Abstract
We recently developed a method for estimating protin dynamics in vivo with 2H2O using MALDI-TOF MS (Rachdaoui N. et al., MCP, 8, 2653-2662, 2009) and we confirmed that 2H-labeling of many hepatic free amino acids rapidly equilibrated with body water. Although this is a reliable method, it required modest sample purification and necessitated the determination of tissue-specific amino acid labeling. Another approach for quantifying protein kinetics is to measure the 2H-enrichments of body water (precursor) and protein-bound amino acid or proteolytic peptide (product) and to estimate how many copies of deuterium are incorporated into a product. In this study we have used nanospray LTQ-FTICR mass spectrometry to simultaneously measure the isotopic enrichment of peptides and protein-bound amino acids. A mathematical algorithm was developed to aid the data processing. The most notable improvement centers on the fact that the precursor:product labeling ratio can be obtained by measuring the labeling of water and a protein(s) (or peptides) of interest, therein minimizing the need to measure the amino acid labeling. As a proof of principle, we demonstrate that this approach can detect the effect of nutritional status on albumin synthesis in rats given 2H2O.
Keywords: albumin, heavy water, protein synthesis, rat, modeling, isotopomers, mass isotopomer distribution, high resolution mass spectrometry
Introduction
Early studies of protein turnover concentrated on whole body and organ specific synthesis [1; 2]. Advances in isotopic tracer methods and improvements in subcellular isolation methods have enabled studies of various protein fractions, i.e. total membrane, mitochondrial, sarcoplasmic or cytosolic proteins [3]. However, understanding of the pathologies related to the regulation of protein metabolism requires methods for studying the synthesis of individual proteins. For example, proteome turnover is differentially regulated and the summation of individual protein fluxes may result in a cancellation of changes in their kinetics [4]. Although recent advances in proteomics yields novel information regarding the static expression of individual proteins, methods to address questions regarding proteome dynamics are starting to emerge [5; 6; 7].
Classical studies of protein turnover relied on precursor-product relationships and involved the administration of a labeled amino acid[8]. In addition to the inconvenience related to long-term oral consumption or intravenous infusion of amino acids and difficulties in determining the true precursor labeling (the intracellular amino acid labeling in a specific organ), these methods also face challenges related to the measurement of the low isotopic labeling of a product and interpretation of the precursor:product labeling ratio.
We and others have recently started to use 2H2O to study protein kinetics in free living organisms [9; 10]. At lower levels , e.g. ~ 0.5% of total body water, 2H2O appears to be well tolerated in human [11; 12] whereas higher levels, e.g. < 10% labeling of water, have been used in a long-term rodent experiments [13]. The oral administration of 2H2O in drinking water results in steady state labeling of body water without perturbing the precursor pool size. In contrast to labeled amino acids, 2H2O rapidly equilibrates with total body water (including intracellular fluids) and it appears to readily label proteogenic amino acids [9; 14; 15]. In fact, steady state [2H]labeling of amino acids was observed in mice after intraperitoneal injection of 2H2O bolus, indicating that amino acids transfer to protein chain is a rate liming step in a protein biosynthesis [16]. Last, incorporation of multiple copies of 2H from 2H2O results in excess isotopic enrichment in a product therein enhancing measurements of labeling.
Previously, isotopic labeling of proteins was measured by GC-MS analysis of amino acids after hydrolysis of a specific protein(s) [10; 14]. In addition to being time consuming, those protocols suffer from potential contamination associated with protein isolation. Recently MALDI-TOF MS was applied to study protein kinetics after metabolic labeling, proteins were isolated and digested with trypsin [16; 17; 18]. Amplification of 2H enrichment in a peptide facilitates measurements of the product labeling by MALDI-TOF MS and the degree of tracer incorporation in a tryptic albumin peptide. Although MALDI-TOF MS is a reasonable choice in certain applications, the absence of a chromatographic inlet compromises broader studies of proteome dynamics. In addition, in some cases these studies necessitate the analysis of amino acids labeling in specific tissues for estimation of true precursor labeling which is needed for calculation of the fractional synthesis rate (FSR) of a protein [16]; clearly, invasive tissue analyses limits the application of this technique mainly to animal protocols and does not readily translate in human studies. In order to circumvent problems related to the measurement of intracellular amino acid labeling and to avoid issues related to sample purification, we improved the heavy water technique for estimating proteome dynamics. Our rationale assumes that the 2H-labeling of body water represents the precursor and the 2H-labeling of a tryptic-peptide(s) can be used to (i) calculate the asymptotic number of deuterium () incorporated into a peptide and/or (ii) measure the labeling of individual protein-bound amino acids via tandem mass spectrometry. The latter point is necessary when calculating the FSR of a protein in cases where there may be questions regarding the kinetics of amino acid labeling, e.g. in cases where the t1/2 of protein may approach the turnover (or labeling constant) of some amino acids. In those scenarios it is best to compare the labeling of a specific (and well characterized) protein-bound amino acid that should have a rapid turnover and undergo extensive labeling (e.g. alanine). We demonstrate the utility of this approach(es) by quantifying the effect of nutritional status on albumin synthesis in rats.
MATERIALS AND METHODS
Materials
HPLC grade solvents for nanospray chromatography and sample preparation were purchased from Fluka (Milwaukee, MO). Pure standards of N-(9-fluoronylmethyloxy-carbonyl-L,D-[2H]-alanine (L,D-[2H]-Fmoc-alanine, was purchased from (Cambridge Isotope Laboratories, Andover, MA, purity > 98%). All other chemicals were from Sigma-Aldrich (St. Louis, MO).
The peptides YLYEIAR and [2-2H]alanyl-YLYEIAR were synthesized using a solid-phase method in the Cleveland Clinic Molecular Biotechnology Core on a 396 52 Peptide Synthesizer (Advanced Chem Tech, Louisville, Kentucky). Stable isotope labeled L,D-[2H]-Fmoc-alanine was coupled in the peptide sequence to give a molecular mass shift of 1 Da from the unlabeled peptide with the average mass of 928.5 Da. Since [2H]-Fmoc-alanine used for labeled peptide synthesis was a mixture of L- and D-alanine derivatives, the synthetic [2-2H]alanyl-YLYEIAR represented the mixture of L- and D- isomers which were resolved and purified on HPLC system. The molecular weights of the purified peptides were verified by MALDI-TOF and ESI-MS and were found to have their expected average masses, the isotopic purity value (98%) was determined by an ESI-MS experiment. The physiological L-isomer of [2-2H]alanyl-YLYEIAR was used in our accuracy experiments.
Stock solutions of unlabeled and labeled peptides were made at concentrations of 13.5 μmol/L and 5 μmol/L in acidic water (pH = 3), respectively. A calibration curve was constructed by adding increasing amount of [2-2H]alanyl-YLYEIAR to constant aliquots of unlabeled YLYEIAR. This solution was divided into 0.05 ml fractions and saved at (−80°C) until analysis.
Animal studies
All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at the Cleveland Clinic and were performed in accordance with NIH guidelines. Male Sprague-Dawley rats (~ 250 g) were purchased from Charles River Laboratories (Wilmington, MA), and were housed in our animal care facility with a 12:12 h light:dark cycle. The animals had free access to food (20% kcal from protein, 70% kcal from carbohydrate and 10% kcal from fat, Harlan Teklad) and water.
