Abstract
Background
Common FTO (fat mass and obesity associated) gene variants have recently been associated with body mass index (BMI) and obesity in several large studies. The role of lifestyle factors (such as physical activity) in those with an underlying FTO genetic predisposition is unknown.
Methods
To determine if FTO variants are associated with BMI in Old Order Amish (OOA) individuals, and to further determine whether the detrimental associations of FTO gene variants can be lessened by increased physical activity, a total of 704 healthy OOA adults were selected from the Heredity and Phenotype Intervention (HAPI) Heart Study, an investigation of gene × environment interactions in cardiovascular disease, for whom objective quantified physical activity measurements were available and for whom 92 single-nucleotide polymorphisms (SNPs) in FTO were genotyped.
Results
Twenty-six FTO SNPs were associated with BMI (P =.04 to <.001), including rs1477196 (P <.001) and rs1861868 (P <.001), 2 SNPs in moderate linkage disequilibrium in the OOA (D′=0.82; r2=0.36). Stratified analyses of rs1861868 revealed its association with BMI to be restricted entirely to those subjects with low sex-and age-adjusted physical activity scores (P <.001); in contrast, the SNP had no effect on those with above-average physical activity scores (P = .29), with the genotype × physical activity interaction achieving statistical significance (P =.01). Similar evidence for interaction was also obtained for rs1477196.
Conclusions
Our results strongly suggest that the increased risk of obesity owing to genetic susceptibility by FTO variants can be blunted through physical activity. These findings emphasize the important role of physical activity in public health efforts to combat obesity, particularly in genetically susceptible individuals.
Obesity and related comorbid conditions represent a global public health burden and account for a growing portion of health care spending in the industrialized world. It is widely acknowledged that there is a substantial genetic contribution to body mass index (BMI), and recently, robust associations of common variants in intron 1 of the fat mass and obesity associated (FTO) gene with BMI, percentage of body fat, and obesity were identified in large studies of white adults and children.1–3 Owing to the high frequency of the obesity-associated FTO variants (about 30% allele frequency for the most strongly associated single-nucleotide polymorphisms [SNPs] in European populations) and their impact (each “risk allele” is associated with a 1.75-kg increase in body weight), these variants carry a population-attributable risk for obesity of greater than 20% in the studied populations.1,3
The function of FTO is incompletely understood, although recent work4 has demonstrated that this gene codes for a protein expressed in the hypothalamus, a center of energy balance, and adipose tissue, where it is localized in cell nuclei and may be involved in demethylation of DNA and perhaps other, as-yet unidentified, functions.
In addition to genetic factors, lifestyle factors, including diet and physical inactivity, are important contributors to weight gain and obesity. Although physical activity has been shown to facilitate weight loss and weight maintenance in obese subjects, there is great interindividual variation in response.5 It is unknown whether lifestyle factors, such as physical activity, can attenuate weight gain and obesity in those with an underlying FTO genetic predisposition. The specific issues addressed in this report are whether FTO variants are associated with BMI in the Old Order Amish (OOA), a population in which moderate to high levels of physical activity are common and, if so, whether the detrimental associations of BMI-associated FTO gene variants can be lessened by increased physical activity.
METHODS
STUDY PARTICIPANTS
Study participants were members of the OOA community in Lancaster County, Pennsylvania. The OOA are a rural-living, closed, founder population of European origin, well known for eschewing many modern conveniences; they do not own cars or use electricity in their homes. Most OOA men are farmers or work in physically demanding occupations such as blacksmithing and carpentry. Women are homemakers, working without the aid of modern appliances and often taking care of many children. Similarities in tradition, formal education, and geographic location among the OOA make for a relatively homogeneous lifestyle, including dietary habits. Individuals included in this report took part in the Heredity and Phenotype Intervention (HAPI) Heart Study, an investigation of gene×environment interactions in cardiovascular disease of 868 adult OOA individuals in generally good health, who were recruited from 2003 to 2007.6 Subjects provided blood samples for DNA analysis and underwent a wide panel of physiological tests, including 7-day measurement of physical activity by accelerometry. The protocol was approved by the institutional review board of the University of Maryland, Baltimore. Subjects gave informed consent before participation.
