Abstract
Background
Vibrio fluvialis is considered to be an emerging foodborne pathogen and has been becoming a high human public health hazard all over the world, especially in coastal areas of developing countries and regions with poor sanitation. The distribution of virulence factors, microbiological and molecular epidemiological features of V. fluvialis isolates in China remains to be examined.
Methods and results
PCR targeted at the virulence determinants and phenotype tests including metabolism, virulence and antibiotic susceptibility were performed. Pulsed-field gel electrophoresis (PFGE) analysis was used to access the relatedness of isolates. A strain with deletion of the arginine dihydrolase system was first reported and proved in molecular level by PCR. Virulence genes vfh, hupO and vfpA were detected in all strains, the ability to produce hemolysin, cytotxin, protease and biofilm formation varied with strains. High resistance rate to β-lactams, azithromycin and sulfamethoxazole were observed. Twenty-seven percent of test strains showed resistant to two and three antibiotics. PFGE analysis demonstrated great genetic heterogeneity of test V. fluvialis strains.
Conclusion
This study evaluated firstly the biological characteristics and molecular epidemiological features of V. fluvialis in China. Some uncommon biochemical characteristics were found. Virulence genes were widely distributed in the isolates from patient and seafood sources, and the occurrence of virulence phenotypes varied with strains. Continued and enhanced laboratory based-surveillance is needed in the future together with systematically collection of the epidemiological information of the cases or the outbreaks.
Keywords: Vibrio fluvialis, VFP, Cytotoxicity, Antibiotic Susceptibility, PFGE, Biofilm, Virulence
Background
V. fluvialis is considered to be an emerging foodborne pathogen and has been implicated in outbreaks and sporadic cases of acute diarrhea [1]–[7]. Besides, V. fluvialis posed a significant economic threat for aquaculture for being pathogenic for cultured fish and lobsters [8]. Gastroenteritis caused by this organism is associated with drinking of contaminated water or consumption of raw or improperly cooked seafood [3,5]. In addition, V. fluvialis-associated extraintestinal infections, such as hemorrhagic cellulites and cerebritis [9], peritonitis [10], acute otitis [11], biliary tract infection [12], bacteraemia [13] and even ocular infections [14] were also reported.
Several toxins that may be important in pathogenesis have been reported in V. fluvialis including a Chinese hamster ovary (CHO) cell elongation factor, CHO cell-killing factor, enterotoxin-like substance, lipase, protease, cytotoxin, and hemolysin [15]–[20]. The cell-free culture filtrates of V. fluvialis strains were capable of evoking distinct cytotoxic and vacuolation effects on HeLa cells [21]. The heme utilization protein gene hupO in V. fluvialis was induced under iron-restricted conditions and is associated with virulence expression through stimulation of hemolysin production and resistance to oxidative stress [22]. In spite of many pathogenic factors were characterized, their precise role in producing the clinical manifestations remains to be known and little definitive information about the pathogenic mechanism of V. fluvialis has been achieved.
The largest outbreak of V. fluvialis infection was reported in Bangladesh between October 1976 and November 1977, with more than 500 patients [2]. In the United States, V. fluvialis accounted for 10% of vibrio-caused clinical cases along the Gulf Coast [23]. Vibrio surveillance data show that it was responsible for 82 of the 1,584 Vibrio infections reported to the Centers for Disease Control and Prevention during 1997–2000 [24]. Srinivasan et al. reported that V. fluvialis is being isolated with an increased frequency from hospitalized patients in Kolkata, India, with cholera-like illnesses [25]. Study of Ramamurthy group revealed the isolation rate of V. fluvialis increased from 0.7% in 2002 to 2.2% in 2009 [4]. Recently, it was reported that 3,529 (91.2%) of 3,871 residents of Pakhirala village of the coastal islands of the Sunderbans, situated in the southern part of West Bengal, eastern India, were affected by watery diarrhea caused by V. fluvialis within a span of six weeks following Cyclone Aila in 2009 [5]. Furthermore, V. fluvialis behaved more aggressively than V. cholerae O1 in an epidemic situation with a higher attack rate and a different clinical picture [5]. In general, the clinical symptoms of the gastroenteritis caused by V. fluvialis are similar to those caused by V. cholera, including mild to moderate dehydration, abdominal pain, vomiting, fever, and diarrhea with presence of blood which is a notable different from cholera [5,26]. The infection of V. fluvialis is generally common in infants, children, and young adults [2,21,27]. Therefore, V. fluvialis has been becoming a high human public health hazard all over the world, especially in coastal areas of developing countries and regions with poor sanitation.
In China, the etiological characteritics of V. fluvialis and its epidemiology of infection were not even fairly known with little information in the literature. 4.8% isolation rate of V. fluvialis was reported in sea products [28]. Considering the occurrence and prevalence of V. fluvialis enteritis in different developed and developing countries [2,6,7,27,29,30], the infection of V. fluvialis in China is probably undetected due to complexity in the identification and less attention in the surveillance. Historically, only toxigenic V. cholerae serogroups O1 and O139 were nationally notifiable. V. fluvialis phenotypically resembles Aeromonas species [26,28], and is closely similar to V. furnissii which is aerogenic in nature [31]. Many questions remain unanswered about its microbiological characteristics, distribution of virulence factors, mechanism of pathogenicity and epidemiology of the infection. The objective of this study was to investigate and compare the virulence determinants, drug resistance profiles of 43 V. fluvialis isolated in China. Main biological characteristics and clonal relationship among the strains were examined.
