1 mM Na3VO4, inhibitor cocktail (Roche), 0.1 mM PMSF, and 2 mM N-ethylmaleimide (NEM; isopeptidase and deubiquitinase inhibitor). Next, 100 μl prewashed Talon magnetic beads (Clontech) were added to the cell extract to a total volume of 6 ml and rotated for 2 hours at 4°C. Beads were washed twice in lysis buffer followed by an additional wash with lysis buffer containing 10 mM imidazole. Next, protein was eluted with 250 mM imidazole in lysis buffer, added to Laemmli sample buffer, and loaded on SDS-PAGE for detection of vimentin by Western blot.
Statistics.
Data represent average ± SD from at least 3 independent experiments. 2-tailed Student’s t tests were carried out to assess the significance of the results. A P value of 0.01 or less was considered significant.
Study approval.
The use and maintenance of mice was in accordance with the Guide of Care and Use of Experimental Animals of the Canadian Council on Animal Care. The present studies were approved by the Comité de Protection des Animaux du Centre Hospitalier Universitaire de Québec (CPA-CHUQ), Québec, Canada.
Supplementary Material
Acknowledgments
We thank Kyunghee Myung for help with EM and Siqing Liu for help quantitating the IF aggregates in patient cells. This work was supported by NIH grants 1P01GM096971 (to R.D. Goldman) and R01 NS062051 (to P. Opal) and a grant from Hannah’s Hope Fund (to R.D. Goldman and P. Opal).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(5):1964–1975. doi:10.1172/JCI66387.
References
- 1.Denk H, Stumptner C, Zatloukal K. Mallory bodies revisited. J Hepatol. 2000;32(4):689–702. doi: 10.1016/S0168-8278(00)80233-0. [DOI] [PubMed] [Google Scholar]
- 2.Li R, Messing A, Goldman JE, Brenner M. GFAP mutations in Alexander disease. Int J Dev Neurosci. 2002;20(3–5):259–268. doi: 10.1016/S0736-5748(02)00019-9. [DOI] [PubMed] [Google Scholar]
- 3.Rudrabhatla P, Jaffe H, Pant HC. Direct evidence of phosphorylated neuronal intermediate filament proteins in neurofibrillary tangles (NFTs): phosphoproteomics of Alzheimer’s NFTs. FASEB J. 2011;25(11):3896–3905. doi: 10.1096/fj.11-181297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asbury AK, Gale MK, Cox SC, Baringer JR, Berg BO. Giant axonal neuropathy — a unique case with segmental neurofilamentous masses. Acta Neuropathol. 1972;20(3):237–247. doi: 10.1007/BF00686905. [DOI] [PubMed] [Google Scholar]
- 5.Berg BO, Rosenberg SH, Asbury AK. Giant axonal neuropathy. Pediatrics. 1972;49(6):894–899. [PubMed] [Google Scholar]
- 6.Pena SD. Giant axonal neuropathy: intermediate filament aggregates in cultured skin fibroblasts. Neurology. 1981;31(11):1470–1473. doi: 10.1212/WNL.31.11.1470. [DOI] [PubMed] [Google Scholar]
- 7.Perrot R, Eyer J. Neuronal intermediate filaments and neurodegenerative disorders. Brain Res Bull. 2009;80(4–5):282–295. doi: 10.1016/j.brainresbull.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Omary MB, Coulombe PA, McLean WH. Intermediate filament proteins and their associated diseases. N Engl J Med. 2004;351(20):2087–2100. doi: 10.1056/NEJMra040319. [DOI] [PubMed] [Google Scholar]
- 9.Bomont P, et al. The gene encoding gigaxonin, a new member of the cytoskeletal BTB/kelch repeat family, is mutated in giant axonal neuropathy. Nat Genet. 2000;26(3):370–374. doi: 10.1038/81701. [DOI] [PubMed] [Google Scholar]
- 10.Bomont P, Koenig M. Intermediate filament aggregation in fibroblasts of giant axonal neuropathy patients is aggravated in non dividing cells and by microtubule destabilization. Hum Mol Genet. 2003;12(8):813–822. doi: 10.1093/hmg/ddg092. [DOI] [PubMed] [Google Scholar]
- 11.Bomont P, et al. Identification of seven novel mutations in the GAN gene. Hum Mutat. 2003;21(4):446. doi: 10.1002/humu.9122. [DOI] [PubMed] [Google Scholar]
- 12.Peiffer J, Schlote W, Bischoff A, Boltshauser E, Muller G. Generalized giant axonal neuropathy: a filament-forming disease of neuronal, endothelial, glial, and schwann cells in a patient without kinky hair. Acta Neuropathol. 1977;40(3):213–218. doi: 10.1007/BF00691956. [DOI] [PubMed] [Google Scholar]