PHENOTYPIC ASSESSMENT
All study subjects underwent a detailed clinical examination at the Amish Research Clinic in Strasburg, Pennsylvania. Height and weight were measured by trained nurses in subjects without shoes and in light clothing using a stadiometer and calibrated scale. Body mass index was calculated as weight in kilograms divided by height in meters squared. Subjects with a BMI of 25 or greater and less than 30 were defined as being overweight, and those with a BMI of 30 or greater were defined as obese. Additional measures of other obesity-related traits were obtained, including waist circumference, measured to the nearest 0.1 cm using an inelastic tape, and in a subset of 355 individuals, body composition (fat mass, lean mass, and derived percentage of body fat) as determined by dual energy x-ray absorptiometry (DEXA).
Objective measurement of physical activity was obtained over 7 consecutive 24-hour days using Actical activity monitors (versions 8.2 and 8.3; Mini Mitter Co Inc, Bend, Oregon) worn on the hip. These devices incorporate an accelerometer, sensitive to 0.01 times gravity in multiple directions, electronic circuitry, and a memory. Acceleration of the device is integrated and expressed as a number—activity counts—for each 15-second recording interval that is stored in memory until the device is returned and data uploaded. Activity can be expressed as raw activity counts, inherently independent of body size, or as its associated energy expenditure (in kilocalories per day) using formulas validated against gas exchange.7 Accelerometry is a well-established method to quantify physical activity.8 Of the 868 HAPI Heart Study participants, physical activity measurements were available for 711 subjects, of whom 704 had valid genotype data.
GENOTYPING
Genomic DNA isolated from whole blood was genotyped with the Affymetrix GeneChip Human Mapping 500 K Array set (Affymetrix, Santa Clara, California). Total genomic DNA (250 ng) was digested with NspI or StyI enzymes and processed according to the Affymetrix protocol. The GeneChip genotyping analysis software (GTYPE version 4.0; Affymetrix) was used for automated genotype calling as part of the GeneChip operating software platform. The GTYPE-generated chip files were reanalyzed using the Bayesian robust linear model with Mahalanobis (BRLMM) genotype calling algorithm (free, downloadable software available at http://www.affymetrix.com/support/technical/whitepapers/brlmm_whitepaper.pdf) and a confidence threshold for call quality of 0.5. As an initial quality control measure, BRLMM-generated chip files with genotype call rates of less than 93% for both enzymes were excluded from further analysis. For this study, SNPs that were either monomorphic or that deviated from Hardy-Weinberg equilibrium using a cut point of P <.001 were excluded from analysis.
The FTO gene, located on chromosome 16q12.2, is approximately 430 kilobases (kb) in length and contains 9 exons. The Affymetrix 500 K arrays contain 92 SNPs within the region of this gene. The mean genotype call rate was 98.6% for these 92 SNPs.
STATISTICAL ANALYSES
Association analyses of FTO SNPs and trait variables were performed using a variance component approach. Non–normally distributed variables were natural log-transformed prior to analyses. We modeled BMI and related traits as a function of measured environmental covariates, additive genetic associations, and a residual error component. The associations of age, age2, sex, and interaction terms for age and sex were estimated jointly with genotype associations. Genotype was scored using an additive model to allow for allele dosage effects. Parameter estimates were obtained by maximum likelihood methods, and the significance of association was tested by a likelihood ratio test. We accounted for the relatedness of OOA study subjects by estimating parameter effects conditional on residual correlations in BMI (or similar trait) between related individuals. The association analyses were performed using the SOLAR software program.9
Pairwise linkage disequilibrium (LD) correlation statistics (r2) were computed using Haploview beta software, version 4.0.10 Multilocus haplotype analyses were performed using HaploStats software,11,12 which estimates haplotype frequencies using an expectation-maximization algorithm in situations in which the haplotype phase is ambiguous. Global and haplotype-specific score statistics and permutation-based P values were calculated after adjustments were made for sex, age, and age2. These analyses did not take into account the relatedness of study subjects.
