Abstract
Adult stem cells reside in specialized niches where they receive environmental cues to maintain tissue homeostasis. In mammals, the stem cell niche within hair follicles is home to epithelial hair follicle stem cells and melanocyte stem cells, which sustain cyclical bouts of hair regeneration and pigmentation1–4. To generate pigmented hairs, synchrony is achieved such that upon initiation of a new hair cycle, stem cells of each type activate lineage commitment2,5. Dissecting the inter-stem-cell crosstalk governing this intricate coordination has been difficult, because mutations affecting one lineage often affect the other. Here we identify transcription factor NFIB as an unanticipated coordinator of stem cell behaviour. Hair follicle stem-cell-specific conditional targeting of Nfib in mice uncouples stem cell synchrony. Remarkably, this happens not by perturbing hair cycle and follicle architecture, but rather by promoting melanocyte stem cell proliferation and differentiation. The early production of melanin is restricted to melanocyte stem cells at the niche base. Melanocyte stem cells more distant from the dermal papilla are unscathed, thereby preventing hair greying typical of melanocyte stem cell differentiation mutants. Furthermore, we pinpoint KIT-ligand as a dermal papilla signal promoting melanocyte stem cell differentiation. Additionally, through chromatin-immunoprecipitation with high-throughput-sequencing and transcriptional profiling, we identify endothelin 2 (Edn2) as an NFIB target aberrantly activated in NFIB-deficient hair follicle stem cells. Ectopically induced Edn2 recapitulates NFIB-deficient phenotypes in wild-type mice. Conversely, endothelin receptor antagonists and/or KIT blocking antibodies prevent precocious melanocyte stem cell differentiation in the NFIB-deficient niche. Our findings reveal how melanocyte and hair follicle stem cell behaviours maintain reliance upon cooperative factors within the niche, and how this can be uncoupled in injury, stress and disease states.
Hair follicle stem cells and melanocyte stem cells remain quiescent within their hair follicle niche for weeks, a period known as telogen phase. With each new hair cycle, these two stem cell populations are coordinately activated. This happens when inhibitory signals are counteracted by activating cues that accumulate from Wnt and BMP/TGFβ (bone morphogenetic protein/transforming growth factor β) crosstalk with dermal papilla at the niche base6–8. Synchronized activity continues throughout the hair cycle. During the growth phase (anagen), melanocytes at the base of the mature hair follicle (‘hair bulb’) produce and transfer pigment to neighbouring committed hair follicle stem cell progeny (‘matrix’) as they differentiate into hair cells2,5.When destruction (catagen) ensues, melanocytes and matrix cells in the hair bulb apoptose, and the dermal papilla (enveloped by the hair bulb during anagen) retracts upward, returning the follicle to telogen. As anagen begins and a new hair bulb emerges, both hair follicle stem cells and melanocyte stem cells contain nuclear β-catenin, implicating canonical Wnt signalling in stem cell coordination6,8. These and several other insights7,9,10 suggest how local environmental signals synchronize proliferation and lineage progression of stem cells during hair cycling.
Uncoupling melanocyte and epithelial stem cell behaviours occurs under transient conditions, that is, in response to ultraviolet radiation, and in various disease and injury states11,12. Given the impact of Wnt and other signals on stem cells and their lineages, and current dogma that matrix cells must differentiate for melanocyte pigment to transfer10, the mechanisms by which melanocyte stem cells can be selectively mobilized from their niche without otherwise disrupting the normal hair cycle remains unknown.
Our venture into this study was prompted by our finding that relative to progeny, hair follicle stem cells express elevated nuclear factor I/B (NFIB)1. NFIB is required for lung and brain development and is often amplified and/or found at oncogenic chromosomal breakpoints in epithelial cancers13–15. NFIB was first detected in epidermis at embryonic day 14.5 (E14.5), concomitant with upregulation of established skin progenitors. Expression intensified as hair follicle stem cells emerged (Fig. 1a and Supplementary Fig. 1a–c).
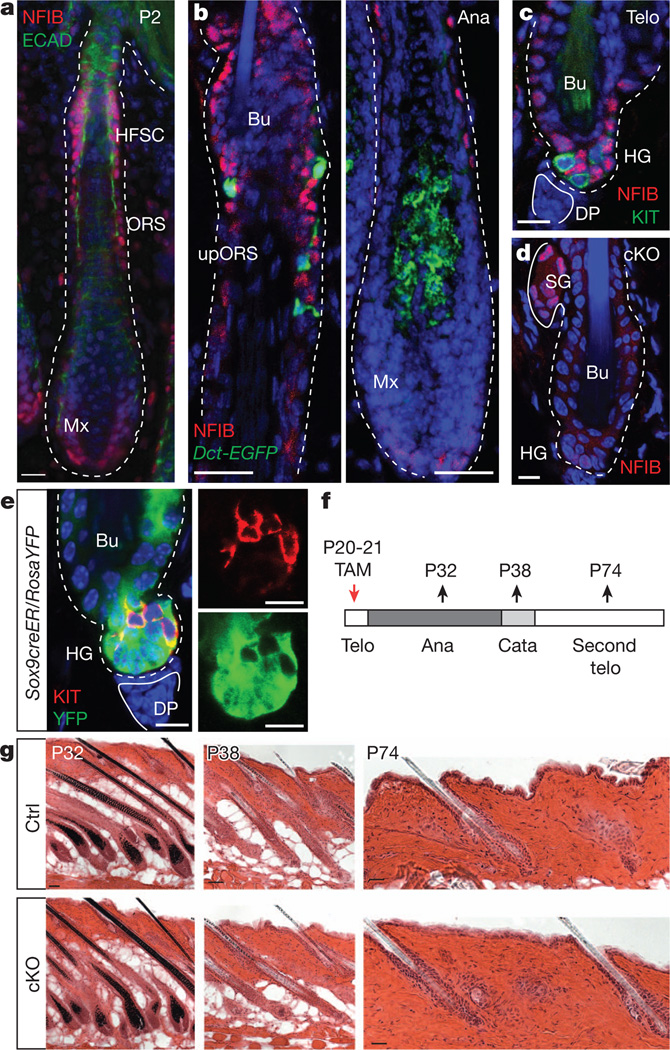
Figure 1. Conditional Nfib targeting in hair follicle stem cells does not perturb hair cycle or follicle architecture.
a–c, Immunofluorescence. a, Enrichment of nuclear NFIB in hair follicle stem cells and ORS of developing hair follicles. ECAD, E-cadherin; HFSC, hair follicle stem cells; Mx, matrix. b, NFIB in anagen hair follicles from adult Dct-EGFP BAC transgenic mice. NFIB is not seen in EGFP+ melanocytes. Ana, anagen; Bu, bulge; upORS, upper ORS. c, Absence of NFIB in KIT+ melanocyte stem cells of telogen hair follicles.DP, dermal papilla;HG, hair germ; Telo, telogen. d–f, Tamoxifen (TAM) was administered to Sox9-CreER/Nfibfl/fl/RosaYFP-cKO mice and analysed at various times thereafter. d, Note hair follicle stem-cell-specific Nfib targeting in bulge and hair germ. SG, sebaceous gland. e, Note YFP reporter activity in hair follicle stem cells but not in KIT+ melanocyte stem cells. f, Schematic. Cata, catagen; P20, postnatal day 20. g, Haematoxylin- and eosin-stained back skins reveal normal hair cycle and follicle architecture upon NFIB loss. Ctrl, control. Scale bars: 50 µm(P32,P38ing); 25 µm(a,b,P74ing); 10 µm(c–e).