Short-term experiment
After 3 days of quarantine, animals were housed in separate cages and their daily food intake was recorded during one week. The experimental variable (feeding versus fasting) implemented at 5:00 PM of the day of 10; rats were randomly divided into fed and fasted groups. Rats from the fed group had full access to food while food was removed from the fasted group. Next day morning at 8:00 AM 2H2O (20 μl of 99% APE 2H-labeled saline per g body weight) was given as an intraperitoneal injection. This priming dose of 2H2O enriches body water to ~ 2.65%. The sham animals (one fed and one fasted) were injected with the same dose of regular (H2O-saline). Animals were returned to their cages and their water was replaced with drinking water enriched with 5% 2H2O. Rats were euthanized (pentabarbital overdose, 120 mg/kg) in one hour intervals during 7 hours. Blood samples were collected through cardiac puncture. The control rats were euthanized after 1 h of H2O-saline injection. The fasting/feeding regimen affects protein turnover via an influx of cold diet derived substrate and allows assessing the effect of nutritional status on protein synthesis.
Long-term experiment
In order to assess the plateau labeling of proteins, one group of animals (5 Sprague-Dawley male rats) received loading dose (intraperitoneal injection) of 2H2O and then they had free access to drinking water enriched with 2H2O (5%) and food. Rats were euthanized after 1, 3, 5, 7 and 10 days. All blood samples were quickly centrifuged at 4 °C, plasma was removed and was stored at −80 °C until analysis.
Analytical
Total body water enrichment
[2H] Enrichment of total body water was measured using a modification [19] of the acetone exchange method [20]. Briefly, 10 μl of plasma was incubated with 2 μl of 10 N NaOH, and 5 μl of pure acetone in a 2 ml glass screw-cap GC vial at room temperature for 4 hours. The acetone from headspace was directly injected (5 μl) for gaschromatography-electron impact mass spectrometry analysis. Acetone was monitored at ions m/z 58 (M0), 59 (M1) and 60 (M2).
Sample preparation for albumin labeling measurement
Total proteins from 25 μl of plasma were precipitated with 100 μl of 10% thrichloroacetic acid (TCA). The pellets were centrifuged and twice washed with 100 μl of 5% TCA. Albumin along with other abundant proteins was extracted with 50 μl of pure ethanol. After evaporation of ethanol the residue was dissolved in 40 μl of sample buffer solution (4% sodium dodecyl sulfate (SDS), 20% glycerol, 10% 2-mercaptoethanol, 0.004% bromophenol blue and 0.125 M Tris HCl, pH 6.8) and purified using a polyacrylamide gel electrophoresis (PAGE).
In-Gel digestion
The albumin spot was exercised from 1D-PAGE gels, protein bands were cut from the gel as closely as possible to minimize excess polyacrylamide, divided into a number of smaller pieces, washed/ destained in 50% ethanol and 5% acetic acid solution in water and reduced with DTT and alkylated with iodoacetamide at 60 °C. Albumin was in-gel digested with an excess of Promega sequencing grade trypsin (5 μL of 20 ng/μL trypsin in 50 mM ammonium bicarbonate) at room temperature, overnight. The peptides that were formed were extracted from the polyacrylamide in two aliquots of 30μ L 50% acetonitrile with 5% formic acid. These extracts were combined and evaporated to <10 μL in a Speedvac and then reconstituted in ~30 μL of 1% acetic acid. 1 μL of the sample solution was injected for the LC-MS analysis.
LC-LTQ-ICFT-MS
Chromatographic separation of the protein digest was performed by an Ultimate 3000 nano HPLC (Dionex, Germering, Germany) with a trapping desalting precolumn (C18, PepMap100, 300 μm ×5 mm, 5 μm particle size, 100 Ǻ, Dionex, Germering, Germany) followed by a reverse phase column (C18, 75 μm×150 mm, 3 μm, Dionex), using a mobile phase A (0.1 % formic acid in a water) and B (80 % acetonitrile, 0.04% formic acid in water) gradient. The run started with 100% of phase A, after 5 min of desalting, the phase B was increased to 13% in 1 min and then linearly to 26% in 40 min. Subsequently the column was washed for 10 min with 100% of mobile phase B and equilibrated for 15min with 100% of A.
Tandem mass spectra were recorded on a Finnigan LTQ FT ULTRA hybrid mass spectrometry (Thermo Electron Corp., Bremen, Germany) equipped with a 7 T superconducting electromagnet and a Packed-Tip nanospray ionization Probe (Thermo Eectron Corp.) operated in a positive ion mode. The peptides were infused at a flow rate of 300 nL/min via the silica noncoated PicoTip emitter (FS360-20-10-C12, New Objective Inc., Woburn, MA) at the voltage of 2.4 kV. The inlet capillary temperature was maintained at 200° C.
The isotopic detection was performed in the ion cyclotron resonance (ICR) cell and the MS analysis was divided into four segments for monitoring selected peptides in a single chromatographic run (Table 1). The parent ions were monitored in the SIM mode (mass interval is 10 Da), while the CID fragments were recorded in the SRM mode (isolation window is 10 Da, mass interval is 30 Da) at a resolution of 12,500. In parallel with ICR measurements, the MS/MS of the selected peptides were produced in LTQ for confirmation of the peptide sequence.
Table 1.
Monitored rat albumin peptides.
| Retention time, min |
Ion | z | Mass Observed |
Mass M+H+ (experimental) |
Mass M+H+ (theoretical) |
Position | Peptide sequence |
|---|---|---|---|---|---|---|---|
| 11.4 | parent | 2 | 763.2983 | 1525.5896 | 1525.5893 | 76-88 | TCVADENAENCDK |
| fragments | 1 | 1165.4383 1094.4029 |
1165.4383 1094.4029 |
1165.4426 1094.4054 |
78-88 79-88 |
VADENAENCDK ADENAENCDK |
|
| 23.7 | parent | 3 | 339.8501 | 1017.5363 | 1017.5364 | 89-97 | SIHTLFGDK |
| fragments | 2 | 465.7553 409.2136 |
930.5036 817.4202 |
930.5043 817.4203 |
90-97 91-97 |
IHTLFGDK HTLFGDK |
|
| 30.5 | parent | 2 | 575.3106 | 1049.6142 | 1049.6150 | 66-75 | LVQEVTDFAK |
| fragments | 1 | 937.4604 809.40131 |
937.4604 809.4031 |
937.4625 809.4040 |
68-75 69-75 |
QEVTDFAK EVTDFAK |
|
| 38.2 | parent | 2 | 633.8221 | 1265.6372 | 1265.6364 | 247-257 | FPNAEFAEITK |
| fragments | 1 | 908.4714 837.4331 |
908.4714 837.4331 |
908.4724 837.4353 |
250-257 251-257 |
AEFAEITK EFAEITK |
Database Searching
For the identification of albumin the data were analyzed using all CID spectra collected in the experiment. Data was searched against NCBI rodent protein database (www.ncbi.nlm.nih.gov, version 2009) using MASCOT software (Matrix Science, London). The search was performed using carbamidomethyl as a fixed modification of cysteine, oxidation as an optional modification of methionine, and one allowed missed cleavage. All matching spectra were verified by manual interpretation.