We evaluated the associations of FTO SNPs on BMI after stratification of the sample according to “high” and “low” physical activity strata. Because the daily number of physical activity accelerometer counts was generally lower in women than in men and decreased with age in both sexes (data not shown), this stratification was performed in a sex- and age-specific fashion. After logarithmic transformation of mean total physical activity counts, subjects were dichotomized into the high- or low-activity stratum depending on whether their age-, age2- and sex-specific residuals were greater than or less than 0. In addition to these stratified analyses, we determined whether the association of genotype on BMI was modified by physical activity levels by constructing a regression model that included the following independent variables: sex, age, age2, age×sex, age2×sex, SNP genotype, ln-transformed physical activity counts, and an interaction term (ln-transformed physical activity counts×genotype). The presence of an interaction between physical activity and SNP genotype on BMI was assessed by a likelihood ratio test, in which we compared a model with the interaction term (full model) with a model without the interaction term (nested model). As in previous analyses, these analyses were performed using variance components analyses in the SOLAR software program to account for the correlations in BMI among family members.
For the conversion of accelerometer counts into activity energy expenditure (in kilocalories per day), we used Actical computer software (version 2.04), provided by the manufacturer of the accelerometers. This software applies a validated regression equation7 to accelerometer counts on a per-epoch basis.
RESULTS
A total of 704 subjects with both physical activity and genotype data were included in our analyses. The mean (SD) age was 43.6 (3.4) years, and the sample included slightly more men than women (53% vs 47%). The mean BMI was higher in women (27.8 [5.7]) than in men (25.7 [3.4]). The prevalence of overweight and obesity in OOA men was 54.0% and 10.1%, respectively, and in OOA women was 63.7% and 30.5%, respectively. Mean physical activity in men and women amounted to 519 (271) and 383 (219)×103 counts/d, respectively.
FTO SNP ASSOCIATIONS WITH OBESITY AND RELATED TRAITS
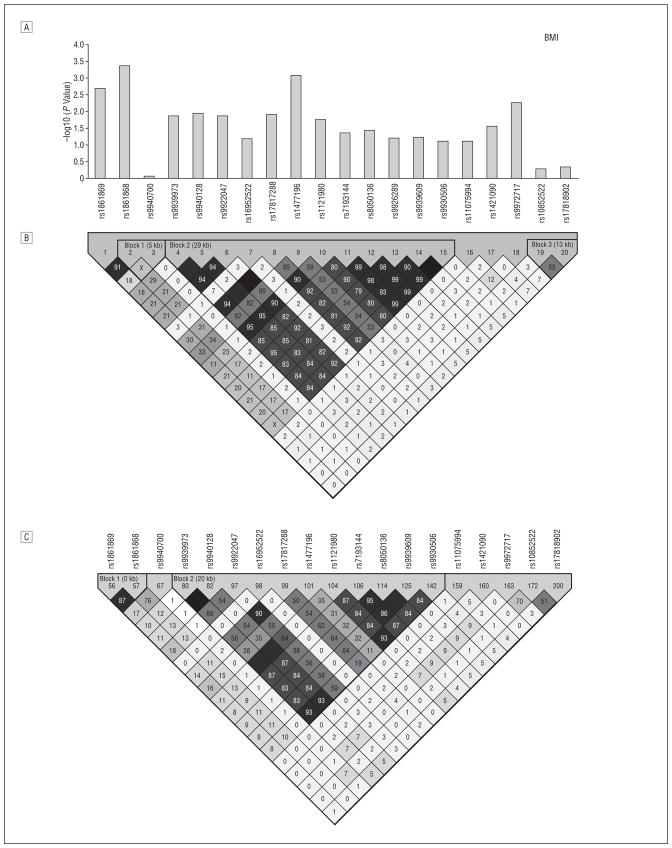
We analyzed 92 SNPs falling in a 347-kb interval that spanned FTO (data are available from the authors at http://medschool.umaryland.edu/endocrinology/afdspublic.asp). Of these SNPs, 26 were associated with BMI (P=.04 to <.001 under an additive genetic model). rs9939609, the associated SNP originally reported by Frayling et al,1 was modestly associated with obesity (P =.03) and not associated with BMI (P=.06) in the OOA, although stronger associations with this SNP were seen with total fat mass (P = .007) and percentage of body fat (P = .01). Figure 1 shows 20 SNPs in the region surrounding rs9939609. These 20 SNPs all fall within an 81-kb region spanning from within intron 1 to within intron 3 of the FTO gene. The SNP most strongly associated with BMI in the OOA was rs1861868, a common SNP with a similar risk allele frequency (0.47) as in HapMap (http://www.hapmap.org) genotype samples of white individuals (0.50). Each A allele of rs1861868 was associated with a 0.75 increase in BMI (P<.001), corresponding to a mean (SD) increase in weight of 2.0 (0.7) kg per allele (Table 1). Individuals with the A allele for rs1861868 were at increased risk of being obese (odds ratio [OR],1.26; P =.006) and of being overweight (OR,1.15; P =.05). The A allele of rs1861868 was also associated with greater waist circumference (P =.003) and weight (P =.002). In a smaller subset of 355 subjects, the same directional associations were seen with total fat mass (P<.001) and with percentage of fat mass (P=.001). Sex-, age-, and age2-adjusted physical activity levels did not differ significantly according to genotype (P =.11).
Figure 1.
Linkage disequilibrium (LD) structure and FTO single-nucleotide polymorphism (SNP) associations with body mass index (BMI). A, For each SNP surrounding rs9939609, we show –log10 (P value) for the additive model in the Old Order Amish (OOA). B, The LD (r 2) of FTO in OOA white subjects. C, The LD (r 2) of FTO in the HapMap (http://www.hapmap.org) genotype of white individuals. The association of rs9939609 with BMI was first reported in European white individuals1 and subsequently in French white individuals2; rs9930506 was the SNP most highly associated with BMI in a Sardinian population3; rs8050136 was associated with type 2 diabetes mellitus (T2DM) in European white subjects.11-13 Zeggini et al13 later found that the association disappeared after adjustment for BMI. In an independent sample of 419 OOA subjects (of whom 124 had T2DM), rs8050136 was associated with T2DM (P = .007), and this association was abolished after adjustment for BMI (P = .53) (data not shown). rs9939609, rs9930506, and rs8050136 all reside in a cluster of SNPs in high LD in HapMap genotypes of white individuals. Black squares indicate r 2>0.80; gray squares, 0.10 ≥r2≤0.80; white squares, r2<0.10. kb Indicates kilobases.
Table 1.
Clinical Characteristics of the Study Population by FTO rs1861868 Genotypesa
| AA (n = 166) | AG (n = 333) | GG (n = 203) | P Value | |
|---|---|---|---|---|
| Characteristic | ||||
| Women, % | 47.0 | 49.6 | 41.9 | .42 |
| Age, y | 43.4 ± 14.9 | 43.2 ± 14.1 | 44.6 ± 13.6 | .38 |
| BMI | 27.4 ± 4.8 | 26.5 ± 4.5 | 25.9 ± 4.1 | <.001 |
| Waist circumference, cm | 89.3 ± 11.4 | 86.9 ± 11.0 | 86.4 ± 10.5 | .003 |
| Weight, kg | 76.5 ± 12.9 | 74.3 ± 13.1 | 73.0 ± 11.8 | .002 |
| Height, cm | 167.5 ± 8.7 | 167.4 ± 9.3 | 167.6 ± 9.7 | .81 |
| Overweight (25 ≤BMI <30), % | 64.5 | 57.1 | 56.7 | .05 |
| Obese (BMI ≥30), % | 27.1 | 18.3 | 15.8 | .006 |
| Total daily accelerometer counts/1000, median (25%, 75% percentiles) | 362 (288, 538) | 392 (273, 572) | 427 (285, 610) | |
| Physical activityb | −0.05 ± 0.43 | 0.002 ± 0.45 | 0.04 ± 0.49 | .11 |
|
| ||||
| Fat and Lean Massc | ||||
| (n = 82) | (n = 169) | (n = 103) | ||
| Total fat mass, kg | 21.8 ± 9.9 | 20.5 ± 9.5 | 18.5 ± 9.2 | <.001 |
| Total lean mass, kg | 55.3 ± 9.8 | 52.6 ± 9.6 | 51.8 ± 10.8 | .02 |
| Total % fat | 27.8 ± 10.1 | 27.5 ± 10.3 | 25.8 ± 10.7 | .001 |
Abbreviation: BMI, body mass index (calculated as weight in kilograms divided by height in meters squared).
