Abstract
The mechanical properties of the extracellular matrix (ECM) are known to influence neuronal differentiation and maturation, though the mechanism by which neuronal cells respond to these biophysical cues is not completely understood. Here we design ECM mimics using self-assembled peptide nanofibers, in which fiber rigidity is tailored by supramolecular interactions, in order to investigate the relationship between matrix stiffness and morphological development of hippocampal neurons. We observe that development of neuronal polarity is accelerated on soft nanofiber substrates, and results from the dynamics of neuronal processes. While the total neurite outgrowth of non-polar neurons remains conserved, weaker adhesion of neurites to soft PA substrate facilitates easier retraction, thus enhancing the frequency of “extension-retraction” events. We hypothesize that higher neurite motility enhances the probability of one neurite to reach a critical length relative to others, thereby initiating the developmental sequence of axon differentiation. Our results suggest that substrate stiffness can influence neuronal development by regulating its dynamics, thus providing useful information on scaffold design for applications in neural regeneration.
Keywords: Self assembly, Peptide amphiphile, Stiffness, Neuronal polarity, Nerve tissue engineering
1. Introduction
Mechanical cues from the extracellular matrix (ECM) are implicated in a variety of biological processes, ranging from cell proliferation, differentiation, and migration to collective cell behaviors such as tubulogenesis [1–4]. Development of neuronal tissue has also been shown to be highly sensitive to the rigidity of the microenvironment. Soft matrices with stiffness matching that of native brain tissue have been found to promote the differentiation of neuronal stem cells into neurons rather than astrocytes and to restrict the growth of glial cell populations [2, 5]. Matrix mechanical properties also influence the maturation of differentiated neurons, with softer substrates shown to promote neurite outgrowth and expedite axon specification [6, 7]. In contrast, more rigid substrates tend to favor a neuron morphology with increased dendrite number and branching [8]. Furthermore, neurons respond in a lower stiffness regime when compared to other tissues [9], but mechanisms for sensing rigidity are thought to be similar to those of other mechano-sensitive cells such as fibroblasts [6].
These studies provide an understanding of the role of matrix stiffness at various stages of neuronal development, and indicate that mechanical properties of biomaterials are an important design parameter to consider in preparing artificial matrices for neural regeneration. However, the materials used in most of these studies to modulate material stiffness (i.e. cross-linked polyacrylamide, polydimethylsiloxane or agarose) often require protein coatings to promote cell adhesion and confounding effects could arise from patterns of tethered proteins [10]; in addition, they are not suitable for use as biomaterials for implantation in brain tissue. Therefore, the development of biodegradable and chemically bioactive materials with tailorable stiffness is an important target to extend the understanding of neuronal response to stiffness and provide material design parameters for future applications in regeneration of brain tissue.
Self-assembling peptide based materials have potential for use in regenerative medicine due to their inherent biocompatibility, ability to display natural signaling epitopes at controlled densities, and their biodegradable nature [11]. One broad class of such materials, peptide amphiphiles (PAs) have been extensively studied by our laboratory and have shown promise as a platform for a wide variety of applications ranging from drug delivery and angiogenesis in ischemic disease to regeneration of neural tissues, bone, cartilage, and enamel, among others [12–19]. PAs contain a peptide segment conjugated to an alkyl tail, which promotes hydrophobic collapse into high-aspect ratio nanofibers in aqueous environments [20]. In the presence of physiological salts, charge screening promotes the formation of self-supporting gels comprised of a network of nanofibers reminiscent of the fibrous structure in native ECM. PA gels that present the laminin-derived epitope IKVAV were demonstrated to selectively promote the differentiation of neural progenitor cells into neurons rather than astrocytes and control both neuron viability and maturation [18, 21]. Furthermore, unlike traditional polymeric approaches that modify cross-linking density or polymer concentration to alter mechanical properties, the mechanical properties of these supramolecular networks can be modified through molecular design [22]. The unique approach to control matrix stiffness through altering the molecular arrangement within individual nanofibers provides the opportunity to integrate both stiffness and biochemical signaling in a single approach. In this work, we used molecular design to prepare two PA nanofiber substrates of different stiffness in order to study the effect of stiffness on development of hippocampal neurons at both early and late stages of culture. We analyzed the distribution of neuronal processes, dynamics of neurite growth and their behavior at the nanofiber interface to investigate a biophysical mechanism underlying the stiffness-mediated regulation of early stages of neuron development.
2. Materials and methods
2.1 PA synthesis
PA molecules were synthesized using standard fluorenylmethyloxycarbonyl-solid phase peptide chemistry with a Rink Amide MBHA resin as previously described [17, 23]. All protected amino acids were purchased from EMD Chemicals and solvents were purchased from Makron. Fmoc deprotection was performed using 30% piperidine in N,N-dimethylformamide (DMF) and amino acid and palmitic acid couplings were performed with 4 eq protected amino acid or palmitic acid, 3.95 eq 2-(1H-benzotriazol-1-yl)-1,1,2,2-tetramethyluronium hexafluorophosphate, and 6 eq of diisopropylethylamine in a solvent mixture of 1:1:1 DMF:dichloromethane (DCM):N-methyl-2pyrrolidone. C-terminal palmitoylation or branching of the peptide headgroup were afforded using Fmoc-Lys(Mtt)-OH where the selective deprotection of the 4-methyltrityl (Mtt) group was performed using 2–3% trifluoroacetic acid, 5% triisopropylsilane (TIPS), and 91% DCM. Continued synthesis on the Fmoc protected α-amine was terminated by the addition of a (Boc)-Lys(Boc)-OH residue. Molecules and protecting groups were cleaved from the resin using a mixture of 95% trifluoroacetic acid (TFA), 2.5% water, and 2.5% TIPS. Molecules containing a tryptophan residue were cleaved with an additional 2.5% ethanedithiol. TFA was removed using rotary evaporation and the product precipitated with cold diethyl ether, dried, and purified using preparative scale reverse phase high performance liquid chromatography on a Varian Prostar Model 210 system equipped with a Phenomenex Jupiter Proteo column (C12 stationary phase, 10 μm, 90 Å pore size, 150 × 30 mm). A gradient of acetonitrile and water with 0.1% TFA was used as the mobile phase for purification. Pure fractions were identified using ESI mass spectrometry, collected, subjected to rotary evaporation to remove excess acetonitrile, lyophilized, and stored at −20 °C until use.
2.2 Rheology
Rheological measurements were performed using a Paar Physica MCR 300 oscillating plate rheometer with a 25 mm diameter cone-plate geometry and a gap of 0.05 mm. PA solutions at 1% (w/v) in water were pipetted in 200 μL volumes onto the rheometer plate and exposed to ammonia vapor for 15 min to form gels. The system was allowed to equilibrate for 10 minutes prior to taking measurements at 0.1% oscillatory strain over an angular frequency range from 100 – 0.1 s−1. The stage temperature was maintained at 25 °C over the entire testing period. Data was plotted as the average of three runs per sample (Error bar represents standard error of the mean).