Methods
Bacterial strains and culture condition
A total of 43 strains of V. fluvialis collected from six different geographical regions in China were included in this study. All strains were maintained in Luria-Bertani (LB) broth supplemented 15% glycerol and stored at −80°C. Of these, 10 were isolated from marine products including shrimp and fish. The rest 33 were isolated from the stool samples of diarrheal patients. Strain CICC21612 was acquired from the National Institutes for food and drug Control (NIFDC) and used as the reference strain of V. fluvialis. In general, V. fluvialis strains were grown in LB broth containing 1% NaCl with shaking at 37°C. The detailed information of every strain was showed in Additional file 1: Table S1.
Polymerase chain reaction (PCR)
PCR was used to confirm the identities of presumptive V. fluvialis by using two species-specific primers toxR-F/toxR-R and VFLU-F/VFLU-R which targeted at the toxR gene and 16S-23S rDNA intergenic sequence [32,33]. PCR assays were also performed to screen the presence of the virulence genes vfh, hupO, vfpA and stn, int IV gene specific for the class IV integron [34], sulII gene encoding for the sulfamethoxazole resistance [35]. Based on sequence of V. furnissii NCTC 11218 [36], primers arc-F/arc-R were designed to amplify the arginine dihydrolase system in V. flluvialis. A water-boiled template of each strain was used in the all PCR assays. Each PCR involved an initial denaturation at 94°C for 4 min, followed by 33 cycles each consisting of an initial denaturation at 94°C for 40 sec followed by annealing and extension steps. Final polymerization was included at 72°C for 6 min. Primers and the corresponding annealing temperatures were listed in Table 1.
Table 1.
Primers and amplification conditions used in this study
| Primer | Sequences (5’-3’) | Target size (bp) | Annealing temp. (°C) | Reference |
|---|---|---|---|---|
|
toxR-F |
GACCAGGGCTTTGAGGTGGACGAC |
217 |
65 |
[32] |
|
toxR-R |
AGGATACGGCACTTGAGTAAGACTC |
|
|
|
| VFLU-F |
ATAAAGTGAAGAGATTCGTACC |
278 |
60 |
[33] |
| VFLU-R |
GTATTCCTGAATGGAATACAC |
|
|
|
|
Int IV-F |
AACACCGCTTGCACCTCTAT |
525 |
53 |
[34] |
|
Int IV-R |
TGTATGCGCTTGAGAGTCC |
|
|
|
|
stn-F |
GGTGCAACATAATAAACAGTCAACAA |
375 |
53 |
[44] |
|
stn-R |
TAGTGGTATGCGTTGCCAGC |
|
|
|
|
sulII-F |
AGGGGGCAGATGTGATCGAC |
606 |
55 |
[35] |
|
sulII-B |
TGTGCGGATGAAGTCAGCTCC |
|
|
|
|
vfh-F |
GCGCGTCAGTGGTGGTGAAG |
800 |
61 |
This study |
|
vfh-R |
TCGGTCGAACCGCTCTCGCTT |
|
|
|
|
hupO-F |
ATTACGCACAACGAGTCGAAC |
600 |
56 |
This study |
|
hupO-R |
ATTGAGATGGT AAACAGCGCC |
|
|
|
|
vfpA -F |
TACAACGTCAAGTTAAAGGC |
1790 |
55 |
This study |
|
vfpA -R |
GTAGGCGCTGTAGCCTTTCA |
|
|
|
|
arc-F |
AGTTTATGCGTCTGGCTTG |
3427 |
56 |
This study |
|
arc-R |
ATGAGTAAGTTATACGTAGG |
|
|
|
|
arc-rev |
GCTTCGGCCCACATAATAA (paired with arc-F) |
2170 |
56 |
This study |
| arc-ck-up | TTACCACCTAATGCGACGA (paired with arc-R) | 1235 | 56 | This study |
Biochemical characteristics of V. Fluvialis
Molecular confirmed V. fluvialis strains were plated on LB agar and thiosulphate citrate bile salts sucrose agar (TCBS) followed by incubation at 37°C overnight. API 20E (bioMérieux) identification strip was used to characterize the biochemical features. The string test was performed using 0.5% sodium deoxycholate solution with fresh colonies grown on LB agar. Cytochrome oxidase was detected using Oxidase Reagent (bioMérieux). Susceptibility to 10 μg of vibriostatic compound O/129 (2, 4-diamino-6, 7-diisopropylpteridine phosphate) was determined in LB agar [38]. Salt tolerance was determined by growing the strains in LB broth overnight with shaking at 37°C without NaCl or with 6% or 7% NaCl.
Haemolysin assay
The ability of V. fluvialis to produce extracellular hemolysin (VFH) was examined on Columbia blood agar containing sheep erythrocytes. Fresh single colonies of each strain from LB agar were spotted onto blood agar plates and incubated at 37°C for 24-72 h. The appearance of hemolytic zone was observed per 24 h.
Haemagglutinin activity
Fresh colonies of each strain from LB agar were suspended in PBS and cell density of the suspension was adjusted to 105-6 cfu/ml. Chicken and human erythrocytes were washed respectively and then diluted to a final concentration of 1.5% (vol/vol) in sterile 10 mM PBS (pH 7.0). 100 μl of the cell suspension was mixed with 100 μl of 1.5% chicken erythrocytes in 8-well 200 μl PCR tubes (Axygen, Germany). The mixture was incubated at 25°C for 45 min, and agglutination was monitored visually. PBS was used as negative control, V. cholerae strain N16961 was used as positive control.
