Abstract
NF-κB is a master regulator of inflammation and has been implicated in the pathogenesis of immune disorders and cancer. Its regulation involves a variety of steps, including the controlled degradation of inhibitory IκB proteins. In addition, the inactivation of DNA-bound NF-κB is essential for its regulation. This step requires a factor known as copper metabolism Murr1 domain–containing 1 (COMMD1), the prototype member of a conserved gene family. While COMMD proteins have been linked to the ubiquitination pathway, little else is known about other family members. Here we demonstrate that all COMMD proteins bind to CCDC22, a factor recently implicated in X-linked intellectual disability (XLID). We showed that an XLID-associated CCDC22 mutation decreased CCDC22 protein expression and impaired its binding to COMMD proteins. Moreover, some affected individuals displayed ectodermal dysplasia, a congenital condition that can result from developmental NF-κB blockade. Indeed, patient-derived cells demonstrated impaired NF-κB activation due to decreased IκB ubiquitination and degradation. In addition, we found that COMMD8 acted in conjunction with CCDC22 to direct the degradation of IκB proteins. Taken together, our results indicate that CCDC22 participates in NF-κB activation and that its deficiency leads to decreased IκB turnover in humans, highlighting an important regulatory component of this pathway.
Introduction
Copper metabolism Murr1 domain–containing (COMMD) proteins are a group of 10 evolutionarily conserved factors present in a wide range of organisms, including plants, protozoa, worms, insects, and vertebrates (1). COMMD1, the prototype member of the family, has been linked to a number of physiologic processes, including copper homeostasis (2–4), sodium balance (5–8), and adaptation to hypoxia (9, 10). COMMD1 has also been found to inhibit NF-κB (11, 12), a proinflammatory transcription factor that regulates close to 400 target genes that play essential roles in immune responses, immune system development, and cell survival and proliferation (13–15).
The underlying mechanism for the diverse functions of COMMD1 has not been fully elucidated, but in several instances, COMMD1 has been shown to promote the ubiquitination of specific cellular proteins (12). Recently, it was shown that COMMD1 and other COMMD family members interact with and regulate the activation of a class of ubiquitin ligases known as Cullin-RING ligases (CRLs) (16). CRLs are multiprotein complexes containing a Cullin family member as the main scaffold protein (Cul1, Cul2, Cul3, Cul4a, Cul4b, Cul5, and Cul7 in humans). To form the active ligase, each Cullin associates with a RING finger protein (Rbx1 or Rbx2) and any of various substrate binding partner proteins specific to each Cullin. This prolific group of enzymes accounts for more than 25% of all ubiquitin ligases in mammals and regulate diverse cellular processes, including cell cycle progression, DNA repair, and many signal transduction pathways, including NF-κB (17).
Activation of NF-κB is controlled by various ubiquitination events, including the critically important degradation of IκB, a constitutive inhibitor of this pathway (18). This step is mediated by Cul1 in association with β-transducin repeat–containing protein (βTrCP), which form the complex CRL1-βTrCP (also known as SCFβTrCP) (19–21). Under basal conditions, so-called “classical” IκB proteins (IκB-α, IκB-β, or IκB-ε) bind to NF-κB dimers and mask their nuclear localization sequence, keeping them inactive in the cytosol (22). IκB phosphorylation by the IκB kinase complex (IKK) generates a phospho-serine motif at the amino termini of classical IκB proteins. This motif is recognized by the F-box proteins βTrCP1 or βTrCP2, the substrate binding subunit of the CRL1-βTrCP ligase, leading to rapid ubiquitination and degradation of IκB (23).
Another CRL-regulated step in the NF-κB pathway is the degradation of chromatin-associated NF-κB subunits such as RelA (also known as p65), which plays an essential role in limiting gene expression (11, 12). This event is triggered by IKK-dependent phosphorylation of RelA (24, 25) and is mediated by a CRL2 complex that depends on COMMD1 for its activation (12, 16). Interestingly, while certain COMMD proteins, such as COMMD8 and COMMD10, bind to Cul1 (16), it was not previously known whether these factors promote the ubiquitination of CRL1 targets such as IκB.
In this study, we demonstrated that coiled-coil domain–containing protein 22 (CCDC22), a highly conserved protein recently implicated in X-linked intellectual disability (XLID) (26), is an associated factor that binds to all COMMD family members. CCDC22 was required for the ubiquitination and subsequent turnover of IκB proteins. Individuals with a hypomorphic mutation in CCDC22 demonstrated IκB stabilization and a blunted NF-κB response. These findings highlight a novel aspect in the activation of IκB ubiquitination and the control of NF-κB through CCDC22.
Results
COMMD proteins associate with CCDC22, a broadly expressed gene.
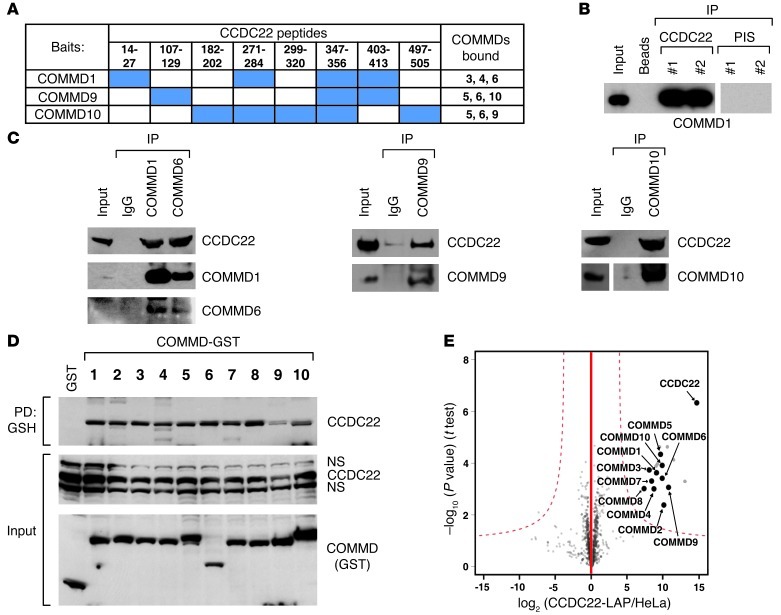
We hypothesized that, given their structural homology, COMMD proteins might assemble similar molecular complexes in vivo and that the identification and characterization of potential protein partners might provide insights into the mechanism of action of COMMD family members in general. In order to accomplish our goal, we began to systematically characterize protein complexes associated with COMMD proteins in vivo using tandem affinity purification (TAP). In these screens, 3 COMMD protein baits were used: COMMD1, COMMD9, and COMMD10. Consistent with the known ability of COMMD proteins to interact with each other (1), the TAP screens identified other endogenous COMMD proteins. Interestingly, these baits interacted with a specific and unique combination of COMMD partners: COMMD1 brought down COMMD3, COMMD4, and COMMD6, whereas COMMD9 and COMMD10 interacted with each other as well as with COMMD5 and COMMD6 (Figure 1A).
Figure 1. Identification of CCDC22 as a COMMD associated factor.
(A) TAP screen identification of CCDC22. CCDC22 peptides identified with high confidence in TAP screens using 3 different COMMD protein baits are indicated by blue shading. The specific COMMD proteins identified with each bait are shown at right. (B and C) Endogenous CCDC22 coimmunoprecipitated with endogenous COMMD proteins. (B) Endogenous CCDC22 was immunoprecipitated (IP) from HEK 293 cell lysates using 2 anti-CCDC22 antisera, and the recovered material was immunoblotted for COMMD1. Preimmune serum (PIS) or beads only were used as negative controls. (C) COMMD1, COMMD6, COMMD9, and COMMD10 were pulled down with polyclonal immune sera, and the precipitated material was immunoblotted for CCDC22. Some input lanes corresponded to different exposures of the same film. (D) CCDC22 associated with all COMMD family members. COMMD proteins fused to GST were expressed in HEK 293 cells and precipitated from Triton X-100 lysates. The recovered material was immunoblotted for endogenous CCDC22. PD, pulldown; NS, nonspecific band. (E) COMMD proteins were the main interaction partners of CCDC22. Volcano plot representation of CCDC22-interacting proteins. LAP-tagged CCDC22 was immunoprecipitated using an antibody directed against the tag. Nontransfected parental HeLa cells served as control. For each protein identified by mass spectrometry, the ratio of the intensities in the CCDC22 IPs over the control was calculated and plotted against the P value (2-tailed t test) calculated from triplicate experiments, both on a logarithmic scale. Dashed curves represent the cutoff, calculated based on a false discovery rate estimation. Specific interactors (top right) are indicated.