- 13.Treiber-Held S, Budjarjo-Welim H, Reimann D, Richter J, Kretzschmar HA, Hanefeld F. Giant axonal neuropathy: a generalized disorder of intermediate filaments with longitudinal grooves in the hair. Neuropediatrics. 1994;25(2):89–93. doi: 10.1055/s-2008-1071592. [DOI] [PubMed] [Google Scholar]
- 14.Kretzschmar HA, Berg BO, Davis RL. Giant axonal neuropathy. A neuropathological study. Acta Neuropathol. 1987;73(2):138–144. doi: 10.1007/BF00693779. [DOI] [PubMed] [Google Scholar]
- 15.Pena SD, Opas M, Turksen K, Kalnins VI, Carpenter S. Immunocytochemical studies of intermediate filament aggregates and their relationship to microtubules in cultured skin fibroblasts from patients with giant axonal neuropathy. Eur J Cell Biol. 1983;31(2):227–234. [PubMed] [Google Scholar]
- 16.Ben Hamida C, et al. Homozygosity mapping of giant axonal neuropathy gene to chromosome 16q24.1. Neurogenetics. 1997;1(2):129–133. doi: 10.1007/s100480050019. [DOI] [PubMed] [Google Scholar]
- 17.Cavalier L, et al. Giant axonal neuropathy locus refinement to a < 590 kb critical interval. Eur J Hum Genet. 2000;8(7):527–534. doi: 10.1038/sj.ejhg.5200476. [DOI] [PubMed] [Google Scholar]
- 18.Igisu H, Ohta M, Tabira T, Hosokawa S, Goto I. Giant axonal neuropathy. A clinical entity affecting the central as well as the peripheral nervous system. Neurology. 1975;25(8):717–721. doi: 10.1212/WNL.25.8.717. [DOI] [PubMed] [Google Scholar]
- 19.Yuan A, et al. Peripherin is a subunit of peripheral nerve neurofilaments: implications for differential vulnerability of CNS and peripheral nervous system axons. J Neurosci. 2012;32(25):8501–8508. doi: 10.1523/JNEUROSCI.1081-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cleveland DW, Yamanaka K, Bomont P. Gigaxonin controls vimentin organization through a tubulin chaperone-independent pathway. Hum Mol Genet. 2009;18(8):1384–1394. doi: 10.1093/hmg/ddp044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klymkowsky MW, Plummer DJ. Giant axonal neuropathy: a conditional mutation affecting cytoskeletal organization. J Cell Biol. 1985;100(1):245–250. doi: 10.1083/jcb.100.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leung CL, Pang Y, Shu C, Goryunov D, Liem RK. Alterations in lipid metabolism gene expression and abnormal lipid accumulation in fibroblast explants from giant axonal neuropathy patients. BMC Genet. 2007;8:6. doi: 10.1186/1471-2156-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabatelli M, et al. Peripheral neuropathy with giant axons and cardiomyopathy associated with desmin type intermediate filaments in skeletal muscle. J Neurol Sci. 1992;109(1):1–10. doi: 10.1016/0022-510X(92)90086-Z. [DOI] [PubMed] [Google Scholar]
- 24.Perez-Torrado R, Yamada D, Defossez PA. Born to bind: the BTB protein-protein interaction domain. Bioessays. 2006;28(12):1194–1202. doi: 10.1002/bies.20500. [DOI] [PubMed] [Google Scholar]
- 25.Xu L, et al. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature. 2003;425(6955):316–321. doi: 10.1038/nature01985. [DOI] [PubMed] [Google Scholar]
- 26.Furukawa M, He YJ, Borchers C, Xiong Y. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat Cell Biol. 2003;5(11):1001–1007. doi: 10.1038/ncb1056. [DOI] [PubMed] [Google Scholar]
- 27.Pintard L, Willems A, Peter M. Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. EMBO J. 2004;23(8):1681–1687. doi: 10.1038/sj.emboj.7600186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dequen F, Bomont P, Gowing G, Cleveland DW, Julien JP. Modest loss of peripheral axons, muscle atrophy and formation of brain inclusions in mice with targeted deletion of gigaxonin exon 1. J Neurochem. 2008;107(1):253–264. doi: 10.1111/j.1471-4159.2008.05601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ganay T, Boizot A, Burrer R, Chauvin JP, Bomont P. Sensory-motor deficits and neurofilament disorganization in gigaxonin-null mice. Mol Neurodegener. 2011;6:25. doi: 10.1186/1750-1326-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen E, et al. Gigaxonin-controlled degradation of MAP1B light chain is critical to neuronal survival. Nature. 2005;438(7065):224–228. doi: 10.1038/nature04256. [DOI] [PubMed] [Google Scholar]