2.3 Small angle X-ray scattering
Small angle X-ray scattering (SAXS) measurements were performed at the Advanced Photon Source using an insertion device station (beamline 5-ID-D Dupont-Northwestern-Dow Collaborative Access team, Argonne National Laboratory, Argonne, IL). Solutions of PA were dissolved at 1% (w/v) in water, diluted to a final concentration of 0.5% (w/v) in a solution of 1:1 water:neurobasal media and were filled into 1.5 mm quartz capillaries (Charles Supper Company). Operating at 15 keV, data was collected using a CCD with a sample to detector distance of 245 cm and an exposure time between 3–6 seconds. The scattering intensity was recorded with a q range of 0.008< q < 0.25 Å−1 where the wave vector q was defined as q = (4 π/λ) sin(θ/2) where θ is the scattering angle. Two-dimensional SAXS images were azimuthally averaged to produce one-dimensional intensity profiles using FIT2D. For background subtraction, scattering profiles were obtained for capillaries filled with solvent. No attempt was made to convert the units to an absolute scale. Detailed methods for SAXS data modeling can be found in Supplementary information.
2.4 X-ray diffraction
X-ray diffraction (XRD) was performed at the Advanced Photon Source using a bending magnet station (beamline 14BM-C, Argonne National Laboratory, Argonne, IL). All data were obtained using a beam energy of 12.67 keV (wavelength: 0.979Å) with a sample to detector distance of 400 mm and exposure time of 30 seconds. Solutions of PA at a concentration of 1% (w/v) in water were dropped on a loop and placed in the path of the X-ray beam. No background subtraction was performed on two-dimensional diffraction patterns.
2.5 Circular dichroism
Circular dichroism (CD) measurements were performed using a J-715 Jasco Circular Dichroism spectrometer at room temperature. PA solutions at a concentration of 0.02% (w/v) in 0.04X neurobasal media diluted with water were used. A quartz cuvette with a 1 mm path length was used for measurement and data was averaged over three scans for each sample.
2.6 Cryogenic transmission electron microscopy
Cryogenic transmission electron microscopy (Cryo-TEM) was performed using a JEOL 1230 microscope equipped with a LaB6 filament at an accelerating voltage of 100kV. For cryo-TEM, 300 mesh copper grids with a lacey carbon support (Electron Microscopy Sciences) were treated with air plasma for 25 seconds (Harrick Plamsa). 5μL of the PA at a concentration of 0.25% (w/v) in 1:1 water: neurobasal media was deposited on a grid. The sample was blotted using a Vitrobot Mark IV (FEI) vitrification robot at 95–100% humidity. The samples were vitrified by plunging them into a liquid ethane reservoir, transferred to liquid nitrogen, placed into a Gatan 626 cryo-holder through a cryo-transfer stage, and imaged using a Gatan 831 CCD camera.
2.7 PA coating procedure
Glass coverslips were coated for PAs following previously described method [23]. Briefly, 12 mm diameter PDL coated sterile glass coverslips were coated with a thin layer of sodium alginate (0.25% w/v), cross-linked by 10 mM CaCl2 solution, and then incubated overnight with 0.05% (w/v) PA 37 °C. Coatings were washed with 2 mM CaCl2 prior to performing experiments.
2.8 Scanning electron microscopy
For scanning electron microscopy (SEM) experiments, PA coatings on glass coverslips (with or without cells plated on it) were fixed with 2.5% glutaraldehyde in PBS (containing 1 mM CaCl2) for1 hour at room temperature (RT) and dehydrated in a graded series of ethanol concentration. Dehydrated samples were dried at the critical point of CO2 using a critical point dryer (Tousimis Samdri-795). Dried samples were then coated with a thin film (14 nm) of osmium metal using an osmium plasma coater (Filgen, OPC-60A). Images were obtained using a Hitachi S-4800 Field Emission Scanning Electron Microscope (FE-SEM) secondary electron detector at an accelerating voltage of 5 kV.
2.9 Fibroblast culture
NIH 3T3 mouse embryonic fibroblasts were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco) with high glucose, supplemented with 10% fetal bovine serum (FBS, Hyclone) and 1% penicillin-streptomycin (P/S) and passaged every 3 days. To study the cell response to PA, cells were seeded at a low density of 62 cells/mm2 on PA coated glass coverslips and incubated for 5 hours (37°C, 5% CO2) under serum free conditions (DMEM and 1% P/S).
2.10 Neuron culture
Dissociated hippocampal cultures were prepared from CD1 mouse embryo (17 day) following a previously described protocol with some modifications [24]. Briefly, the embryos were collected from anaesthetized timed pregnant mice and the hippocampal region of the brain was dissected out under ice cold calcium and magnesium-free Hank’s Balanced Salt Solution. Harvested brain tissues were dissociated into single cells by trypsinization (0.25%, 20 min at 37°C) followed by mechanical trituration, and suspended in medium containing DMEM F-12 and 10% FBS. The concentration of cells in suspension was adjusted to 0.25×106 cells/ml by adding DMEM F-12 (final FBS concentration <1%) before plating on PA coated surfaces and cultured in serum free neurobasal media (supplemented with B-27 supplement, GlutaMAX™ and P/S). Cell plating densities were 180 cells/mm2 and 166 cells/mm2 for 8 well chambered-slides (BD Bioscience; used for immunostaining) and glass coverslips (used for SEM), respectively. All saline solutions and culture medium supplements were obtained from Gibco Life Technology, while reagents and enzymes were obtained from Sigma-Aldrich. Dissection and animal handling were carried out in accordance with Public Health Service Policy on Humane Care and Use of Laboratory Animals and in compliance with protocol approved by Institutional Animal Care and Use Committee.
2.11 Immunostaining
All solutions used for staining and washing were supplemented with 1 mM CaCl2 to prevent disruption of the PA coating (anchored to the glass surface by Ca2+ cross-linked alginate). Fixed cell samples (4% paraformaldehyde in PBS, 15 min, RT) were permeabilized and blocked with 0.4% Triton X-100, 10% normal goat serum and 2% BSA in PBS. Incubation with primary antibody solution was done for 2 hours at RT or overnight at 4°C. Following primary antibody conditions were used in this work: Rabbit monoclonal Anti-β-tubulin III, 1:1000 dilution, Covance; mouse monoclonal anti-microtubule associated protein 2 (MAP2), 1:500 dilution, Sigma; chicken polyclonal anti-glial fibrillary acidic protein (GFAP), 1:1000 dilution, Abcam; rabbit monoclonal anti-synaptophysin, 1:1000 dilution, Millipore; mouse monoclonal anti-vinculin, 1:400 dilution, Sigma. Primary antibodies were detected with Alexa Fluor® conjugated secondary antibodies (Life Technologies; 1 hour incubation at RT). Actin filaments were visualized by staining with rhodamine conjugated phalloidin (Life Technologies; 1:200 dilution, 1 hour at RT). Cell nuclei were counterstained with DAPI (Life Technologies; 5 μg/ml).
