Abstract
Background
Canine distemper virus (CDV) infects a variety of carnivores, including wild and domestic Canidae. In this study, we sequenced and phylogenetic analyses of the hemagglutinin (H) genes from eight canine distemper virus (CDV) isolates obtained from seven raccoon dogs (Nyctereutes procyonoides) and a giant panda (Ailuropoda melanoleuca) in China.
Results
Phylogenetic analysis of the partial hemagglutinin gene sequences showed close clustering for geographic lineages, clearly distinct from vaccine strains and other wild-type foreign CDV strains, all the CDV strains were characterized as Asia-1 genotype and were highly similar to each other (91.5-99.8% nt and 94.4-99.8% aa). The giant panda and raccoon dogs all were 549Y on the HA protein in this study, irrespective of the host species.
Conclusions
These findings enhance our knowledge of the genetic characteristics of Chinese CDV isolates, and may facilitate the development of effective strategies for monitoring and controlling CDV for wild canids and non-cainds in China.
Keywords: Canine distemper virus, Haemagglutinin (H) gene, Genotype, Phylogenetic analysis
Background
CD is caused by the canine distemper virus (CDV) which belongs to the Morbillivirus genus of the Paramyxoviridae virus family. This virus infects a broad range of animals, such as Mustelidae (ferrets, minks, skunks, weasels, and badgers), Procyonidae (raccoons), Ursidae (bears and pandas), Viverridae (civets, genets, and linsangs), hyaenidae (hyenas), and Felidae (lions and tigers) [1-7]. The genome of CDV encodes the following genes: matrix (M), fusion (F), hemagglutinin (H), nucleocapsid (N), polymerase (L), and phosphoprotein (P). The H gene protein is responsible for viral attachment to the cell host [8] and is the most variable protein described for all members of the genus Morbillivirus [9]. Experimental investigation showed that changing one amino acid (Y549H) in the CDV-H protein of one dog strain can decreased expression of specialist traits and increased expression of generalist traits, thereby confirming its functional importance [10].
The H gene has been used to investigate the genetic relationships among the various strains [11-13]. On the basis of the full-length sequence of the H gene of CDV strains identified globally, at least seven main geographic groups (genotypes) appear to circulate among the various susceptible hosts, namely America-1, America-2, Asia-1 and Asia-2, Europe, Europe wildlife and Arctic [14-17]. In addition, some CDV strains identified in Africa, Asia and Argentina [18-20] appear to diverge substantially and might represent separate geographic groups.
N glycosylation has been shown to be important for the correct folding, transport, and function of other paramyxovirus fusion and attachment glycoproteins [21]. There are significant molecular differences between the glycoproteins of wild-type and vaccine CDV strains. It has been hypothesized that reduced N glycosylation is an important attenuating factor and that an increase in N glycosylation may eventually result in vaccine failure [15,22].
Although the use of live attenuated vaccines has greatly helped to prevent CDV infections in susceptible animals, the dental abnormalities and enamel erosion that are found in many young and adult giant pandas [23,24] may be related to early infection with CDV [25,26]. Several episodes also have been reported in raccoon dogs [20]. So in this work we analyzed a total of 7 samples from raccoon dogs with clinical signs of canine distemper and 1 sample from giant panda which were submitted to our laboratories and the viruses were analyzed using a fragment of the H gene.
Results
CDV detection by NP-based RT-PCR
We analyzed a total number of 108 samples taken from raccoon dogs with suspect of CDV infection. CDV RNA was detected in 56 out of 108 (51.8%) blood samples, most of the raccoon dogs, between the age of 15–30 weeks. Negative samples were assayed twice to confirm the results. Forty-one of the 56 positive clinical specimens (73.2%) were obtained from raccoon dogs which had been vaccinated against CDV at least once. Twelve samples (21.4%) were obtained from unvaccinated raccoon dogs, and the vaccination status of the three remaining raccoon dogs was unknown. Forty-nine of the 56 positive animals (87.5%), showed symptoms typical of CDV, such as respiratory disease and neurological dysfunction, two raccoon dogs were asymptomatic, while no records were available for the other five raccoon dogs. The giant panda showed a mild fever (anal temperature 39.0°C).
Sequence and phylogenetic analysis of the H gene
Amplification of fragments of the expected size was obtained with H gene and the sequences determined. After removing primer-derived sequences and some sequences with inconsistencies at the 5’ and 3’ ends, a 594 bp-long fragment of the H gene were obtained.