- 31.Ding J, et al. Gene targeting of GAN in mouse causes a toxic accumulation of microtubule-associated protein 8 and impaired retrograde axonal transport. Hum Mol Genet. 2006;15(9):1451–1463. doi: 10.1093/hmg/ddl069. [DOI] [PubMed] [Google Scholar]
- 32.Wang W, et al. Gigaxonin interacts with tubulin folding cofactor B and controls its degradation through the ubiquitin-proteasome pathway. Curr Biol. 2005;15(22):2050–2055. doi: 10.1016/j.cub.2005.10.052. [DOI] [PubMed] [Google Scholar]
- 33.Yang HY, Lieska N, Goldman AE, Goldman RD. A 300,000-mol-wt intermediate filament-associated protein in baby hamster kidney (BHK-21) cells. J Cell Biol. 1985;100(2):620–631. doi: 10.1083/jcb.100.2.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prahlad V, Yoon M, Moir RD, Vale RD, Goldman RD. Rapid movements of vimentin on microtubule tracks: kinesin-dependent assembly of intermediate filament networks. J Cell Biol. 1998;143(1):159–170. doi: 10.1083/jcb.143.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herrmann H, et al. Structure and assembly properties of the intermediate filament protein vimentin: the role of its head, rod and tail domains. J Mol Biol. 1996;264(5):933–953. doi: 10.1006/jmbi.1996.0688. [DOI] [PubMed] [Google Scholar]
- 36.Rogers KR, et al. Truncation mutagenesis of the non-alpha-helical carboxyterminal tail domain of vimentin reveals contributions to cellular localization but not to filament assembly. Eur J Cell Biol. 1995;66(2):136–150. [PubMed] [Google Scholar]
- 37.Zanotto-Filho A, Braganhol E, Battastini AM, Moreira JC. Proteasome inhibitor MG132 induces selective apoptosis in glioblastoma cells through inhibition of PI3K/Akt and NFkappaB pathways, mitochondrial dysfunction, and activation of p38-JNK1/2 signaling. Invest New Drugs. 2012;30(6):2252–2262. doi: 10.1007/s10637-012-9804-z. [DOI] [PubMed] [Google Scholar]
- 38.Tsui CC, Pierchala BA. The differential axonal degradation of Ret accounts for cell-type-specific function of glial cell line-derived neurotrophic factor as a retrograde survival factor. J Neurosci. 2010;30(15):5149–5158. doi: 10.1523/JNEUROSCI.5246-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiu FC, Goldman JE. Synthesis and turnover of cytoskeletal proteins in cultured astrocytes. J Neurochem. 1984;42(1):166–174. doi: 10.1111/j.1471-4159.1984.tb09713.x. [DOI] [PubMed] [Google Scholar]
- 40.McTavish CF, Nelson WJ, Traub P. The turnover of vimentin in Ehrlich ascites tumour cells. FEBS Lett. 1983;154(2):251–256. doi: 10.1016/0014-5793(83)80159-8. [DOI] [PubMed] [Google Scholar]
- 41.Nixon RA, Logvinenko KB. Multiple fates of newly synthesized neurofilament proteins: evidence for a stationary neurofilament network distributed nonuniformly along axons of retinal ganglion cell neurons. J Cell Biol. 1986;102(2):647–659. doi: 10.1083/jcb.102.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barry DM, Millecamps S, Julien JP, Garcia ML. New movements in neurofilament transport, turnover and disease. Exp Cell Res. 2007;313(10):2110–2120. doi: 10.1016/j.yexcr.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 43.Brown A. Live-cell imaging of slow axonal transport in cultured neurons. Methods Cell Biol. 2003;71:305–323. doi: 10.1016/s0091-679x(03)01014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lasek RJ, Paggi P, Katz MJ. Slow axonal transport mechanisms move neurofilaments relentlessly in mouse optic axons. J Cell Biol. 1992;117(3):607–616. doi: 10.1083/jcb.117.