In addition, in all 3 screens, mass spectrometry analysis identified peptides that matched with high confidence to a protein of previously unknown function, termed CCDC22 (Figure 1A). Using deposited data available from BioGPS (27), we found that CCDC22 is broadly expressed in human tissues (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI66466DS1). Moreover, we examined the pattern of expression for the Ccdc22 ortholog in mouse tissues and confirmed similar findings at the mRNA level by quantitative real-time RT-PCR (qRT-PCR) and at the protein level by Western blot analysis (Supplemental Figure 1, B and C).
CCDC22-COMMD interactions were readily validated in endogenous coimmunoprecipitations using 2 antisera raised against CCDC22, which coprecipitated endogenous COMMD1 (Figure 1B). Reciprocal precipitations using antisera against COMMD1, COMMD6, COMMD9, and COMMD10 also coprecipitated endogenous CCDC22 (Figure 1C). Consistent with prior reports (1, 28), COMMD1 and COMMD6 were present in the same complex (Figure 1C). The interaction of CCDC22 with 4 COMMD family members suggested the possibility that other COMMD proteins might also interact with this factor. Given the paucity of characterized antibodies to other COMMD family members, we expressed COMMD proteins fused to glutathione S-transferase (GST) in mammalian cells and subsequently precipitated these proteins from cell lysates. All 10 COMMD proteins were able to coprecipitate endogenous CCDC22 to a similar extent (Figure 1D). Finally, a quantitative BAC-GFP interactomics experiment (29) using GFP-tagged CCDC22 as bait was also performed. For these experiments, CCDC22 was stably expressed in HeLa cells through the introduction of a BAC encoding GFP-tagged CCDC22 from its native locus (30). Again, all COMMD proteins were identified with high confidence as interacting partners of CCDC22 using 2 distinct affinity tag combinations (Figure 1E and Supplemental Figure 2), which confirmed that this protein is an associated factor of all COMMD family members.
CCDC22 regulates the cellular localization of COMMD proteins.
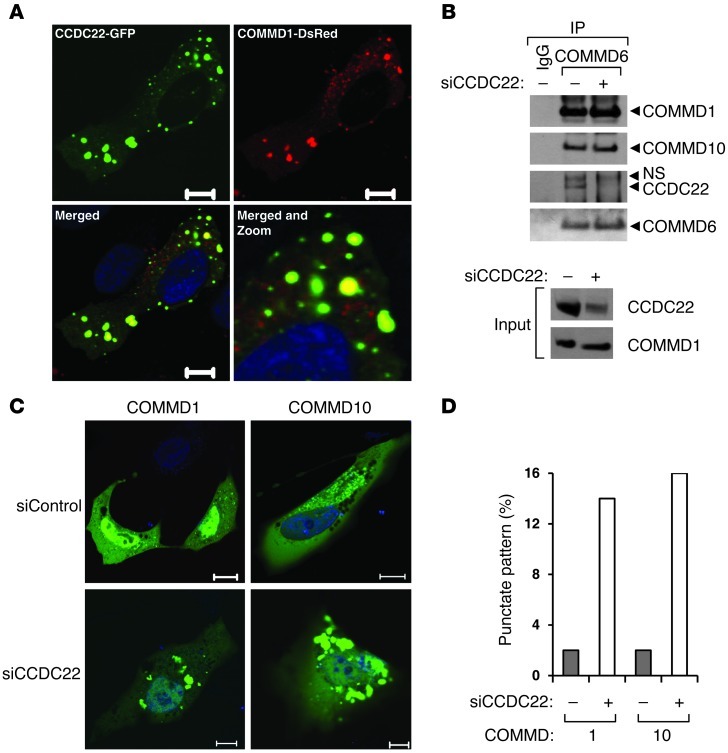
Next, we examined the cellular distribution of CCDC22. Using a yellow fluorescent protein (YFP) fusion protein, we found CCDC22 to be localized primarily in the cytosol, with a punctate perinuclear distribution, and with minor presence in the nucleus (Supplemental Figure 1D), in agreement with what is reported for the endogenous protein by the Protein Atlas project (31). Since the cellular distribution of CCDC22 was similar to the pattern displayed by several COMMD proteins (16), we also analyzed whether CCDC22 and COMMD1 could colocalize. Using GFP- and DsRed2-tagged versions of CCDC22 and COMMD1, respectively, we identified clear colocalization, while at the same time, some fraction of these proteins displayed exclusive cellular localization (Figure 2A). This colocalization is in agreement with the protein-protein interaction identified biochemically.
Figure 2. CCDC22 is required for proper cellular distribution of COMMD family proteins.
(A) Colocalization of CCDC22 with COMMD1. GFP-tagged CCDC22 and DsRed2-tagged COMMD1 were overexpressed in U2OS cells. Cells were counterstained with Hoechst and imaged by confocal microscopy. Scale bars: 10 μm. The merged view is also shown enlarged (merged and zoom; enlarged ×3-fold). (B) CCDC22 was not required for COMMD-COMMD interaction. HEK 293 cells were transfected with an siRNA targeting CCDC22 (siCCDC22) or a control duplex. After 48 hours, endogenous COMMD6 was immunoprecipitated, and the recovered material was immunoblotted for endogenous COMMD1 and COMMD10. (C and D) CCDC22 deficiency led to COMMD mislocalization. U2OS cells were transfected with YFP-tagged COMMD1 or COMMD10 together with siRNA against CCDC22 or a control duplex (siControl). (C) Nuclear counterstaining with Hoechst was performed just prior to confocal imaging. Scale bars: 10 μm. (D) The proportion of cells with a large perinuclear punctate pattern was determined by examining more than 100 cells per dish in a blinded manner.
In order to clarify the role of CCDC22 binding to all COMMD proteins, we examined whether CCDC22 regulates the ability of COMMD proteins to interact with each other. After CCDC22 silencing, the amount of COMMD1 or COMMD10 bound to endogenous COMMD6 was unaffected (Figure 2B), which suggests that these key protein-protein interactions are not controlled by CCDC22. Next, we evaluated whether CCDC22 plays a role in the cellular distribution of COMMD proteins. Silencing of CCDC22 dramatically changed the cellular localization of fluorescently tagged COMMD family members, inducing redistribution to large perinuclear punctate foci (Figure 2C). Quantitatively, CCDC22 deficiency led to a nearly 5-fold higher number of cells with YFP-tagged COMMD1 or COMMD10 aggregates (Figure 2D). These data indicate that CCDC22 regulates the cellular distribution of COMMD proteins, but is not required for COMMD-COMMD interactions.
The amino terminus of CCDC22 and the COMM domain are necessary and sufficient for interaction.
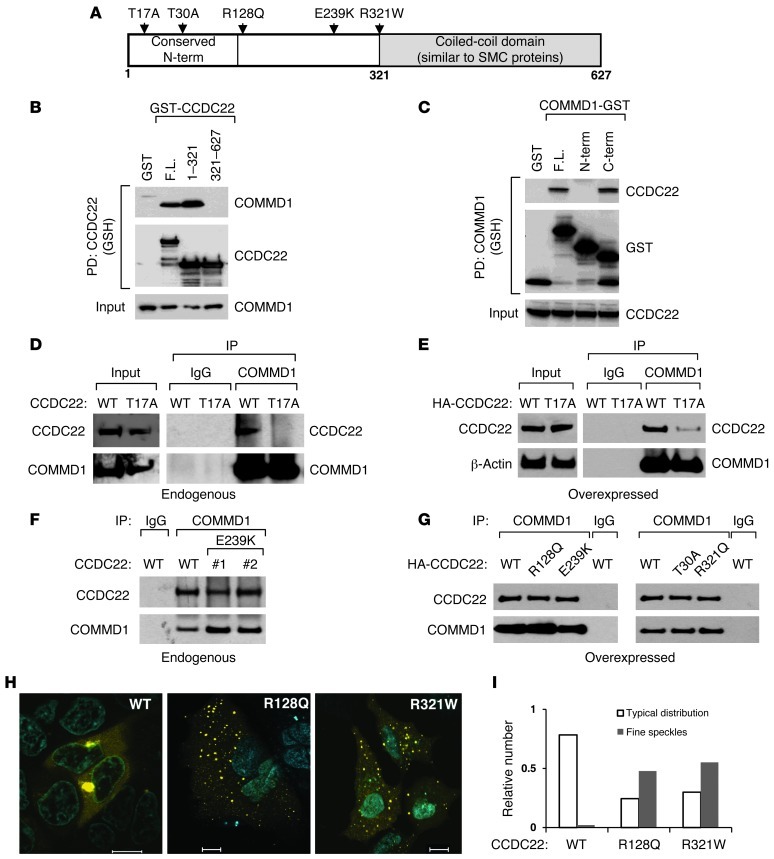
Homology analysis using the Conserved Domain Database (32) indicates that CCDC22 is highly conserved and a single ortholog is present in plants, protozoa, worms, insects, and vertebrates, a range of organisms similar to that in which COMMD genes are found. Notably, CCDC22 and COMMD orthologs are not evident in yeast or bacteria. The areas of highest homology among CCDC22 orthologs (Figure 3A) are their extreme amino termini (first ∼150 amino acids) and their carboxyl termini (∼350 amino acids). This latter region corresponds to a coil-coiled domain and bears similarity to structural maintenance of chromosomes (SMC) proteins, factors that bind to chromatin and are involved in meiosis and DNA repair (33). Further characterization of the CCDC22-COMMD1 interaction showed that COMMD1 bound to the amino terminus of CCDC22 (Figure 3B). In addition, the COMM domain of COMMD1, a carboxyterminal homology domain present in all COMMD proteins, was necessary and sufficient for CCDC22 binding (Figure 3C), in agreement with the binding of all COMM domain–containing proteins to CCDC22.