Metalloprotease activity
V. fuvialis protease (VFP) activity was measured using an azocasein assay [39]. Briefly, V. fuvialis strains were cultured overnight in LB broth at 37°C with agitation. 100 μl of azocasein (5 mg/ml) in 100 mM Tris (pH 8.0) was incubated with 100 μl of cell culture supernatants for 1 h at 37°C. The reaction was stopped by adding 400 μl of 10% trichloroacetic acid solution. After centrifugation for 15 minutes at 13000 rpm, the trichloroacetic acid supernatant was transferred to a new tube containing 700 μl of 525 mM NaOH, and the optical density was determined at 442 nm (OD442). One azocasein unit is the amount of enzyme that produces an increase of 0.01 optical density units per h. Three independent cultures for each strain were tested and LB broth was used as blanks.
Biofilm formation
Biofilm formation was measured by the crystal violet staining method [40]. Overnight cultures of each strain were diluted 1:50 in fresh medium and 100 μl of dilution transferred to 96-well flat-bottom microtitre plates. The plates were incubated statically for 24 h at 30°C for biofilm development. At the desired end-point, OD600 was determined and the plates were rinsed with PBS buffer for to remove the non-adherent cells. Biofilms were stained with 120 μl 0.1% crystal violet for 30 min at 30°C followed by rinsing four times with PBS. The cell-associated dye was solubilized in 120 μl of dimethyl sulfoxide (DMSO) and quantified by measuring the OD570 of the resulting solution. Final results were normalized for growth and expressed as the A570/OD600 ratio. Each assay was performed in triplicate.
Antibiotic susceptibility test
Antibiotic susceptibility test was performed using the microbroth dilution method according to the guidelines of the current Clinical and Laboratory Standards Institute (CLSI). All V. fluvialis strains were tested for susceptibility to 15 antibiotics which include ampicillin, amoxicillin/clavulanic acid, cefotaxime, ceftriaxone, ceftazidime, chloramphenicol, ciprofloxacin, gentamicin, nalidixic acid, streptomycin, sulfamethoxazole, trimethoprim, co-trimoxazole, tetracycline, and azithromycin. Multidrug resistance was defined as a presence of resistance to two or more classes of antibiotics. Escherichia coli ATCC 25922 was used for quality control. No interpretive criteria for V. fluvialis were available based on CLSI guidelines, the minimum inhibitory concentrations (MICs) of antibiotics were determined by referring to the CLSI standards for V. cholerae if available; otherwise breakpoints for Enterobacteriaceae were applied.
Tissue culture assay
Human laryngeal carcinoma Hep-2 cells were grown in RPMI Medium 1640 (Gibco) supplemented with 10% fetal bovine serum at 37°C in a humidified 5% CO2 atmosphere (Thermo Scientific, USA). Around 2×105 cells were seeded in each well of 24-well plates and cultured overnight. Tissue culture medium was removed and cells were washed with 1640 medium three times before treatment. V. fluvialis strains were grown in Brain Heart Infusion broth (BHI, OXOID) supplemented with 0.5% NaCl at 37°C for 18 h in a rotary shaker. The culture supernatant, collected by centrifugation at 8000 rpm for 2 min, was filter-sterilized using 0.22 μm filter units (Millipore, USA). The resultant cell-free culture filtrate was serially diluted and aliquots of each test dilution were added in triplicate to the cell culture plate and incubated for 24 h. Morphological changes and cytotoxic effects were observed after 24 h using an inverted microscope (Nikon ECLIPSE Ti-SR, JAPAN). The toxin titer was expressed as the highest dilution that affected 50% of the cells in a well [21]. The cell pellet was washed and suspended in 1640 medium. Approximately 107 bacterial cells were then added in triplicate to the cell culture plate and incubated for 8 h at 37°C with 5% CO2. Cytotoxic activity was detected by Lactate dehydrogenase (LDH) cytotoxicity assays Kit (Promega) according to the instruction of manufacture. 2% Triton X-100 served as positive controls and BHI, RPMI Medium 1640 as negative controls. Cytotoxic activity was calculated according to the following formula: LDH(%)=[OD490(sample) − OD490 (negative control)/OD490(positive control) − OD490(negative control)] ×100%.
PFGE
PFGE was performed according to the PulseNet standardized PFGE protocol for V. cholerae subtyping [41]. Genomic DNA of V. fluvialis strains was prepared in agarose plugs and digested with the restriction enzyme NotI, separated in a 1% agarose gel in 0.5×TBE buffer at 14°C using a CHEF-DRIII apparatus (Bio-Rad, Hercules, CA, USA). The pulse time ranged from 2 to 10 s for 13 h, and then from 20 to 25 s for 6 h, both at 6 v/cm. Gels were stained in distilled water containing 1.0 μg ethidium bromide ml-1 for 30 min, destained several times and photographed under UV light using the Gel Doc 2000 (Bio-Rad, Hercules, CA, USA). After visualization, the PFGE patterns were analyzed using BioNumerics. Dendrograms were clustered and constructed by using the UPGMA with a tolerance of 1.5%.
Nucleotide sequence accession number
The nucleotide sequences encoding the enzymes involved in the arginine dihydrolase system and vfh gene have been deposited in the GenBank database under accession number KC569550 and KC569551, respectively.