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nekrasova OE, et al. Vimentin intermediate filaments modulate the motility of mitochondria. Mol Biol Cell. 2011;22(13):2282–2289. doi: 10.1091/mbc.E10-09-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner OI, Lifshitz J, Janmey PA, Linden M, McIntosh TK, Leterrier JF. Mechanisms of mitochondria-neurofilament interactions. J Neurosci. 2003;23(27):9046–9058. doi: 10.1523/JNEUROSCI.23-27-09046.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schnitzer J, Franke WW, Schachner M. Immunocytochemical demonstration of vimentin in astrocytes and ependymal cells of developing and adult mouse nervous system. J Cell Biol. 1981;90(2):435–447. doi: 10.1083/jcb.90.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gyoeva FK, Gelfand VI. Coalignment of vimentin intermediate filaments with microtubules depends on kinesin. Nature. 1991;353(6343):445–448. doi: 10.1038/353445a0. [DOI] [PubMed] [Google Scholar]
- 49.Helfand BT, Chang L, Goldman RD. Intermediate filaments are dynamic and motile elements of cellular architecture. J Cell Sci. 2004;117(pt 2):133–141. doi: 10.1242/jcs.00936. [DOI] [PubMed] [Google Scholar]
- 50.Yoon M, Moir RD, Prahlad V, Goldman RD. Motile properties of vimentin intermediate filament networks in living cells. J Cell Biol. 1998;143(1):147–157. doi: 10.1083/jcb.143.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Herrmann H, Strelkov SV, Burkhard P, Aebi U. Intermediate filaments: primary determinants of cell architecture and plasticity. J Clin Invest. 2009;119(7):1772–1783. doi: 10.1172/JCI38214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Herrmann H, Aebi U. Intermediate filaments: molecular structure, assembly mechanism, and integration into functionally distinct intracellular Scaffolds. Annu Rev Biochem. 2004;73:749–789. doi: 10.1146/annurev.biochem.73.011303.073823. [DOI] [PubMed] [Google Scholar]
- 53.Strelkov SV, et al. Conserved segments 1A and 2B of the intermediate filament dimer: their atomic structures and role in filament assembly. EMBO J. 2002;21(6):1255–1266. doi: 10.1093/emboj/21.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Helfand BT, et al. Vimentin organization modulates the formation of lamellipodia. Mol Biol Cell. 2011;22(8):1274–1289. doi: 10.1091/mbc.E10-08-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goto H, et al. Phosphorylation of vimentin by Rho-associated kinase at a unique amino-terminal site that is specifically phosphorylated during cytokinesis. J Biol Chem. 1998;273(19):11728–11736. doi: 10.1074/jbc.273.19.11728. [DOI] [PubMed] [Google Scholar]
- 56.Sihag RK, Inagaki M, Yamaguchi T, Shea TB, Pant HC. Role of phosphorylation on the structural dynamics and function of types III and IV intermediate filaments. Exp Cell Res. 2007;313(10):2098–2109. doi: 10.1016/j.yexcr.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cohen S, Zhai B, Gygi SP, Goldberg AL. Ubiquitylation by Trim32 causes coupled loss of desmin, Z-bands, and thin filaments in muscle atrophy. J Cell Biol. 2012;198(4):575–589. doi: 10.1083/jcb.201110067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Verma R, Deshaies RJ. A proteasome howdunit: the case of the missing signal. Cell. 2000;101(4):341–344. doi: 10.1016/S0092-8674(00)80843-0. [DOI] [PubMed] [Google Scholar]
- 59.Hoyt MA, Coffino P. Ubiquitin-free routes into the proteasome. Cell Mol Life Sci. 2004;61(13):1596–1600. doi: 10.1007/s00018-004-4133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jariel-Encontre I, Bossis G, Piechaczyk M. Ubiquitin-independent degradation of proteins by the proteasome. Biochim Biophys Acta. 2008;1786(2):153–177. doi: 10.1016/j.bbcan.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 61.Schrader EK, Harstad KG, Matouschek A. Targeting proteins for degradation. Nat Chem Biol. 2009;5(11):815–822. doi: 10.1038/nchembio.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gonzalez SL, Stremlau M, He X, Basile JR, Munger K. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J Virol. 2001;75(16):7583–7591. doi: 10.1128/JVI.75.16.7583-7591.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berezutskaya E, Bagchi S. The human papillomavirus E7 oncoprotein functionally interacts with the S4 subunit of the 26 S proteasome. J Biol Chem. 1997;272(48):30135–30140. doi: 10.1074/jbc.272.48.30135. [DOI] [PubMed] [Google Scholar]