In adult hair follicles, NFIB co-localized with hair follicle stem cells, whose niche in telogen is subdivided into an upper ‘bulge’ compartment and lower ‘hair germ’ (or secondary hair germ) adjacent to dermal papilla (Supplementary Fig. 1d). During anagen, NFIB-positive cells were also found within the upper outer root sheath (ORS), which in early catagen will form the new niche (bulge and hair germ) for the next hair cycle16 (Fig. 1b). NFIB waned in transit-amplifying (TA) matrix progenitors (Fig. 1b and Supplementary Fig. 1e). NFIB was not detected in melanocyte stem cells marked by dopachrome tautomerase (DCT) and tyrosine kinase receptor KIT in the upper ORS and bulge/hair germ, nor in differentiated melanocytes within the hair bulb17 (Fig. 1b, c). Overall, both inside and outside the niche, hair follicle stem cells and melanocyte stem cells showed synchronized behaviours but distinct expression patterns.
To address the function of NFIB, we conditionally induced Nfib ablation (cKO) in mouse hair follicle stem cells by using Sox9-CreER and K15-CrePGR. Unless specified, data are fromSox9-CreER mice, but both gave similar results. NFIB immunofluorescence verified that Nfib was efficiently targeted in hair follicle stem cells, consistent with K15 and Sox9 expression primarily in ORS/bulge/hair germ; furthermore, when Rosa26floxSTOPfloxYFP (RosaYFP; yellow fluorescent protein (YFP)) was used to mark and trace the progeny of Cre-induced cells, only hair follicle stem cells and their subsequent progeny, and not KIT+ melanocytes or dermal cells, were fluorescently labelled18 (Figs 1d, e and Supplementary Fig. 2a–c). Despite efficient targeting, however, Nfib-cKO hair coats and hair cycling appeared normal (Fig. 1f, g).
Equally surprising to the absence of hair follicle stem cell lineage defects were melanocyte lineage abnormalities. Fontana–Masson staining revealed atypical melanogenic cells at the niche base of telogen hair follicles (Fig. 2a and Supplementary Fig. 2d). Immunostaining showed increases in melanocytes (KIT+DCT+) throughout hair germ and bulge, and ectopic presence of differentiated melanocytes (KIT+DCT+TYRP1+MITF+TYR+) within hair germ (Fig. 2b, c and Supplementary Fig. 3a–c).
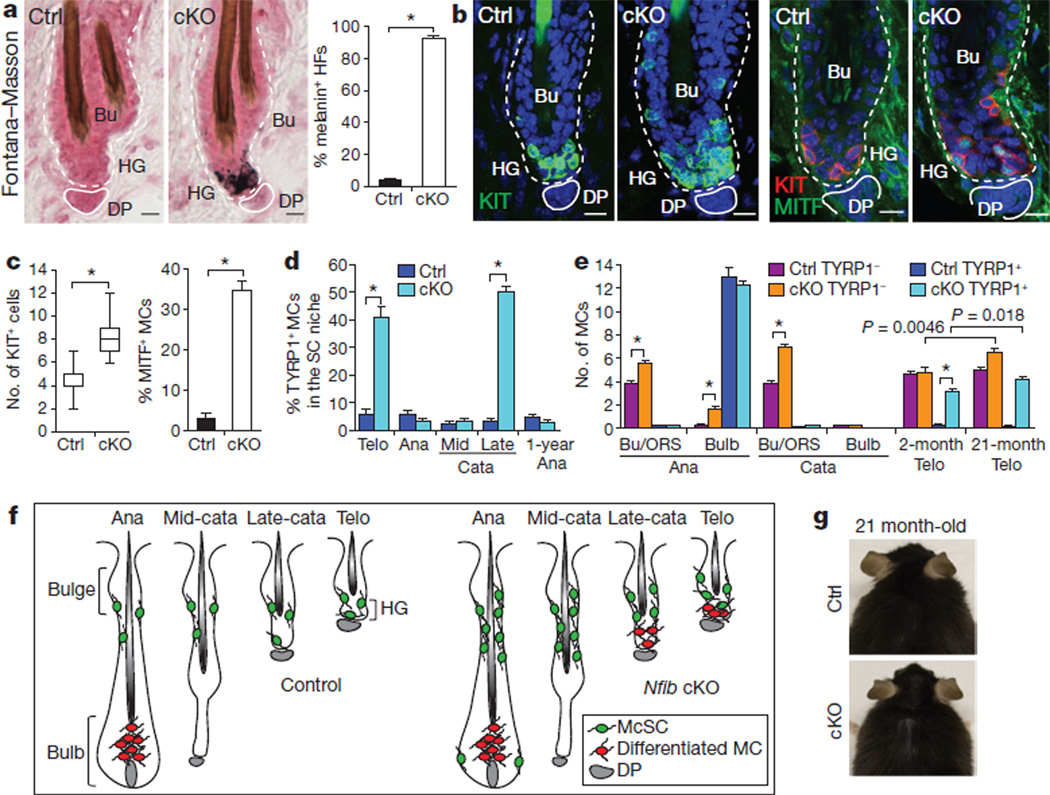
Figure 2. NFIB loss enhances melanocyte stem-cell self-renewal and perturbs melanocyte stem cells activity in the hair follicle stem cell niche.
a, Ectopic pigmentation in telogen-phase cKO hair germ as detected by Fontana–Masson melanin staining and quantifications (n=3 mice per experiment, >40 hair follicles per mouse). b–e, Immunofluorescence and quantifications. b, c, Increased melanocytes and ectopic differentiation in telogen cKO hair follicles (30–50 hair follicles; 2 mice per experiment). KIT marks melanocyte stem cells and differentiated melanocytes; MITF andTYRP1 are differentiation-specific melanocyte markers. MCs, melanocytes. d, Precocious melanocyte differentiation begins in the niche at late catagen. Quantifications of TYRP1+ among KIT+ or Dct-EGFP+ melanocytes (>40 hair follicles; 2 mice per experiment). Analyses shown start at second telogen. SC, stem cell. e, Quantifications of melanocyte stem cells (TYRP1−) and differentiated melanocytes (TYRP1+) at different hair follicle stages (>30 hair follicles; 2 mice per experiment). f, Summary of data. McSC, melanocyte stemcell;MC, melanocyte. g, Lack of hair greying in ageing Nfib-cKO mice. Scale bars, 10 µm (a, b). *P<0.001. Error bars indicate s.e.m.
A priori, ectopic differentiated melanocytes in cKO hair germs might reflect hair bulb melanocytes that somehow escaped apoptosis during catagen. Alternatively, they could arise from precocious differentiation of niche melanocyte stem cells. To distinguish between these possibilities, we analysed melanocyte behaviour at each hair cycle stage (Fig. 2d and Supplementary Fig. 4). During anagen, TYRP1+ cells dropped to wild-type levels in the bulge/upper ORS. As dermal papilla returned to the niche during late catagen, TYRP1+ numbers again soared, persisting until the next anagen. By contrast, control stem cell niches displayed much more modest melanocyte stem cell differentiation, which occurred at telogen→anagen rather than catagenRtelogen. The difference in this timing was >3 weeks for young adult mice.
Accompanying the disappearance of differentiated melanocytes within the anagen Nfib-cKO bulge/upper ORS, was their appearance in the hair bulb (upper matrix) (Fig. 2e and Supplementary Fig. 4). Within the bulb, pigment transfer to differentiating hair cell recipients seemed normal. Additionally, at the start of catagen, TYRP1+ melanocytes underwent apoptosis and were eliminated by mid-catagen in both control and cKO hair bulbs. In contrast to differentiated melanocytes, TYRP1− melanocyte stem cells remained elevated throughout cKO hair cycles, where they resided in the bulge/upper ORS. The only time at which melanocyte stem cells dropped transiently was during late catagen/telogen, when melanocyte stem cells near dermal papilla precociously differentiated.
These results pinpointed defects to the stem cell niche; furthermore, dermal papilla proximity seemed to affect primarily the uncoupling and premature differentiation of melanocyte stem cells rather than self-renewal (Fig. 2f). In addition, melanocyte stem cells in anagen stem cell niches were still negative for TYRP1 even in 1-year-old Nfib-cKO hair follicles (Fig. 2d), indicating that ectopic differentiated melanocytes from telogen do not accumulate within the niche over hair cycles.