Results and discussion
Biological features and biochemical characterizatio
Considering that V. fluvialis shares biochemical properties with Aeromonas species and the API 20E system gave ambiguous identity sometimes [26], the identities of V. fluvialis were first confirmed by using two sets of species-specific primers which targeted at the conserved transcriptional activation and variable membrane tether regions of the toxR gene and 16S-23S rDNA intergenic sequence, respectively [32,33]. Excluding three strains with negative amplifications, 43 strains with both the expected amplicons size were molecular-confirmed as V. fluvialis and included in this study. All 44 strains including the reference strain CICC21612 grew as yellow colonies on the TCBS plates and grew well in the presence of 6% salt; however, 36 strains (81.8%) did not grow in LB without NaCl. And 8 strains (18.2%) grew very poorly. Salt tolerance test is very important in distinguishing V. fluvialis from the Aeromonas species as Aeromonas species cannot grow in the presence of 6% NaCl [42]. Variable results were observed in LB with 7% salt: 31 strains (70.5%) grew and 13 strain (29.5%) did not grow. 28 strains (63.6%) were resistant to 10 μg of vibriostatic agent. All the strains were positive in Oxidase and String test, negative in Voges–Proskauer (VP) test, H2S production, urease and tryptophane deaminase. They were positive in mannitol, sucrose and arabinose fermentation, negative in inositol, rhamnose and melibiose fermentation. o-nitrophenyl-β-D-galactopyranoside (ONPG) was positive except for strain JS38. Except strains JS50 and Ma-2531 which were arginine dihydrolase-negative, all strains were lysine decarboxylase-negative, ornithine decarboxylase-negative and arginine dihydrolase-positive. ONPG positivity is a generally believed biochemical trait in V. fluvialis, but ONPG-negative V. fluvialis had been reported [21]. The results of V. fluvialis strains in a variety of biological features and API 20E profile numbers are shown in Additional file 1: Table S1.
Arginine dihydrolase -negative V. fluvialis strain was not reported in the literature before. The arginine dihydrolase system which converts arginine to ornithine, ammonia and carbon dioxide via citrulline consists of three enzyme reactions catalysed by arginine deiminase, ornithine carbamoyltransferase and carbamate kinase [43,44] and facilitates acid tolerance. Arginine deiminase, i.e. arginine dihydrolase is the first enzyme involved in this system catalysing the chemical reaction: L-arginine + H2O ↮ L-citrulline + NH3 [43]. PCR amplification revealed that nucleotide sequences encoding arginine deiminase, ornithine carbamoyltransferase and carbamate kinase are absent in strain Ma-2531. Though strain JS50 displayed negative phenotype of arginine dihydrolase, PCR assay gave the similar-size of amplicon as other strains and sequence analysis revealed no mutation in the enzyme’s coding region, suggesting somehow the expression of the arginine dihydrolase system was affected in JS50. To further exclude the possibility of negative amplification due to sequence variation of the primer annealing regions in strain Ma-2531, additional two primers arc-rev and arc-ck-up were designed based on the sequence of arc operon of V. fluvialis strain JS50. Primer pairs arc-rev/arc-F and arc-ck-up/arc-R gave the expected size of amplicons in all tested V. fluvialis strains except Ma-2531. Sequence analysis of vfh gene of Ma-2531 displayed 98% identity to the reference sequence of V. fluvialis hemolysin gene in GenBank (AF348455.1), which further confirmed Ma-2531 was an arginine dihydrolase-negative V. fluvialis. The corresponding gene sequences encoding the enzymes involved in the arginine dihydrolase system in JS50 and vfh gene in Ma-2531 were deposited in the Genbank under accession number KC569550 and KC569551, respectively. Lysine decarboxylase, ornithine decarboxylase, arginine didydrolase, and L-arabinose are often used as the species-specific minimal biochemical tests to identify V. fluvialis from V. cholerae and nonagglutinating (NAG) vibrios [4]. The appearance of arginine didydrolase-negative V. fluvialis increased the complexity of the identification through biochemical tests.
Identification of virulence genes and phenotypes
Several virulence factors important in pathogenesis have been reported in V. fluvialis[15]–[22]. Though the precise role in producing the clinical manifestations remains unclear, these factors may increase the pathogenicity of V. fluvialis and contribute to diarrhea. We screened the presence of the virulence genes vfh, hupO, vfpA and stn by PCR. All strains were positive for genes vfh, hupO and vfpA, negative for gene stn encoding the toxin NAG-ST enterotoxin [37].
Azocasein assay was used to determine the product of VFP protease (Figure 1). 17 (39.5%) strains had medium to high expression of VFP with the asocasein unit values ranging from 10.95 to 26.87, the others showed lower VFP productions with the asocasein unit values below 10. Among the above 17 strains, 13 were isolated from stool samples. In contrast to the prevalence of the vfp genes in the all tested strains, the corresponding phenotypes were not detected in an equal rate, suggesting the defective expression of VFP in some strains and needs further study. VFP is 70% homologous to precursor proteins of metalloproteases from other human pathogenic vibrios such as V. cholerae[45] and V. vulnificus[46]. It shows haemagglutinating, permeability-enhancing and haemorrhagic activities in addition to proteolytic activity like V. vulnificus protease [47]. Metalloprotease of V. cholerae, also called haemagglutinin/protease (Hap), plays an important role in cholera pathogenesis by proteolytically activating cholera toxin A subunit [48] and the El Tor cytolysin/haemolysin [49], hydrolysing physiologically important proteins [50] and promoting mucin gel penetration, detachment and spread of infection along the gastrointestinal tract [51]. Based on the similar biological activities and high homology to metalloproteases of V. cholerae and V. vulnificus, VFP may also function as an important pathogenic factor in V. fluvialis. Strains with higher expression of VFP could be potentially more virulent in the pathogenesis than those with lower expression or no expression, which was consistent with our observation that clinical isolations were predominant in those with medium to high VFP production.
Figure 1.