2.12 Image acquisition and analysis
Images from fluorescently stained samples were acquired using an inverted fluorescence microscope (Nikon, Eclipse TE2000U) attached with a cooled CCD camera (Coolsnap, Photometrics) or an inverted confocal laser scanning microscope (Zeiss LSM 510 META). Morphological quantification was performed on β-tubulin III stained images of the neurons obtained at 10X objective magnification. Neurons were classified into developmental stages by using criteria previously described by Dotti et al [25]. Neurite tracing and their length measurement was done using the NeuronJ plugin in ImageJ software [26]. Measurements were made from 50 neurons for each condition (10 neurons each from 5 fields) and were repeated for 3 independent sets of cultures (total sample size: 150 neurons for each condition); higher morphological maturation was used as a selection criteria within one field, since the inclusion of neurons with little neurite growth could disproportionately affect the average and confound the difference between various conditions. Neuronal processes were only counted during the quantification of primary neurite number if it was >8 μmin length. For quantification of spine density, confocal image stacks of synaptophysin and MAP2 stained neurons (acquired under 40X objective and 3X digital zoom) was projected, and the number of spines present on proximal 40 μm of major dendrite was counted. Astrocytes and neuron number at 7 days were quantified by GFAP and β-tubulin III stained images. All experiments were repeated in three independent culture experiments unless mentioned otherwise.
2.13 Live cell imaging
PAs were coated as described above on a 4-chambered Hi-Q4 culture dish (Nikon). Cells were seeded on the coatings and imaged using a Nikon Biostation IM for a total of 5 or 18 hours at 37° C, 5% CO2.
2.14 Statistical analysis
Data were tested for normality by the Shapiro-Wilk test. The nonparametric Mann-Whitney U test was performed to compare between two groups of data, and the Kruskal-Wallis test was carried out to compare between multiple groups, followed by a pair-wise comparison by the Steel-Dwass test. Bar graphs represent mean ± standard error of the mean.
3. Results
The amino acids immediately adjacent to the hydrophobic alkyl tail of PA molecules can direct the degree of β-sheet formation and influence the stiffness of nanofiber networks [22, 27]. When amino acids with a strong propensity to form β-sheet secondary structures are incorporated, the resulting assembled nanofibers have increased stiffness compared to PAs incorporating amino acids that less readily form β-sheet hydrogen bonds. Following this strategy, stiff nanofibers were designed using a peptide backbone containing valine and alanine residues (“stiff” PA: Fig. 1a). Lysine residues were incorporated to enhance water solubility and promote nanofiber network formation in the presence of physiological salts through charge screening by counterions. To decrease the rigidity of PA nanofibers, we sought to reduce both packing density of molecules within the nanofiber and also hydrogen bonding close to the aliphatic core. To accomplish this, the soft nanofiber design includes both a branched headgroup by using a lysine dendron moiety in the peptide chain [28, 29] along with a weaker β-sheet region consisting of alanine and glycine residues (“soft” PA, Fig. 1a). Palmitoylation was afforded on the ε-amine of the C-terminal lysine residue, effectively reversing the polarity of the molecule [28]. For both “stiff” and “soft” PAs, we further added the laminin γ1 chain-derived bioactive peptide sequence KDI, which has previously been shown to promote neurite outgrowth and axon guidance [30, 31].
Fig. 1.
Design and characterization of PA nanofibers with variable stiffness.(a) Chemical structure of stiff (left) and soft (right) PA molecules. (b) CryoTEM images of both PAs in aqueous environment show long cylindrical nanofibers formed by both PAs. Scale bar 200 nm. (c) Animation illustrating the differences in molecular arrangement within the stiff and soft PA nanofibers. (d) CD (circular dichroism) spectra of the stiff PA and the soft PA. (d, Inset) X-ray diffraction pattern of the same PAs; arrowhead indicates a Bragg reflection at 4.68 Å in the stiff PA. (e) Rheological measurements of stiff or soft PA gels formed at 1% (w/v) (n=3).
The morphology of the PA assemblies was observed using cryogenic transmission electron microscopy (cryoTEM), and both stiff and soft PAs were found to form high-aspect-ratio cylindrical nanofibers (Fig. 1b). Small angle X-ray scattering (SAXS) further confirmed a cylindrical geometry for both PAs based on the power-law dependence of q−1 in the low q region, and a fit to cylindrical core-shell model with estimated diameters of 6.5 nm and 10 nm for stiff and soft PAs, respectively (Fig. S1). The slightly larger diameter of soft PA nanofibers can be attributed to the larger peptide segment of the individual molecule. Fig. 1c illustrates the expected arrangement of molecules within the assemblies of stiff and soft PA. The ordered arrangement of PA molecules in the stiff PA nanofiber is due to β-sheet formation between the peptide backbone segments, evidenced by both circular dichroism (CD) spectroscopy and X-ray diffraction (XRD): The CD spectrum of stiff PA revealeda β-sheet signature with a minimum at 218 nm, and XRD exhibited a reflection at 4.68 Å (Fig. 1d), the characteristic spacing of β-sheet hydrogen bonding between two β-strands [32]. Soft PA molecules were more disordered within the assembled nanofiber, as the CD demonstrated a random coil character and the XRD pattern was amorphous (Fig. 1d).
To measure the mechanical properties of both soft and stiff PAs, rheological measurements were performed using 1% (w/v) PA gels. The storage modulus (G′) and the loss modulus (G′) of the gels were measured over an angular frequency range of 1–100 s−1 using a cone-plate rheometer (Fig. 1e). The storage modulus (a measure of the elastic modulus or stiffness) of gels formed by stiff PA was found to be three times higher in magnitude than gels made of soft PA (22.9 ± 5 kPa vs. 7.3 ± 0.9 kPa, at an angular frequency of 10 s−1 and 0.1% strain). It is important to note that while trends in the bulk moduli for PA gels are expected to correlate with the stiffness of individual fibers, the absolute values should be different.