The neighbor joining tree (Figure 1) generated using a 594 base pair (bp) long partial sequences of the H gene (amplified with primers “B”), evidenced geographic patterns (genotypes) resembling the patterns described previously using the full-length H gene (Asia 1, Asia 2, Europe, Arctic, America1 and America 2). In the phylogenetic tree (Figure 1) all the Chinese wild-type viruses (8/8) grouped together in one branch in the Asia-1 genotype. In this genotype, other Chinese CDV strains identified from domestic dogs or other wildlife species after the 1990s were also included, along with some Japanese and Korean strains detected before 1998. The 8 Asia-1 strains displayed identity to each other in the H gene (91.5-99.8% nt and 94.4-99.8% aa), while identity to the vaccine strains CDV3, Onderstepoort and Lederle was lower (85.4-89.2% nt and 84.4-88.3% at aa level) (Table 1). The 4 strains (Sichuan 02–05) displayed high identity to each other (97.4-99.8% nt and 98.4-99.8% aa). On the other hand, although strain Sichuan06 appeared to belong to the main Asia 1 clade, it is more distant from the group (Figure 1).
Figure 1.
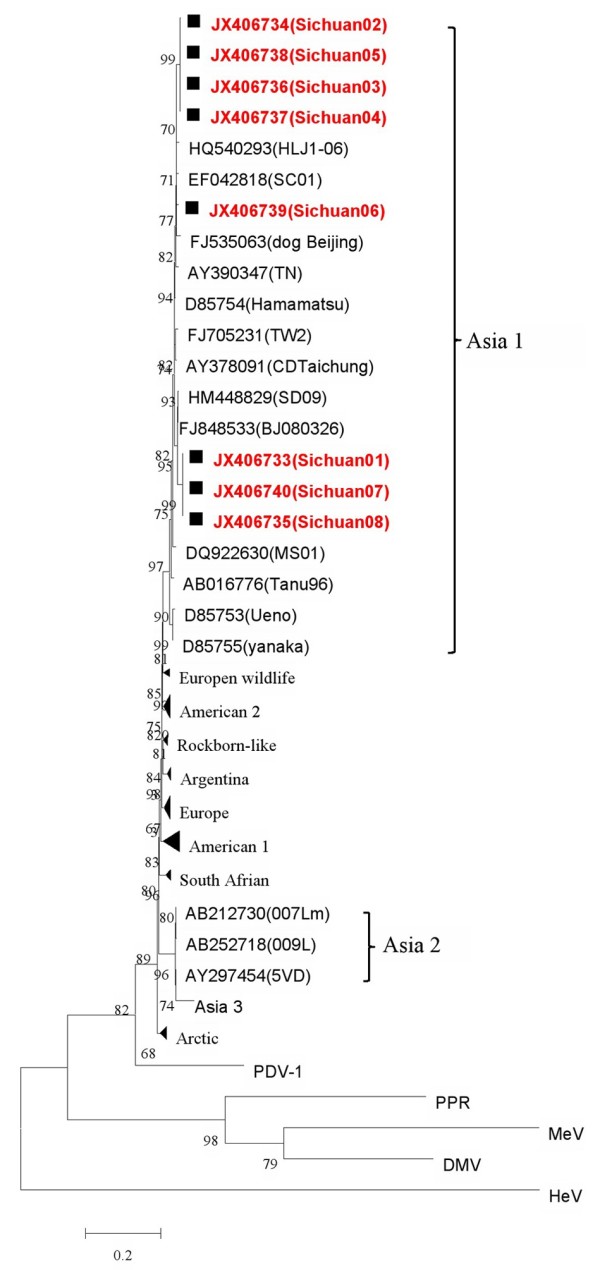
Phylogenetic relationships among 73 CDV isolates based on the amino acid sequences of the H gene. Distance values were calculated by the ClustalW program within the MEGA 5.0 software package. The Neighbor-Joining algorithm was used to generate the tree. Bootstrap values were calculated on 1000 replicates. Only bootstrap values >75% were shown. Filled quadrate (■) indicates the 8 Chinese wild-type CDV strains analyzed in this study.
Table 1.