- 64.Kaiser P, Tagwerker C. Is this protein ubiquitinated? Methods Enzymol. 2005;399:243–248. doi: 10.1016/S0076-6879(05)99016-2. [DOI] [PubMed] [Google Scholar]
- 65.Goldman RD, Grin B, Mendez MG, Kuczmarski ER. Intermediate filaments: versatile building blocks of cell structure. Curr Opin Cell Biol. 2008;20(1):28–34. doi: 10.1016/j.ceb.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soellner P, Quinlan RA, Franke WW. Identification of a distinct soluble subunit of an intermediate filament protein: tetrameric vimentin from living cells. Proc Natl Acad Sci U S A. 1985;82(23):7929–7933. doi: 10.1073/pnas.82.23.7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Helfand BT, Mendez MG, Pugh J, Delsert C, Goldman RD. A role for intermediate filaments in determining and maintaining the shape of nerve cells. Mol Biol Cell. 2003;14(12):5069–5081. doi: 10.1091/mbc.E03-06-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Helfand BT, Chang L, Goldman RD. The dynamic and motile properties of intermediate filaments. Annu Rev Cell Dev Biol. 2003;19:445–467. doi: 10.1146/annurev.cellbio.19.111401.092306. [DOI] [PubMed] [Google Scholar]
- 69.Kular RK, Cvetanovic M, Siferd S, Kini AR, Opal P. Neuronal differentiation is regulated by leucine-rich acidic nuclear protein (LANP), a member of the inhibitor of histone acetyltransferase complex. J Biol Chem. 2009;284(12):7783–7792. doi: 10.1074/jbc.M806150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parysek LM, Goldman RD. Characterization of intermediate filaments in PC12 cells. J Neurosci. 1987;7(3):781–791. doi: 10.1523/JNEUROSCI.07-03-00781.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Starger JM, Brown WE, Goldman AE, Goldman RD. Biochemical and immunological analysis of rapidly purified 10-nm filaments from baby hamster kidney (BHK-21) cells. J Cell Biol. 1978;78(1):93–109. doi: 10.1083/jcb.78.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Helfand BT, Mikami A, Vallee RB, Goldman RD. A requirement for cytoplasmic dynein and dynactin in intermediate filament network assembly and organization. J Cell Biol. 2002;157(5):795–806. doi: 10.1083/jcb.200202027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cvetanovic M, Rooney RJ, Garcia JJ, Toporovskaya N, Zoghbi HY, Opal P. The role of LANP and ataxin 1 in E4F-mediated transcriptional repression. EMBO Rep. 2007;8(7):671–677. doi: 10.1038/sj.embor.7400983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Correia I, Chu D, Chou YH, Goldman RD, Matsudaira P. Integrating the actin and vimentin cytoskeletons. adhesion-dependent formation of fimbrin-vimentin complexes in macrophages. J Cell Biol. 1999;146(4):831–842. doi: 10.1083/jcb.146.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Murthy SN, Lomasney JW, Mak EC, Lorand L. Interactions of G(h)/transglutaminase with phospholipase Cdelta1 and with GTP. Proc Natl Acad Sci U S A. 1999;96(21):11815–11819. doi: 10.1073/pnas.96.21.11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ohta T, Michel JJ, Schottelius AJ, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell. 1999;3(4):535–541. doi: 10.1016/S1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- 77.Kraut DA, et al. Sequence- and species-dependence of proteasomal processivity. ACS Chem Biol. 2012;7(8):1444–1453. doi: 10.1021/cb3001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24(19):8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bloom J, Pagano M. Experimental tests to definitively determine ubiquitylation of a substrate. Methods Enzymol. 2005;399:249–266. doi: 10.1016/S0076-6879(05)99017-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.