To demonstrate the difference in stiffness between the two PA nanofibers on a cellular level, we evaluated the spreading of 3T3 fibroblasts on two-dimensional PA substrates prepared on glass surfaces. It is well-known that fibroblast morphology changes in response to substrate stiffness [33, 34]. Uniform and stable coating of PA nanofibers were prepared on glass surfaces by sequential treatment of positively charged poly-D-lysine (PDL), negatively charged alginate, and positively charged PA nanofibers (Fig. S2). The purpose of the alginate layer was twofold—to enable the PA nanofibers to adhere to the alginate through electrostatic complexation and to prevent non-specific cell adhesion below the PA layer, as alginate is unable to facilitate cell adhesion [35]. 3T3 fibroblasts grown on a substrate of stiff PA exhibited intermediate spreading, with actin-rich lamellipodial extensions (Fig. 2), but both fibrillar adhesions (visualized by vinculin staining) and actin stress fiber formation that are hallmarks of fibroblast adhesion to stiffer tissue culture plastic or glass surfaces (Fig. S3) were absent. However, on soft PA substrates fibroblasts had a rounded shape with occasional small protrusions, a morphology consistent with fibroblast adhesion to softer substrates, suggesting that this PA is too soft to support fibroblast spreading [33]. Time-lapse imaging showed dynamic lamellipodial and filopodial extensions when cultured on stiff PA, while cells cultured on the soft PA were unable to spread and maintained their initial round shape (Movie 1 and 2). Overall, the differences in fibroblast morphology when cultured on substrates of stiff and soft PA confirms that differences in the bulk mechanical properties translate to differences in stiffness experienced at the cellular level.
Fig. 2.

Fibroblast morphology on stiff (left) and soft (right) PA coated surfaces. Fluorescent micrographs of 3T3 fibroblasts 5 hours after plating and stained with phalloidin (red), anti-vinculin antibody (green) and DAPI (blue). Scale bar 10 μm.
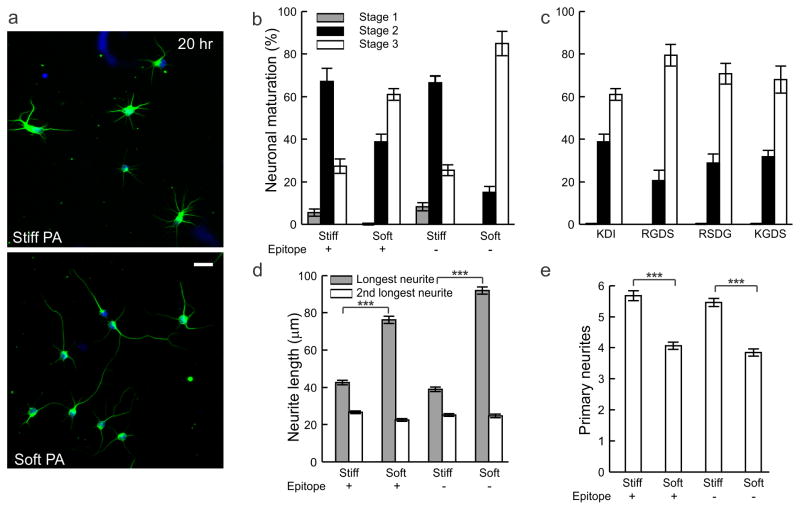
Next, we evaluated the effects of PA substrate stiffness on the development of primary mouse hippocampal neurons. Interestingly, we found a ten-fold decrease in astrocyte number (stained positive for glial fibrillary acidic protein, GFAP) on soft PA substrates compared with the stiff PA after 7 days in culture (Fig. 3). However, a corresponding reduction in the neuron density (visualized by β-tubulin III staining) was not observed. Our finding of substrate stiffness-dependent effects on neuron and glial population are consistent with those from a previous study [5]. While long-term culture revealed prominent difference in astrocyte number based on stiffness, differences in neuron morphology on these two substrates were apparent as early as 20 hours after plating. On stiff PA substrates, multiple short neurites were formed, while culture on soft PA substrates favored a single long neurite with a few short neurites (Fig. 4a). Neuronal maturation can be classified based on established morphological criteria [25], with immature neurons lacking neurites classified as stage 1, neurons with multiple short neurites (minor processes) without established polarity classified as stage 2, and neurons that have achieved polarity and possess one neurite considerably longer than the rest (major process) classified as stage 3. Using this classification revealed that most neurons on stiff PA reached developmental stage 2 (67.1 ± 6.2%) in the same time (20 hours following plating) that most neurons cultured on soft PA substrates advanced to stage 3 of development (60.9 ± 2.6%), thus indicating faster maturation on soft PA (Fig. 4b). When cultured on PA substrates of stiff and soft PAs with the same peptide backbones but without the KDI epitope (Fig. S4), a similar trend in neuronal development was observed (Fig. 4b), suggesting that the KDI epitope is not a critical signal for early neuronal maturation. To further probe the effect of peptide epitopes on early neuronal maturation, we also cultured neurons on PAs with a soft peptide backbone presenting the RGDS epitope known to promote neurite outgrowth [36]. When cultured on substrates of soft PA bearing the RGDS epitope, the trend in neuron development was similar to that seen for the soft PA with KDI (Fig. 4c); moreover, this trend persisted in cultures with a scrambled or mutated control of the RGDS epitope. These data suggest that epitope bioactivity is less critical in regulating neuronal maturation than substrate stiffness, emphasizing the potency of mechano-signaling in the early stages of neuronal development.
Fig. 3.

Influence of PA stiffness on astrocyte and neuron density in hippocampal culture.(a ) Micrographs from 7 day old hippocampal culture showing the distribution of neurons (TuBIII staining, green) and astrocytes (GFAP staining, red) on stiff (left) and soft (right) PA substrates (DAPI, blue). Scale bar 50 μm. (b) Quantification of astrocyte and neuron density (number per mm2) on stiff and soft PA substrates (***p<0.001, triplicate).
Fig. 4.
Effects of PA stiffness on early stages of neuron development. (a) Hippocampal neurons were cultured on stiff (top) and soft (bottom) PA coated surfaces and stained after 20 hours for β-tubulin III (green) and DAPI (blue). Scale bar 20 μm. (b) Distribution of developmental stages for neurons after 20 hours culture on stiff and soft PAs with and without the KDI epitope (n>450 neurons for each condition). (c) Distribution of developmental stages for soft PA with the KDI epitope compared to that for the same PA backbone presenting the RGDS epitope along with scrambled and mutated control sequences for the RGDS epitope (n>700 neurons). (d) Quantification of the length of the longest and the second longest neurites when cultured in conditions identical to those for (b) (***p<0.001, n=150 neurons). (e) Quantification of the total number of primary neurites of the same set of neurons in (d) (***p<0.001).