Calculation of the fractional synthesis rate (FSR)
The FSR in a long-term experiment can be determined based on a single compartmental model by fitting a time course of total labeling of a peptide (Epeptide (t)) to an exponential rise curve equation:
| (1) |
This equation allows determining of asymptotical total labeling (E0), the rate constant (k) and the half-life (t1/2 = ln2/k) of a protein. Total labeling of a peptide was calculated using the formula:
| (2) |
where MPE Mi is the molar percent enrichment of isotopomer and calculated as
| (3) |
Although our approach does not require the background correction for natural enrichment, the total isotopic excess due to 2H incorporation was calculated via subtraction of the total baseline enrichment calculated for the control rat.
The rational for calculation of protein fractional synthesis rate in short-term experiments is based on the precursor:product relationship, assuming that 2H2O is the precursor and a proteolitic peptide is the product. Thus the FSR for the protein of interest is calculated using the equation:
| (4) |
where slope is the rate of the increase in 2H-labeling of peptide during 2H2O administration and Ewater is the steady state enrichment of total body water. is the asymptotic number of deuterium incorporated into a peptide, which is calculated using a mathematical algorithm (see below) based on the plateau labeling of a peptide and steady state labeling of total body water. Thus, estimation of the FSR in a short-term experiment requires measurements of peptide labeling by LC-MS/MS, water labeling (typically by GC-MS) and calculation of the asymptotic number of deuterium incorporated into the peptide, .
The FSR of a protein can also be calculated based on an isotopic excess of a peptide-bound amino acid. Note that estimation of the FSR based on a peptide-bound amino acid may require long-term labeling studied in order to incorporate sufficient quantity of 2H into a single non essential amino acid.
Mathematical Algorithm
The algorithm presented in this study is similar to the probability based models reported by MacCoss [21] and Cabral [22] where the asymptotic labeling of a peptide is a function of total body water and number of exchangeable hydrogen atoms:
| (5) |
Thus, when two of the three parameters are known the third one can be calculated. Since body water labeling can be easily determined by a simple protocol using GC-MS, here we consider application of the algorithm for the simulation of isotopic distribution and for calculation of .
Prediction of asymptotic labeling of a peptide
In our model an isotopomer distribution of a peptide was constructed using combinatorial linear equations based on a molecular formula for each peptide accounting for the number of exchangeable H atoms and the enrichment of body water. For this purpose H atoms in a peptide are divided into two groups, i.e. non exchangeable and exchangeable H atoms. Thus for a peptide molecule with C, H, N, O and S atoms the molecular formula for a peptide can be stated as
where c, n, o and s are number of C, H, N, O and S, respectively; we denote h as a number of non exchangeable hydrogen atoms (1H) and as a number of the C-H sites that can be exchanged with 2H as a result of metabolic labeling with 2H2O. The goal is to determine the number of incorporated hydrogen atoms () into a peptide as a function of total body 2H2O enrichment and amino acid composition of a peptide.
Experimentally the maximum number of exchangeable hydrogen atoms for each peptide of interest could be determined based on amino acid composition and the number of C-H sites in each amino acid that exchange with 2H as a result of equilibrium with total body water. It is known that the equilibrium of 2H incorporation from total body water into C-H sites of amino acids is not complete and the value of equilibrium constant depends on amino acid’s metabolic origin. Theoretically, all amino acids, including essential amino acids could exchange one H atom as a consequence of a transamination reaction. In practice, much lower values (0.04-0. 47) of deuterium incorporation were observed for essential amino acids [16]. For non essential amino acids the asymptotical numbers of exchangeable hydrogen atoms varies depending on their structure and their biosynthetic origin. Note that in this approach we ignore N-H, O-H and S-H sites, because hydrogen atoms at these sites spontaneously exchange with 2H2O and they easily back exchange with H2O during the sample preparation procedure.
Simulations of the mass isotopomer distribution profile of a peptide is based on probabilities of enriched isotopes of each atom. We compute theoretical isotopic distributions from the elemental composition of the sequence, first using self-convolution for every chemical element, and then convolutions between elements. This approach is has been implemented in the software, MassXplorer developed by Sadigov [23], for quantification using 18O-water labeling. When considering an element enriched with a specific isotope (for example, a mixture with enriched 2H), we create a “dummy” element. For this dummy element the isotope distribution is determined by the enrichment distribution. For example, if the 2H concentration in water is 5%, the isotope distribution of the dummy element will be 95% and 5% for 1H and 2H, respectively. Once the isotope distribution of the element is computed via self-convolution (given the assumed number of exchangeable hydrogens), we mix it with the other elemental distributions to obtain the final isotope distribution.
Calculation of asymptotical number of incorporated deuterium atoms ()
The inverse application of this software uses the measured isotopic enrichments of both the peptide and body water and to determine the asymptotic maximum number of incorporated deuterium atoms () into a peptide. Briefly, to calculate , a selected narrow m/z range of high resolution precursor SIM scans of a peptide from the ten day labeling experiment is extracted from the Xcalibur RAW file. The software generates all predicated isotopic distributions with different at specific precursor (total body water) labeling. Each predicted isotope distribution is then correlated against the measured isotopic distribution, the mean sum of square errors (SSE) is used to fit the experimental result to the predicted data. Thus, for a specific peptide, the best fit of is determined based on the minimum error between the theoretical isotopic distribution simulated by the program and the experimentally measured isotope distribution. The algorithm was applied to study albumin kinetics in rats whose body water was enriched with 2H2O during short-term experiment, the resulting calculated by the algorithm was subsequently used to estimate albumin FSR using formula (4). The algorithm for calculation of asymptotical number of incorporated deuterium atoms, as well as for simulations of the mass isotopomer distribution profile is implemented in a program written in the C/C++ language of Visual Studio 9. The program is freely available and can be requested from the corresponding author.
Data Presentation and Statistics
Data shown in figures are the total excess labeling in a given rat experiment. Each symbol in the figures corresponds to one rat experiment. The statistical significance of differences between the profiles of [2H]-enrichments of a selected peptide in fed and fasted experiments was tested using two-tailed t test with equal variance. The best fit for the number of exchangeable hydrogens was calculated as the sum of squares of the differences between the observed and theoretical. The exchange number with the smallest sum of squares is assumed the best fit to the experimental data, and the corresponding optimal value for the number of exchangeable hydrogens.
RESULTS
Prior to administering the tracer, rats consumed ~ 20 to 25 g food per day and exhibited normal growth; no adverse effect(s) was observed during 2H2O treatment in long-term experiments (rats consumed the same amount of food and maintained their normal growth). Samples from control rats (sham, H2O treated) were analyzed by an LTQ system using data dependent survey analysis to identify albumin-derived tryptic peptides that could be used for isotopic distribution analysis in subsequent SRM and SIM experiments. The sequence of rat albumin was conformed by the presence of 41 peptides of which 4 were selected for isotopic distribution analysis. Our selection criteria was based on (i) high frequency of alanine, glutamine, glutamate and glycine in the peptide sequence, (ii) high intensity of a peptide and its fragment ions with and without selected nonessential amino acids and (iii) good chromatographic properties including separation from isobaric peptides, elution time and chromatographic peak shape. Table 1 represents characteristic retention times, ion charge, mass to charge ratio, amino acid composition and position in protein sequence for each peptide and their selected fragments for mass isotopomer distribution analysis.