To test melanocyte stem cell activity further, we mated Nfib-cKO to Dct-EGFP mice and isolated EGFP+ bulge cells from telogen hair follicles. Melanocyte differentiation markers, for example, Kit, Mitf, Tyrp1, Tyr and Sox10, were upregulated in EGFP+ cells, whereas genes expressed by both melanocyte stem cells and differentiated melanocytes showed no change (Supplementary Fig. 3d). Finally, hair coats of ageing cKO mice were still pigmented, and melanocyte stem cell levels were sustained (Fig. 2e, g and Supplementary Fig. 3e). These data provided compelling evidence that NFIB-deficiency in hair follicle stem cells affects the timing of melanocyte differentiation without compromising melanocyte stem cell biology and/or function, which results in hair greying6,7,9,11.
Ultrastructural analysis unveiled new defects within the stem cell niche (Fig. 3a and Supplementary Fig. 5). Niche melanocytes closest to dermal papilla had pigment granules and immature melanosomes. Surprisingly, however, adjacent hair germ cells also had pigment granules. Their epithelial identity was shown by keratin filaments, desmosomes and hemidesmosomes. Although poorly understood, hair cell uptake of pigment from differentiated melanocytes is thought to depend on FOXN110, expressed by differentiating matrix cells but not by hair follicle stem cells19.
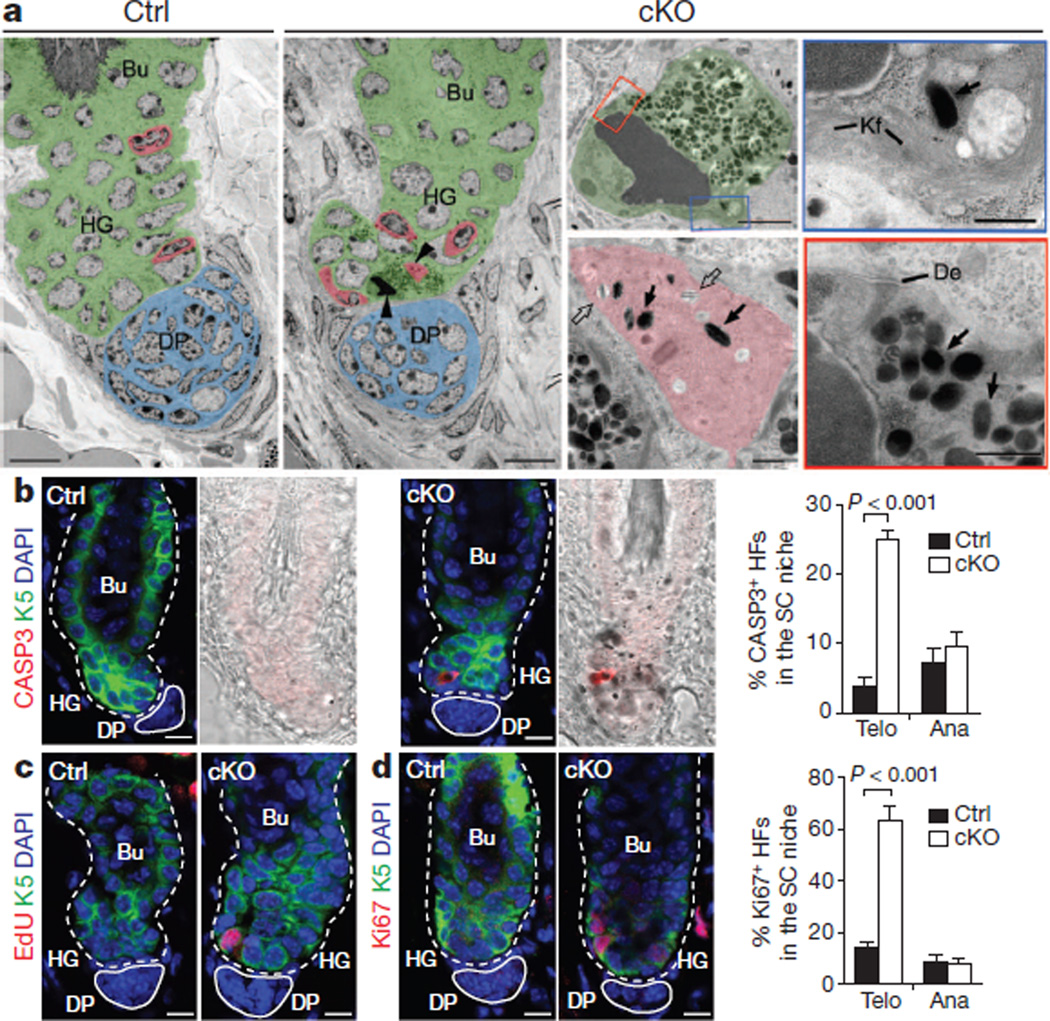
Figure 3. Premature transfer of pigment promotes apoptotic cell death in hair follicle stem cells in the NFIB-deficient niche.
a, Hair follicle ultrastructure. Pseudo-colouring highlights distinct stem cell niche cell types: green, hair follicle stem cell; red, melanocyte; blue, dermal papilla. Arrowheads mark cells shown at higher magnification. Open and solid arrows denote immature and mature melanosomes, respectively. Note pigment-laden apoptotic hair follicle stem cells whose boxed regions at higher magnification reveal condensed chromatin, degenerating mitochondrion, keratin filaments (Kf), desmosomes (De) and melanosomes. b, Activated caspase 3 antibody marks K5+ apoptotic cKO hair follicle stem cells, seen in telogen but not anagen (n=3 mice; >30 hair follicles per mouse). HFs, hair follicles; SC, stem cell. c, d, Increased hair follicle stem cell proliferation in telogen but not anagen NfibcKO hair follicles. Assessment was by EdU (5-ethynyl-2′-deoxyuridine) incorporation (administered twice, 24 h before analysis) and Ki67 immunostaining (n=3 mice; >30 hair follicles per mouse). Scale bars: 10 µm (low magnifications in a and b–d); 2 µm (apoptotic hair follicle stem cell in a); 0.5 µm(cytoplasmic segments and melanocyte in a). Error bars indicate s.e.m.
Inappropriate accumulation of pigment proved calamitous for quiescent hair follicle stem cells in the telogen hair germ: it elicited their apoptotic death, typified by condensed chromatin, mitochondrial destruction, and cleaved caspase-3-immunolabelling (Fig. 3a, b and Supplementary Fig. 6). In addition, unaffected neighbouring K5-positive hair germ cells proliferated (Fig. 3c, d and Supplementary Fig. 6). Although hair germ proliferation is a normal sign of telogen→anagen8, precocious hair cycle entry was not observed.
Apoptotic and proliferative defects within the niche disappeared in anagen (Fig. 3b, d), concomitant with movement of differentiated melanocytes from niche to hair bulb (Fig. 2d–f). These results indicate that apoptosis and hyperproliferation of Nfib-null hair follicle stem cells depend upon precocious differentiation of neighbouring melanocyte stem cells. They also indicate that when differentiated melanocytes inappropriately bequeath pigment to hair follicle stem cells rather than their customary differentiated progeny (hair cells), pigment-laden hair follicle stem cells are unable to cope, whereas healthy hair follicle stem cell neighbours proliferate to restock the niche.
Notably, in older Nfib-cKO mice, black-pigmented dermal cells swarmed hair follicles and were even visible from the skin surface (Supplementary Fig. 7). Although dermal melanoblasts exist in certain skin regions such as the ear, and can differentiate under some conditions20, fewTYRP1+melanocytes were found in cKO dermis. Rather, a number of cells encompassing this pigment were Mac1+ with features of macrophages. Irrespective of whether pigment-laden vacuoles within dermal cells reflected engulfment of dying, pigmented cells, or direct pigment transfer from melanocytes, these defects made the normalcy of the hair coat of ageing cKO mice all the more remarkable.