Data are given as mean ± SD except where noted; “n” indicates the number of samples.
Group means for age- and sex-residualized daily physical activity counts.
Measures of body composition as determined by dual-energy x-ray absorptiometry (DEXA).
Similar associations were observed with the C allele of rs1477196, the second most significant SNP in the OOA. In the OOA, each risk allele for rs1477196 was associated with a mean (SD) increase in BMI of 0.84 (0.25) (P <.001). The SNPs rs1861868 and rs1477196 are 17.9 kb apart in intron 1 and are in moderate LD in the OOA (D′=0.82; r2=0.36).
We examined the relationship between the 2 best SNPs identified from our analyses with rs9939609. In the OOA, this SNP is in strong LD with rs1477196 (D′=1; r2=0.54) but not with rs1861868 (D′=0.44; r2=0.17) (Figure 1). Together, the 3 SNPs (ordered from 5′ to 3′ rs1861868-rs1477196-rs9939609) defined 6 possible haplotypes (2 with frequencies <5%), 3 of which were associated with BMI (global permuted P=.004) (Table 2). The GTA haplotype had a frequency of 33.0% and was associated with decreased BMI (permuted P<.001), whereas the complementary ACT haplotype had a frequency of 35.0% and was associated with increased BMI (permuted P =.08). The less frequent ACA haplotype (10.4%) was also associated with increased BMI (permuted P =.006). These results suggest that in the OOA, regardless of rs9939609 allele (A or T), rs1861868 and rs1477196 define a haplotype that is associated with increased BMI.
Table 2.
Haplotypes for FTO SNPs rs1861868, rs1477196, and rs9939609
| Haplotype | Frequency | Haplotype Score | P Valuea |
|---|---|---|---|
| GTAf | 0.330 | −3.49 | <.001 |
| GCA | 0.039 | −0.25 | .80 |
| GCT | 0.147 | −0.13 | .90 |
| ATA | 0.030 | 0.20 | .84 |
| ACT | 0.349 | 1.77 | .08 |
| ACA | 0.104 | 2.73 | .006 |
ANALYSES STRATIFIED BY PHYSICAL ACTIVITY LEVELS
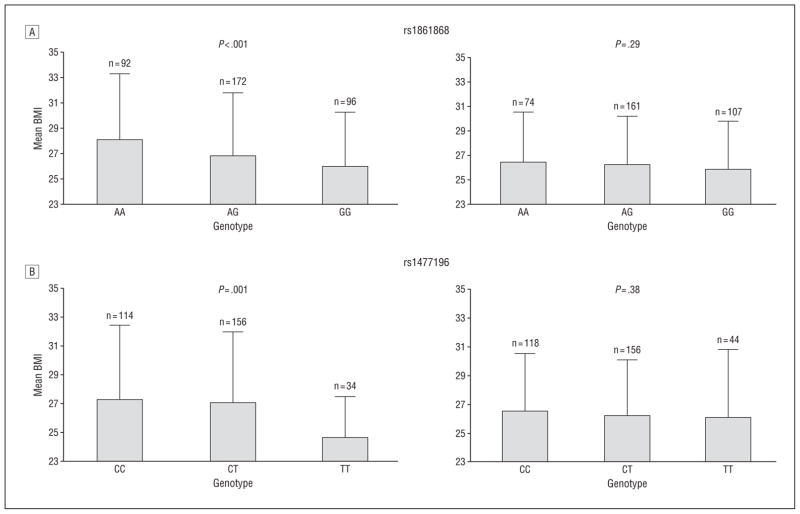
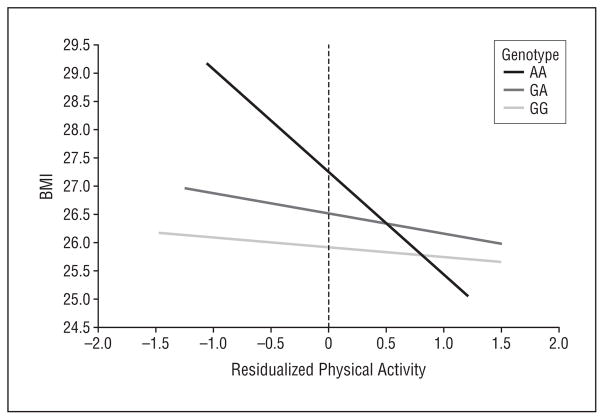
We next assessed the relationship of rs1861868 with BMI and obesity, stratified according to physical activity level (Figure 2). Among those within the lower half of the physical activity distribution (n=361), the rs1861868 A allele was strongly associated with greater BMI (a mean [SD] increase of 1.12 [0.33] for each risk allele; P<.001). In contrast, BMI was not significantly associated with the rs1861868 genotype in subjects in the upper half of the physical activity distribution (n=341) (0.30 [0.28] increase in BMI per risk allele; P =.29). rs1861868 was also associated with being obese in the low-activity group (OR, 1.28 for each risk allele; P =.03) but not in the