Mass Isotopomer Distribution Analysis of Peptides
One technical parameter that is crucial for mass isotopomer distribution analysis is the spectral accuracy of the mass spectrometer [24]. Although ICR mass spectrometry is capable of achieving unsurpassed resolution and mass accuracy with high sensitivity, our initial observations revealed that the highest resolving power (~ 1,000,000) results in greatest spectral error (up to 4%) when compared with the theoretical isotopomer distribution of a given peptide. Presumably this observation is largely attributed to the space charge effect observed in the ICR cell of these instruments [25], for example, similar errors in isotopic abundance measurements have been observed using LTQ/Orbitrap instruments and found to vary with the resolving power [26]. Rather than adopt additional methods to address issues regarding space charge effects, etc. [25], we determined that acquiring data at ~ 12,500 resolution would result in a reasonably low spectral error (i.e. ~ 1.0 %). Note that we previously reported the use of a modified data processing method that could achieve a minimal spectral error (i.e. <0.5 %)[27], however, we found that further improvement in spectral accuracy did not have a substantial affect on the biological kinetic parameters considering the effort that would be required to further modify the algorithm for processing the spectra to be practical at this time.
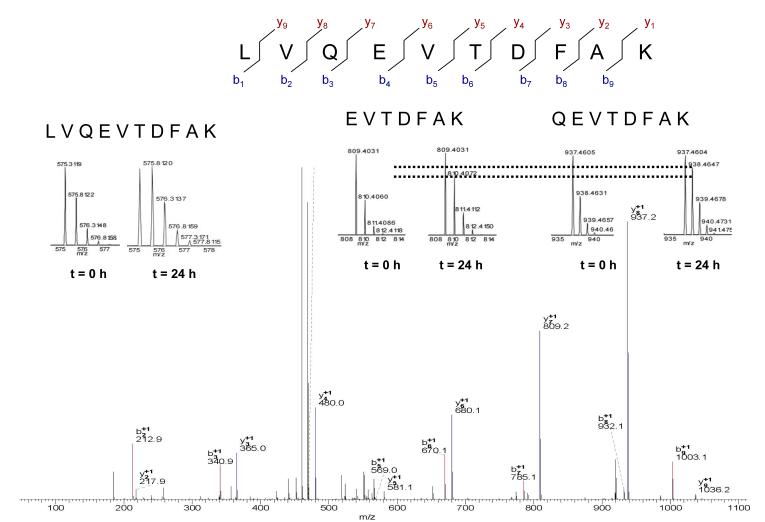
Figure 1 shows the FT MS/MS mass spectrum of the doubly charged precursor ion (575.312 Da) from the albumin peptide LVQEVTDFAK. The natural isotopic abundance of LVQEVTDFAK (from M0 to Mi), measured in plasma from sham treated rats, approximated the theoretical values (calculated using web-based Protein Prospector application, http://prospector.ucsf.edu/prospector/mshome.htm). Collision induced dissociation (CID) of this peptide yields multiple, and highly abundant, fragment ions. To determine whether ICR-FT MS would allow us to estimate the labeling of a specific peptide-bound amino acid we identified fragment ions with and without specific amino acids. Since glutamine (Q) is one candidate non-essential amino acid that may incorporate substantial amount of deuterium, QEVTDFAK and EVTDFAK were selected for SIM analysis. The inserts in Fig. 2 show the high resolution spectra of the parent ion (LVQEVTDFAK) and fragment ions (QEVTDFAK and EVTDFAK) before and after 24 h of 2H2O treatment, as expected, the precursor ion of LVQEVTDFAK peptide incorporated more 2H than product ions. The dashed line is added to aid in visualizing differences in the isotopic abundance, the data demonstrate that the daughter ion QEVTDFAK containing glutamine (m/z 937.460) has a higher M1/M0 ratio than the daughter ion EVTDFAK (m/z 809.403), the difference reflecting the labeling of glutamine.
Figure 1.
FT MS/MS CID spectra of LVQEVTDFAK from the doubly charged precursor ion identified at 575.31 Da in positive ion mode. The inserts show the high resolution spectra of the parent and two consecutive fragment ions (QEVTDFAK and EVTDFAK) before and after 24 h of 2H2O treatment, the dashed line is add to aid in visualizing the changes.
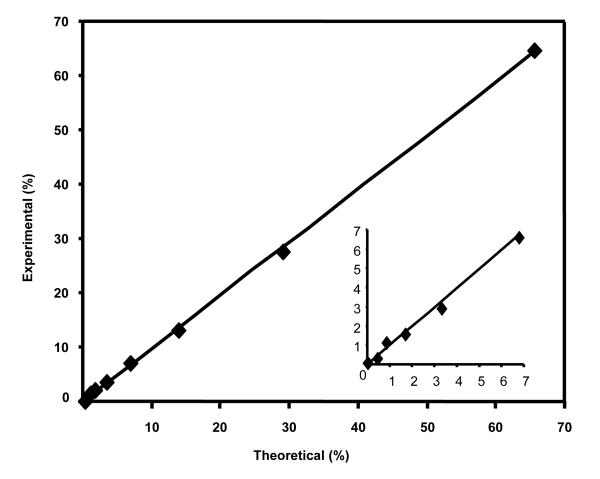
Figure 2.
Calibration curve of [2-2H]alanyl-YLYEIAR enrichment (1 to 68%), y = 0.9817x – 0.1703,r2 = 0.9994. The insert demonstrates the ability to quantify low levels of enrichment (e.g. < 7%), y = 09901x – 0.0052, r2 = 0.9909).
Figure 2 shows a calibration curve of [2H]alanyl-YLYEIAR enrichment. The curve is linear over a large range of labeling, i.e. we observed an r2 = 0.98 when analyzing samples containing from 1 to 68% enriched peptide mixtures.
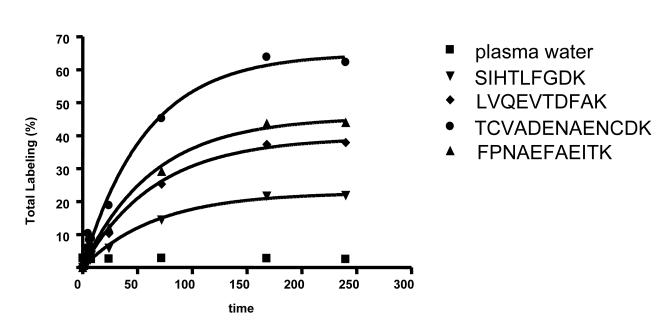
Figure 3 represents the time dependent labeling of body water and four albumin peptides from plasma obtained from fed rats. Consistent with other reports [10; 15; 16], total body water labeling rapidly reaches the steady state labeling and stabilizes at ~2.5-2.7%. The peptide labeling data were fit to a single pool model therein yielding the asymptotic labeling of each tryptic peptide and the rate constants describing albumin turnover. Note that the peptides reach different plateau labeling which is consistent with the fact that each contains different amino acids, however we observed similar rate constants (and half-lives) for each of the four peptides (Table 2). The latter parameter is comparable to previously reported values [28; 29].
Figure 3.
Time course labeling of body water and rat plasma albumin. ICR-FT MS analysis of albumin demonstrated an exponential increase in the labeling of four different albumin peptides. The total labeling of each peptide is different because of the amino acid sequence, however, all peptides reach plateau labeling after ~ 10 days of 2H2O administration. The fractional synthetic rate constants (k), the asymptotic total labeling (E0) of each peptide and the half-live (t1/2) of albumin calculated based on the best fit to an exponential curve for each peptide are presented in Table 2.