During neuronal development, the transition from stage 2 to stage 3 requires neurons to break symmetry and establish polarity, where one of the neurites (major process) extends to be much longer than the others (minor processes) and eventually forms the axon [25]. Since neuronal development and neurite distribution are closely related at this early stage of development, we measured the length of the longest neurites when grown on PA substrates (Fig. 4d). The average length of the longest neurite on soft PA (76.3 ± 1.9 μm) was almost double compared with stiff PA (42.5 ± 1.3 μm, p<0.001); the difference in neurite length was maintained on soft and stiff PA substrates without KDI epitopes. Meanwhile, the length of the second longest neurite (a measure of the minor processes) was similar among all conditions (ca. 22–26 μm), demonstrating that soft PA substrates did not promote the growth of all neurites. This observation supports the finding that neurons grown on soft PA substrates develop polarity more rapidly. Interestingly, the total number of primary neurites when cultured on soft PA was significantly less than that for culture on stiff PA substrates (4.0 ± 0.1 vs. 5.7 ± 0.1, p<0.001; Fig. 4e). Taken together, these data for neurite length and neurite number suggest that the combined total length of all neurites is roughly conserved regardless of substrate stiffness, and therefore the observed phenotypic difference results from an altered distribution of neuronal processes.
To establish a mechanism by which substrate stiffness controls the distribution of neuronal processes, cell dynamics were evaluated using time-lapse imaging. The processes from symmetric neurons were dynamic with frequent extensions and retractions (Fig. 5a, movie 3 and 4), as has been previously reported [25, 37]. However, a distinct difference in neurite motility was observed between culture on stiff and soft PA substrates. On stiff PA, retraction was less frequent and rarely completed, resulting in an increased number of shorter primary neurites over time (Fig. 5a). Meanwhile, neurites on soft PA substrates demonstrated several retraction events, and complete retractions were observed. These differences in neurite motility could arise from the influence of nanofiber stiffness at the cell-material interface. Scanning electron microscopy (SEM) was used to visualize the interface between neurons and PA nanofibers (Fig 5b). On stiff PA, there was a dense network of nanofibers, with neurites attached predominantly at focal points where multiple nanofibers clustered together (arrows). In contrast, soft PA nanofibers were uniformly attached along the length of the neurites. Moreover, on soft PA substrates fibers were oriented parallel to each other and perpendicular to the neurite length, perhaps resulting from the tension applied to the flexible PA nanofibers during cell attachment. The observations from SEM and time-lapse imaging suggest that differences in nanofiber stiffness influence interactions between nanofibers and neurites, the dynamics of which result in a distinct pattern of neurite distribution (illustrated in Fig. 5c).
Fig. 5.
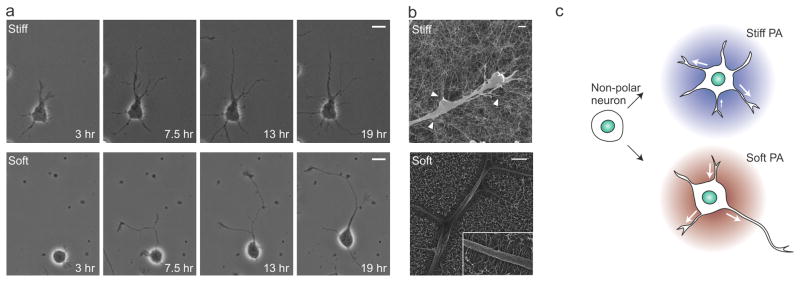
Dynamics of the neurite growth on different PA nanofiber substrates. (a) Individual frames from time-lapse imaging (with time noted on image) showing neurite growth on stiff (top) and soft (bottom) PA substrates. (b) Scanning electron micrographs showing the interaction between neurites and nanofibers, with stiff PA nanofibers (left) attached to neurites in bundles (arrowheads) and soft nanofibers (right) attached as individual fibers (inset). (c) Schematic representation of the effect of PA stiffness on neurite motility and development of polarity (Arrows indicate the extension and retraction events at a given time-point). Scale bars: a, 10 μm; b, 1 μm.
After 3–4 days in culture, minor neuronal processes, which remain a constant average length in earlier days, begin to grow rapidly and form the dendrites [25]. Therefore, we next evaluated the effects of PA substrate stiffness on dendrite development. Following 7 days in culture, dendrites (visualized by staining for microtubule associated protein 2, MAP2) exhibited distinct patterns depending on nanofiber stiffness (Fig. 6a). On soft PA substrates, the majority of neurons had one major apical dendrite at one side of the soma and a few minor dendrites appearing from the opposite side, similar to the structure of pyramidal neurons. On stiff PA substrates, however, multiple short dendrites extended from the soma, and often more than one major dendrite was present. The difference in the dendrite arborization was further reflected in the synaptic connectivity. Quantification of the total synapse number (visualized by presynaptic axon terminal marker synaptophysin) at the proximal segment of the main dendrites revealed a two-fold increase in synapse density when cultured on soft PA substrates (31.7 ± 1.3/100 μm, Fig. 6b) compared with culture on stiff PA substrates (15.0 ± 1.2/100 μm, p<0.001). However, this difference does not necessarily suggest an absolute increase in synaptic connections on soft PA substrates, as multiple smaller dendrites around the soma when cultured on stiff PA (Fig. 6a, arrowhead) provided the site for additional synapse formation. Differences in dendrite morphology resulting from substrate stiffness have been previously reported and attributed to indirect mechanisms such as soluble factors released by astrocytes [8]. To test whether there is a contact-mediated contribution of stiffness in these findings neurons were cultured on both stiff and soft PA substrates with shared culture medium. Within 3 days of culture, neurons on stiff and soft PA substrates exhibited distinct dendrite morphologies similar to that observed without shared culture medium (Fig. 6c). This observation suggests that PA substrate stiffness has a robust contact-guided influence in shaping dendrite morphology.
Fig. 6.
Effects of PA stiffness on dendrite morphology and synapse distribution. (a) Representative dendrite morphology (MAP2 staining, red) and distribution of synapses (synaptophysin staining, green), observed in hippocampal neurons after 7 days in culture on stiff (left) and soft (right) PA substrates; arrowheads indicate multiple thin dendrites extending from the neuronal soma on the stiff PA substrate. (b) Quantification of synapse density on the major dendrite process (***p<0.001, n=40 neurons). (c) Schematic of the experimental setup where colored spots represent three different PA coatings made on a PDL coated glass slide. Representative dendrite morphology (MAP2, red; DAPI, blue) of neurons with all of them sharing same culture medium after 3 days of culture on the stiff PA substrate without an epitope (1), stiff PA substrate presenting a KDI epitope (2), and soft PA substrate presenting the RGDS epitope (3). Scale bars: a, 10 μm; c, 20 μm.
4. Discussion
In this work, we used molecular design to prepare PA molecules which self-assemble into nanofibers of different stiffness to probe the effects of substrate stiffness on the development of primary hippocampal neurons. Culture on these substrates revealed that hippocampal neuron development, including achievement of polarity, dendrite maturation and astrocyte growth, was strongly dependent on substrate stiffness. The substrate stiffness altered cell-material interactions, leading to distinct motility patterns of neurites, and eventually resulting in a specific neurite distribution with differences in the temporal course for neuron development.