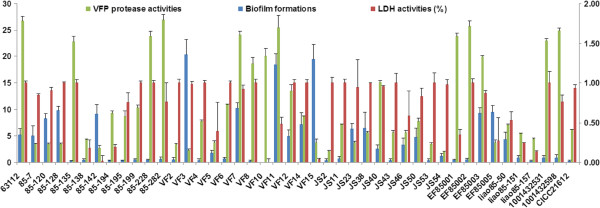
The virulence phenotypes of V. fluvialis including VFP protease production, cytotoxic activity and biofilm formation.
All strains were agglutination-negative to both the chicken and human erythrocytes. On Columbia blood agar, the colonies of all strains were medium to large, mucoid, gray and β-hemolytic, indicating that VFH was routinely produced. Except JS 23 and liao85-157, all strains showed obvious hemolytic zone after 24 h incubation. Longer incubation (more than 72 h) was needed to develop clear hemolytic phenotype for JS23 and liao85-157. The role and biological properties of hemolysin from V. fluvialis have been independently studied by two groups [18,20]. In addition to lysing erythrocytes from different animal species, this enterotoxigenic El Tor-like hemolysin was cytotoxic towards CHO cells and induced fluid accumulation in mouse [18]. Pores formed by VFH in erythrocyte membrane seem larger than those formed by other Vibrio hemolysins such as V. cholera, V. parahaemolyticus and V.vulnificus[20], which may be somehow related with the bloody diarrhea [2,5,26] occurred in some patients.
Different with V. cholerae and V. parahaemolyticus, which isolates from clinical sources are predominately toxigenic by containing virulence genes such as ctxAB or tdh and trh, no correlation between the prevalence of virulence genes and isolation source was observed in V. fluvialis. Virulence genes vfh, hupO and vfpA were equally detected in patient isolates and seafood isolates. All test V. fluvialis were haemolysis-positive irrespective of the isolation sources. Our results indicate the higher risk and potential public health threaten of seafood contaminated by V. fluvialis.
Cytotoxic activity
The supernatants from all the V. fluvialis strains were capable of causing cytotoxic to Hep-2 cell. Cell morphological changes including cell rounding and destruction of the monolayer were readily observed after 24 h treatment. The end-point titres showing cytotoxic effects on 50% Hep-2 cells in a well mainly ranged from 2 to 8, only four (VF7, VF2, Ma-2598 and Ma-2531) showed high titres of 16, 32, 16 and 64, respectively. In contrast to the report of Chakraborty et al.[21], vacuolating effect was not clearly observed, maybe due to the variation of strains and the difference of tissue cell line used.
Cytotoxocity was also detected by measuring lactate dehydrogenase (LDH) activities of Hep-2 cells after 8 h treatment with V. fluvialis. Cytotoxocity was expressed as LDH released into the medium as a percentage of total cellular LDH, obtained by treatment with 2% Triton X-100. LDH activities varied with strains (Figure 1). 33 strains (75%) displayed media to high cytotoxocity with LDH value between 53.12% and 100%, the rest 11 strains (25%) showed low cytotoxocity with LDH value below 48.5%. The average LDH activity evoked by 10 seafood-isolated strains was 68%, which was no significantly different from average value of 76% caused by 34 clinical isolates. It was found that for the above four strains (VF7, VF2, Ma-2598 and Ma-2531) which had the highest end-point titres of supernatants showing cytotoxic effects, LDH values were also high; for the strains with LDH below 50%, the end-point titres of supernatants showing cytotoxic effects were all between 2 and 4.
Biofilm formation
V. fluvialis was found to be the most predominant species among Vibrio isolated from both the suburban and urban community effluents in South Africa [52], suggesting its high capacity of survival and persistence in the environment. Amel et al. reported a long-term survival (6 years) of V. fluvialis in marine sediment [53]. Therefore the ability to form biofilm in V. fluvialis was tested. The biofilm formation varied greatly with different strains (Figure 1), 20 (45.5%) strains could form biofilm in vitro, among which three strains VF3, VF11 and VF 15 isolated from stool samples made very thick biofilms. Microbes in biofilm communities are more resistant to environmental stresses and protozoan predation. Strains with the capacity to form higher biofilms could survive better in the infecting host and estuarine system than those forming less biofilms and thus contribute to the pathogenesis.
Antibiotic susceptibility test
All V. fluvialis strains were found to be sensitive to ceftazidime, chloramphenicol, ciprofloxacin, gentamicin, nalidixic acid, trimethoprim and co-trimoxazole (Table 2). Except JS2, all strains were sensitive to tetracycline. And all strains were sensitive to streptomycin except Ma-2531. JS43 is the only strain showed resistant to the third generation of cephalosporins cefotaxime and ceftriaxone with MIC of 4 μg/ml. Additionally, there were six strains showed intermediate to cefotaxime and ceftriaxone with MIC of 2 μg/ml. 45.5%, 38.6%, 25.0% and 38.7% resistance (including intermediate resistance) to ampicillin, azithromycin, sulfamethoxazole and amoxicillin/clavulanic acid were observed with the corresponding MIC ranged from 16 to 64 μg/ml, 4 to 16 μg/ml, more than 1024 μg/ml, and 16/8 to 64/32 μg/ml, respectively. 12 (27.3%) strains were resistant to two and three antibiotics, if strains showing intermediate were included, the rate was up to 34.0%. Strain Ma-2531 isolated from clinical sample in 2010 was resistant to five antibiotics. Antibiotic susceptibility comparison of strains from Fujian revealed that patient isolates showed higher resistant rate (91.7%) and broader resistant spectrum than the seafood-isolates (30%), which suggests patient isolates may bring a more severe medical and public health concern.
Table 2.