Table 2.
Calculated kinetic parameters and maximum number of incorporated deuterium into tryptic albumin peptides. The total plateau labeling, the turnover constant and half-life for each peptide were calculated using the best fit to a single exponential rise Equations (data are shown as mean ± se). The algorithm described in the text was used to calculate the best fit for the number of exchangeable hydrogens based on minimum of the sum of squares of error (SSE) differences between the observed and theoretical isotopic distribution.
| Peptide sequence and molecular formula |
Calculated Total Plateau labeling (%) |
Observed Fractional Synthetic Rate (day−1) |
Half-life (day) | Maximum number of incorporated deuterium |
|
|---|---|---|---|---|---|
| N | SSE | ||||
| TCVADENAENCDK C57 H93 N18 O27 S2 |
64.88 ± 0.13 | 0.41 ± 0.04 | 1.70 ± 0.17 | 24 | 0.00018 |
| SIHTLFGDK C46 H73 N12 O14 |
22.75 ± 0.94 | 0.36 ± 0.04 | 1.96 ± 0.24 | 9 | 0.00011 |
| LVQEVTDFAK C52 H85 N12 O17 |
39.41 ± 0.18 | 0.36 ± 0.03 | 1.93 ± 0.17 | 15 | 0.00016 |
| FPNAEFAEITK C59 H88 N13 O18 |
45.58 ± 0.76 | 0.38 ± 0.04 | 1.86 ± 0.2 | 20 | 0.00011 |
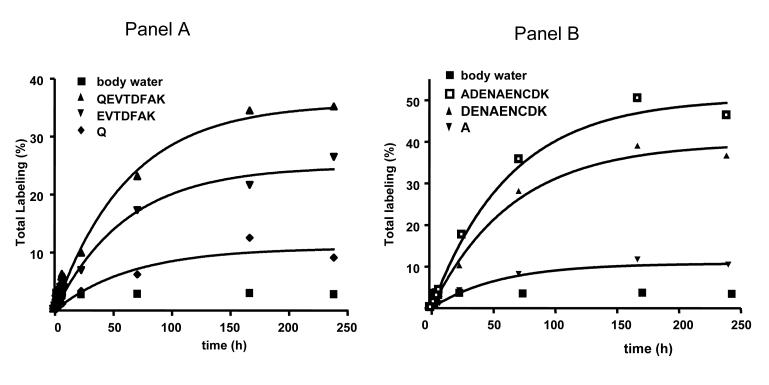
Figure 4 demonstrates the time dependent labeling observed in daughter ions derived from two peptides (LVQEVTDFAK (Panel A) and TCVADENAENCDK (Panel B)) presented in Figure 3. The difference between the labeling of two consecutive fragment ions, i.e. QEVTDFAK vs EVTDFAK (Panel A) and ADENAENCDK vs DENAENCDK (Panel B), represents the labeling of peptide-bound glutamine and alanine, respectively. As expected, glutamine and alanine, which each have > 1 exchangeable hydrogen atom, incorporates a substantial quantity of deuterium during long-term experiment and allowing a reliable estimation of albumin kinetic parameters (Table 3) when compared with data obtained using the precursor ion (Table 2) and, as noted earlier, agree with data in the literature [28; 29].
Figure 4.
Time course labeling of measured peptide fragments and calculated labeling of peptide-bound amino acid. Panel A demonstrates labeling profiles of QEVTDFAK and EVTDFAK fragments of QEVTDFAK peptide and calculated time-dependent labeling of glutamine (Q). Panel B demonstrates the labeling profiles of ADENAENCDK and DENAENCDK fragments of TCVDENAENCDK. The labeling of protein-bound alanine is estimated as a difference of the labeling of ADENAENCDK and DENAENCDK fragments.
Table 3.
Albumin kinetic parameters based on fragment peptides and protein-bound amino acid labeling (data are expressed as mean ± se).
| Fragments | Total Plateau Labeling, E0 (%) |
FSR, k (day−1) |
|---|---|---|
|
| ||
| ADENAENCDK | 50.1 ± 2.7 | 0.40 ± 0.07 |
| DENAENCDK | 39.4 ± 1.5 | 0.39 ± 0.07 |
| A | 10.7 ± 0.5 | 0.44 ± 0.09 |
| QEVTDFAK | 36.4 ± 1.0 | 0.33 ± 0.04 |
| EVTDFAK | 27.5 ± 1.1 | 0.36 ± 0.06 |
| Q | 10.2 ± 0.8 | 0.37 ± 0.05 |
Calculation of asymptotical number of 2H in C-H sites
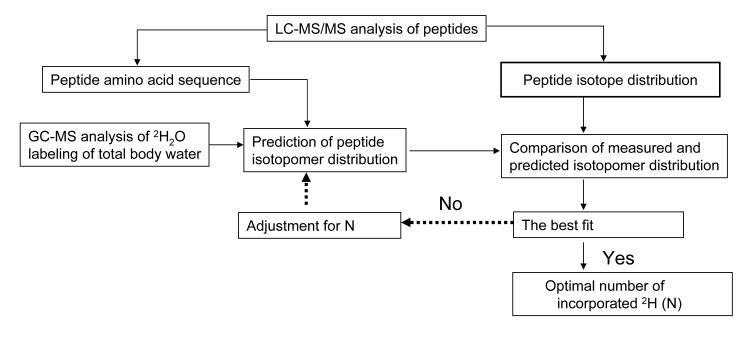
We used our algorithm to calculate the number of exchanged hydrogen atoms in each analyzed peptide (Figure 5). The software generates the mass isotopomer distribution of a peptide based on plasma 2H2O labeling and the different number of incorporated 2H atoms and compares that with the experimentally measured plateau labeling of a peptide (Figure 4). For the plateau labeling of albumin we used the data from our 10 day 2H2O experiment. Since the half-life of rat albumin is ~ 1.8 day, the total pool of rat albumin is close to completely turned over during the 10 day labeling experiment indicating that the number of incorporated deuterium atoms reflects the maximum possible incorporation. The minimum of square root of error was used to fit the experimental data to the theoretical model, the best fit for asymptotical number of exchanged hydrogen atoms represented in the last column of Table 2 for each analyzed peptide.
Figure 5.
A flow diagram for the analysis of metabolic protein labeling with 2H2O.
Effect of rat’s nutritional status on albumin synthesis
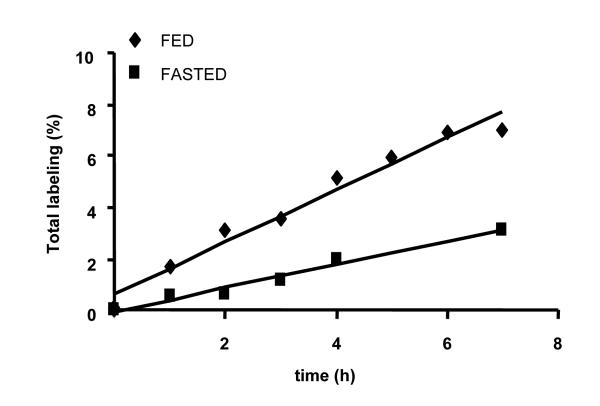
We applied the technique to assess the affect of nutritional status on albumin synthesis during a short-term (7 h) experiment in rats. Figure 6 shows that feeding stimulates 2H incorporation into albumin. Note that the data on fed rats represents the semi-linear part of the exponential curve from the long-term experiment. This technique allows a reliable measurement of the label incorporated into a tryptic peptide after one hour of 2H2O administration both in fed and fasted rats. The FSR of albumin calculated using equation 4 suggests that albumin synthesis in the fed state is ~2 fold higher than in the fasted state. The calculated rate of albumin synthesis was found equal to 13.3% versus 6.3% newly synthesized albumin over 7 h in fed versus fasted rats. This is also in a good agreement with earlier finding that in fasting state the total hepatic protein synthesis rate is reduced to 60-70% of the fed values [17].