PA materials provide a unique approach to study the role of matrix stiffness on cell response and development. Unlike conventional materials used to study the effects of stiffness on cells, where cross-linking density between polymer chains is varied to change the stiffness, PA stiffness results from supramolecular interactions, especially molecular packing and β-sheet hydrogen bonding. While defects in covalently cross-linked materials can propagate and lead to failure due to irreversibility of bond breakage, materials prepared using non-covalent interactions do not have such limitation, as bonds can spontaneously reform under physiological conditions. Although individual hydrogen bonds do not possess the strength of a covalent bond, when arranged in ordered β-sheet structure these bonds can produce materials with extraordinary stiffness and resilience. Examples from nature of such materials with a high degree of hydrogen bonding include amyloid fibrils and spider silk [38, 39]. Many self-assembling peptide materials, including PAs, incorporate similar hydrogen bonding in their design in order to prepare stiff one-dimensional nanofibers [40]. PA molecules have the additional design component of an alkyl tail which facilitates hydrophobic collapse and serves as a driving force for molecular self-assembly. Thus, compared to other self-assembling peptides, PA self-assembly is not completely reliant on β-sheet hydrogen bonds for nanofiber formation and stability, which allows more freedom to tune the β-sheet strength without compromising nanofiber assembly. In this work, nanofiber stiffness was further modulated by introduction of a branched head-group in the peptide chain, which effectively reduced the number of β-sheets per cross-section by decreasing the packing density [29].
Using molecular design, PAs were prepared with different nanofiber stiffness to study the effect of stiffness on the development of primary hippocampal neurons. Consistent with previous reports [6], neurons developed polarity at a faster rate on soft substrates than on stiff substrates. It has been shown that in the non-polar stage neurons exhibit multiple short neurites that undergo random extensions and retractions, while in the polar stage, the length of one neurite surpasses the others and continues to grow at a faster rate to form the axon [25]. It is shown here that the growth of one major neurite on the soft PA substrate was accompanied by fewer total primary neurites per cell, implying that neurite distribution is dependent on substrate stiffness. Microtubule assembly is suggested to be critical for the growth of neuronal processes [41], and its availability could explain the observation that the total length of all neurites was conserved, in spite of striking differences in the neurite distribution. Non-neuronal cells are known to contain similar levels of polymerized microtubules at early time-points in culture, independent of their degree of spreading [42]. Since the neurite extensions are physically supported by bundles of microtubules running along their core, if each immature neuron also have an equivalent amount of polymerized microtubules, then each should be capable of producing approximately an equivalent length of total neurites, independent of substrate stiffness. Though all neuronal processes initially have equal probability of becoming the axon, the probability of a specific process becoming the axon is greatly increased once its length exceeds all others by a critical length of 10 μm [37]. Asymmetry in the distribution of neuronal processes accelerates the development of polarity, as demonstrated previously in the classical axotomy experiment [37, 43]. In this experiment, if the remaining length of a severed axon is shorter than the rest of the processes, the neuron takes substantially longer to reestablish polarity. Once polarity is achieved, the major process specified to be the axon grows at a much faster rate than the others, a process facilitated by the accumulation of neurite growth promoting molecules at the growth cone [44, 45]. Our results suggest that physical cues from substrate stiffness impose a bias on the distribution of neuronal processes, thereby controlling the temporal development of neuronal polarity.
While the distribution pattern of neuronal processes favors axon specification, the dynamics of these processes have also been suggested to play a critical role in the development of neuronal polarity [46]. It is shown here that PA substrate stiffness strongly influenced the dynamics of neurite growth, and the retraction of early neuronal processes was dramatically enhanced on soft PA substrates. Increased neurite retraction on soft PA substrates can be explained by the difference in interactions at the neurite-nanofiber interface. In general, stiff substrates are known to afford stable focal adhesions, while adhesions to soft substrates are less stable and more transient [9]. The uniformly distributed attachment of individual soft PA nanofibers along the neuronal processes suggests the formation of transient adhesions. Conversely, stiff PA nanofibers attach to cells at specific points along the projecting neurite. It is known that stable focal adhesions induce localized stabilization of microtubules through integrins and focal adhesion kinases [47, 48]; in turn, enhanced microtubule stability leads to fewer retraction events [49]. Therefore, in the presence of weaker adhesions on soft substrates, microtubules will be less stable and allow for larger retractions. The neurons can then redistribute the microtubule resources gained by retraction in order to extend additional neurites, effectively enhancing the overall dynamics. Consistent with this hypothesis, only partial retractions were observed in studies where hippocampal neurons were grown on PDL coated glass surfaces [25]. We speculate that higher motility, ormore frequent “extension-retraction” events, of the uncommitted neuronal processes increase the random chance that one process will extend to a critical length (Fig. 5c), initiating the sequence of developmental events for axon differentiation.
We also demonstrated that later stages of neuronal development, such as dendrite morphology and synaptic connections, were strongly influenced by substrate stiffness. For neurons, both of these characteristics are critical to integrate signals from other neurons. Although indirect mechanisms, such as astrocyte secreted factors and activation of ionotropic glutamate receptors, have been suggested as the means by which substrate stiffness controls dendrite development [8], our results demonstrate that physical cues from the substrate directly influence this process. While fewer primary neurites on soft PA substrates could facilitate a characteristic pyramidal neuron morphology with a distinct apical dendrite and few basal dendrites, some of the excess primary neurites on stiff PA possibly exist as multiple smaller dendrites in the mature neurons. Recent reports have shown substantial differences in tissue stiffness between different regions within the hippocampus (i.e. CA1 vs. CA3) [50]. Taken together with our findings here, this suggests that stiffness could be an important factor influencing the difference in dendrite morphology observed in pyramidal neurons in these hippocampal regions.
5. Conclusions
We have reported here the design of self-assembled PA materials with varying stiffness, and demonstrated their use as substrates to probe the effects of matrix stiffness on the development of hippocampal neurons. This material enables us to better understand the biophysical basis for stiffness-mediated neuronal maturation during the early phases of culture. The ability to use PA molecular design to control the development and maturation of neurons, coupled with the suppression of astrocyte proliferation, through tuning nanofiber stiffness is expected to enhance the therapeutic potential of these materials in applications for regeneration of the nervous system.