Homology of nucleotide and amino acid sequences of CDV H gene
| Sichuan01 | Sichuan02 | Sichuan03 | Sichuan04 | Sichuan05 | Sichuan06 | Sichuan07 | Sichuan08 | CDV3 | Onderstepoot | Lederle | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| |
Nucleotides(%) |
|
|
|
|
|
|
|
|
||
| Sichuan01 |
|
93.1 |
94.1 |
94.9 |
94.9 |
94.6 |
98.3 |
98.0 |
86.5 |
85.4 |
86.7 |
| Sichuan02 |
94.4 |
|
98.5 |
98.3 |
98.3 |
96.6 |
94.9 |
94.9 |
89.1 |
87.7 |
89.2 |
| Sichuan03 |
95.0 |
98.4 |
|
97.4 |
97.4 |
94.6 |
95.2 |
92.1 |
87.7 |
86.5 |
88.0 |
| Sichuan04 |
95.0 |
99.4 |
99.7 |
|
99.8 |
97.3 |
97.5 |
91.7 |
88.0 |
86.5 |
88.2 |
| Sichuan05 |
95.0 |
99.5 |
99.6 |
99.8 |
|
97.5 |
97.7 |
91.9 |
88.2 |
86.6 |
88.5 |
| Sichuan06 |
95.5 |
98.9 |
99.4 |
99.5 |
99.4 |
|
95.9 |
91.5 |
88.0 |
86.5 |
88.2 |
| Sichuan07 |
99.8 |
94.4 |
95.0 |
95.0 |
95.0 |
95.5 |
|
93.9 |
88.0 |
86.4 |
88.3 |
| Sichuan08 |
99.7 |
94.5 |
95.3 |
95.0 |
95.0 |
95.5 |
99.5 |
|
88.4 |
87.2 |
88.6 |
| CDV3 |
86.0 |
86.6 |
87.2 |
87.2 |
87.5 |
87.7 |
86.0 |
86.4 |
|
96.1 |
99.0 |
| Onderstepoot |
84.4 |
84.9 |
85.5 |
85.5 |
85.7 |
86.0 |
84.4 |
84.7 |
93.9 |
|
96.0 |
| Lederle |
86.6 |
87.2 |
87.7 |
87.7 |
87.9 |
88.3 |
86.6 |
86.8 |
97.2 |
93.9 |
|
| Amino acids(%) | |||||||||||
Analysis of the amino acid sequences of the H gene of wild-type CDV strains
The inferred aa sequences of the partial H protein of the 8 wild-type Chinese CDV strains and of vaccine (America-1 genotype) strains were aligned. The giant panda and raccoon dogs all were 549Y on the HA protein from the current study (Table 2). Inspection of the H protein alignment (Table 3) revealed that all the 7 Asia-1 strains possessed 9 potential N-linked glycosylation sites (N-X-S/T) at aa positions 19–21, 149–151, 309–311, 391–393,422–424, 456–458, 584–586, 587–589 and 603–605. In the H protein of strain Sichuan01, the sites at positions 584–586 and 603–605 were disrupted. The strain Onderstepoort had only four glycosylation sites, while strains CDV3, Lederle and Convac displayed seven glycosylation sites.
Table 2.
CDV isolates from China, organs from which they were isolated and their GenBank accession numbers
| Serial no | Isolate | Host | Foster mode | Vaccinated | Organ source | GenBank accession no. | 549 |
|---|---|---|---|---|---|---|---|
| 1 |
Sichuan01 |
Raccoon dog |
Captivity |
Yes |
Blood |
JX406733 |
Y |
| 2 |
Sichuan02 |
Raccoon dog |
Captivity |
Yes |
Blood |
JX406734 |
Y |
| 3 |
Sichuan03 |
Raccoon dog |
Captivity |
Yes |
Blood |
JX406736 |
Y |
| 4 |
Sichuan04 |
Raccoon dog |
Captivity |
Yes |
Blood |
JX406737 |
Y |
| 5 |
Sichuan05 |
Raccoon dog |
Captivity |
Yes |
Blood |
JX406738 |
Y |
| 6 |
Sichuan06 |
Raccoon dog |
Captivity |
Yes |
Blood |
JX406739 |
Y |
| 7 |
Sichuan07 |
Raccoon dog |
Captivity |
Yes |
Blood |
JX406740 |
Y |
| 8 | Sichuan08 | Giant Panda | Captivity | Yes | Blood | JX406735 | Y |
Table 3.
Distribution status of N-glycosylation sites of H gene in different genetypes of CDV
| Genetype | CDV isolate |
N-glycosylation sites in each sequence |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 19 | 149 | 309 | 391 | 421 | 456 | 584 | 587 | 603 | ||
| Asia-1 |
Sichuan01 |
N |
N |
N |
N |
N |
N |
N |
N |
N |
| Sichuan02 |
N |
N |
N |
N |
N |
N |
N |
N |
N |
|
| Sichuan03 |
N |
N |
N |
N |
N |
N |
N |
N |
N |
|
| Sichuan04 |
N |
N |
N |
N |
N |
N |
N |
N |
N |
|
| Sichuan05 |
N |
N |
N |
N |
N |
N |
N |
N |
N |
|
| Sichuan06 |
N |
N |
N |
N |
N |
N |
N |
N |
N |
|
| Sichuan07 |
N |
N |
N |
N |
N |
N |
N |
N |
N |
|
| Sichuan08 |
N |
N |
N |
N |
N |
N |
N |
N |
N |
|
| Vaccines | Onderstepoort |
N |
N |
- |
- |
N |
- |
- |
N |
- |
| Convac |
N |
N |
- |
N |
N |
N |
- |
N |
N |
|
| CDV3 |
N |
N |
- |
N |
N |
N |
- |
N |
N |
|
| Lederle | N | N | - | N | N | N | - | N | N | |
Note:“N”: N-glycosylation Sites;“-”: negative.