Figure 6.
Influence of feeding on albumin synthesis. Rats were either given access to food or were fasted overnight before and during 2H2O administration. The slope of total labeling of FPNAEFAEITK (1.01 vs 0.46), body water labeling (2.79 ± 0.05 vs 2.6 ± 0.16) and asymptotical number of incorporated deuterium atoms (N) (20 ± 0.0001, calculated based on 10 days labeling experiment in a fed rat) were used for calculation of the fraction synthesis rate of albumin in fed vs fasted rats, respectively.
We also tested whether a high resolution mass spectrometry permits measurement of 2H incorporation into individual protein-bound amino acid in a short-term experiment. However, because of the lower enrichment of individual protein-bound amino acids, 2H incorporation could be measured only after ~4 hours of 2H2O administration. The relatively lower spectral accuracy of protein-bound amino acid labeling does not permit a reliable estimation of FSR based on protein-bound alanine and glutamine labeling in short-term experiments.
DISCUSSION
We present a novel and simple technique to study in vivo protein dynamics using metabolic heavy water labeling, high-resolution ICR-FT mass spectrometry and a mathematical algorithm for calculation of a protein FSR. This approach is based on measurement of enrichments of total body water, tryptic peptide or protein-bound amino acid and estimation of asymptotical number of deuterium incorporated into a peptide. As a proof of principle we applied this technique for estimation of the FSR of rat plasma albumin where we were able to quantify changes in protein synthesis that are consistent with expectations based on nutritional status.
One of the major challenges in protein turnover studies employing amino acid tracers is determination of intracellular precursor labeling for turnover calculations. In contrast to amino acids, 2H2O rapidly equilibrates with total body water and exchangeable hydrogen atoms of intracellular amino acids. This should allow investigator to consider 2H2O as a tracer studying for protein synthesis. Indeed, oral administration of heavy water after a bolus injection easily maintains a steady state labeling of total body water and results in a substantial enrichment of albumin. In contrast to difficulties related to measurement of the labeling of an intracellular amino acid, the enrichment of body water is easily and accurately assessed by simple head-space GC-MS analysis [30].
Although some protein turnover studies have used the average mass shift approach for kinetic calculations [17; 31], in our 2H2O labeling experiments we have quantified a shift in the relative mass isotopomer distribution (i.e. an increase in M1, M2, M3,.. isotopomers). This necessitates an accurate measurement of mass isotopomer distribution of a peptide which is achieved using nano-spray chromatography coupled with high resolution mass spectrometry of proteolytic-peptides. We believe that the SRM experiment improves the specificity of the assay and allows selective identification of peptide and its fragments for isotopomer analysis. Using SIM and SRM experiments in the same chromatographic run allow reliable quantification of the isotopic distribution of both the parent peptide and its fragment ions. Most importantly, an accurate measurement of two consecutive peptide fragments allowed us to calculate the labeling a specific protein-bound amino acid (Figure 4, Table 3).
According to a precursor:product relationship the fraction of protein that was newly synthesized is calculated as a ratio of the measured and calculated maximum asymptotic molar percent enrichment of peptide or peptide-bound amino acid [10]. The measured molar percent enrichment of a peptide is the combination of natural enrichments of atoms (13C, 2H, 17O, 18O, 33S and 34S) composing the peptide and 2H isotopic enrichment (excess mole fraction) during protein synthesis. The excess mole fractions of each isotopomer could be calculated after data reduction for natural baseline abundances [32], the asymptotical number of incorporated hydrogen atoms could be then be calculated from the ratio of two consecutive mass isotopomers. Recently Xiao et al. developed a mathematical algorithm for extraction of deuterium isotopic distribution from observed MALDI-TOF spectra [18]. In this study the isotopomer distribution is considered as a concatenation of groups of isotopomers from the elements composing a molecule. According to this rational, the inverse concatenation operation generates the deuterium isotopomers based on observed isotopomer distribution and theoretical carbon isotopomer distribution. However, for a baseline enrichment calculation Xiao et al. simplified their experimental model to carbon isotopes (12C and 13C) and ignored other elements (H, O, N and S) isotopes. They demonstrated that modeling of the isotope envelope of selected peptides based on carbon and deuterium is in reasonably good agreement with the predicted isotopic distribution, indicating that the approach allows one to resolve the distribution of heavier (newly made) protein from the lighter (natural) protein. However, ignoring isotopic abundance of 34S (5.6%) for cysteine and methionine containing peptides may give erroneous results. Similar to the mass isotopomer distribution analysis (MIDA) approach developed by Hellerstein and colleagues [10; 33] we use the theoretical asymptotical labeling of a peptide to calculate the fraction of newly synthesized protein.
Cabral et al. recently applied an alternative strategy for estimation of number of C-H bonds in glutathione that equilibrates with 2H2O[22]. In contrast to the methods discussed above, the latter technique does not require correction for natural abundances of carbon and other atoms. In this approach the natural abundances are included in the equations that are used for the model. Instead this approach requires a long-term experiment for achievement of the plateau labeling of the product. Our method expands on the work of Cabral et al., we used the data from a long-term experiment where that was complete turnover for calculation of . The calculated is then used in short-term experiments when protein turnover is not complete. The duration of 2H2O administration (i.e. defining “short” versus “long” term labeling) has to be determined based on the half-life of a specific protein. Collection of multiple samples at early hours of the study enables the estimation of turnover rates of multiple proteins with short half-life, while extending the experiment for several days or weeks allows the estimation of the kinetics for proteins with slow turnover rates. Rat albumin, which has a half-life of ~1.8 days and fractional synthetic rate constant of 0.38 × day−1, would approximately reach steady-state labeling in ~ 10 days. Using our algorithm based on data obtained following 10 days of labeling, we found the number of deuterium incorporated into the peptides, TCVADENAENCDK, SIHTLFGDK, LVQEVTDFAK, and FPNAEFAETK to be 24, 9, 15, and 20 respectively. These calculated values are in the range of expected number of exchanged C-H sites for specific amino acid labeling determined in our previous study and other data in the literature [16; 34]. Both essential and non-essential amino acids incorporate 2H as a result of reverse transamination reaction, labeling of non-essential amino acids with 2H2O also occurs during their synthesis and through intermediary metabolism involving intracellular water as a co-substrate. In addition, amino acids having β-hydrogen atom(s) could be labeled via keto-enol tautomerization (simple chemical H/D exchange with water at physiological pH) of the related α-keto acid. Our previous study demonstrated distinct equilibrium constants for 2H incorporation into different amino acids which is typically less than one [16]. The incomplete equilibrium of H/D exchange or C-H incorporation could be the result of an isotopic effect and/or incomplete reversibility of metabolic pathways involved in the synthesis of non-essential amino acids. Earlier studies on the estimation of lipogenesis found similar values for palmitate and stearate with low and high 2H2O enrichment, suggesting that there is negligible isotopic effect in conditions where the 2H-labeling of water is generally a few percent excess [35]. Our previous and current study demonstrates that only a few amino acids, namely glutamine, glutamate and alanine incorporate substantial number of deuterium atoms. These non-essential amino acids get extensively labeled through labeling of their precursors (oxaloacetate for alanine and α-ketoglutarate for glutamine and glutamate) in citric acid cycle and through keto-enol tautomerization. The observations are consistent with the recent report of DeRiva et al. [33]. The mathematical algorithm developed in this study enable calculations of the maximum number of 2H atoms incorporated into a peptide molecule. Since represents the number of 2H atoms in a peptide when the total protein pool has turned over completely and FSR of protein is a ratio of the measured and calculated maximum asymptotic molar percent enrichment of peptide, the determination of in combination with the steady state labeling of body water and peptide labe ling allows calculation of the FSR of a protein in short-term experiment.