high-activity group (P =.15). A formal test for interaction revealed significant evidence that increased physical activity levels blunted the association between the rs1861868 A allele on BMI (interaction P =.01). In Figure 3, we graphically illustrate, using the predicted values from the linear regression of residualized physical activity on BMI and rs1861868 genotype, that the difference in BMI across rs1861868 genotypes is large in less physically active individuals but small and not statistically significant in more physically active individuals. The impact of increased physical activity on BMI reduction is most apparent for individuals with the rs1861868 AA genotype.
Figure 2.
Mean body mass index (BMI), calculated as weight in kilograms divided by height in meters squared, in high and low physical activity groups according to FTO single-nucleotide polymorphism genotypes for rs1861868 (A) and rs1477196 (B). High activity was defined as age-, age2-, and sex-specific residuals greater than 0, and low activity was defined by residuals less than 0. Interaction P values: rs1861868, P = .01; rs1477196, P = .004.
Figure 3.
Predicted body mass index (BMI), calculated as weight in kilograms divided by height in meters squared, as a function of residualized age- and sex-specific ln-transformed physical activity accelerometer counts according to FTO rs1861868 genotypes. On the left side of the plot (low physical activity), BMI levels are strikingly dissimilar between rs1861868 genotypes. In contrast, on the right side of the plot, similar BMI levels can be seen across genotypes, particularly in subjects with very high levels of physical activity.
Similarly, the rs1477196 C allele was associated with a 1.22 (0.38) increase in BMI per risk allele in the low-activity group (P =.001) but only a 0.27 (0.31) increase in BMI in the high-activity group (P =.38) (interaction P =.004) (Figure 2). The rs1477196 C allele was associated with obesity in the low-activity group (OR, 1.58 for each risk allele; P=.001) but not in the high-activity group (P =.11).
The level of physical activity in the 2 strata can be described by the energy expenditure at the 25th and 75th centiles of the distribution, respectively. For women, energy expenditure was 2610 and 3590 kcal/d at the 25th and 75th centiles, respectively, whereas for men energy expenditure was 3130 and 3990 kcal/d at the same centiles. Thus, in this population, a mean activity level of 860 calories for women and 980 calories for men separates the high- and low-activity strata, which differ in their phenotypic expression of the at-risk FTO genotypes.