Discussion
Despite the vaccination procedures adopted in China in the last decades, CDV is still a serious threat to breeding raccoon dogs as well as to domestic dogs and it is responsible for relevant economic losses in Chinese fur industry [20]. In order to determine the frequency of this infection, we analyzed the samples taken from raccoon dogs with suspect of CDV infection. Our result revealed a relatively high (51.8%) prevalence of CDV infection animals sampled with clinical signs consistent with CDV infection. This indicated that CDV still represents a high risk to the canine population in China. Furthermore, all samples were tested twice and the results were reproducible. One possible explanation for the 52 animals thought to be infected with CDV were found negative by PCR screening is the ability of detecting low copy numbers of RNA. Another possible explanation for the negative is the presence of Taq polymerase inhibitors on blood samples, since it is known that blood material contains nucleases that degrades RNA, and substances, such as heparin and hemoglobin were found to inhibit the activity of Taq DNA polymerase [27].
The molecular analysis of the amplifications obtained by the use of primer pair “B” helped to clarify the relationship of the China strains with other CDV strains reported in other parts of the world. The analysis has shown that the strains of giant panda and raccoon dogs in this study are placed on one branches of the phylogenetic tree, were characterized as Asia-1 genotype. The H gene-based neighbor-joining phylogenetic tree analysis using selected sequences retrieved from GenBank revealed geographic-related patterns of segregation. Our findings are consistent with previous studies demonstrating the occurrence of strains belonging to genetically distinct CDV lineages in Asia [20,28,29]. The Chinese Asia-1 strains displayed a high genetic conservation with other Asia-1 CDVs, which detected from raccoon dogs or other carnivores in China over nearly two decades (Figure 1). The strain Sichuan08 was obtained from a mild fever giant panda in the Mar of 2012, and it showed the identity between Sichuan02-06 were 94.5-95.5%. Because there were few reports of giant panda clinical onset of CDV infection up to now, so we couldn't determine whether or not the fever is the clinical manifestations of the giant panda.
The observed diversity between the vaccine strains and recent wild-type CDVs may be accounted for by several mechanisms, such as adaptation to new host species [30,31], antigenic escape [32-35] and/or genetic recombination between wild-type strains [36], variously driving the evolution of the virus. It has been described that changes in N-glycosylation of the H protein may affect neutralization by antibodies and replication in vitro [15,37,38]. So, the connected aspartic amide N glycosylation site potentially is a spotlight in H proteins between vaccine and wild strains of CDV. Usually, there are four (Onderstepoort strain) or seven (CDV3, Lederle and Convac strain) potential sites in the vaccine strains. However, seven or nine sites have been detected in all wild CDV strains, of which 309–311 N-connected amide asparagine glycosylation sites are specific to CDV field strains. Some studies believed that the variants from H protein glycosylation played a crucial role in the antigenic differences and increase in N glycosylation may eventually result in vaccine failure [15,39]. So, we conjectured that the giant panda and raccoon dogs infected CDV maybe due to this reason in this time.
Based on phylogenetic analyses of complete sequences data from CDV strains retrieved from domestic dog and non-dog species it was hypothesized that positive selection drives residues 530 and 549 of the HA gene, and that the presence of specific residues at these positions resulted in the emergence of CDV as a disease of novel host species [31]. Recently, it has been suggested that there no evidence that amino acid 530 was strongly affected by host species, while wild canids there was a nearly significant trend towards a 549Y bias, non-canid strains showed no significant bias towards either H or Y at site 549 [9]. In this study, We found that amino acid 549Y was conserved within CDV lineages, regardless of host species. CDV transmission in wild carnivore and non-canids species may also most often occur between individuals within a species, and will be influenced by a range of factors such as population size and ranging patterns [40,41].
Conclusions
In conclusion, the results presented by this study from a relatively small number of samples obtained from two host species conform to information obtained by other studies in China, that information currently available on CDV strains in China, including the results from the current study, is insufficient to determine whether or not the Asia 1 genotype is the predominant genotype in China. Analysis of CDV strains from a variety of host species will provide a more in-depth understanding of the global ecology of CDV, So, detect these more research on a broader range of potential carnivore hosts is required.
Materials and methods
Viruses and clinical specimens
Ethics statement
The animal from which specimens were collected, was handled in accordance with animal protection law of the People’s Republic of China (a draft of an animal protection law in China released on September 18, 2009). This study was approved by the National Institute of Animal Health Animal Care and Use Committee at Sichuan Agricultural University (approval number 2010–020).