Figure 3. CCDC22-COMMD interactions and the effects of XLID-associated variants.
(A) Schematic representation of CCDC22. Conserved regions and the location of nonrecurrent sequence variants identified in XLID patients are displayed. (B and C) The amino terminus of CCDC22 and the COMM domain of COMMD1 were necessary and sufficient for binding. (B) Full-length (F.L.) and indicated domains of CCDC22 were expressed fused to GST, and their binding to endogenous COMMD1 was examined by coprecipitation. (C) Similar experiments were performed to detect coprecipitation of endogenous CCDC22 with full-length COMMD1 or its aminoterminal (N-term; amino acids 1–118) or carboxyterminal (C-term; amino acids 119–190) domains. (D and E) The XLID-associated mutation CCDC22 T17A impaired COMMD1 binding. (D) Coimmunoprecipitation between endogenous CCDC22 and COMMD1 was examined in LCLs derived from the kindred with the T17A mutation and a healthy control subject (WT). (E) Endogenous COMMD1 was similarly immunoprecipitated from HEK 293 cells expressing CCDC22 T17A or the WT. (F and G) Interaction of COMMD1 with other XLID-associated variants of CCDC22. (F) The ability of endogenous COMMD1 and CCDC22 E239K to interact was examined using available LCLs. (G) Interactions were examined by expressing HA-tagged CCDC22 proteins in HEK 293 cells. Immunoprecipitation of endogenous COMMD1 was followed by immunoblotting for HA-tagged CCDC22. (H and I) Abnormal cellular distribution of CCDC22 variants. (H) Distribution of YFP-tagged CCDC22 variants, determined by confocal microscopy. Scale bars: 10 μm. (I) Cellular distribution after examination of more than 100 cells per group in a blinded manner.
An XLID-associated CCDC22 mutation impairs COMMD binding.
Recently, a mutation in CCDC22 was identified in a family with XLID (OMIM 300859) and other developmental abnormalities (26). Affected members carry a single point mutation in CCDC22 (c.49A>G/p.T17A) that changes codon 17 from Thr to Ala, a highly conserved residue among vertebrates. It has been shown that this T17A mutation, by virtue of its close proximity to a splice site, results in abnormal splicing and diminished mRNA levels (26). Indeed, we found that EBV-immortalized B lymphocytes (lymphoblastoid cell lines; LCLs) from affected individuals had reduced CCDC22 transcript levels, whereas expression of forkhead box P3 (FOXP3), a gene in close proximity to CCDC22, was not affected (Supplemental Figure 3, A and B). Interestingly, the corresponding reduction in CCDC22 protein levels was relatively modest in LCLs (Figure 3D, input), and we therefore speculated that the T17A mutation may also result in functional alterations of the CCDC22 protein. Indeed, endogenous T17A was unable to bind to COMMD1 in lymphoid cells from affected individuals, a finding that was disproportionate to the small decrement in CCDC22 expression noted in these cells (Figure 3D). Similarly, T17A expressed in HEK 293 cells was defective in its ability to bind to endogenous COMMD1, even when both the WT and mutant CCDC22 proteins were expressed at comparable levels (Figure 3E).
Several other rare variants of CCDC22 were identified in additional families (Figure 3A). Although some of them, such as the E239K variant, have subsequently been found at a low frequency in the NHLBI exome sequencing project (12 of 8,627 genomes), the other variants appear to be unique to the XLID kindred. Therefore, we examined how these CCDC22 variants bind to COMMD1 using a coimmunoprecipitation approach. None of these variants seemed to affect COMMD1 binding when tested in LCLs or after expression in HEK 293 cells (Figure 3, F and G). Similarly, quantitative BAC-GFP interactomics experiments with T30A, R128Q, E239K, and R321W failed to disclose any changes in their binding to COMMD proteins (data not shown). However, 2 of the variants, R128Q and R321W, displayed substantial changes in their cellular localization patterns, demonstrating a fine speckled distribution in the cell that was different from the coarse dots noted when expressing the WT protein (Figure 3, H and I). Thus, independent of their binding to COMMD proteins, these 2 mutants demonstrated a functional impairment.
The T17A mutation or CCDC22 deficiency leads to blunted NF-κB activation.
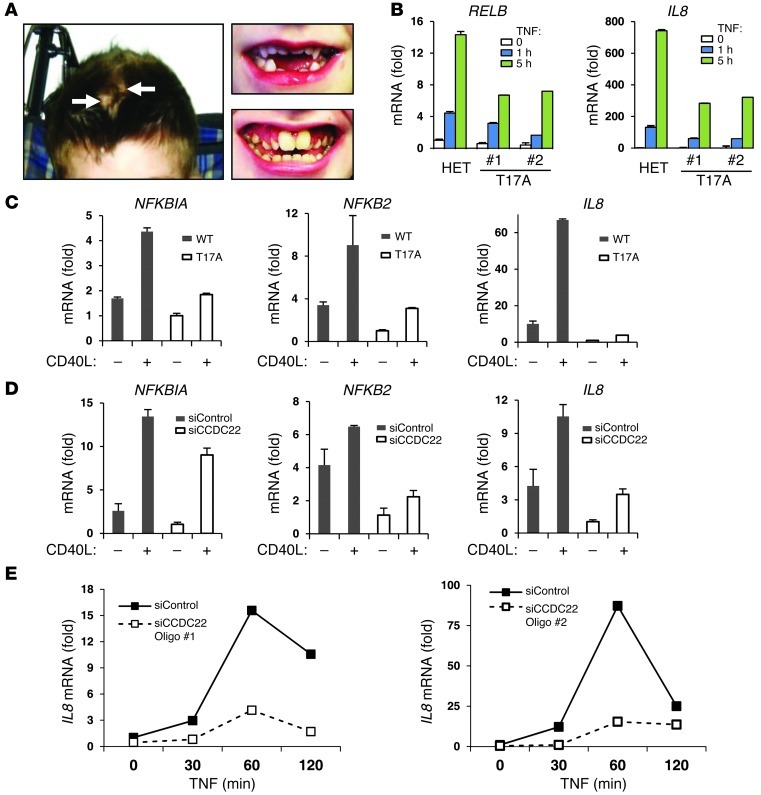
Given the role of COMMD1 in the regulation of NF-κB, we turned our attention to a possible role for CCDC22 in this pathway. Careful examination of the XLID kindred carrying the T17A mutation revealed that some affected individuals had aplasia cutis and markedly abnormal dentition (Figure 4A), 2 manifestations of ectodermal dysplasia. This congenital change can be found in individuals with hypomorphic mutations in NEMO (34, 35), which encodes an essential scaffold protein in the IKK complex. Ectodermal dysplasia can be similarly observed as a result of IκB-α mutations that alter its degradation (36), or in individuals with mutations in the ectodysplasin (EDA) pathway, a TNF-related signaling cascade that is involved in ectodermal development (37). Therefore, this clinical phenotype suggested a potential blockade of NF-κB activation during development (38).
Figure 4. CCDC22 is required for NF-κB activation.
(A) Aplasia cutis (left, arrows) and examples of abnormal dentition (right) in patients with the T17A mutation. (B) Fibroblasts from XLID patients displayed blunted activation of NF-κB–dependent genes. Primary dermal fibroblasts from patients demonstrated decreased TNF-induced activation of IL8 and RELB compared with their mother, a heterozygote mutation carrier (HET). (C) Response of NF-κB genes to CD40L activation was decreased in a LCL derived from an XLID patient. LCLs derived from an XLID patient or a healthy control were stimulated with CD40L. (D) CCDC22 deficiency phenocopied the T17A mutation. LCLs derived from the healthy control in C were transfected with the indicated siRNA oligonucleotides and subsequently stimulated with CD40L. (E) CCDC22 was required for activation of NF-κB–responsive genes in HEK 293 cells. 2 siRNA oligonucleotides targeting CCDC22 were used, and cells were subsequently stimulated with TNF. (B–E) Gene induction was evaluated in triplicate experiments by qRT-PCR; data represent mean and SEM.