To dissect the molecular miscommunication between hair follicle stem cells and melanocyte stem cells in the niche, we used high throughput RNA-seq to transcriptionally profile bulge and hair germ hair follicle stem cells. Fluorescence activated cell sorting (FACS) of skins from K15-CrePGR/RosaYFP/Nfibfl/fl (cKO) and Nfibfl/+ (het) mice were used for purifying CD34+-bulge andCD34−-hair germ cells (both YFP+Sca1−α6highCD200+) (Supplementary Figs 2a and 8a).
Of 800–1,000 messenger RNAs changed by≥twofold in NFIB-deficient hair follicle stem cells relative to control, 145 were upregulated and 99 were downregulated (Supplementary Figs 8b and 9 and Supplementary Table 1). Quantitative PCR (qPCR) of independently purified samples validated the differences (Supplementary Fig. 8c, d). Notably absent from the list were Foxn1, Fgf2, Nog, Egfr, F2rl1 and derivatives of Pomc—all previously implicated in melanocyte differentiation and/or pigment transfer10,12,21. Also absent were genes involved in BMP/TGF-β signalling, known to function in stem cell niche quiescence. Similarly, NFIB loss did not seem to affect canonical Wnt signalling, a key stimulus for stem cell activity and fate commitment: Wnt-sensitive target gene Axin2 was unchanged in NFIB-deficient hair follicle stem cells, and upregulated genes included both negative and positive Wnt regulators.
These results were consistent with the normal hair cycle displayed by Nfib-cKO skin (Fig. 1g). Had any of these signalling pathways been perturbed, both stem cell populations—not just melanocyte stem cells—should have been affected. Moreover, hair follicle stem cells from mice genetically defective for Wnt, BMP and TGF-β signalling still expressed NFIB protein and mRNA (Supplementary Fig. 10a, b). Thus at least within the quiescent stem cell niche, these signalling pathways seemed to be refractory to loss of NFIB, and NFIB seemed to be refractory to these signalling pathways.
To identify direct NFIB target genes, we performed chromatin-immunoprecipitation with high-throughput-sequencing (ChIP-seq) analysis on 10–15 million bulge hair follicle stem cells FACS-purified from 15–20 mice19. Applying immunoprecipitation-grade NFIB antibody to chromatin, we identified 1,449 genes that were directly and reproducibly bound by NFIB (Fig. 4a and Supplementary Table 2). NFIB-bound genes included Krt5 (Supplementary Fig. 11a). Intriguingly, like Krt5 and its transcription factor-AP2 family regulators, NFIB was absent in most areas of Trp63-null skin (Supplementary Fig. 10c). Additionally, Nfib is bound by p63 (ref. 22) and harbours TFAP2 binding motifs (data not shown), suggesting possible connections to these early stem cell markers.
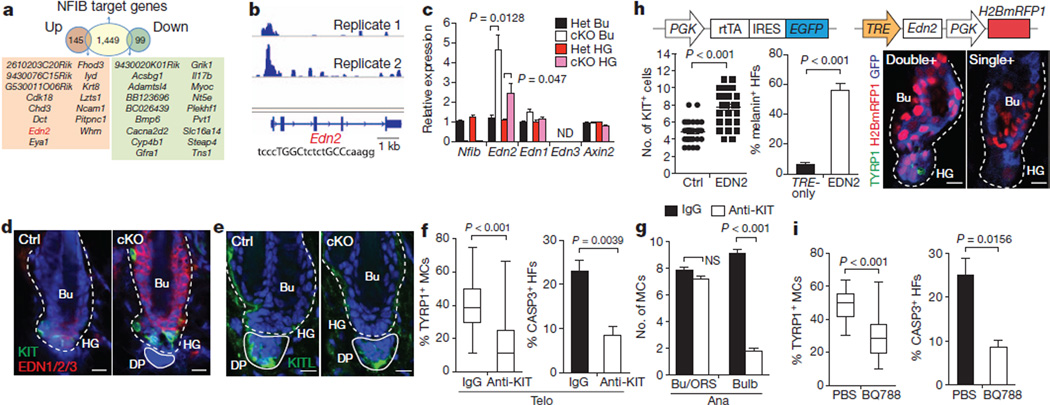
Figure 4. RNA-seq and ChIP-seq analyses identify Edn2 as a direct NFIB regulated gene mediating inter-stem cell crosstalk.
a, Identification of direct NFIB-regulated genes as those that bind NFIB (from ChIP-seq) and are up/down regulated in Nfib-cKO hair follicle stem cells (from RNA-seq). b, NFIB ChIP-seq profiles of Edn2 promoter. NFIB palindromic-binding-sequence exists within the peak. c, qPCR on FACS-isolated populations confirm RNA-seq results (Het, normalizations to heterozygote values). d, Immunofluorescence shows ectopic endothelins in Nfib-cKO hair follicles. e, Immunofluorescence reveals KITL in dermal papilla. f, Reductions in TYRP1+ melanocytes (>60 hair follicles; 3mice) and apoptosis (n = 4 mice;>30 hair follicles permouse) in cKO hair follicles after injecting anti-KIT blocking antibody (anti-KIT) during catagen/telogen. IgG, control. g, Anti-KIT-mediated inhibition of KIT signalling during anagen (>45 hair follicles in two mice). h, Overhead schematic: lentiviral constructs. Quantifications show increased KIT+ melanocytes (32 hair follicles; two mice per experiment) and melanin+ hair follicles (n=3 mice;>60 hair follicles per mouse) upon EDN2 induction. Immunofluorescence showsTYRP1+ melanocyte differentiation in doubly- (Edn2) and not singly-transduced/uninfected (Ctrl) hair follicle stem cells. i, Reduced TYRP1+ melanocytes (>46 hair follicles in two mice) and apoptosis (n=3 mice; >25 hair follicles per mouse) in Nfib-cKO hair follicles after injection of EDNRB inhibitor BQ788. Scale bars: 10 µm (d, e, h). ND, not determined; NS, not significant. Error bars indicate s.e.m.
NFIB peaks were enriched in±2 kilobases (kb; 12%) and 2–50 kb (29%) sequences proximal to gene transcription initiation sites (Supplementary Fig. 11b, c). A de novo motif search identified five sequences within these peaks (Supplementary Fig. 11d, e). Most common were TGGCA/T and A/TGCCA, which when combined, comprised a palindromic motif. Notably, NFIB protein self-dimerizes, and its preferred binding motif is a TTGGCANNNTGCCAA palindrome23. Moreover, in response to NFIB loss, 201 (~14%) NFIB target genes were differentially expressed relative to control hair follicle stem cells (Supplementary Fig. 12). Of these, 44% were upregulated whereas 56% were downregulated in NFIB-deficient hair follicle stem cells. NFIB’s lack of apparent bias for hair follicle stem cell gene activation differed from the role of NFIC in cultured fibroblasts23.
Searching for candidates whose altered expression might enhance melanocyte stem cell proliferation and differentiation, we focused on the 33NFIB-binding genes≥twofold up- or downregulated in bulge and hair germ hair follicle stem cells when Nfib is ablated (Fig. 4a). Notable was the gene Edn2, encoding endothelin-2. Within the Edn2 promoter was an NFI-binding palindrome sequence containing an optimal spacer (Fig. 4b). In both ChIP-seq replicates, NFIB bound to this site.
Endothelins are secreted factors with the ability to mediate intercellular crosstalk. All three endothelins stimulate cultured melanocyte stem cells5. Although Edn3 is required for neural crest migration and melanocyte specification during embryogenesis24, and Edn1 has been implicated in Wnt-mediated melanocyte proliferation6, neither Edn3 nor Edn1 appeared on our list of hair follicle stem cell genes bound by NFIB. Moreover, as assessed by RNA-seq and qPCR, Edn3 was not expressed in hair follicle stem cells, and Edn1 showed low expression and little change upon NFIB loss (Fig. 4c and Supplementary Table 1). By contrast, at telogen→anagen, Edn2 was transiently activated inwild-type hair follicle stem cells19, and in Nfib-null hair follicle stem cells, Edn2 was upregulated more than twofold relative to controls. Pan anti-endothelin immunolabelling was also stronger in Nfib-null relative to control hair follicle stem cells (Fig. 4d). Finally, hair follicle stem cells express endothelial converting enzyme (ECE1) necessary to process/activate EDN2.