From 2011 to 2012, 108 samples (whole blood) of raccoon dogs with suspect of CDV infection were sent to our laboratory for diagnostic confirmation and were identified only in 56 samples (51.8%) by using a RT-PCR, which amplifies a 242 bp-long fragment of the nucleoprotein (NP) gene with primers “A” ( Table 4). Out of 56 samples, 7 could be included in this study based on the amount of the sample and the amount of RNA extracts available for additional analysis. The samples were submitted to our laboratory from 3 different private fur farms within ya’an, Sichuan province. The organs of the giant panda (whole blood) were submitted to our laboratory by China Conservation and Research Center for the Giant Panda, Ya’an, China. Detailed information on the origin and the accession numbers of the CDV-positive samples is shown in Table 2.
Table 4.
Description of the primer pairs
| Primer pair | Sequence 5’–3’ | Target | Sense | Purpose | Amplicon size (bp) |
|---|---|---|---|---|---|
| A |
CGGAAATCAACGGACCTAAAT |
N |
+ |
Diagnostic |
242 |
| |
TCCTTGAGCTTTCGACCCTT |
|
_ |
|
|
| B |
TGGTTCACAAGATGGTATTC |
H |
+ |
Phylogenetic |
613 |
| CAACACCAC TAAATTGGACT | _ |
Extraction of nucleic acids
Viral RNA was extracted from leucocytes obtained by washing with nuclease-free water. Briefly, 500 μl of nuclease-free water were added to 500 μl of whole blood in a 1.5 microcentrifuge tube, and centrifuged 7 min at 5,000 g. A total of 500 μl of the supernatant was discarded, then 500 μl of nuclease-free water were added, repeating this step 3 times. Total RNA was extracted with TRI Reagent (Sigma®, USA), following the manufacturer’s instructions.
RT-PCR
Reverse transcription was carried out by PrimeScript™ RT reagent Kit (TaKaRa, Dalian, China) according to the manufacturer’s instruction: 37°C for 15 min, 85°C for 5 sec, using 7 μl of RNA, 2 μl 5 × PrimeScript buffer, 0.5 μl random primer and 0.5 μl PrimeScript RT Enzyme. The CDV-positive samples were tested with primers “B” (Table 4), which amplifying a 613 bp fragment (corresponding to positions 8006–8619 of the Onderstepoort strain genome). The thermo-cycling condition were as follows: 5 min at 94°C (initial denaturation), 30 cycles of 30 sec at 94°C, 30 sec at 51°C, 30 sec at 72°C, and a final extension of 10 min at 72°C.
Cloning and sequencing
PCR products of the correct size (613 bp in length) were amplified and cloned into the pMD 19-T vector (TaKaRa, Dalian, China). For each CDV strain, 3–5 positive recombinant plasmids were sequenced in both directions using primer M13 (Shanghai Invitrogen Biotechnology Company, Shanghai, China) and additional primers selected on the basis of the obtained sequences. The accession numbers are shown in Table 2.
Amino acid analysis
To investigate the differences between the Chinese wild-type CDV strains and the vaccine strains available in the market in China, the deduced amino acid (aa) sequences of the H protein were aligned. Multiple sequence alignment was carried out using the software package MegAlign (Lasergene7.0, DNAStar Inc., Madison, WI). The aa present at sites 549 on the HA protein was determined for the eight strains from the current study. The potential N-linked glycosylation sites of the H protein were determined using the software NetNGlyc1.0.
Phylogenetic analysis
The eight China CDV sequences were compared to those of 64 CDV strains that were obtained from various part of the world up until November 2010. The Accession numbers for these H gene nucleotide sequences were obtained from the GenBank nucleotide database at the National Center for Biotechnology Information (NCBI). The nucleotide sequences were analyzed preliminary with cognate sequences available in GenBank using BLAST software (http://www.ncbi.nlm.nih.gov/). Multiple sequence alignment was made with ClustalW software [42] included in MEGA 5.0 software [43].
Sequences for comparison were obtained corresponded to nucleotide sequences of the H gene. Phylogenetic analysis of nucleotide were computed using the Kimura two parameter algorithm and the tree was constructed with the Neighbor Joining method with a 1000 bootstrap repeats using the MEGA 5.0 Software [43]. The measles virus (MeV) (NC001498), phocine (seal) distemper virus (PDV) (AF479274), peste-des- petits-ruminants virus (PPRV) (NC006383), hendra virus (HeV) (NC001906) and dolphin morbillivirus (DMV) (NC005283) were used as outgroup in the neighbor joining trees.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Conceived and designed the experiments: LG QGY SJC. Performed the experiments: LG SLY ZXH JL HBP. Analyzed the data: LG SLY MLZ. Contributed reagents/materials /analysis tools: SJC CDW RH XNY . Wrote the paper: LG SLY. All of the authors read and approved the final version of the manuscript.