Consistent with this, primary fibroblasts from 2 probands demonstrated decreased TNF-dependent activation of NF-κB target genes, such as IL8 and TNF-α–induced protein 3 (TNFAIP3; also known as A20), when compared with their heterozygous carrier mother (Figure 4B). Moreover, NF-κB–dependent gene expression in response to CD40 ligand (CD40L) stimulation was impaired in an LCL from an individual with the T17A mutation compared with a normal control LCL (Figure 4C). Similar to the effect of the T17A mutation, CCDC22 silencing led to impaired activation of NF-κB target genes in the control LCL (Figure 4D and Supplemental Figure 4). Finally, to exclude the possibility of an off-target effect of the siRNA experiments, these were repeated using 2 separate siRNA duplexes in HEK 293 cells. Again, in both instances, CCDC22 silencing led to decreased TNF-induced activation of NF-κB–dependent genes (Figure 4E and Supplemental Figure 5A). Conversely, CCDC22 overexpression led to greater IL8 activation in this system (Supplemental Figure 5B).
Given these effects on NF-κB activity, we examined whether CCDC22 was itself regulated by NF-κB in any way. We found that CCDC22 expression was not inducible by NF-κB activation, and its binding to COMMD1 was constitutive and not induced by TNF stimulation (Supplemental Figure 6, A and B). Together, these data indicated that CCDC22 deficiency, or a hypomorphic mutation in this gene that impairs the interaction with COMMD proteins, leads to blunted activation of NF-κB.
CCDC22 is required for IκB turnover.
The positive role that CCDC22 plays in NF-κB activation stands in contrast with the inhibitory function of COMMD1 in this pathway (1, 11, 12). Nevertheless, consistent with the interaction between these 2 proteins, we found that CCDC22 was present in a complex containing both Cul2 and COMMD1 (Supplemental Figure 7A), and, like COMMD1, CCDC22 could be inducibly bound to the NF-κB–responsive promoter of baculoviral IAP repeat containing 3 (BIRC3; also known as c-IAP2; Supplemental Figure 7B). Although these findings were consistent with the interaction between CCDC22 and COMMD1, they failed to explain CCDC22’s inhibitory role on NF-κB. Therefore, we hypothesized that this factor must have an additional site of action that is COMMD1 independent.
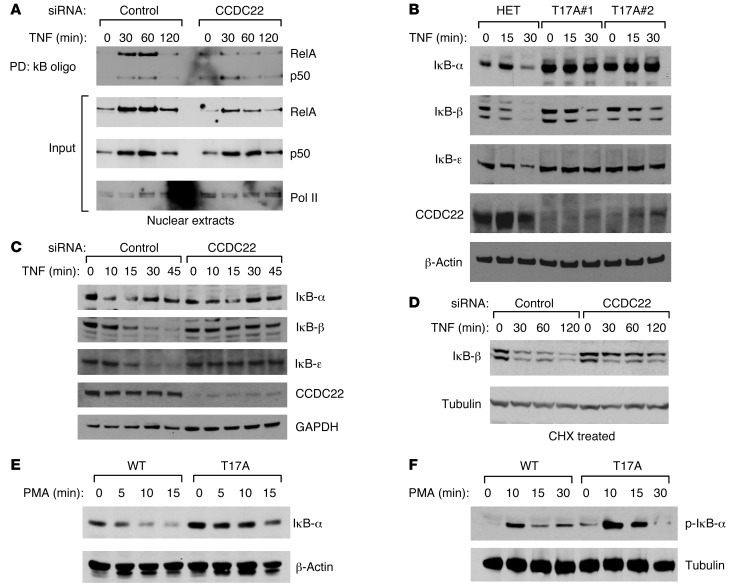
To test this possibility, we examined the effects of CCDC22 silencing on NF-κB nuclear accumulation. CCDC22 silencing led to a reduction in nuclear translocation of NF-κB after cell stimulation, as shown by DNA-protein coprecipitation using oligonucleotides containing tandem κB sites or by direct Western blotting of nuclear extracts (Figure 5A). This finding suggested a possible blockade in IκB degradation, akin to that observed in hypomorphic NEMO mutations (35). This possibility was examined in a variety of systems. First, primary dermal fibroblasts from individuals with the T17A mutation displayed markedly delayed or absent IκB degradation compared with cells derived from their mother, a heterozygous carrier (Figure 5B). All 3 classical IκB proteins were affected, and the basal level of IκB-α was also notably elevated. In addition, CCDC22 protein levels were more significantly reduced in mutant fibroblasts than in LCLs (compare Figure 3D and Figure 5B), highlighting the variable effect of the mutation on mRNA splicing and gene expression (26). Experiments using siRNA in HEK 293 cells demonstrated a similar result, although in these cells, the effects were most dramatic for IκB-β and IκB-ε (Figure 5C). Moreover, the changes in IκB-β turnover proved to result from altered protein stability, not from alterations in mRNA expression. Using cycloheximide (CHX) to block new protein synthesis, we found that TNF-induced degradation of IκB-β was markedly delayed by RNAi against CCDC22 (Figure 5D). Finally, LCLs from individuals with the T17A mutation also displayed reduced degradation of IκB-α after PMA or CD40L stimulation (Figure 5E and Supplemental Figure 8A). Less dramatic changes in IκB-β degradation at later time points were also evident in this LCL model (Supplemental Figure 8B).
Figure 5. CCDC22 is required for RelA nuclear transport and IκB degradation.
(A) CCDC22 deficiency resulted in depressed nuclear accumulation of active NF-κB. HEK 293 cells were transfected with the indicated siRNAs and stimulated with TNF. The presence of active NF-κB complexes in nuclear extracts was assessed by a DNA-protein coprecipitation assay (top) or by direct immunoblotting (input, bottom). RNA polymerase II (Pol II) served as a loading control. (B) TNF-induced degradation of classical IκB proteins was impaired in primary fibroblasts bearing the T17A mutation. Primary dermal fibroblasts from 2 patients demonstrate decreased TNF-induced degradation of IκB-α, IκB-β, and IκB-ε compared with fibroblasts from their mother, a heterozygote mutation carrier. (C) CCDC22 deficiency impaired TNF-induced IκB degradation. HEK 293 cells were transfected with the indicated siRNA oligonucleotides and treated with TNF. IκB degradation was determined by Western blot analysis. (D) CCDC22 deficiency affected IκB-β stability. Cells transfected with the indicated siRNA were subsequently treated with cycloheximide (CHX) to inhibit new protein synthesis. IκB-β stability after TNF stimulation was examined by immunoblotting. (E) IκB-α degradation was impaired in XLID-derived LCLs. LCLs derived from a healthy control subject and an XLID patient with the T17A mutation were stimulated with PMA, and IκB-α degradation was examined by immunoblotting. (F) IκB-α phosphorylation was not affected in XLID-derived LCLs. LCLs derived from a healthy control and an XLID patient were stimulated with PMA for indicated times. Phosphorylation of IκB-α at serines 32 and 36 was determined by immunoblotting using a phosphospecific antibody.
In all instances, the impaired degradation of IκB-α was not associated with reduced IκB-α phosphorylation (Figure 5F and Supplemental Figure 8C), which indicates that the blockade in degradation was not caused by impaired IKK function. In fact, phospho–IκB-α levels were higher in mutant cells, which indicates that the reduced IκB degradation is caused by an event that occurs after phosphorylation. Thus, these data suggest that impaired CCDC22 activity leads to decreased degradation of phosphorylated IκB.
CCDC22 is required for IκB ubiquitination.