The Edn2 changes were cell-autonomous, because Edn2 mRNA was also upregulated in cultured Nfib-null keratinocytes (Supplementary Fig. 13). Moreover, this difference depended upon NFIB, as it was rescued by expressing the main keratinocyte isoform, NFIB3. By contrast, even though KIT ligand (Kitl)mRNA seemed to be modestly induced in Nfib-null hair follicle stem cells (Supplementary Table 1), the levels were not influenced by NFIB3-rescue (Supplementary Fig. 13), nor did we observe NFIB binding to the Kitl promoter/enhancer in ChIP-seq analyses. Interestingly, however, whereas KITL was not detected in hair follicle stem cells, it was seen in dermal papilla17 (Fig. 4e).
Together, these results indicated a model whereby Edn2 induction by Nfib-null hair follicle stem cells enhances proliferation of neighbouring melanocyte stem cells and sensitizes them to precociously differentiate when they encounter KITL and possibly additional signals from dermal papilla. If true, then blocking KIT signalling should ameliorate precocious melanocyte stem cell differentiation in cKO hair follicles, whereas elevating EDN2 in wild-type hair follicle stem cells should generate phenotypic features of NFIB-deficiency.
We first tested this hypothesis by injecting a KIT-receptor-blocking antibody into the skins of cKO mice beginning in late catagen. By telogen, marked reductions were seen in TYRP1+ melanocytes and in activated-CASP3+ hair follicle stem cells (Fig. 4f). Importantly and in agreement with previous findings17,25, KIT inhibition in anagen hair follicles only affected lineage-committed proliferation and differentiation: undifferentiated melanocyte stem cells remained elevated in the cKO niche (Fig. 4g and Supplementary Fig. 14).
We next induced EDN2 in wild-type adult skin by co-transducing E9.5 mouse embryos in utero with high-titre lentiviruses26 harbouring: (1) a tetracycline-regulatable transactivator (rtTA) coupled in a bicistronic transcript to EGFP; and (2) H2BmRFP1 and a tetracycline-inducible promoter driving either Edn2 (TRE-Edn2) or nothing (TRE-only) (Fig. 4h). Following selective EDN2 induction (Supplementary Fig. 15), rtTA/TRE-Edn2-transduced hair follicle stem cell niches contained increased KIT+ melanocytes, melanin+ cells and TYRP1+ differentiated melanocytes (Fig. 4h). Notably, de novo melanogenesis was not detected in similarly transduced epidermis, indicating that EDN2’s effects were confined to locations where preexisting melanocyte stem cells reside (data not shown). Finally, premature melanocyte stem cell differentiation in Nfib-cKO niches depended upon EDN2, because marked reductions in TYRP1+ melanocytes and activated-CASP3+ hair follicle stem cells were observed when an endothelin receptor inhibitor (BQ788) was injected intradermally into cKO skin from late-catagen (Fig. 4i).
In summary, our findings expose an unexpected gatekeeper, NFIB, which governs activity within the quiescent stem cell niche of hair follicles. Upregulation of a singleNFIB target, Edn2, seemed to be sufficient to uncouple coordinated stem cell behaviour. In contrast to all known genetic pathways perturbing melanocyte stem cell function within hair follicles, precocious melanocyte differentiation and ensuing chaos was cyclical and did not compromise stem cell pools, hair pigmentation or growth, even in ageing animals. The mechanism underlying this unprecedented phenotype lends new importance to the two-tiered structure of the hair follicle stem cell niche, and to dermal papilla/KITL-independent and dependent steps in controlling the effects of EDN2.
Endothelin 2 has not been implicated hitherto in normal cutaneous melanocyte physiology. Moreover, whereas melanocyte specification during embryogenesis fails to occur without Edn3 (ref. 27), melano-genesis is seemingly normal in Edn1 knockout mice28. This has left it unclear as to whether endothelins function postnatally. Similarly to Edn3, elevating Edn2 in epidermis did not promote melanogenesis20; however, our results show that when provided with the proper microenvironment and additional stimulatory signals, endothelins can influence adult melanocyte stem cells and their differentiation.
In closing, our findings add new understanding to how melanocyte stem cell and hair follicle stem cell behaviours maintain reliance upon cooperative factors within the niche. They also reveal how this communication might be selectively uncoupled in injury and disease states. Notably, Edn2 is induced upon ultraviolet irradiation and other stress conditions associated with increased pigmentation29,30. Although beyond the scope of the present study, testing the possible role of NFIB in skin cancers, wound repair and stress responses merit investigation, as does removing possible redundancy from other NFI members expressed by hair follicle stem cells. In the future, it will be interesting to see the extent to which Nfib downregulation will tip the balance from coupled to uncoupled states in health and disease. Our studies here emphasize the importance of endothelins as important messengers to uncouple melanocyte and hair follicle stem cell synchrony.
METHODS
Mice
P52–60 CD1 mice from Charles River laboratories were used for NFIB ChIP-seq experiments. Nfibfl/fl and Nfib−/− mice have been described15,31, as have transgenic K15-crePGR, knock-in Sox9-creERT2 and Rosa26floxSTOPfloxYFP (RosaYFP) mice32–34. Dct-EGFP animals were generated by GENSAT project35. CreER was activated by intraperitoneal injection of P20–21 mice with 20mgml−1 tamoxifen (Sigma) in corn oil (Sigma) (1mg per 10 g body weight per day). Cre-PGR was induced by RU486 treatment: 5-day intraperitoneal injection of 10 mg ml−1 RU486 (VWR) in sesame oil (sigma) (0.75mg10 g body weight per day), together with 2-week topical application with 4% RU486 in ethanol.
All animals were maintained in an AAALAC-approved Comparative Bioscience Center (CBC) at The Rockefeller University and procedures were performed using IACUC-approved protocols that adhere to the standards of the National Institutes of Health.
Histology and immunofluorescence
Embryos (<E16.5) or back skins were embedded in OCT (Tissue Tek), frozen on dry ice and stored at −80 °C. Only in the case of RosaYFP reporter or lentiviral transduced mice, tissues were prefixed in4% paraformaldehyde (PFA) for 30 min at room temperature before embedding in OCT in order to preserve the fluorescence signals. Un-prefixed frozen sections (10–20 µm) were fixed in4%PFAfor 10 min at room temperature. For histological analysis, sections were stained with haematoxylin and eosin. Melanin staining was performed using a Fontana–Masson stain kit (MarketLab Inc.) according to the manufacturer’s directions. For immunofluorescence, sections were permeabilized in 0.3%Triton X-100 in PBS for 20 min and blocked for 1 h at room temperature in blocking buffer including 2.5%normal donkey serum, 2.5%normal goat serum(or 5% normal donkey serum alone when goat primary antibodies were used), 0.5% BSA and 0.1% Triton X-100 in PBS. For melanocyte markers, including DCT, TYRP1 and TYR, sections were permeabilized with 0.3% H2O2 in cold methanol for 30 min at-20 °C36.MOM Basic kit (Vector Laboratories) was used for blocking when primary antibodies were generated from mouse. Primary antibodies were diluted in blocking buffer and sections were incubated overnight at 4 °C. After washing with PBS for 30 min at room temperature, sections were incubated for 1–2 h at room temperature with secondary antibodies conjugated to Alexa-488, Alexa-546, Alexa-647 (Molecular Probes), or RRX (Jackson Laboratories). EdU staining was performed using Click-iT EdU Alexa Fluor 594 Imaging Kit (Life Technologies) and following manufacturer’s directions. Nuclei were stained using 4′6′-diamidino-2-phenylindole (DAPI). Imaging was performed on a Zeiss Axioplan 2, Zeiss Apotome, Zeiss Inverted LSM 510 laser scanning confocal microscope, or Zeiss Inverted LSM 780 laser scanning confocal microscope. Figures were prepared using ImageJ, Adobe Photoshop and Illustrator CS3.