Contributor Information
Ling Guo, Email: yaanguoling@163.com.
Shao-lin Yang, Email: 280598792@qq.com.
Cheng-dong Wang, Email: wangchengdong@yahoo.cn.
Rong Hou, Email: hourong2000@yahoo.com.
Shi-jie Chen, Email: cworld@126.com.
Xiao-nong Yang, Email: yangxn058@yahoo.com.cn.
Jie Liu, Email: 375254375@qq.com.
Hai-bo Pan, Email: 1359995115@qq.com.
Zhong-xiang Hao, Email: 1205230776@qq.com.
Man-li Zhang, Email: 505069467@qq.com.
San-jie Cao, Email: csjie@sicau.edu.cn.
Qi-gui Yan, Email: yanqigui@126.com.
Acknowledgments
This work was supported by Program for Changjiang Scholars and Innovative Research Team in University, China (IRT0848), Specific projects of public welfare of the Ministry of Science and Technology of the P.R. China (200910188), Program of Science and Technology Department of Sichuan Province (2010SZ0050), National Key Technology R&D Program (2012BAC01B06) and the “973 project” of prophase special program (2012CB722207).
References
- Appel MJG, Yates RA, Foley GL, Bernstein JJ, Santinelli S, Spelman LH, Miller LD, Arp LH, Anderson M, Barr M. Canine distemper epizootic in lions, tigers, and leopards in North America. J Vet Diagn Invest. 1994;6:277. doi: 10.1177/104063879400600301. [DOI] [PubMed] [Google Scholar]
- Deem SL, Spelman LH, Yates RA, Montali RJ. Canine distemper in terrestrial carnivores: a review. J Zoo Wildlife Med. 2000;31:441–451. doi: 10.1638/1042-7260(2000)031[0441:CDITCA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Evermann J, Leathers C, Gorham J, McKeirnan A, Appel M. Pathogenesis of two strains of lion (Panthera leo) morbillivirus in ferrets (Mustela putorius furo) Vet Pathol. 2001;38:311. doi: 10.1354/vp.38-3-311. [DOI] [PubMed] [Google Scholar]
- Pearson R, Gorham J. Canine distemper virus. Virus. 1987;1:371–373. [Google Scholar]
- Van De Bildt MWG, Kuiken T, Visee AM, Lema S, Fitzjohn TR, Osterhaus ADME. Distemper outbreak and its effect on African wild dog conservation. Emerg Infect Dis. 2002;8:212. doi: 10.3201/eid0802.010314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel MJ, Summers BA. Pathogenicity of morbilliviruses for terrestrial carnivores. Book Pathogenicity of morbilliviruses for terrestrial carnivores. 1995;4:187–191. doi: 10.1016/0378-1135(95)00011-x. [DOI] [PubMed] [Google Scholar]
- Haas L, Hofer H, East M, Wohlsein P, Liess B, Barrett T. Canine distemper virus infection in Serengeti spotted hyenas. Vet Microbiol. 1996;48:147–152. doi: 10.1016/0378-1135(95)00180-8. [DOI] [PubMed] [Google Scholar]
- Von Messling V, Zimmer G, Herrler G, Haas L, Cattaneo R. The hemagglutinin of canine distemper virus determines tropism and cytopathogenicity. J Virol. 2001;75:6418–6427. doi: 10.1128/JVI.75.14.6418-6427.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolin VM, Wibbelt G, Michler FU, Wolf P, East ML. Susceptibility of carnivore hosts to strains of canine distemper virus from distinct genetic lineages. Vet Microbiol. 2012;156:45–53. doi: 10.1016/j.vetmic.2011.10.009. [DOI] [PubMed] [Google Scholar]
- Nikolin VM, Osterrieder K, von Messling V, Hofer H, Anderson D, Dubovi E, Brunner E, East ML. Antagonistic pleiotropy and fitness trade-offs reveal specialist and generalist traits in strains of canine distemper virus. PLoS One. 2012;7:e50955. doi: 10.1371/journal.pone.0050955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeter Z, Lakatos B, Palade EA, Kozma T, Forgách P, Rusvai M. Genetic diversity of Hungarian canine distemper virus strains. Vet Microbiol. 2007;122:258–269. doi: 10.1016/j.vetmic.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas L, Martens W, Greiser-Wilke I, Mamaev L, Butina T, Maack D, Barrett T. Analysis of the haemagglutinin gene of current wild-type canine distemper virus isolates from Germany. Virus Res. 1997;48:165–171. doi: 10.1016/S0168-1702(97)01449-4. [DOI] [PubMed] [Google Scholar]