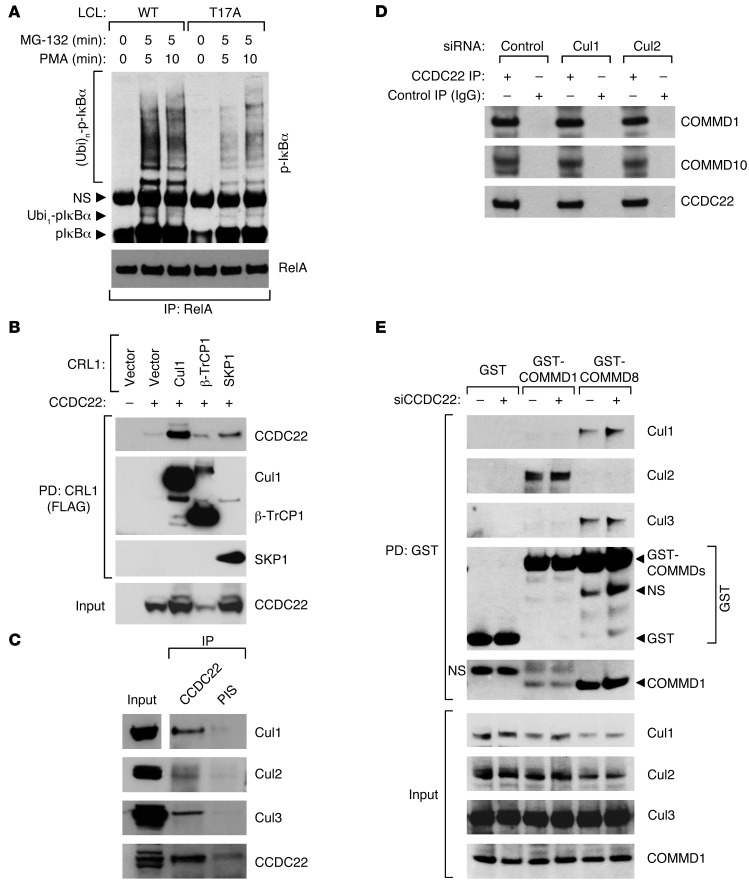
Based on the above findings, we reasoned that the ubiquitination of phosphorylated IκB proteins might be regulated by CCDC22. Indeed, in an LCL from a T17A-affected patient, we noted marked reductions in endogenous IκB-α ubiquitination after PMA stimulation (Figure 6A). IκB ubiquitination in response to this signal is carried out by CRL1-βTrCP, a Cul1-containing complex. As noted previously, certain COMMD proteins — most notably COMMD8 and COMMD10 (16) — can bind to Cul1, which suggests the possibility that CCDC22 might interact with this complex as well. Indeed, CCDC22 coprecipitated various components of this multimeric ligase when expressed in HEK 293 cells (Figure 6B). Moreover, precipitation of endogenous CCDC22 led to the recovery of endogenous Cul1, Cul2, and Cul3 (Figure 6C). Finally, CCDC22 was able to interact with other Cullin family members (Supplemental Figure 9A), in line with similar observations for COMMD family members (16). Together, these data demonstrated that CCDC22 interacts with the ligase that targets IκB proteins for degradation and is required for its ability to ubiquitinate this substrate in vivo.
Figure 6. CCDC22 is required for IκB ubiquitination.
(A) IκB-α ubiquitination is reduced in lymphoid cells bearing the T17A mutation. LCLs derived from a healthy control subject and an XLID patient with the T17A mutation were stimulated with PMA. The protease inhibitor MG-132 was concurrently administered. Ubiquitinated phospho–IκB-α levels were determined by immunoprecipitating NF-κB complexes with a RelA antibody, followed by phospho–IκB-α immunoblotting. (B) CCDC22 interacted with various CRL1-βTrCP components. FLAG-tagged Cul1, βTrCP1, or SKP1 were expressed along with CCDC22 in HEK 293 cells. CRL components were immunoprecipitated using a FLAG antibody, and CCDC22 was detected in the recovered material by immunoblotting. (C) Endogenous CCDC22 interacted with Cul1, Cul2, and Cul3. CCDC22 was immunoprecipitated, and the recovered material was immunoblotted for endogenous Cul1, Cul2, and Cul3. Some input lanes corresponded to different exposures of the same film. (D) Cul1 and Cul2 were not required for CCDC22-COMMD interaction. HEK 293 cells were treated with siRNA against Cul1, Cul2, or an irrelevant control. Endogenous CCDC22 was subsequently immunoprecipitated, and the recovered material was immunoblotted for endogenous COMMD1 and COMMD10. (E) CCDC22 was not required for Cullin-COMMD interaction. HEK 293 cells were transfected with GST, GST-COMMD1, or GST-COMMD8 along with control or anti-CCDC22 siRNA. After 48 hours, cell lysates were purified on GST-agarose, and the recovered material was immunoblotted for endogenous Cul1, Cul2, Cul3, and COMMD1.
The ability of both CCDC22 and COMMD family members to interact with CRLs led us to examine whether CRLs play a role in the interactions between CCDC22 and COMMD proteins. Silencing of Cul1 or Cul2 did not affect CCDC22-COMMD1 or CCDC22-COMMD10 interactions in coimmunoprecipitation experiments (Figure 6D and Supplemental Figure 9B). Conversely, we next evaluated whether CCDC22 could play a role in the interaction between COMMD proteins and CRLs. The binding between COMMD1 or COMMD8 and their respective preferential Cullin partners was not substantially affected by CCDC22 silencing (Figure 6E and Supplemental Figure 9C). This experiment also served as a further demonstration that COMMD1 binds preferentially to Cul2, whereas COMMD8 binds better to Cul1 and Cul3. Thus, these data suggested that despite the ability of COMMDs, CCDC22, and Cullin to form a triple complex, none of them seems to play a scaffold role.
COMMD8 is also required for IκB turnover and NF-κB activation.
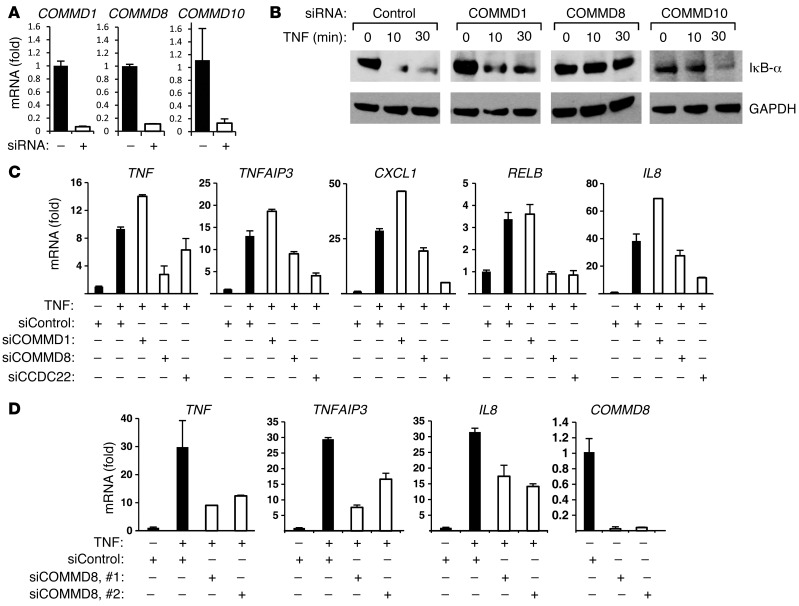
Our results indicated that CCDC22 promotes IκB ubiquitination and degradation, and we reasoned that this effect might be mediated by CCDC22’s interaction with another COMMD family member or might be COMMD independent. To identify a possible COMMD protein that might be similarly involved in the degradation of IκB, we focused our attention on COMMD family members that interact with Cul1 (the central subunit in the CRL1-βTrCP complex), such as COMMD8 and COMMD10 (16). COMMD8 repression led to decreased IκB-α degradation, an effect not seen after COMMD1 or COMMD10 RNAi (Figure 7, A and B). Similarly, COMMD8 or CCDC22 silencing led to decreased activation of several NF-κB target genes, while an opposite effect was seen after COMMD1 RNAi (Figure 7C). These observations were recapitulated using 2 independent RNAi duplexes targeting COMMD8 (Figure 7D), which indicates that this is unlikely to be an off-target effect. Together, these data suggested that a CCDC22-COMMD8 complex participates in the activation of CRL1-βTrCP and is required for optimal IκB degradation and subsequent activation of NF-κB target genes.
Figure 7. COMMD8, a partner of CCDC22, is also required for IκB degradation.
(A and B) COMMD8 deficiency impaired TNF-induced IκB-α degradation. (A) HEK 293 cells were transfected with the indicated siRNA oligonucleotides, and the effectiveness of the silencing was determined by qRT-PCR. (B) In parallel, cells were stimulated with TNF, and IκB-α degradation was examined by immunoblotting. (C and D) COMMD8 was required for NF-κB–responsive gene expression. (C) HEK 293 cells were transfected with the indicated siRNA oligonucleotides and subsequently treated with TNF. Induction of several NF-κB–responsive genes was examined. (D) The findings in C were recapitulated using 2 distinct siRNA oligonucleotides against COMMD8. (A, C, and D) Gene induction was evaluated in triplicate experiments by qRT-PCR; data represent mean and SEM.
Discussion
In this study, we identified CCDC22 as a novel interacting protein that binds avidly to COMMD family members. Our data indicate that each COMMD protein, from COMMD1 to COMMD10, interacts with CCDC22. However, it is unlikely that this occurs in a single complex containing CCDC22 and all 10 COMMD family members at the same time. Rather, we postulate that different and distinct CCDC22-COMMD complexes exist in vivo. This is based on previously published studies indicating that COMMD proteins form dimers (1, 39) and on our mass spectrometry data demonstrating that only certain COMMD combinations were present in vivo (Figure 1A). Thus, based on the available data, we conclude that CCDC22 plays a critical role in controlling the cellular distribution of these complexes (Figure 2), and in so doing, it seems to be necessary for their normal function. We postulate that the specific composition of COMMD proteins present in any given CCDC22-COMMD complex, as well as the specific CRLs to which they bind, ultimately determine their unique functions. In addition, we speculate that CCDC22 and COMMD proteins work in concert to regulate these ligases, probably acting as a complex to displace the CRL inhibitor CAND1, as recently reported in the case of COMMD1 (16).