Antibiotic susceptibility patterns ofV. fluvialisstrains
|
Antibiotic |
Breakpoints (mg/ml) |
Resistance |
Sensitivity |
|
|---|---|---|---|---|
| R (%) | I (%) | S (%) | ||
| Ampicillin |
S<=8 I=16 R>=32a |
12 (27.3) |
8 (18.2) |
24 (54.5) |
| Amoxicillin/Clavulanic acid |
S<=8/4 I=16/8 R>=32/16b |
10 (22.7) |
7 (15.9) |
27 (61.4) |
| Cefotaxime |
S<=1 I=2 R>=4b |
1 (2.3) |
6 (13.6) |
37 (84.1) |
| Ceftriaxone |
S<=1 I=2 R>=4b |
1 (2.3) |
6 (13.6) |
37 (84.1) |
| Ceftazidime |
S<=4 I=8 R>=16b |
0 (0) |
0 (0) |
44 (100) |
| Chloramphenicol |
S<=8 I=16 R>=32a |
0 (0) |
0 (0) |
44 (100) |
| Ciprofloxacin |
S<=1 I=2 R>=4b |
0 (0) |
0 (0) |
44 (100) |
| Gentamicin |
S<=4 I=8 R>=16b |
0 (0) |
0 (0) |
44 (100) |
| Nalidixic acid |
S<=16 R>=32b |
0 (0) |
- |
44 (100) |
| Streptomycin |
S<16 R>=16d |
1 (2.3) |
- |
43 (97.7) |
| Sulfamethoxazole |
S<=256 R>=512a |
11 (25.0) |
- |
33 (75.0) |
| Trimethoprim |
S<=8 R>=16b |
0 (0) |
- |
44 (100) |
| Co-trimoxazole |
S<=2/38 R>=4/76a |
0 (0) |
- |
44 (100) |
| Tetracycline |
S<=4 I=8 R>=16a |
1 (2.3) |
0 (0) |
43 (97.7) |
| Azithromycin | S<=2 I=4 R>=8c | 12 (27.3) | 5 (11.4) | 27 (61.3) |
aBreakpoints are based on the CLSI standards for V. cholerae.
bBreakpoints refer to the CLSI criteria for Enterobacteriaceae.
cBreakpoint is based on the CLSI standards for C. jejuni.
dBreakpoint refer to the reference [61].
R, resistance; I, intermediate; S, sensitivity.
Compared to the high sensitivity (99.4%) of V. cholerae to the ampicillin during the similar time period (1977–1989) [55], V. fluvialis exhibited much higher resistant to β- lactams. In regard to sulfamethoxazole resistance, sulII gene was detected in 85–142, JS2 and Ma-2531 strains. Resistance to the azithromycin (38.7%, including intermediate resistance) is the unique feature of the V. fluvialis tested in this study, strain 63112 isolated in 1963 from a diarrheal patient also showed azithromycin resistance with MIC of 8 μg/ml. Wang et al. reported that the earliest azithromycin-resistant V. cholerae strains appeared in 1965 in China [55]. Considering the isolation time of the strains and the time-to-market of the second generation macrolides antibiotics, we reasoned that azithromycin-resistant phenotype of V. fluvialis in this study was due to the cross-resistance caused by the first generation macrolides antibiotics which had been widely used in the clinical since 1952.
In general, the antibiotic resistant conditions of our V. fluvialis strains were not as serious as those reported in the literatures where the SXT element, plasmid and integrons mediated MDRs were quite common in V. fluvialis[3,25,56]–[58]. And V. fluvialis was reported to be the most abundant strain harboring most of the antibiotic resistance genes and SXT element among the Vibrio strains isolated from wasterwater final effluents [59]. The probable reason was the earlier isolation time. The majority of the tested strains were isolated in 1980s. It was reported that integrons appeared in V. fluvialis after 1998 [25] and SXT was detected for the first time in O139 V. cholerae isolated in 1992 [60]. Consistent to the phenotypes of lacking of SXT specific MDR pattern and sensitivity to the aminoglycosides, PCR screening of the SXT integrase and 3’ conserved sequence (3’CS) and 5’CS of class I integron gave no amplicons (data not shown). From the other side, our results further suggest the rapid increasing and spreading of antibiotic resistance in the V. fluvialis in recent 20 years, the appearance of MDR strains will be a severe medical and public health problem due to its epidemic-causing potential. It's worth noting that int IV gene specific for the class IV integron [34] was negative in all strains, thus suggesting there are maybe no superintegron in the V. fluvialis or sequence variation occurred in the int IV gene.
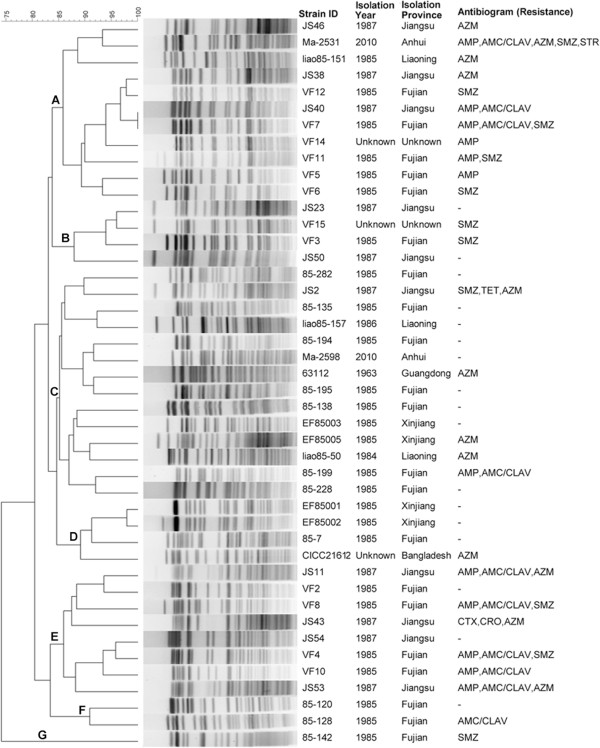
PFGE
PFGE was used to analyze the genetic relatedness, molecular-subtyping characteristics of V. fluvialis strains, especially to determine whether some strains from the same location during the same year were clonal. PFGE of the NotI-digested DNA of 44 strains generated 43 distinct patterns, only two (JS40 and VF7) possessed the same pattern (Figure 2). All strains formed 7 clusters at the 85% similarity breakpoint. The top two clusters contained 14 and 11 strains respectively. The smallest cluster only contained one strain. Five out of 11 strains in cluster A were patient isolates from Fujian in 1985, and 7 out of 14 strains in cluster C were seafood isolates from Fujian. Cluster C exhibited less antibiotic resistance than Cluster A, the most strains in cluster E displayed multiple antibiotic resistance. According to the interpretive criteria proposed by Tenover [61], strain EF85001 and EF85002 were considered to be closely related by showing one band difference which were isolated from same location at same time, strain 85–199 and 85–228, VF5 and VF6, 85–120 and 85–128 were possibly related.