COMMENT
Our study replicates the association between variants in the FTO gene and obesity-related traits (eg, BMI, body weight, waist circumference, and percentage of body fat) recently reported by others.1–3,13–15 Furthermore, we have shown that the association of genotype on body composition is much smaller and not statistically significant in subjects having higher physical activity levels. The SNPs most strongly associated with BMI in our study were rs1861868 and rs1477196, both of which are common in the OOA (0.49 and 0.35, respectively) and in non-OOA white individuals (0.48 and 0.28, respectively). These SNPs are in higher LD in the OOA (D′= 0.84; r2=0.36) than in non-OOA European white individuals (HapMap genotypes from white individuals, D′=0.61, r2=0.15). Greater LD in the OOA compared with the general white population might be expected because the OOA are a relatively young founder population. Although an association of rs1477196 with obesity (P=6.0×10−9 based on an additive model) has previously been reported in a study of French adults,2 to our knowledge, the association of rs1861868 with BMI or obesity has not been described. In our analyses, a common haplotype defined by both SNPs (rs1861868, A allele; rs1477196, C allele) seems to jointly confer risk to increased BMI. This seems to be independent of rs9939609, the SNP originally found to be associated with BMI in multiple European white populations1,3 (Table 2). At least in the OOA, these SNPs may both be marking a common, yet unknown functional allele or may themselves have functional effects on body size through an as-yet undetermined mechanism. Further fine mapping with additional genotyping to help localize the true functional variant was beyond the scope of the current study.
A primary finding of this study is that of a gene×environment interaction between variants in the FTO gene and physical activity. Using objective-, age-, and sex-adjusted physical activity measures obtained for 7 consecutive 24-hour days, we were able to investigate the genotype associations of FTO variants in OOA subjects with high and low physical activity levels. Unlike questionnaires, the Actical device provides an objective estimate of physical activity that is unaffected by study participant recall bias. We found that SNP associations with BMI and related measures were limited to less physically active individuals. In the less active group, being AA homozygous for the rs1861868 FTO variant was associated with an increase in BMI of 2.1, a slightly higher estimate than that obtained by Frayling et al1 for rs9939609 in United Kingdom (UK) Wellcome Trust Case Control subjects with type 2 diabetes mellitus, and much higher than the same group’s estimate for rs9939609 in UK type 2 diabetes mellitus genetics consortium controls.1 By contrast, in the more physically active OOA stratum, the associations of the FTO variants were much smaller and not statistically significant. Because the FTO genotype was not associated with physical activity, this finding suggests a strong moderating effect of physical activity on the deleterious effects of FTO variants, consistent with published findings16 based on self-reported physical activity data. When men and women were evaluated separately without consideration of covariates, the association of rs1861868 genotype reached nominal statistical significance in women only, not in men. However, there was no evidence that the effect of genotype on BMI differed substantially between men and women, particularly when the level of physical activity was included in the regression model (ie, the genotype by sex interaction term was not statistically significant; P =.17).
Recent work4,17 has demonstrated that FTO codes for a protein expressed in the hypothalamus, the function of which is impacted in many obesity-related genetic defects in humans. Because FTO is down-regulated by Krebs cycle intermediates,4 it is conceivable that this protein is involved in incompletely understood nutrient sensing pathways, which are pivotal to central regulation of energy intake. Thus, a mechanism whereby increased physical activity can negate the association of FTO variants with fat accretion could be through pertubation of energy flux resulting in alterations in expression of FTO.
Activity levels in the “high-activity” stratum were approximately 900 kcal higher than in the “low-activity” stratum, which, depending on body size, corresponds to about 3 to 4 hours of moderately intensive physical activity, such as brisk walking, house cleaning, or gardening.18 Although this seems to be a large amount of physical activity, the OOA demonstrate that this level of activity was typical of an agrarian lifestyle without modern machinery. Of course, our cross-sectional study is unsuited to determine the amount of activity required to negate the effect of an FTO-related genetic predisposition to weight gain; however, in a retrospective analysis in which weight regain was measured as a function of physical activity energy expenditure, Schoeller et al18 found that the addition of 80 min/d of moderate activity or 35 min/d of vigorous activity to a sedentary lifestyle was sufficient for weight maintenance. Prospective intervention studies will be necessary to define these parameters more accurately.