Since CRLs are involved in a vast array of cellular processes, we anticipate that CCDC22 will have pleiotropic effects in multiple pathways besides the effects on NF-κB transcription observed here. Indeed, the complex phenotype noted in individuals carrying the hypomorphic T17A mutation indicates that CCDC22 plays an important developmental role in the nervous system and beyond. In this regard, it is noteworthy that Cul4b has been similarly linked to XLID, and CCDC22 may therefore also be involved in the regulation of CRL4B targets that are important for neuronal biology (40). Other rare variants in this gene were also noted in families with XLID; in the case of 2 of these variants, R128Q and R321W, we demonstrated a clear functional alteration of the mutant proteins, which were mislocalized in the cell. The mechanism for this abnormal localization was not mediated by altered COMMD binding, but may involve other protein-protein interactions that remain to be elucidated. With regard to the T30A and E239K variants, no functional effects were identified by our studies; with respect to E239K, we speculate that this may be a rare but functionally normal protein, since it can be found in the NHLBI exome database. Ultimately, we anticipate that additional mutations in this gene will be uncovered in the context of XLID and other X-linked developmental disorders.
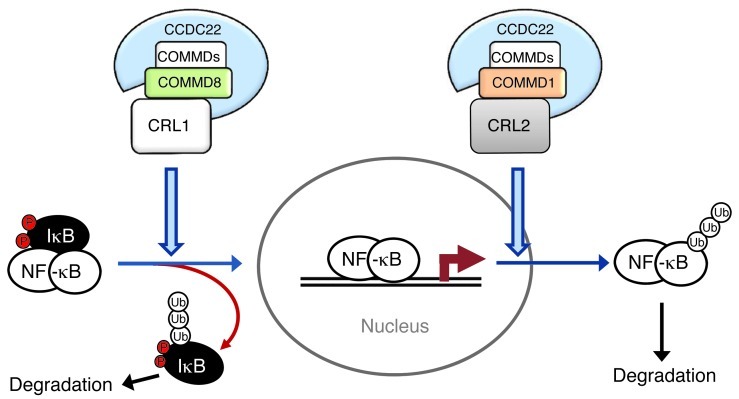
Our data suggested that in the NF-κB pathway, a CCDC22-COMMD8 complex plays an important role in IκB turnover and NF-κB activation through its interaction with Cul1, the ligase that targets IκB (21). However, we conversely found that CCDC22-COMMD1 complexes bound preferentially to Cul2, a ligase involved in NF-κB/RelA ubiquitination and removal of chromatin-bound NF-κB (12, 25). With these dual functions, CCDC22 deficiency or mutation would impair the activity of both COMMD1 and COMMD8 complexes, but the lack of IκB degradation, an upstream and initial step in this pathway, had a dominant role and was responsible for the impaired NF-κB activation described herein (Figure 8). The role of CCDC22 in IκB degradation is supported by ample analysis of cells derived from individuals with the hypomorphic T17A mutation. Moreover, our concordant findings after RNAi-induced silencing of CCDC22 confirmed the role of this factor in NF-κB activation. In agreement with these observations, some of the individuals with XLID affected by the T17A mutation displayed ectodermal dysplasia, a congenital change that can result from blunted NF-κB activation downstream of EDAR, a member of the TNF receptor superfamily (37). Moreover, NF-κB plays an important role in neuronal function and learning processes (41, 42) and is similarly important for myelination and Schwann cell function (43). Therefore, the alterations in the NF-κB pathway seen in these patients may contribute to their neurologic phenotype.
Figure 8. Role of CCDC22-COMMD complexes in NF-κB pathway regulation.
With respect to the role of CCDC22 in immune function, our observations were mainly restricted to 6 individuals with the T17A mutation, with only 2 of them being of adult age at this point. Nevertheless, increased infections, autoimmunity, or unusual malignancies have not been noted thus far. This may indicate that chronic CCDC22 deficiency is better compensated in vivo than in isolated culture systems or that this hypomorphic mutation is not severe enough to result in an obvious immune phenotype in children. Alternatively, the immune defects in vivo may be restricted to selective microorganisms; this has been observed with important immune regulators, such as TRIF, for which inactivating mutations lead to a narrow and specific susceptibility to herpes encephalitis (44). If this were the case here, a larger patient cohort and longer-term follow-up may be needed to fully comprehend the immune phenotypes of CCDC22 mutations. Nevertheless, it is interesting to note that a genetic study has indicated that single nucleotide polymorphisms in the CCDC22 gene affect the risk for allergic rhinitis (45). However, because the CCDC22 gene is located in the complementary strand and in close proximity to FOXP3, a major regulator of immune function, it is possible that such polymorphisms may be more relevant to the function of FOXP3. In any event, it is important to note that the functional analysis of CCDC22 presented here, including various RNAi experiments, did not reflect FOXP3 function, and that the T17A mutation did not affect FOXP3 expression (Supplemental Figure 3B). Finally, the high level of CCDC22 expression observed in the immune system, particularly in myeloid and T cells (Supplemental Figure 1A), suggests an important immunological role, in keeping with the NF-κB regulatory function reported here.
The involvement of CCDC22 and COMMD8 in the ubiquitination of IκB represents a novel aspect in the regulation of this critical pathway that might be amenable to therapeutic manipulation. Disrupting CCDC22-COMMD interactions in a manner akin to the effect of the T17A mutation should result in impaired IκB degradation and NF-κB blockade, an effect that would be desirable in certain contexts, such as chronic inflammatory disorders or specific cancers.
Methods
Plasmids and siRNAs.
Expression vectors for CCDC22 (pEBB-FLAG-CCDC22, pEBB-HA-CCDC22, pEBB-YFP-CCDC22, pEBB-GFP-CCDC22, and pEBG-CCDC22) were generated by PCR amplification of the coding sequence using IMAGE clone 3449051 as template. Expression vector for βTrCP1 (pEBB-FLAG-βTrCP1) was generated by PCR amplification of the coding sequence using as template a plasmid obtained from Y. Ben-Neriah (Hebrew University of Jerusalem, Jerusalem, Israel). Deletion constructs for CCDC22 were similarly generated by PCR, with the amino acid boundaries of the encoded mutant proteins being 1–320 and 321–627. Point mutations T17A, T30A, R128Q, E239K, and R321W were introduced into pEBB-HA-CCDC22 by site-directed mutagenesis (Stratagene). The pEBB-2×HA-TB vector used for TAP screening was constructed using complementary oligonucleotides coding tandem HA tags, which were inserted into the BamHI site of pEBB-TB. Subsequently, pEBB-2×HA-COMMD9-TB and pEBB-2×HA-COMMD10-TB were constructed by subcloning the COMMD9 and COMMD10 sequences from the corresponding pEBB-FLAG vectors. All other plasmids used were described previously (1, 3, 16, 25, 28, 46–48). See Supplemental Table 1 for target sequences of the siRNA duplexes used.
Cell culture and transfection.
HEK 293, HeLa, and U2OS cell lines were obtained from ATCC and cultured in DMEM supplemented with 10% FBS and l-glutamine. A standard calcium phosphate transfection protocol was used to transfect plasmids and siRNA in HEK 293 cells (48); the Fugene transfection system (Invitrogen) was used for HeLa and U2OS cells. Patient-derived cells were obtained after IRB approval and informed consent. LCLs were generated by immortalization of peripheral lymphocytes with EBNA as previously described (49). Primary human fibroblasts of XLID patients and their mother, a heterozygote carrier of the CCDC22 mutation, were obtained from skin punch biopsies. LCLs and primary fibroblasts were cultured in RPMI 1640 (Cellgro, 10-040-CM) supplemented with 10% FBS. For siRNA transfection of LCLs, electroporation was performed using the Neon Transfection System (Invitrogen) following the manufacturer’s recommendations. Briefly, 1 × 108 cells in 10 ml RPMI 1640 supplemented with 10% FBS and l-glutamine were pulsed 2 times (pulse voltage, 1,100 V; pulse width, 30 ms each). In certain experiments, TNF (1,000 U/ml; Roche), cycloheximide (60 μg/ml; BioVision), PMA (250 nM; Fisher BioReagents), CD40L (50 ng/ml; Enzo Life Science), and/or MG-132 (40 μM; Boston Biochem) were applied to the growth media.
qRT-PCR.
Total RNA was extracted from cells using the RNeasy procedure (Qiagen) according to the manufacturer’s instructions. An RT reaction with 5 μg total RNA in 20 μl was performed, using random hexamers and reverse transcription reagents according to the manufacturer’s instructions (Invitrogen). This was followed by qRT-PCR performed using a Mastercycler ep realplex2 system (Eppendorf). Oligonucleotides and internal probes for NFKBIA, CUL1, and IL8 transcripts were obtained from Applied Biosystems, and a Taqman PCR Master Mix with GAPDH mRNA quantitation was duplexed in the same well as an internal control. See Supplemental Table 2 for primers used for Sybr green–based qRT-PCR of other genes.