Figure 2.
The results of the PFGE analysis using No tI digestion of V. fluvialis strains and the MDR patterns. The dendrogram was produced using the Dice coefficient and the unweighted-pair group method with an arithmetic mean algorithm (UPGMA) with a position tolerance of 1.5%. Abbreviation: AMP, ampicillin; AMC/CLAV, amoxicillin/clavulanic acid; CTX, cefotaxime; CRO, ceftriaxone; STR, streptomycin; SMZ, sulfamethoxazole; TET, tetracycline; AZM, azithromycin.
Conclusions
In this study, we examined the main biological characteristics, virulence phenotypes and their correlation with genetic factors, drug resistance profiles of V. fluvialis isolated from patients and environment in China. One strain was found to be negative in arginine dihydrolase system. There was no significant correlation between the prevalence of virulence phenotypes and isolation source. Virulence genes vfh, hupO and vfpA were widely distributed, the ability to produce hemolysin, cytotxin and protease varied with strains. Resistance to β-lactams and Sulfamethoxazole were prevalence. Azithromycin resistance is a unique feature of the V. fluvialis tested in this study. PFGE-based comparative molecular analysis of isolates demonstrated great genetic heterogeneity of V. fluvialis in China. To our knowledge, this is the first study that specifically evaluated etiological characteristics and molecular relatedness of V. fluvialis isolated in China. The obtained information contributed to the understanding of pathogenicity and the epidemiological features of V. fluvialis and it’s necessary to enhance surveillance in the future due to the increasing appearance of MDR strains and its epidemic-causing potential.
Abbreviations
PCR: Polymerase chain reaction; TCBS: Thiosulphate citrate bile salts sucrose agar; VFP: V. fuvialis protease; CLSI: Clinical and Laboratory Standards Institute; MICs: Minimum inhibitory concentrations; LDH: Lactate dehydrogenase; PFGE: Pulsed-field gel electrophoresis; ONPG: O-nitrophenyl-β-D-galactopyranoside; VFH: V. fluvialis hemolysin.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
PL carried out the molecular genetic studies and phenotypes test, participated in the sequence submission and drafted the manuscript. XC carried out the tissue culture. XD participated in the PFGE analysis. BK participated in the design of the study and helped to draft the manuscript. WL conceived of the study, and participated in its design and coordination and drafted the manuscript. All authors read and approved the final manuscript.
Authors' information
State Key Laboratory for Infectious Disease Prevention and Control, and National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention. Beijing 102206, People’s Republic of China.
Supplementary Material
Information and biological features of V. fluvialis strains used in this study.
Contributor Information
Pu Liang, Email: liangpu8802@163.com.
Xiaoying Cui, Email: cuixiaoying@icdc.cn.
Xiaoli Du, Email: duxiaoli@icdc.cn.
Biao Kan, Email: kanbiao@icdc.cn.
Weili Liang, Email: liangweili@icdc.cn.
Acknowledgements
This work was partially supported by the Project of the National Natural Science Foundation of China (81071410). We are very grateful to Dr. Jingyun Zhang for the help in preparing the figures and formatting the bibliography.