- Martella V, Cirone F, Elia G, Lorusso E, Decaro N, Campolo M, Desario C, Lucente M, Bellacicco A, Blixenkrone-Møller M. Heterogeneity within the hemagglutinin genes of canine distemper virus (CDV) strains detected in Italy. Vet Microbiol. 2006;116:301–309. doi: 10.1016/j.vetmic.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Bolt G, Jensen TD, Gottschalck E, Arctander P, Appel MJ, Buckland R, Blixenkrone-Moller M. Genetic diversity of the attachment (H) protein gene of current field isolates of canine distemper virus. J Gen Virol. 1997;78:367–372. doi: 10.1099/0022-1317-78-2-367. [DOI] [PubMed] [Google Scholar]
- Iwatsuki K, Miyashita N, Yoshida E, Gemma T, Shin YS, Mori T, Hirayama N, Kai C, Mikami T. Molecular and phylogenetic analyses of the haemagglutinin (H) proteins of field isolates of canine distemper virus from naturally infected dogs. J Gen Virol. 1997;78:373–380. doi: 10.1099/0022-1317-78-2-373. [DOI] [PubMed] [Google Scholar]
- Martella V, Elia G, Lucente MS, Decaro N, Lorusso E, Banyai K, Blixenkrone-Møller M, Lan NT, Yamaguchi R, Cirone F. Genotyping canine distemper virus (CDV) by a hemi-nested multiplex PCR provides a rapid approach for investigation of CDV outbreaks. Vet Microbiol. 2007;122:32–42. doi: 10.1016/j.vetmic.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Pardo ID, Johnson GC, Kleiboeker SB. Phylogenetic characterization of canine distemper viruses detected in naturally infected dogs in North America. J Clin Microbiol. 2005;43:5009–5017. doi: 10.1128/JCM.43.10.5009-5017.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon MG, Remorini P, Periolo O, Iglesias M, Mattion N, La Torre J. Detection by RT-PCR and genetic characterization of canine distemper virus from vaccinated and non-vaccinated dogs in Argentina. Vet Microbiol. 2007;125:341–349. doi: 10.1016/j.vetmic.2007.05.020. [DOI] [PubMed] [Google Scholar]
- Woma TY, van Vuuren M, Bosman AM, Quan M, Oosthuizen M. Phylogenetic analysis of the haemagglutinin gene of current wild-type canine distemper viruses from South Africa: lineage Africa. Vet Microbiol. 2010;143:126–132. doi: 10.1016/j.vetmic.2009.11.013. [DOI] [PubMed] [Google Scholar]
- Zhao JJ, Yan XJ, Chai XL, Martella V, Luo GL, Zhang HL, Gao H, Liu YX, Bai X, Zhang L. Phylogenetic analysis of the haemagglutinin gene of canine distemper virus strains detected from breeding foxes, raccoon dogs and minks in China. Vet Microbiol. 2010;140:34–42. doi: 10.1016/j.vetmic.2009.07.010. [DOI] [PubMed] [Google Scholar]
- Hu A, Cattaneo R, Schwartz S, Norrby E. Role of N-linked oligosaccharide chains in the processing and antigenicity of measles virus haemagglutinin protein. J Gen Virol. 1994;75:1043–1052. doi: 10.1099/0022-1317-75-5-1043. [DOI] [PubMed] [Google Scholar]
- Sawatsky B, von Messling V. Canine distemper viruses expressing a hemagglutinin without N-glycans lose virulence but retain immunosuppression. J Virol. 2010;84:2753–2761. doi: 10.1128/JVI.01813-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen DL, Edwards MS, Sutherland-Smith M, Yu J, Li D, Zhong G, Wei R, Chenglin Z, Miller R, Phillips L. Giant pandas: biology, veterinary medicine, and management. Cambridge, U.K: Cambridge University Press; 2006. Significant medical issues and biological reference values for giant pandas from the biomedical survey; pp. 59–86. [Google Scholar]
- Janssen DL, Morris P, Sutherland-Smith M, Greenberg M, Li D, Hu D, Mauroo N, Spelman L. Giant pandas: biology, veterinary medicine, and management. Cambridge, U.K: Cambridge University Press; 2006. Medical management of captive adult and geriatric giant pandas; pp. 353–376. [Google Scholar]
- Bittegeko S, Arnbjerg J, Nkya R, Tevik A. Multiple dental developmental abnormalities following canine distemper infection. J Am Anim Hosp Assoc. 1995;31:42–45. doi: 10.5326/15473317-31-1-42. [DOI] [PubMed] [Google Scholar]
- Dubielzig RR, Higgins RJ, Krakowka S. Lesions of the enamel organ of developing dog teeth following experimental inoculation of gnotobiotic puppies with canine distemper virus. Vet Pathol. 1981;18:684–689. doi: 10.1177/030098588101800513. [DOI] [PubMed] [Google Scholar]