Immunoblotting and precipitation.
Whole cell lysates were prepared by adding Triton lysis buffer (25 mM HEPES, 100 mM NaCl, 10 mM DTT, 1 mM EDTA, 10% glycerol, 1% Triton X-100) or RIPA buffer (PBS, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 10 mM DTT) supplemented with 1 mM sodium orthovanadate and protease inhibitors (Roche), as indicated in each experiment. Cytosolic and nuclear extracts were prepared as previously described (1). Native immunoprecipitation, denatured immunoprecipitation, GSH precipitation, and immunoblotting were performed as previously described (1, 3, 50). Antiserum to COMMD1 has been previously described (1). Antisera against human CCDC22, COMMD6, COMMD9, and COMMD10 were generated by serial immunization of rabbits with purified full-length recombinant proteins prepared in E. coli. These antibodies were validated by Western blot for their ability to detect overexpressed protein after transient transfection, as well as the loss of a specific band of the correct molecular weight after RNAi (silencing confirmed by qRT-PCR). Antiserum to Cul3 was provided by M. Peter (ETH Zurich, Zurich, Switzerland). See Supplemental Table 3 for all commercial antibodies used.
TAP and bimolecular affinity purification.
TAP screening for COMMD1 has been previously reported (1). TAP screening for COMMD9 and COMMD10 were performed by sequential purification through HA binding resin (Roche) and streptavidin agarose (Pierce). Briefly, 10 plates of seeded HEK 293 cells (15 cm each) were transfected with pEBB-2×HA-COMMD9-TB or pEBB-2×HA-COMMD10-TB. 2 days later, nuclear and cytosolic extracts were prepared as previously described (1) and pooled as a single lysate prior to purification. The lysate was incubated with HA binding resin at 4°C for 2 hours. At that point, the bait was eluted 3 times using excess HA peptide (1 mg/ml in 20 mM Tris-HCl [pH 7.4], 100 mM NaCl, 0.1 mM EDTA). This pooled eluate was applied to a streptavidin agarose column to bind the bait at 4°C for 2 hours. The column was washed with detergent-free buffer, and beads were then submitted to the proteomic facility for trypsin digestion and LC/MS-MS analysis. Quantitative BAC-GFP interactomics (QUBIC) for CCDC22 was performed as described previously (29). A BAC harboring the locus for CCDC22 was modified to include a carboxyterminal (LAP) or aminoterminal (NFLAP) GFP tag by BAC transgenomic method (30). Engineered BACs were used as transgenes to generate HeLa cells stably expressing GFP-tagged CCDC22 under endogenous control. Affinity purifications were performed and analyzed as described previously (29). Finally, bimolecular affinity purification of the Cul2-COMMD1 complex (GST-Cul2, COMMD1-TB) was performed as described previously (51).
κB pulldown assay.
Nuclear extracts (100 μg) were incubated with a biotinylated 2κB oligonucleotide and subsequently precipitated using streptavidin agarose beads (Thermo Scientific), as described previously (52).
Confocal microscopy.
HeLa or U2OS cells were plated in chambered coverglass plates and transfected with the indicated plasmids. Cells were counterstained with Hoechst 33342 (8 μM) for 30 minutes, and images were obtained with a Zeiss LSM 510 META confocal microscope equipped with a Chameleon XR NIR laser. All confocal microscopy experiments were repeated at least twice; representative results are shown.
ChIP.
ChIP was performed as previously reported (50). Briefly, HEK 293 cells were cultured overnight in low serum media and stimulated with TNF (1,000 U/ml for 30 minutes) prior to ChIP. The antibodies used included RelA (Santa Cruz, sc-372), COMMD1 (1), CCDC22 (generated as described above), and normal rabbit IgG (Cell Signaling, 2729). After immunoprecipitation, DNA was extracted and used as template for PCR. The primers used for amplification of the BIRC3 promoter were 5′-GCATGCTTACCAATACGTGC-3′ and 5′-ATTGCGCAATTGTAGCGGTA-3′.
Statistics.
In all gene expression experiments, data represent mean ± SEM. A P value less than 0.05 was considered significant.
Study approval.
All patient-related evaluation was performed with the written consent of the participants or their legal guardians, after review and approval of the study protocols by the human research ethics committee of the Women’s and Children’s Health Network (Adelaide, South Australia, Australia). Animal studies were approved by the IACUC at UT Southwestern (protocol no. 2008-0327).
Note added in proof.
Recent data from a genome-wide siRNA screen for regulators of NOD2-regulated pathways also identified that COMMD8 is required for NF-κB activation (53).
Supplementary Material
Acknowledgments
The authors are grateful to the patients and family members that participated in this study. This work was supported by NIH R01 grant DK073639, a CCFA Senior Research Award, and the Disease Oriented Clinical Scholars’ Program at UT Southwestern to E. Burstein; by German medical genome research grant FKZ01GS0861 to M.Y. Hein; by Australian NH&MRC Project Grant 1008077 to J. Gecz, who is supported by a NH&MRC Principal Research Fellowship; and by EURO-MRX support to A.W. Kuss. The authors thank Irit Alkalai, Yinon Ben-Neriah, Denis Guttridge, Michele Pagano, Ina Poser, and Anthony A. Hyman for providing reagents; Marit Leuschner, Andrea Ssykor, Ina Poser, and Anthony A. Hyman for generating stable BAC transgenic HeLa cell lines expressing the GFP-tagged mutated and nonmutated CCDC22; Matthias Peter for providing the Cul3 antibody; Marie Shaw and Renee Carroll for handling patient cell lines; and Andrew Syvyk for help with electroporation of lymphocytes with siRNA. The authors are also grateful to Colin Duckett for his support and assistance during the transition of the Burstein lab to UT Southwestern.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(5):2244–2256. doi:10.1172/JCI66466.
References
- 1.Burstein E, et al. COMMD proteins: A novel family of structural and functional homologs of MURR1. J Biol Chem. 2005;280(23):22222–22232. doi: 10.1074/jbc.M501928200. [DOI] [PubMed] [Google Scholar]
- 2.van de Sluis B, Rothuizen J, Pearson PL, van Oost BA, Wijmenga C. Identification of a new copper metabolism gene by positional cloning in a purebred dog population. Hum Mol Genet. 2002;11(2):165–173. doi: 10.1093/hmg/11.2.165. [DOI] [PubMed] [Google Scholar]
- 3.Burstein E, et al. A novel role for XIAP in copper homeostasis through regulation of MURR1. EMBO J. 2004;23(1):244–254. doi: 10.1038/sj.emboj.7600031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klomp AE, van de Sluis B, Klomp LW, Wijmenga C. The ubiquitously expressed MURR1 protein is absent in canine copper toxicosis. J Hepatol. 2003;39(5):703–709. doi: 10.1016/S0168-8278(03)00380-5. [DOI] [PubMed] [Google Scholar]
- 5.Biasio W, Chang T, McIntosh CJ, McDonald FJ. Identification of Murr1 as a regulator of the human δ epithelial sodium channel. J Biol Chem. 2004;279(7):5429–5434. doi: 10.1074/jbc.M311155200. [DOI] [PubMed] [Google Scholar]
- 6.Ke Y, Butt AG, Swart M, Liu YF, McDonald FJ. COMMD1 downregulates the epithelial sodium channel through Nedd4-2. Am J Physiol Renal Physiol. 2010;298(6):F1445–F1456. doi: 10.1152/ajprenal.00257.2009. [DOI] [PubMed] [Google Scholar]
- 7.Chang T, Ke Y, Ly K, McDonald FJ. COMMD1 regulates the delta epithelial sodium channel (δENaC) through trafficking and ubiquitination. Biochem Biophys Res Commun. 2011;411(3):506–511. doi: 10.1016/j.bbrc.2011.06.149. [DOI] [PubMed] [Google Scholar]
- 8.Drevillon L, et al. COMMD1-mediated ubiquitination regulates CFTR trafficking. PLoS One. 2011;6(3):e18334. doi: 10.1371/journal.pone.0018334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van de Sluis B, et al. Increased activity of hypoxia-inducible factor 1 is associated with early embryonic lethality in Commd1 null mice. Mol Cell Biol. 2007;27(11):4142–4156. doi: 10.1128/MCB.01932-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van de Sluis B, et al. COMMD1 disrupts HIF-1α/β dimerization and inhibits human tumor cell invasion. J Clin Invest. 2010;120(6):2119–2130. doi: 10.1172/JCI40583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganesh L, et al. The gene product Murr1 restricts HIV-1 replication in resting CD4+ lymphocytes. . Nature. 2003;426(6968):853–857. doi: 10.1038/nature02171. [DOI] [PubMed] [Google Scholar]
- 12.Maine GN, Mao X, Komarck CM, Burstein E. COMMD1 promotes the ubiquitination of NF-κB subunits through a Cullin-containing ubiquitin ligase. EMBO J. 2007;26(2):436–447. doi: 10.1038/sj.emboj.7601489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burkly L, et al. Expression of relB is required for the development of thymic medulla and dendritic cells. Nature. 1995;373(6514):531–536. doi: 10.1038/373531a0. [DOI] [PubMed] [Google Scholar]