The following antibodies and dilutions were used: NFIB (rabbit, 1:1,000, Active Motif); KIT (rat, 1:1,000, BD Pharmingen); P-cadherin (goat, 1:100, R&D); E-cadherin (rat, 1:500, Fuchs laboratory); K5 (guinea-pig, 1:500, Fuchs laboratory); CD34 (rat, 1:100, BD Pharmingen); Ki67 (rabbit, 1:300, Novocastra); cleaved-caspases 3 (rabbit, 1:300, R&D); GFP (chicken, 1:2,000, Abcam); EDN 1/2/3 (rabbit, 1:100, Santa Cruz); KITL (rat, 1:300, R&D); Mac-1 (rat, 1:100, eBioscience) MITF (mouse,1:100, Abcam); DCT (rabbit, 1:500, gift from V. J. Hearing); TYRP1 (rabbit, 1:1,000, gift from V. J. Hearing); TYR (rabbit, 1:1,000, gift from V. J. Hearing).
Electron microscopy
Back skins were fixed in 2% glutaraldehyde, 4% PFA and 2mMCaCl2 in 0.05M sodium cacodylate buffer, pH7.2, at room temperature for >1 h, post-fixed in 1% osmium tetroxide, and processed for Epon embedding. Ultrathin sections (60–70 nm) were counterstained with uranyl acetate and lead citrate. Images were taken with a transmission electron microscope (Tecnai G2–12; FEI) equipped with a digital camera (model XR60; Advanced Microscopy Techniques).
Isolation of hair follicle stem cells and FACS
Subcutaneous fat was removed from skins with a scalpel. To isolate bulge hair follicle stem cells for NFIB ChIP-seq experiment, skins were placed on 0.25% trypsin (GIBCO) at 37 °C for 30 min with dermis side down. For isolation of bulge and hair germ hair follicle stem cells, skins were first incubated in 0.25% collagenase (Sigma) in HBSS (GIBCO) at 37 °C for 1 h to remove dermis. After gentle PBS washing, skins were treated with trypsin as described above. Single-cell suspensions were obtained by scrapping the skin gently and filtering through 70-µm strainers, followed by 40-µm strainers. Staining buffer (PBS with 5% FBS treated with BioRad Chelex to remove calcium) was added to inactivate Trypsin, and cells were collected by centrifugation for 5 min at 300g. Cell suspensions were incubated with the appropriate antibodies diluted in staining buffer for 30 min at 4 °C. The following antibodies were used: CD34–Alexa 660 (1:200, eBioscience); α6–phycoerythrin (1:500, eBioscience); SCA1–Alexa 700 (1:200, eBioscience); CD200–biotin (1:200, AbD Serotec); streptavidin– phycoerythrin–Cy7 (1:200, eBioscience). DAPI (100 ng ml−1) was used for death cell exclusion. Cell isolations were performed on BD FACSAria II sorters equipped with BD FACSDiva software. For RNA extraction, cells were sorted directly into TRIzol LS Reagent (Life Technologies).
RNA extraction and RNA-seq analysis
Cells were isolated from Nfibfl/+/K15-crePGR/RosaYFP (Het) and Nfib fl/fl/K15-crePGR/RosaYFP (cKO) mice. Nfib ablation was induced from second telogen (P50–60) with RU486 treatment, and mice were taken during third telogen (P120–130).The protocol of cell sorting was described above with antibodies: CD34–Alexa 660, CD200–biotin, α6–phycoerythrin, SCA1– Alexa 700 and streptavidin– phycoerythrin–Cy7. Cells were lysed with TRIzol LS Reagent (Life Technologies) and RNA was extracted using Direct-zol RNA MiniPrep (Zymo Research) with DNase treatment to remove residual genomic DNA. Isolated RNA samples were provided to the Genomics Resources Core Facility at Weill Cornell Medical College for quality control, library preparation, clustering and sequencing. RNA-seq reads were aligned to the mouse genome (mm9,NCBI Build 37) using TopHat37 (http://tophat.cbcb.umd.edu/). Cufflinks38 (http://cufflinks.cbcb.umd.edu/) was subsequently used to assemble the aligned reads into transcripts and then estimate the transcript abundances. Pathway analysis was performed using iPAGE39 algorithm, included in the ChIPseeqer framework40.
Quantitative real-time PCR with reverse transcription
RNA isolation was described above. Complementary DNA was synthesized from isolated RNA using SuperScript III First-Strand Synthesis System with oligo-dT primers (Invitrogen). cDNAs were mixed with indicated primers and Power SYBR Green PCR Master Mix (Applied Biosystems), and quantitative PCR (qPCR) was performed on a Applied Biosystems 7900HT Fast Real-Time PCR system for 40 cycles. Specificity was confirmed by subsequent melting curve analysis or gel electrophoresis. Levels of PCR product were expressed as a function of peptidylprolyl isomerase B (Ppib). Primers were designed through Primer 3 and amplified products encompassed exon/intron boundaries. The following primer sequences were used: Nfib forward 5′-ATGACCCATCCAGTCCTCAA-3′, reverse 5′-TTGAAGGAAAGG CTCTCCAA-3′; Dct forward 5′-AGAGAAACAACCCTTCCACAGA-3′, reverse 5′-CCAATGACCACTGAGAGAGTTG-3′; Kit forward 5′-GGGCTAGCCA GAGACATCAG-3′, reverse 5′-AGGAGAAGAGCTCCCAGAGG-3′; Mitf forward 5′-TTGAAAACCGACAGAAGAAGC-3′, reverse 5′-TGGATGGGATA AGGGAAAGTC-3′; Tyrp1 forward 5′-GCCTCCAGTTACCAACACAGA-3′, reverse 5′-AACGGATCAGACAAGAAGCAA-3′; Tyr forward 5′-CCAGAAG CCAATGCACCTAT-3′, reverse 5′-ATAACAGCTCCCACCAGTGC-3′; Sox10 forward 5′-GACCAGTACCCTCACCTCCA-3′, reverse 5′-AGCCTCTCAGC CTCCTCAAT-3′; Ednrb forward 5′-CTCTGTTGGCTTCCCCTTC-3′, reverse 5′-CGATTGGATTGATGCAGGA-3′; Pax3 forward 5′-GCGAGAAAAAGG CTAAACACA-3′, reverse 5′-CGGAGCCTTCATCTGACTG-3′; S100a1 forward 5′-CAAGGAAGGGGACAAATATAAGC-3′, reverse 5′-CGTTTTCATCCAG TTCCTTCA-3′; Edn1 forward 5′-TACTTCTGCCACCTGGACATC-3′, reverse 5′-CCCTGAGTTCTTTTCCTGCTT-3′; Edn2 forward 5′-TTCTGCCATCGAA GACACTG-3′, reverse 5′-TGGCCTTTCTTGTCACCTCT-3′; Edn3 forward 5′- TGGGAAACAAGAGGACAAGG-3′, reverse 5′-CTGGGAGCTTTCTGGAAC TG-3′; Axin2 forward 5′-ACTGACCGACGATTCCATGT-3′, reverse 5′-CTGC GATGCATCTCTCTCTG-3′; Gpnmb forward 5′-TGCCTGCTGTCTGTGAG AAG-3′, reverse 5′-GGCAGTTTCCTATTGGCTTG-3′; Kitl forward 5′-CAAG TCTTACAAGGGCAGTTGA-3′, reverse 5′-ACAAGGTCACGGGTAGCAAG-3′; Chd3 forward 5′-ACTTTGATGAGCGTCCTGAAG-3′, reverse 5′-GGCTT GTCCTTCTCATTTCG-3′; Whrn forward 5′-TGGCTTATAGACCTGATGGA GAA-3′, reverse 5′-CTTCTGAGGGGGATTTGACAT-3′; Acsbg1 forward 5′-AAGTTCCTGTCCATGCTGCT-3′, reverse 5′-TGGAGAAGTCACGTTGGAG A-3′; Adamtsl4 forward 5′-GGCAACCAGACTCTCAGCTC-3′, reverse 5′- CGGCAGCAGGTAGTTGTGTA-3′; Cyp4b1 forward 5′-GCCTGATCTCTCTG CACATCTA-3′, reverse 5′-CACCTTCATCTCGTTCATAGCA-3′; Gfra1 forward 5′-TGCTGGCCCTCTAGATCCATAAC-3′, reverse 5′-ACAGCGCTTCT GGCAGTTGATA-3′; Myoc forward 5′-TCGGCTTTAGAGGAAGAGAAGA-3′, reverse 5′-CATACTTGCCAGCGATTGTTT-3′; Nt5e forward 5′-ATGAA CATCCTGGGCTACGA-3′, reverse 5′-GTCCTTCCACACCGTTATCAA-3′; Steap4 forward 5′-TGATTCCTATCCGTTACTATGTTCG-3′, reverse 5′-ATG GGCTGTCTTTATTAGTTAGGG-3′; Tns1 forward 5′-TCTTACCATTGCC CTCAATCC-3′, reverse 5′-CCACGGACTCCACATAGCTC-3′; Ppib forward 5′-GTGAGCGCTTCCCAGATGAGA-3′, reverse 5′-TGCCGGAGTCGACAAT GATG-3′.