After validation and optimization we applied this technique for estimation of albumin synthesis in rat plasma. Relatively accurate measurements of isotopic excess enabled the estimation of the FSR based on tryptic peptides and protein-bound amino acids. Although, albumin kinetics could be accurately measured using peptide labeling in both long-term and short-term experiments, protein-bound amino acid labeling permits only the measurement of long-term experiments when body water labeling is at ~2.5-2.7 %. However, future advances in high resolution mass spectrometry (with improvements in spectral accuracy measurements) may enable the utilization of protein-bound amino acid labeling for protein turnover studies in short-term experiments.
The technique was verified by comparing our results of albumin turnover rate and half-life with values reported in the literature using different approaches. We also demonstrated that the technique presented here detects decrease in albumin synthesis as a result of fasting. Since plasma albumin originates from liver[4], it may be possible to establish a link between the synthetic rate of plasma albumin and liver disease. In contrast to traditional studies of whole-body and/or tissue-specific protein synthesis, this approach could be used to investigate the turnover of individual proteins. The fact that 2H2O can be administered via a simple oral route in the drinking water may enable the application in basic and clinical research routines. As well, since 2H from 2H2O is incorporated into other important end-products, such as lipids, one could envision comprehensive studies of lipoprotein kinetics.
In conclusion, we have developed a new technique and algorithm for estimating protein turnover using 2H2O. This technique could be used to study the regulation of protein homeostasis and its relation to diseases associated with altered amino acid and protein metabolism. The algorithm presented in this report could be applied to global proteome turnover studies involving 2H2O.
Acknowledgement
We thank Hazel Huang and the ICUC staff for their help with the animal studies and Dr John Kirwan and Dr Visvanathan Chandramouli for helpful discussions during preparation of the manuscript. This work was supported in part by National Institutes of Health grant 5R21RR025346-02.
Abbreviations used
- 2H2O
heavy water
- Fmoc
9-fluorenylmethoxycarbonyl derivative
- μLC-LTQ-FTICR-MS
nanospray liquid chromatography linear trap Fourier transform ion cyclotron resonance mass spectrometry
- MALDI TOF MS
matrix-assisted laser desorption ionization time of flight mass spectrometry
- GC-MS
gas chromatography mass spectrometry
- SIM
selected ion monitoring
- SRM
selected reaction monitoring
- MIDA
mass isotopomer distribution analysis
- FSR
fractional synthesis rate
- SSE
sum of squares of error
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Mass isotopomers are molecules that differ by the presence of different isotopes resulting in isotopomer pattern in a mass spectrum. Mi represents the mass of the ith isotopomer above than mass of monoisotopic ion M0.
Reference
- [1].Smith K, Rennie MJ. The measurement of tissue protein turnover. Baillieres clinical endocrinology and metabolism. 1996;10:469–95. doi: 10.1016/s0950-351x(96)80651-3. [DOI] [PubMed] [Google Scholar]
- [2].Fukagawa NK, Minaker KL, Rowe JW, Goodman MN, Matthews DE, Bier DM, Young VR. Insulin-mediated reduction of whole body protein breakdown. Dose-response effects on leucine metabolism in postabsorptive men. The Journal of clinical investigation. 1985;76:2306–11. doi: 10.1172/JCI112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Guillet C, Boirie Y, Walrand S. An integrative approach to in-vivo protein synthesis measurement: from whole tissue to specific proteins. Current opinion in clinical nutrition and metabolic care. 2004;7:531–8. doi: 10.1097/00075197-200409000-00005. [DOI] [PubMed] [Google Scholar]
- [4].De Feo P, Gaisano MG, Haymond MW. Differential effects of insulin deficiency on albumin and fibrinogen synthesis in humans. The Journal of clinical investigation. 1991;88:833–40. doi: 10.1172/JCI115384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Price JC, Guan S, Burlingame A, Prusiner SB, Ghaemmaghami S. Analysis of proteome dynamics in the mouse brain. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14508–13. doi: 10.1073/pnas.1006551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Soufi B, Kumar C, Gnad F, Mann M, Mijakovic I, Macek B. Stable isotope labeling by amino acids in cell culture (SILAC) applied to quantitative proteomics of Bacillus subtilis. Journal of proteome research. 2010;9:3638–46. doi: 10.1021/pr100150w. [DOI] [PubMed] [Google Scholar]
- [7].Beynon RJ, Pratt JM. Metabolic labeling of proteins for proteomics. Molecular & cellular proteomics : MCP. 2005;4:857–72. doi: 10.1074/mcp.R400010-MCP200. [DOI] [PubMed] [Google Scholar]
- [8].Wolfe RR, Chinkes DL. Isotope Tracers in metabolic Research: Principles and Practice of Kinetic Analyses. Wiley-Liss; New York: 2004. [Google Scholar]
- [9].Previs SF, Fatica R, Chandramouli V, Alexander JC, Brunengraber H, Landau BR. Quantifying rates of protein synthesis in humans by use of 2H2O: application to patients with end-stage renal disease. American journal of physiology. 2004;286:E665–72. doi: 10.1152/ajpendo.00271.2003. [DOI] [PubMed] [Google Scholar]
- [10].Busch R, Kim YK, Neese RA, Schade-Serin V, Collins M, Awada M, Gardner JL, Beysen C, Marino ME, Misell LM, Hellerstein MK. Measurement of protein turnover rates by heavy water labeling of nonessential amino acids. Biochimica et biophysica acta. 2006;1760:730–44. doi: 10.1016/j.bbagen.2005.12.023. [DOI] [PubMed] [Google Scholar]
- [11].Peng SK, Ho KJ, Taylor CB. Biologic effects of prolonged exposure to deuterium oxide. A behavioral, metabolic, and morphologic study. Archives of pathology. 1972;94:81–9. [PubMed] [Google Scholar]
- [12].Hellerstein MK, Hoh RA, Hanley MB, Cesar D, Lee D, Neese RA, McCune JM. Subpopulations of long-lived and short-lived T cells in advanced HIV-1 infection. The Journal of clinical investigation. 2003;112:956–66. doi: 10.1172/JCI17533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Jones PJ, Leatherdale ST. Stable isotopes in clinical research: safety reaffirmed. Clinical science (London, England : 1979) 1991;80:277–80. doi: 10.1042/cs0800277. [DOI] [PubMed] [Google Scholar]