- De Paula SO, Lopes da Fonseca BA. Optimizing dengue diagnosis by RT-PCR in IgM-positive samples: comparison of whole blood, buffy-coat and serum as clinical samples. J Virol Methods. 2002;102:113–117. doi: 10.1016/S0166-0934(02)00005-8. [DOI] [PubMed] [Google Scholar]
- Wang F, Yan X, Chai X, Zhang H, Zhao J, Wen Y, Wu W. Differentiation of canine distemper virus isolates in fur animals from various vaccine strains by reverse transcription-polymerase chain reaction-restriction fragment length polymorphism according to phylogenetic relations in china. J Virol. 2011;8:85. doi: 10.1186/1743-422X-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B, Wen Y-J, Wang F-X, Zhang S-Q, Wang X-D, Hu J-X, Shi X-C, Yang B-C, Chen L-Z, Cheng S-P. Pathogenesis and phylogenetic analyses of canine distemper virus strain ZJ7 isolate from domestic dogs in China. J Virol. 2011;8:520. doi: 10.1186/1743-422X-8-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo E, Holland JJ. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- McCarthy AJ, Shaw MA, Goodman SJ. Pathogen evolution and disease emergence in carnivores. Proc Biol Sci. 2007;274:3165–3174. doi: 10.1098/rspb.2007.0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatsuki K, Tokiyoshi S, Hirayama N, Nakamura K, Ohashi K, Wakasa C, Mikami T, Kai C. Antigenic differences in the H proteins of canine distemper viruses. Vet Microbiol. 2000;71:281–286. doi: 10.1016/S0378-1135(99)00172-8. [DOI] [PubMed] [Google Scholar]
- Lan NT, Yamaguchi R, Furuya Y, Inomata A, Ngamkala S, Naganobu K, Kai K, Mochizuki M, Kobayashi Y, Uchida K, Tateyama S. Pathogenesis and phylogenetic analyses of canine distemper virus strain 007Lm, a new isolate in dogs. Vet Microbiol. 2005;110:197–207. doi: 10.1016/j.vetmic.2005.07.016. [DOI] [PubMed] [Google Scholar]
- Lan NT, Yamaguchi R, Inomata A, Furuya Y, Uchida K, Sugano S, Tateyama S. Comparative analyses of canine distemper viral isolates from clinical cases of canine distemper in vaccinated dogs. Vet Microbiol. 2006;115:32–42. doi: 10.1016/j.vetmic.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Martella V, Elia G, Lucente MS, Decaro N, Lorusso E, Banyai K, Blixenkrone-Moller M, Lan NT, Yamaguchi R, Cirone F. Genotyping canine distemper virus (CDV) by a hemi-nested multiplex PCR provides a rapid approach for investigation of CDV outbreaks. Vet Microbiol. 2007;122:32–42. doi: 10.1016/j.vetmic.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Han GZ, Liu XP, Li SS. Cross-species recombination in the haemagglutinin gene of canine distemper virus. Virus Res. 2008;136:198–201. doi: 10.1016/j.virusres.2008.04.022. [DOI] [PubMed] [Google Scholar]
- Harder TC, Kenter M, Vos H, Siebelink K, Huisman W, van Amerongen G, Orvell C, Barrett T, Appel MJ, Osterhaus AD. Canine distemper virus from diseased large felids: biological properties and phylogenetic relationships. J Gen Virol. 1996;77:397–405. doi: 10.1099/0022-1317-77-3-397. [DOI] [PubMed] [Google Scholar]
- Lan NT, Yamaguchi R, Kawabata A, Uchida K, Sugano S, Tateyama S. Comparison of molecular and growth properties for two different canine distemper virus clusters, Asia 1 and 2, in Japan. J Vet Med Sci. 2007;69:739–744. doi: 10.1292/jvms.69.739. [DOI] [PubMed] [Google Scholar]
- Zhao J, Yan X, Wu W. Genetic variations and cellular receptors of Canine distemper virus–a review. Acta Microbiol Sin. 2008;48:986. [PubMed] [Google Scholar]
- Guiserix M, Bahi-Jaber N, Fouchet D, Sauvage F, Pontier D. The canine distemper epidemic in Serengeti: are lions victims of a new highly virulent canine distemper virus strain, or is pathogen circulation stochasticity to blame? J R Soc Interface. 2007;4:1127–1134. doi: 10.1098/rsif.2007.0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begon M, Townsend CR, Harper JL. Ecology: From Individuals to Ecosystems. Oxford, UK: Blackwell; 2006. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]