NFIB ChIP-seq analysis
All materials and methods for ChIP-seq experiments have been described19. For each independent NFIB ChIP experiment, 10–15 million bulge hair follicle stem cells were isolated by FACS (CD34–Alexa 660 and α6–phycoerythrin antibodies) from 15–20 CD-1 mice, and then used to isolate chromatin and incubated with anti-NFIB antibody (Active Motif) for ChIP-seq analysis. ChIP-seq reads were aligned to the mouse genome (mm9, build 37) with the Bowtie program41 and the Illumina Analyzer Pipeline. ChIP-seq peak calling, genomic annotation ofNFIB peaks, and comparison between NFIB targets and gene expression signature genes were performed using ChIPseeqer40. Motif analysis was performed using FIRE42 algorithm, included in the ChIPseeqer framework.
Plasmid DNA constructions
Plasmids encoding full-length Nfib1 cDNA were obtained from ATCC (clone MGC-13959). Since NFIB3 is the major isoform present in epidermal keratinocytes, Nfib3 cDNA was used in this study. Nfib3 DNA fragment was amplified from Nfib1 cDNA clone by PCR with primers: forward 5′-GTTGCGAGCTCTCATGATGTATTCTC-3′, reverse 5′-GTCAACC CGGGCTAGCCCAGGTACCAGGACTGGCTCGTTTGAGGA-3′.
For expression of EGFP-NFIB3, Nfib3 DNA fragment was then inserted into pEGFP-C1 between SacI and XmaI restriction sites. All lentiviral vectors (LV) were present in the pLKO lentiviral backbone (pLKO-no-stuffer-PGK-MCS, Addgene 10879). For construction of LV-rtTA-IRES-EGFP, DNA fragment of rtTA-IRESEGFP was excised from pMSCV-rtTA2S-M2-IRES-EGFP with BglII and SalI restriction enzymes (RE) and cloned between BamHI and SalI sites of pLKO backbone. rtTA2S-M2 (Tet-On Advanced) was cloned from pUHD-rtTA2S-M2. This was excised by digesting with BamHI and SacII, and blunting with Klenow. The blunted fragment was subcloned into pMSCV-IRES-EGFP digested with HpaI, and dephosphorylated with SAP. For construction of LV-TRE-PGKH2BmRFP1, TRE promoter was first inserted into ClaI and AgeI sites of pLKO vector to replace U6 promoter (pLKO-TRE-PGK). The TRE promoter from Clontech’s pTRE2 was amplified by PCR using primers flanked with ClaI and AgeI sites: forward 5′-CGTATATCGATGCCCTTTCGTCTCGA-3′, reverse 5′-GAATTACCGGTCGCGGAGGCTGGAT-3′.
H2BmRFP1 DNA fragment was obtained from pCR-H2BmRFP1 by BglII and NsiI restriction enzyme digestion26, which was then inserted between BamHI and NsiI sites of pLKO-TRE-PGK. Edn2 cDNA were amplified from keratinocyte cDNA library with primers flanked with EcoRI site: forward 5′-CGCCAGAA TTCATGGTCTCCGCCTGGT-3′, reverse 5′-CGCCAGAATTCGGTGTTATC TCTTCCTCCATC -3′.
Afterwards, DNA fragment was inserted into EcoRI site of LV-H2BmRFP1-TRE vector. All insertions were verified by DNA sequencing.
A detailed description of the ultrasound-guided lentiviral injection procedure and production of high-titre lentiviruses is described elsewhere26. Transduced mice were confirmed by genotyping with EGFP primers: forward 5′-AATGG CCACAAGTTCAGC-3′, reverse 5′-TCGCCGATGGGGGTATTCT-3′. Positive mice were fed with doxycycline-containing chow from P21.
Cell culture and in vitro Nfib overexpression
Primary Nfib-null (KO) keratinocytes were isolated from epidermis of E18.5 Nfib knockout embryos (Nfib−/2) and control keratinocytes were from heterozygous Nfib−/2 embryos. Keratinocytes were maintained in E-media supplemented with 15% serum and a final concentration of 0.05mM Ca2. Experiments were performed using primary cells with fewer than 15 passages. Plasmids expressing EGFP or EGFP–NFIB3 were introduced into control and KO keratinocytes with FuGENE 6 Transfection Reagent (Roche). After 24 h transfection, EGFP-positive cells were isolated by FACS for qPCR analysis.
Intradermal injection of EDNRB receptor inhibitor BQ788 and anti-KIT blocking antibody
To inhibit EDNRB receptor from late catagen, 100 µl BQ788 dissolved in PBS (1mgml−1, Tocris Bioscience) or PBS alone was injected intradermally into the back skin of mice on first catagen when skin colour turned from black to pink6. To inhibit KIT signalling, 100 µl anti-KIT antibody (BD Pharmingen, 1.0 mgml−1, clone: ACK45 NA/LE) or control Rat IgG was used17. Injections were conducted every other day for 3 days (day 0, 2, 4), and mice were taken and analysed on day 5. For inhibition of KIT signalling during anagen, 100 µl anti-KIT antibody or control IgG was injected intradermally into back skin 1 day after depilation. Injections were conducted every other day for 5 days (day 0, 2, 4, 6, 8) and mice were taken and analysed on day 10.
Statistics
To determine significance between two groups indicated in figures, comparisons were performed in Prism 5 (GraphPad Software) with unpaired two-tailed student’s t-test.
Supplementary Material
Acknowledgements
We thank Y.-C. Hsu, T. Chen, B. Keyes, S. E. Williams, A. Rodriguez-Folgueras and other Fuchs laboratory members for intellectual input and suggestions; L. Polak and N. Stokes for breeding of mouse lines and conducting in utero lentiviral injections; V. J.Hearing for providing anti-DCT, TYRP1 and TYR antibodies. We also thank Rockefeller facilities: Comparative Bioscience Center (AAALAC accredited) for care and husbandry care of mice in accordance with National Institutes of Health (NIH) guidelines; Bioimaging Center for advice on image acquisition; Flow Cytometry facility for FACS sorting. E.F. is an investigator of the Howard Hughes Medical Institute. This work was supported by grants from the NIH to E.F. (R01-AR050452 and R01-AR31737) and R.M.G. (R01-HL080624), and a CAREER grant to O.E. from the National Science Foundation (DB1054964).
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions C.-Y.C. performed all experiments; H.A.P. performed the ultrastructural analyses; E.G.G. and O.E. performed the bioinformatic analyses; G.G. carried out the initial characterization of NFIB expression during mouse development; R.M.G. provided the conditional Nfibfl/fl mice; E.F. supervised the project; E.F. and C.-Y.C. wrote the manuscript.