In conclusion, we have replicated the associations of common SNPs in the FTO gene with increased BMI and risk to obesity in the OOA. Furthermore, we provide quantitative data to show that the weight increase resulting from the presence of these SNPs is much smaller and not statistically significant in subjects who are very physically active. This finding offers some clues to the mechanism by which FTO influences changes in BMI and may have important implications in targeting personalized lifestyle recommendations to prevent obesity in genetically susceptible individuals.
Footnotes
Financial Disclosure: None reported.
Author Contributions: Study concept and design:Rampersaud, Mitchell, Shen, Shuldiner, and Snitker. Acquisition of data: Fu, Ducharme, Hines, Sack, Naglieri, Shuldiner, and Snitker. Analysis and interpretation of data:Rampersaud, Mitchell, Pollin, Fu, O’Connell, Shuldiner, and Snitker. Drafting of the manuscript: Rampersaud, Mitchell, and Snitker. Critical revision of the manuscript for important intellectual content: Rampersaud, Mitchell, Pollin, Fu, Shen, O’Connell, Ducharme, Hines, Sack, Naglieri, Shuldiner, and Snitker. Statistical analysis: Rampersaud, Mitchell, Pollin, Shen, O’Connell, and Snitker. Obtained funding: Shuldiner. Administrative, technical, and material support: Fu, Ducharme, and Sack. Study supervision:Mitchell, Shuldiner, and Snitker.
References
- 1.Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dina C, Meyre D, Gallina S, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39(6):724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 3.Scuteri A, Sanna S, Chen WM, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3(7):e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerken T, Girard CA, Tung YC, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate dependent nucleic acid demethylase. Science. 2007;318(5855):1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pérusse L, Rice T, Province MA, et al. Familial aggregation of amount and distribution of subcutaneous fat and their responses to exercise training in the HERITAGE family study. Obes Res. 2000;8(2):140–150. doi: 10.1038/oby.2000.15. [DOI] [PubMed] [Google Scholar]
- 6.Mitchell BD, McArdle PF, Shen H, et al. The genetic response to short-term interventions affecting cardiovascular function: rationale and design of the Heredity and Phenotype Intervention (HAPI) Heart Study. Am Heart J. 2008;155(5):823–828. doi: 10.1016/j.ahj.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heil DP. Predicting activity energy expenditure using the Actical activity monitor. Res Q Exerc Sport. 2006;77(1):64–80. doi: 10.1080/02701367.2006.10599333. [DOI] [PubMed] [Google Scholar]
- 8.Westerterp KR. Physical activity assessment with accelerometers. Int J Obes Relat Metab Disord. 1999;23(suppl 3):S45–S49. doi: 10.1038/sj.ijo.0800883. [DOI] [PubMed] [Google Scholar]
- 9.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62(5):1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 11.Lake SL, Lyon H, Tantisira K, et al. Estimation and tests of haplotype-environment interaction when linkage phase is ambiguous. Hum Hered. 2003;55(1):56–65. doi: 10.1159/000071811. [DOI] [PubMed] [Google Scholar]
- 12.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70(2):425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeggini E, Weedon MN, Lindgren CM, et al. Wellcome Trust Case Control Consortium. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316(5829):1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316(5829):1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andreasen CH, Stender-Petersen KL, Mogensen MS, et al. Low physical activity accentuates the association of the FTO rs9939609 polymorphism on body fat accumulation. Diabetes. 2008;57(1):95–101. doi: 10.2337/db07-0910. [DOI] [PubMed] [Google Scholar]
- 17.Fletcher GF, Balady GJ, Amsterdam EA, et al. Exercise standards for testing and training: a statement for healthcare professionals from the American Heart Association. Circulation. 2001;104(14):1694–1740. doi: 10.1161/hc3901.095960. [DOI] [PubMed] [Google Scholar]
- 18.Schoeller DA, Shay K, Kushner RF. How much physical activity is needed to minimize weight gain in previously obese women? Am J Clin Nutr. 1997;66(3):551–556. doi: 10.1093/ajcn/66.3.551. [DOI] [PubMed] [Google Scholar]