Supplementary Material
Time lapse observation of fibroblast cell morphology on stiff PA coated surface. Recording started 1 hour after cell plating and total elapsed time is 5 hours. [Scale bar, 20 μm]
Time lapse observation of fibroblast cell morphology on soft PA coated surface. Recording started 1 hour after cell plating and total elapsed time is 5 hours. [Scale bar, 20 μm]
Time lapse observation of neurite growth from hippocampal neuron on stiff PA coated surface. Recording started 3 hours after cell plating and total elapsed time is 18 hours. [Scale bar, 10 μm]
Time lapse observation of neurite growth from hippocampal neuron on stiff PA coated surface. Recording started 3 hours after cell plating and total elapsed time is 18 hours. [Scale bar, 10 μm]
Acknowledgments
This work was supported by NIBIB, grant No. 2R01EB003806-06A2. We are grateful to the following core facilities at Northwestern University: the Peptide Synthesis Core at the Institute for BioNanotechnology in Medicine (IBNAM), the Biological Imaging Facility (BIF), the Cell Imaging Facility (CIF) and Keck Biophysics Facility for instrument use. We are grateful to Prof. Wes Burghardt for the use of rheological equipment. We acknowledge Dr. Steven Weigand for assistance with SAXS experiments and Guy Macha and Vukica Srajer for assistance with XRD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 2.Saha K, Keung AJ, Irwin EF, Li Y, Little L, Schaffer DV, et al. Substrate modulus directs neural stem cell behavior. Biophys J. 2008;95:4426–38. doi: 10.1529/biophysj.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci U S A. 1997;94:13661–5. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo CL, Ouyang M, Yu JY, Maslov J, Price A, Shen CY. Long-range mechanical force enables self-assembly of epithelial tubular patterns. Proc Natl Acad Sci U S A. 2012;109:5576–82. doi: 10.1073/pnas.1114781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Georges PC, Miller WJ, Meaney DF, Sawyer ES, Janmey PA. Matrices with compliance comparable to that of brain tissue select neuronal over glial growth in mixed cortical cultures. Biophys J. 2006;90:3012–8. doi: 10.1529/biophysj.105.073114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kostic A, Sap J, Sheetz MP. RPTPalpha is required for rigidity-dependent inhibition of extension and differentiation of hippocampal neurons. J Cell Sci. 2007;120:3895–904. doi: 10.1242/jcs.009852. [DOI] [PubMed] [Google Scholar]
- 7.Balgude AP, Yu X, Szymanski A, Bellamkonda RV. Agarose gel stiffness determines rate of DRG neurite extension in 3D cultures. Biomaterials. 2001;22:1077–84. doi: 10.1016/s0142-9612(00)00350-1. [DOI] [PubMed] [Google Scholar]
- 8.Previtera ML, Langhammer CG, Langrana NA, Firestein BL. Regulation of dendrite arborization by substrate stiffness is mediated by glutamate receptors. Ann Biomed Eng. 2010;38:3733–43. doi: 10.1007/s10439-010-0112-5. [DOI] [PubMed] [Google Scholar]
- 9.Discher DE, Mooney DJ, Zandstra PW. Growth factors, matrices, and forces combine and control stem cells. Science. 2009;324:1673–7. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trappmann B, Gautrot JE, Connelly JT, Strange DG, Li Y, Oyen ML, et al. Extracellular-matrix tethering regulates stem-cell fate. Nat Mater. 2012;11:642–9. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- 11.Matson JB, Stupp SI. Self-assembling peptide scaffolds for regenerative medicine. Chem Commun (Camb) 2012;48:26–33. doi: 10.1039/c1cc15551b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartgerink JD, Beniash E, Stupp SI. Self-assembly and mineralization of peptide-amphiphile nanofibers. Science. 2001;294:1684–8. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 13.Webber MJ, Matson JB, Tamboli VK, Stupp SI. Controlled release of dexamethasone from peptide nanofiber gels to modulate inflammatory response. Biomaterials. 2012;33:6823–32. doi: 10.1016/j.biomaterials.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajangam K, Behanna HA, Hui MJ, Han X, Hulvat JF, Lomasney JW, et al. Heparin binding nanostructures to promote growth of blood vessels. Nano Lett. 2006;6:2086–90. doi: 10.1021/nl0613555. [DOI] [PubMed] [Google Scholar]
- 15.Webber MJ, Tongers J, Newcomb CJ, Marquardt KT, Bauersachs J, Losordo DW, et al. Supramolecular nanostructures that mimic VEGF as a strategy for ischemic tissue repair. Proc Natl Acad Sci U S A. 2011;108:13438–43. doi: 10.1073/pnas.1016546108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah RN, Shah NA, Del Rosario Lim MM, Hsieh C, Nuber G, Stupp SI. Supramolecular design of self-assembling nanofibers for cartilage regeneration. Proc Natl Acad Sci U S A. 2010;107:3293–8. doi: 10.1073/pnas.0906501107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Z, Newcomb CJ, Bringas P, Jr, Stupp SI, Snead ML. Biological synthesis of tooth enamel instructed by an artificial matrix. Biomaterials. 2010;31:9202–11. doi: 10.1016/j.biomaterials.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, et al. Selective differentiation of neural progenitor cells by high-epitope density nanofibers. Science. 2004;303:1352–5. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 19.Tysseling-Mattiace VM, Sahni V, Niece KL, Birch D, Czeisler C, Fehlings MG, et al. Self-Assembling Nanofibers Inhibit Glial Scar Formation and Promote Axon Elongation after Spinal Cord Injury. J Neurosci. 2008;28:3814–23. doi: 10.1523/JNEUROSCI.0143-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui H, Webber MJ, Stupp SI. Self-assembly of peptide amphiphiles: from molecules to nanostructures to biomaterials. Biopolymers. 2010;94:1–18. doi: 10.1002/bip.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sur S, Pashuck ET, Guler MO, Ito M, Stupp SI, Launey T. A hybrid nanofiber matrix to control the survival and maturation of brain neurons. Biomaterials. 2012;33:545–55. doi: 10.1016/j.biomaterials.2011.09.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pashuck ET, Cui H, Stupp SI. Tuning supramolecular rigidity of peptide fibers through molecular structure. J Am Chem Soc. 2010;132:6041–6. doi: 10.1021/ja908560n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sur S, Matson JB, Webber MJ, Newcomb CJ, Stupp SI. Photodynamic control of bioactivity in a nanofiber matrix. ACS nano. 2012;6:10776–85. doi: 10.1021/nn304101x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goslin K, Asmussen H, Banker G. Rat hippocampal neurons in low-density culture. In: Goslin K, Banker G, editors. Culturing Nerve Cells. 2. Cambridge, Massachusetts: The MIT Press; 1998. pp. 339–70. [Google Scholar]