- 14.Kontgen F, et al. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 1995;9(16):1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- 15.Sha WC, Liou HC, Tuomanen EI, Baltimore D. Targeted disruption of the p50 subunit of NF-κB leads to multifocal defects in immune responses. Cell. 1995;80(2):321–330. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- 16.Mao X, et al. Copper metabolism MURR1 domain containing 1 (COMMD1) regulates Cullin-RING ligases by preventing Cullin-associated NEDD8-dissociated (CAND1) binding. J Biol Chem. 2011;286(37):32355–32365. doi: 10.1074/jbc.M111.278408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6(1):9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 18.Hayden MS, Ghosh S. Shared principles in NF-κB signaling. Cell. 2008;132(3):344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 19.Chen Z, et al. Signal-induced site-specific phosphorylation targets IκBα to the ubiquitin-proteasome pathway. Genes Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 20.Henkel T, Machleidt T, Alkalay I, Krönke M, Ben-Neriah Y, Baeuerle PA. Rapid proteolysis of IκB-α is necessary for activation of transcription factor NF-κB. Nature. 1993;365(6442):182–185. doi: 10.1038/365182a0. [DOI] [PubMed] [Google Scholar]
- 21.Yaron A, et al. Identification of the receptor component of the IκBα-ubiquitin ligase. Nature. 1998;396(6711):590–594. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- 22.Baeuerle PA, Baltimore D. IκB: a specific inhibitor of the NF-κB transcription factor. Science. 1988;242(4878):540–546. doi: 10.1126/science.3140380. [DOI] [PubMed] [Google Scholar]
- 23.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and β-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8(6):438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geng H, Wittwer T, Dittrich-Breiholz O, Kracht M, Schmitz ML. Phosphorylation of NF-κB p65 at Ser468 controls its COMMD1-dependent ubiquitination and target gene-specific proteasomal elimination. EMBO Rep. 2009;10(4):381–386. doi: 10.1038/embor.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mao X, et al. GCN5 is a required cofactor for a ubiquitin ligase that targets NF-κB/RelA. Genes Dev. 2009;23(7):849–861. doi: 10.1101/gad.1748409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voineagu I, et al. CCDC22: a novel candidate gene for syndromic X-linked intellectual disability. Mol Psychiatry. 2012;17(1):4–7. doi: 10.1038/mp.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu C, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10(11):R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Bie P, et al. Characterization of COMMD protein-protein interactions in NF-κB signalling. Biochem J. 2006;398(1):63–71. doi: 10.1042/BJ20051664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hubner NC, et al. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J Cell Biol. 2010;189(4):739–754. doi: 10.1083/jcb.200911091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poser I, et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat Methods. 2008;5(5):409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barbe L, et al. Toward a confocal subcellular atlas of the human proteome. Mol Cell Proteomics. 2008;7(3):499–508. doi: 10.1074/mcp.M700325-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Marchler-Bauer A, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39(Database issue):D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schleiffer A, Kaitna S, Maurer-Stroh S, Glotzer M, Nasmyth K, Eisenhaber F. Kleisins: a superfamily of bacterial and eukaryotic SMC protein partners. Mol Cell. 2003;11(3):571–575. doi: 10.1016/S1097-2765(03)00108-4. [DOI] [PubMed] [Google Scholar]
- 34.Doffinger R, et al. X-linked anhidrotic ectodermal dysplasia with immunodeficiency is caused by impaired NF-κB signaling. Nat Genet. 2001;27(3):277–285. doi: 10.1038/85837. [DOI] [PubMed] [Google Scholar]
- 35.Jain A, Ma CA, Liu S, Brown M, Cohen J, Strober W. Specific missense mutations in NEMO result in hyper-IgM syndrome with hypohydrotic ectodermal dysplasia. Nat Immunol. 2001;2(3):223–228. doi: 10.1038/85277. [DOI] [PubMed] [Google Scholar]
- 36.Picard C, Casanova JL, Puel A. Infectious diseases in patients with IRAK-4, MyD88, NEMO, or IκBα deficiency. Clin Microbiol Rev. 2011;24(3):490–497. doi: 10.1128/CMR.00001-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mikkola ML. Molecular aspects of hypohidrotic ectodermal dysplasia. Am J Med Genet A. 2009;149A(9):2031–2036. doi: 10.1002/ajmg.a.32855. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt-Ullrich R, Aebischer T, Hulsken J, Birchmeier W, Klemm U, Scheidereit C. Requirement of NF-κB/Rel for the development of hair follicles and other epidermal appendices. Development. 2001;128(19):3843–3853. doi: 10.1242/dev.128.19.3843. [DOI] [PubMed] [Google Scholar]
- 39.Narindrasorasak S, Kulkarni P, Deschamps P, She YM, Sarkar B. Characterization and copper binding properties of human COMMD1 (MURR1). Biochemistry. 2007;46(11):3116–3128. doi: 10.1021/bi0620656. [DOI] [PubMed] [Google Scholar]
- 40.Nakagawa T, Xiong Y. X-linked mental retardation gene CUL4B targets ubiquitylation of H3K4 methyltransferase component WDR5 and regulates neuronal gene expression. Mol Cell. 2011;43(3):381–391. doi: 10.1016/j.molcel.2011.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boersma MC, Dresselhaus EC, De Biase LM, Mihalas AB, Bergles DE, Meffert MK. A requirement for nuclear factor-κB in developmental and plasticity-associated synaptogenesis. J Neurosci. 2011;31(14):5414–5425. doi: 10.1523/JNEUROSCI.2456-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, Baltimore D. NF-κB functions in synaptic signaling and behavior. Nat Neurosci. 2003;6(10):1072–1078. doi: 10.1038/nn1110. [DOI] [PubMed] [Google Scholar]
- 43.Chen Y, et al. HDAC-mediated deacetylation of NF-κB is critical for Schwann cell myelination. Nat Neurosci. 2011;14(4):437–441. doi: 10.1038/nn.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sancho-Shimizu V, et al. Herpes simplex encephalitis in children with autosomal recessive and dominant TRIF deficiency. J Clin Invest. 2011;121(12):4889–4902. doi: 10.1172/JCI59259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suttner K, et al. Genetic variants harbored in the forkhead box protein 3 locus increase hay fever risk. J Allergy Clin Immunol. 2010;125(6):1395–1399. doi: 10.1016/j.jaci.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 46.Duckett CS, Li F, Wang Y, Tomaselli KJ, Thompson CB, Armstrong RC. Human IAP-like protein regulates programmed cell death downstream of Bcl-xL and cytochrome c. Mol Cell Biol. 1998;18(1):608–615. doi: 10.1128/mcb.18.1.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richter BW, et al. Molecular cloning of ILP-2, a novel member of the inhibitor of apoptosis protein family. Mol Cell Biol. 2001;21(13):4292–4301. doi: 10.1128/MCB.21.13.4292-4301.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duckett CS, Gedrich RW, Gilfillan MC, Thompson CB. Induction of nuclear factor κB by the CD30 receptor is mediated by TRAF1 and TRAF2. Mol Cell Biol. 1997;17(3):1535–1542. doi: 10.1128/mcb.17.3.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neitzel H. A routine method for the establishment of permanent growing lymphoblastoid cell lines. Hum Genet. 1986;73(4):320–326. doi: 10.1007/BF00279094. [DOI] [PubMed] [Google Scholar]
- 50.Li H, et al. Regulation of NF-κB activity by competition between RelA acetylation and ubiquitination. Oncogene. 2012;31(5):611–623. doi: 10.1038/onc.2011.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maine GN, Gluck N, Zaidi IW, Burstein E. Bimolecular Affinity Purification (BAP): Tandem affinity purification using two protein baits. Cold Spring Harb Protoc. 2009;2009(11):pdb.prot5318. doi: 10.1101/pdb.prot5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vaira S, Alhawagri M, Anwisye I, Kitaura H, Faccio R, Novack DV. RelA/p65 promotes osteoclast differentiation by blocking a RANKL-induced apoptotic JNK pathway in mice. J Clin Invest. 2008;118(6):2088–2097. doi: 10.1172/JCI33392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Warner N, et al. A genome-wide siRNA screen reveals positive and negative regulators of the NOD2 and NF-κB signaling pathways. Sci Signal. 2013;6(258):rs3. doi: 10.1126/scisignal.2003305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.