ChIP-seq data have been deposited in the Gene Expression Omnibus database under accession number GSE42900.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
References
- 1.Tumbar T, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishimura EK, et al. Dominant role of the niche in melanocyte stem cell fate determination. Nature. 2002;416:854–860. doi: 10.1038/416854a. [DOI] [PubMed] [Google Scholar]
- 3.Blanpain C, Fuchs E. Epidermal homeostasis: a balancing act of stem cells in the skin. Nature Rev. Mol. Cell Biol. 2009;10:207–217. doi: 10.1038/nrm2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cotsarelis G. Gene expression profiling gets to the root of human hair follicle stem cells. J. Clin. Invest. 2006;116:19–22. doi: 10.1172/JCI27490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirobe T. How are proliferation and differentiation of melanocytes regulated? Pigment Cell Melanoma Res. 2011;24:462–478. doi: 10.1111/j.1755-148X.2011.00845.x. [DOI] [PubMed] [Google Scholar]
- 6.Rabbani P, et al. Coordinated activation of Wnt in epithelial and melanocyte stem cells initiates pigmented hair regeneration. Cell. 2011;145:941–955. doi: 10.1016/j.cell.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishimura EK, et al. Key roles for transforming growth factor βin melanocyte stem cell maintenance. Cell Stem Cell. 2010;6:130–140. doi: 10.1016/j.stem.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greco V, et al. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell. 2009;4:155–169. doi: 10.1016/j.stem.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanimura S, et al. Hair follicle stem cells provide a functional niche formelanocyte stem cells. Cell Stem Cell. 2011;8:177–187. doi: 10.1016/j.stem.2010.11.029. [DOI] [PubMed] [Google Scholar]
- 10.Weiner L, et al. Dedicated epithelial recipient cells determine pigmentation patterns. Cell. 2007;130:932–942. doi: 10.1016/j.cell.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 11.Inomata K, et al. Genotoxic stress abrogates renewal of melanocyte stem cells by triggering their differentiation. Cell. 2009;137:1088–1099. doi: 10.1016/j.cell.2009.03.037. [DOI] [PubMed] [Google Scholar]
- 12.Fitch KR, et al. Genetics of dark skin in mice. Genes Dev. 2003;17:214–228. doi: 10.1101/gad.1023703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gründer A, et al. Nuclear factor I-B (Nfib) deficient mice have severe lung hypoplasia. Mech. Dev. 2002;112:69–77. doi: 10.1016/s0925-4773(01)00640-2. [DOI] [PubMed] [Google Scholar]
- 14.Dooley AL, et al. Nuclear factor I/B is an oncogene in small cell lung cancer. Genes Dev. 2011;25:1470–1475. doi: 10.1101/gad.2046711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steele-Perkins G, et al. The transcription factor gene Nfib is essential for both lung maturation and brain development. Mol. Cell. Biol. 2005;25:685–698. doi: 10.1128/MCB.25.2.685-698.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu YC, Pasolli HA, Fuchs E. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell. 2011;144:92–105. doi: 10.1016/j.cell.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Botchkareva NV, Khlgatian M, Longley BJ, Botchkarev VA, Gilchrest BA. SCF/c-kit signaling is required for cyclic regeneration of the hair pigmentation unit. FASEB J. 2001;15:645–658. doi: 10.1096/fj.00-0368com. [DOI] [PubMed] [Google Scholar]
- 18.Rendl M, Lewis L, Fuchs E. Molecular dissection of mesenchymal-epithelial interactions in the hair follicle. PLoS Biol. 2005;3:e331. doi: 10.1371/journal.pbio.0030331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lien WH, et al. Genome-wide maps of histone modifications unwind in vivo chromatin states of the hair follicle lineage. Cell Stem Cell. 2011;9:219–232. doi: 10.1016/j.stem.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aoki H, Yamada Y, Hara A, Kunisada T. Two distinct types of mouse melanocyte: differential signaling requirement for the maintenance of non-cutaneous and dermal versus epidermal melanocytes. Development. 2009;136:2511–2521. doi: 10.1242/dev.037168. [DOI] [PubMed] [Google Scholar]
- 21.D’Orazio JA, et al. Topical drug rescue strategy and skin protection based on the role of Mc1r in UV-induced tanning. Nature. 2006;443:340–344. doi: 10.1038/nature05098. [DOI] [PubMed] [Google Scholar]
- 22.McDade SS, et al. Genome-wide analysis ofp63binding sites identifiesAP-2factors as co-regulators of epidermal differentiation. Nucleic Acids Res. 2012;40:7190–7206. doi: 10.1093/nar/gks389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pjanic M, et al. Nuclear factor I revealed as family of promoter binding transcription activators. BMC Genomics. 2011;12:181. doi: 10.1186/1471-2164-12-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pla P, Larue L. Involvement of endothelin receptors in normal and pathological development of neural crest cells. Int. J. Dev. Biol. 2003;47:315–325. [PubMed] [Google Scholar]
- 25.Nishikawa S, et al. In utero manipulation of coat color formation by a monoclonal anti-c-kit antibody: two distinct waves of c-kit-dependency during melanocyte development. EMBO J. 1991;10:2111–2118. doi: 10.1002/j.1460-2075.1991.tb07744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beronja S, Livshits G, Williams S, Fuchs E. Rapid functional dissection of genetic networks via tissue-specific transduction and RNAi in mouse embryos. Nature Med. 2010;16:821–827. doi: 10.1038/nm.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baynash AG, et al. Interaction of endothelin-3 with endothelin-B receptor is essential for development of epidermal melanocytes and enteric neurons. Cell. 1994;79:1277–1285. doi: 10.1016/0092-8674(94)90018-3. [DOI] [PubMed] [Google Scholar]
- 28.Reid K, et al. Multiple roles for endothelin in melanocyte development: regulation of progenitor number and stimulation of differentiation. Development. 1996;122:3911–3919. doi: 10.1242/dev.122.12.3911. [DOI] [PubMed] [Google Scholar]
- 29.Adur J, Takizawa S, Uchide T, Casco V, Saida K. High doses of ultraviolet-C irradiation increases vasoactive intestinal contractor/endothelin-2 expression in keratinocytes of the newborn mouse epidermis. Peptides. 2007;28:1083–1094. doi: 10.1016/j.peptides.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 30.Klipper E, et al. Induction of endothelin-2 expression by luteinizing hormone and hypoxia: possible role in bovine corpus luteum formation. Endocrinology. 2010;151:1914–1922. doi: 10.1210/en.2009-0767. [DOI] [PubMed] [Google Scholar]
- 31.Hsu YC, et al. Mesenchymal nuclear factor I B regulates cell proliferation and epithelial differentiation during lung maturation. Dev. Biol. 2011;354:242–252. doi: 10.1016/j.ydbio.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soeda T, et al. Sox9-expressing precursors are the cellular origin of the cruciate ligament of the knee joint and the limb tendons. Genesis. 2010;48:635–644. doi: 10.1002/dvg.20667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris RJ, et al. Capturing and profiling adult hair follicle stem cells. Nature Biotechnol. 2004;22:411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- 34.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heintz N. Gene expression nervous system atlas (GENSAT) Nature Neurosci. 2004;7:483. doi: 10.1038/nn0504-483. [DOI] [PubMed] [Google Scholar]
- 36.Govender D, Davids LM, Kidson SH. Immunofluorescent identification of melanocytes in murine hair follicles. J. Mol. Histol. 2006;37:1–3. doi: 10.1007/s10735-005-9011-8. [DOI] [PubMed] [Google Scholar]
- 37.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodarzi H, Elemento O, Tavazoie S. Revealing global regulatory perturbations across human cancers. Mol. Cell. 2009;36:900–911. doi: 10.1016/j.molcel.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giannopoulou EG, Elemento O. An integrated ChIP-seq analysis platform with customizable workflows. BMC Bioinformatics. 2011;12:277. doi: 10.1186/1471-2105-12-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elemento O, Slonim N, Tavazoie S. Auniversal framework for regulatory element discovery across all genomes and data types. Mol. Cell. 2007;28:337–350. doi: 10.1016/j.molcel.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.