- [14].Dufner DA, Bederman IR, Brunengraber DZ, Rachdaoui N, Ismail-Beigi F, Siegfried BA, Kimball SR, Previs SF. Using 2H2O to study the influence of feeding on protein synthesis: effect of isotope equilibration in vivo vs. in cell culture. American journal of physiology. 2005;288:E1277–83. doi: 10.1152/ajpendo.00580.2004. [DOI] [PubMed] [Google Scholar]
- [15].Belloto E, Diraison F, Basset A, Allain G, Abdallah P, Beylot M. Determination of protein replacement rates by deutera ted water: validation of underlying assumptions. American journal of physiology. Endocrinology and metabolism. 2007;292:E1340–7. doi: 10.1152/ajpendo.00488.2006. [DOI] [PubMed] [Google Scholar]
- [16].Rachdaoui N, Austin L, Kramer E, Previs MJ, Anderson VE, Kasumov T, Previs SF. Measuring proteome dynamics in vivo: as easy as adding water? Molecular & cellular proteomics : MCP. 2009;8:2653–63. doi: 10.1074/mcp.M900026-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Vogt JA, Hunzinger C, Schroer K, Hèolzer K, Bauer A, Schrattenholz A, Cahill MA, Schillo S, Schwall G, Stegmann W, Albuszies G. Determination of fractional synthesis rates of mouse hepatic proteins via metabolic 13C-labeling, MALDI-TOF MS and analysis of relative isotopologue abundances using average masses. Analytical chemistry. 2005;77:2034–42. doi: 10.1021/ac048722m. [DOI] [PubMed] [Google Scholar]
- [18].Xiao GG, Garg M, Lim S, Wong D, Go VL, Lee WN. Determination of protein synthesis in vivo using labeling from deuterated water and analysis of MALDI-TOF spectrum. Journal of applied physiology (Bethesda, Md. : 1985) 2008;104:828–36. doi: 10.1152/japplphysiol.00976.2007. [DOI] [PubMed] [Google Scholar]
- [19].Shah V, Herath K, Previs SF, Hubbard BK, Roddy TP. Headspace analyses of acetone: a rapid method for measuring the 2H-labeling of water. Analytical biochemistry. :404235–7. doi: 10.1016/j.ab.2010.05.010. [DOI] [PubMed] [Google Scholar]
- [20].Yang D, Diraison F, Beylot M, Brunengraber DZ, Samols MA, Anderson VE, Brunengraber H. Assay of low deuterium enrichment of water by isotopic exchange with [U-13C3]acetone and gas chromatography-mass spectrometry. Analytical biochemistry. 1998;258:315–21. doi: 10.1006/abio.1998.2632. [DOI] [PubMed] [Google Scholar]
- [21].MacCoss MJ, Wu CC, Matthews DE, Yates JR., 3rd Measurement of the isotope enrichment of stable isotope-labeled proteins using high-resolution mass spectra of peptides. Analytical chemistry. 2005;77:7646–53. doi: 10.1021/ac0508393. [DOI] [PubMed] [Google Scholar]
- [22].Cabral CB, Bullock KH, Bischoff DJ, Tompkins RG, Yu YM, Kelleher JK. Estimating glutathione synthesis with deuterated water: a model for peptide biosynthesis. Analytical biochemistry. 2008;379:40–4. doi: 10.1016/j.ab.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sadygov RG, Zhao Y, Haidacher SJ, Starkey JM, Tilton RG, Denner L. Using power spectrum analysis to evaluate (18)O-water labeling data acquired from low resolution mass spectrometers. Journal of proteome research. 2010;9:4306–12. doi: 10.1021/pr100642q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cassano AG, Wang B, Anderson DR, Previs S, Harris ME, Anderson VE. Inaccuracies in selected ion monitoring determination of isotope ratios obviated by profile acquisition: nucleotide 18O/16O measurements. Analytical biochemistry. 2007;367:28–39. doi: 10.1016/j.ab.2007.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bresson JA AG, Bruce JE, Smith RD. Improved isotopic abundance measurments for high resolution fourier transform ion cyclotron resonance mass spectra via time-domain data extraction. Journal of American Society for Mass Spectrometry. 1998;9:799–804. [Google Scholar]
- [26].Erve JC, Gu M, Wang Y, DeMaio W, Talaat RE. Spectral accuracy of molecular ions in an LTQ/Orbitrap mass spectrometer and implications for elemental composition determination. Journal of the American Society for Mass Spectrometry. 2009;20:2058–69. doi: 10.1016/j.jasms.2009.07.014. [DOI] [PubMed] [Google Scholar]
- [27].Ilchenko S, Li L, Rachdaoui N, Willard B, McCullough A, Kasumov T. A mathematical algorithm for estimation of protein dynamics using mass spectrometry. Journal of the American Society for Mass Spectrometry. 2010:S93. [Google Scholar]
- [28].Papageorgopoulos C, Caldwell K, Shackleton C, Schweingrubber H, Hellerstein MK. Measuring protein synthesis by mass isotopomer distribution analysis (MIDA) Analytical biochemistry. 1999;267:1–16. doi: 10.1006/abio.1998.2958. [DOI] [PubMed] [Google Scholar]
- [29].Stephen JM, Waterlow JC. Use of carbon-14-labelled arginine to measure the catabolic rate of serum and liver proteins and the extent of amino-acid recycling. Nature. 1966;211:978–80. doi: 10.1038/211978a0. [DOI] [PubMed] [Google Scholar]
- [30].Shah V, Herath K, Previs SF, Hubbard BK, Roddy TP. Headspace analyses of acetone: a rapid method for measuring the 2H-labeling of water. Analytical biochemistry. 2010;404:235–7. doi: 10.1016/j.ab.2010.05.010. [DOI] [PubMed] [Google Scholar]
- [31].Zhao Y, Lee WN, Lim S, Go VL, Xiao J, Cao R, Zhang H, Recker RR, Xiao GG. Quantitative proteomics: measuring protein synthesis using 15N amino acid labeling in pancreatic cancer cells. Analytical chemistry. 2009;81:764–71. doi: 10.1021/ac801905g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jennings ME, 2nd, Matthews DE. Determination of complex isotopomer patterns in isotopically labeled compounds by mass spectrometry. Analytical chemistry. 2005;77:6435–44. doi: 10.1021/ac0509354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].De Riva A, Deery MJ, McDonald S, Lund T, Busch R. Measurement of protein synthesis using heavy water labeling and peptide mass spectrometry: Discrimination between major histocompatibility complex allotypes. Analytical biochemistry. 2010;403:1–12. doi: 10.1016/j.ab.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Commerford SL, Carsten AL, Cronkite EP. The distribution of tritium among the amino acids of proteins obtained from mice exposed to tritiated water. Radiation research. 1983;94:151–5. [PubMed] [Google Scholar]
- [35].Lee WN, Bassilian S, Ajie HO, Schoeller DA, Bergner J. Edmond, E. A., Byerley LO. In vivo measurement of fatty acids and cholesterol synthesis using D2O and mass isotopomer analysis. The American journal of physiology. 1994;266:E699–708. doi: 10.1152/ajpendo.1994.266.5.E699. [DOI] [PubMed] [Google Scholar]