- 25.Dotti CG, Sullivan CA, Banker GA. The establishment of polarity by hippocampal neurons in culture. J Neurosci. 1988;8:1454–68. doi: 10.1523/JNEUROSCI.08-04-01454.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meijering E, Jacob M, Sarria JCF, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry Part A. 2004;58:167–76. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- 27.Paramonov SE, Jun HW, Hartgerink JD. Self-assembly of peptide-amphiphile nanofibers: the roles of hydrogen bonding and amphiphilic packing. J Am Chem Soc. 2006;128:7291–8. doi: 10.1021/ja060573x. [DOI] [PubMed] [Google Scholar]
- 28.Jiang H, Guler MO, Stupp SI. The internal structure of self-assembled peptide amphiphile nanofibers. Soft Matter. 2007;3:454–62. doi: 10.1039/b614426h. [DOI] [PubMed] [Google Scholar]
- 29.Storrie H, Guler MO, Abu-Amara SN, Volberg T, Rao M, Geiger B, et al. Supramolecular crafting of cell adhesion. Biomaterials. 2007;28:4608–18. doi: 10.1016/j.biomaterials.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 30.Liesi P, Laatikainen T, Wright JM. Biologically active sequence (KDI) mediates the neurite outgrowth function of the gamma-1 chain of laminin-1. J Neurosci Res. 2001;66:1047–53. doi: 10.1002/jnr.1250. [DOI] [PubMed] [Google Scholar]
- 31.Wiksten M, Liebkind R, Laatikainen T, Liesi P. Gamma 1 laminin and its biologically active KDI-domain may guide axons in the floor plate of human embryonic spinal cord. J Neurosci Res. 2003;71:338–52. doi: 10.1002/jnr.10495. [DOI] [PubMed] [Google Scholar]
- 32.Sunde M, Blake C. The structure of amyloid fibrils by electron microscopy and X-ray diffraction. Adv Protein Chem. 1997;50:123–59. doi: 10.1016/s0065-3233(08)60320-4. [DOI] [PubMed] [Google Scholar]
- 33.Georges PC, Janmey PA. Cell type-specific response to growth on soft materials. J Appl Physiol. 2005;98:1547–53. doi: 10.1152/japplphysiol.01121.2004. [DOI] [PubMed] [Google Scholar]
- 34.Rhee S, Jiang H, Ho CH, Grinnell F. Microtubule function in fibroblast spreading is modulated according to the tension state of cell-matrix interactions. Proc Natl Acad Sci U S A. 2007;104:5425–30. doi: 10.1073/pnas.0608030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rowley JA, Madlambayan G, Mooney DJ. Alginate hydrogels as synthetic extracellular matrix materials. Biomaterials. 1999;20:45–53. doi: 10.1016/s0142-9612(98)00107-0. [DOI] [PubMed] [Google Scholar]
- 36.Schense JC, Bloch J, Aebischer P, Hubbell JA. Enzymatic incorporation of bioactive peptides into fibrin matrices enhances neurite extension. Nat Biotechnol. 2000;18:415–9. doi: 10.1038/74473. [DOI] [PubMed] [Google Scholar]
- 37.Goslin K, Banker G. Experimental observations on the development of polarity by hippocampal neurons in culture. J Cell Biol. 1989;108:1507–16. doi: 10.1083/jcb.108.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buehler MJ. Strength in numbers. Nat Nanotechnol. 2010;5:172–4. doi: 10.1038/nnano.2010.28. [DOI] [PubMed] [Google Scholar]
- 39.Keten S, Buehler MJ. Geometric confinement governs the rupture strength of H-bond assemblies at a critical length scale. Nano Lett. 2008;8:743–8. doi: 10.1021/nl0731670. [DOI] [PubMed] [Google Scholar]
- 40.Wu EC, Zhang SG, Hauser CAE. Self-Assembling Peptides as Cell-Interactive Scaffolds. Adv Funct Mater. 2012;22:456–68. [Google Scholar]
- 41.Conde C, Caceres A. Microtubule assembly, organization and dynamics in axons and dendrites. Nat Rev Neurosci. 2009;10:319–32. doi: 10.1038/nrn2631. [DOI] [PubMed] [Google Scholar]
- 42.Mooney DJ, Hansen LK, Langer R, Vacanti JP, Ingber DE. Extracellular matrix controls tubulin monomer levels in hepatocytes by regulating protein turnover. Mol Biol Cell. 1994;5:1281–8. doi: 10.1091/mbc.5.12.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dotti CG, Banker GA. Experimentally induced alteration in the polarity of developing neurons. Nature. 1987;330:254–6. doi: 10.1038/330254a0. [DOI] [PubMed] [Google Scholar]
- 44.Goslin K, Schreyer DJ, Skene JH, Banker G. Development of neuronal polarity: GAP-43 distinguishes axonal from dendritic growth cones. Nature. 1988;336:672–4. doi: 10.1038/336672a0. [DOI] [PubMed] [Google Scholar]
- 45.Toriyama M, Sakumura Y, Shimada T, Ishii S, Inagaki N. A diffusion-based neurite length-sensing mechanism involved in neuronal symmetry breaking. Mol Syst Biol. 2010;6:394. doi: 10.1038/msb.2010.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kollins KM, Hu J, Bridgman PC, Huang YQ, Gallo G. Myosin-II negatively regulates minor process extension and the temporal development of neuronal polarity. Dev Neurobiol. 2009;69:279–98. doi: 10.1002/dneu.20704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palazzo AF, Eng CH, Schlaepfer DD, Marcantonio EE, Gundersen GG. Localized stabilization of microtubules by integrin- and FAK-facilitated Rho signaling. Science. 2004;303:836–9. doi: 10.1126/science.1091325. [DOI] [PubMed] [Google Scholar]
- 48.Kaverina I, Rottner K, Small JV. Targeting, capture, and stabilization of microtubules at early focal adhesions. J Cell Biol. 1998;142:181–90. doi: 10.1083/jcb.142.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Solomon F. Neuroblastoma cells recapitulate their detailed neurite morphologies after reversible microtubule disassembly. Cell. 1980;21:333–8. doi: 10.1016/0092-8674(80)90469-9. [DOI] [PubMed] [Google Scholar]
- 50.Elkin BS, Azeloglu EU, Costa KD, Morrison B., 3rd Mechanical heterogeneity of the rat hippocampus measured by atomic force microscope indentation. J Neurotrauma. 2007;24:812–22. doi: 10.1089/neu.2006.0169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Time lapse observation of fibroblast cell morphology on stiff PA coated surface. Recording started 1 hour after cell plating and total elapsed time is 5 hours. [Scale bar, 20 μm]
Time lapse observation of fibroblast cell morphology on soft PA coated surface. Recording started 1 hour after cell plating and total elapsed time is 5 hours. [Scale bar, 20 μm]
Time lapse observation of neurite growth from hippocampal neuron on stiff PA coated surface. Recording started 3 hours after cell plating and total elapsed time is 18 hours. [Scale bar, 10 μm]
Time lapse observation of neurite growth from hippocampal neuron on stiff PA coated surface. Recording started 3 hours after cell plating and total elapsed time is 18 hours. [Scale bar, 10 μm]