References
- Thekdi R, Lakhani AG, Vachha SM, Chandrakapure MR. Vibrio fluvialis (group F vibrio) in Maharashtra. Indian J Med Res. 1982;76:80–85. [PubMed] [Google Scholar]
- Huq MI, Alam AK, Brenner DJ, Morris GK. Isolation of Vibrio-like group, EF-6, from patients with diarrhea. J Clin Microbiol. 1980;11(6):621–624. doi: 10.1128/jcm.11.6.621-624.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury G, Pazhani GP, Nair GB, Ghosh A, Ramamurthy T. Transferable plasmid-mediated quinolone resistance in association with extended-spectrum beta-lactamases and fluoroquinolone-acetylating aminoglycoside-6'-N-acetyltransferase in clinical isolates of Vibrio fluvialis. Int J Antimicrob Agents. 2011;38(2):169–173. doi: 10.1016/j.ijantimicag.2011.04.013. [DOI] [PubMed] [Google Scholar]
- Chowdhury G, Pazhani GP, Dutta D, Guin S, Dutta S, Ghosh S, Izumiya H, Asakura M, Yamasaki S, Takeda Y, Arakawa E, Watanabe H, Mukhopadhyay AK, Bhattacharya MK, Rajendran K, Nair GB, Ramamurthy T. Vibrio fluvialis in Patients with Diarrhea, Kolkata, India. Emerg Infect Dis. 2012;18(11):1868–1871. doi: 10.3201/eid1811.120520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S, Bhattacharjee S, Bal B, Pal R, Niyogi SK, Sarkar K. Is Vibrio fluvialis emerging as a pathogen with epidemic potential in coastal region of eastern India following cyclone Aila? J Health Popul Nutr. 2010;28(4):311–317. doi: 10.3329/jhpn.v28i4.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allton DR, Forgione MA Jr, Gros SP. Cholera-like presentation in Vibrio fluvialis enteritis. South Med J. 2006;99(7):765–767. doi: 10.1097/01.smj.0000223657.22296.e6. [DOI] [PubMed] [Google Scholar]
- Klontz KC, Desenclos JC. Clinical and epidemiological features of sporadic infections with Vibrio fluvialis in Florida, USA. J Diarrhoeal Dis Res. 1990;8(1–2):24–26. [PubMed] [Google Scholar]
- Tall BD, Fall S, Pereira MR, Ramos-Valle M, Curtis SK, Kothary MH, Chu DM, Monday SR, Kornegay L, Donkar T, Prince D, Thunberg RL, Shangraw KA, Hanes DE, Khambaty FM, Lampel KA, Bier JW, Bayer RC. Characterization of Vibrio fluvialis-like strains implicated in limp lobster disease. Appl Environ Microbiol. 2003;69(12):7435–7446. doi: 10.1128/AEM.69.12.7435-7446.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KC, Hsu RW. Vibrio fluvialis hemorrhagic cellulitis and cerebritis. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2005;40(9):e75–77. doi: 10.1086/429328. [DOI] [PubMed] [Google Scholar]
- Ratnaraja N, Blackmore T, Byrne J, Shi S. Vibrio fluvialis peritonitis in a patient receiving continuous ambulatory peritoneal dialysis. J Clin Microbiol. 2005;43(1):514–515. doi: 10.1128/JCM.43.1.514-515.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera Rodriguez LE, Monroy SP, Morier L, Ramirez Alvarez MM, Fernandez Abreu A, Castro Escarpulli G, Longa Briceno A, Bravo Farinas L. Severe otitis due to Vibrio fluvialis in a patient with AIDs: first report in the world. Rev Cubana Med Trop. 2005;57(2):154–155. [PubMed] [Google Scholar]
- Liu WL, Chiu YH, Chao CM, Hou CC, Lai CC. Biliary tract infection caused by Vibrio fluvialis in an immunocompromised patient. Infection. 2011;39(5):495–496. doi: 10.1007/s15010-011-0146-0. [DOI] [PubMed] [Google Scholar]
- Lai CH, Hwang CK, Chin C, Lin HH, Wong WW, Liu CY. Severe watery diarrhoea and bacteraemia caused by Vibrio fluvialis. J Infect. 2006;52(3):e95–98. doi: 10.1016/j.jinf.2005.05.023. [DOI] [PubMed] [Google Scholar]
- Penland RL, Boniuk M, Wilhelmus KR. Vibrio ocular infections on the U.S. Gulf Coast. Cornea. 2000;19(1):26–29. doi: 10.1097/00003226-200001000-00006. [DOI] [PubMed] [Google Scholar]
- Chikahira M, Hamada K. Enterotoxigenic substance and other toxins produced by Vibrio fluvialis and Vibrio furnissii. Nihon juigaku zasshi. 1988;50(4):865–873. doi: 10.1292/jvms1939.50.865. [DOI] [PubMed] [Google Scholar]
- Wong HC, Ting SH, Shieh WR. Incidence of toxigenic vibrios in foods available in Taiwan. J Appl Bacteriol. 1992;73(3):197–202. doi: 10.1111/j.1365-2672.1992.tb02978.x. [DOI] [PubMed] [Google Scholar]
- Rahim Z, Aziz KM. Factors affecting production of haemolysin by strains of Vibrio fluvialis. J Diarrhoeal Dis Res. 1996;14(2):113–116. [PubMed] [Google Scholar]
- Kothary MH, Lowman H, McCardell BA, Tall BD. Purification and characterization of enterotoxigenic El Tor-like hemolysin produced by Vibrio fluvialis. Infect Immun. 2003;71(6):3213–3220. doi: 10.1128/IAI.71.6.3213-3220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockwood DE, Kreger AS, Richardson SH. Detection of toxins produced by vibrio fluvialis. Infect Immun. 1982;35(2):702–708. doi: 10.1128/iai.35.2.702-708.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JH, Lee JH, Choi YH, Park JH, Choi TJ, Kong IS. Purification, characterization and molecular cloning of Vibrio fluvialis hemolysin. Biochim Biophys Acta. 2002;1599(1–2):106–114. doi: 10.1016/s1570-9639(02)00407-7. [DOI] [PubMed] [Google Scholar]
- Chakraborty R, Chakraborty S, De K, Sinha S, Mukhopadhyay AK, Khanam J, Ramamurthy T, Takeda Y, Bhattacharya SK, Nair GB. Cytotoxic and cell vacuolating activity of Vibrio fluvialis isolated from paediatric patients with diarrhoea. J Med Microbiol. 2005;54(Pt 8):707–716. doi: 10.1099/jmm.0.45820-0. [DOI] [PubMed] [Google Scholar]
- Ahn SH, Han JH, Lee JH, Park KJ, Kong IS. Identification of an iron-regulated hemin-binding outer membrane protein, HupO, in Vibrio fluvialis: effects on hemolytic activity and the oxidative stress response. Infect Immun. 2005;73(2):722–729. doi: 10.1128/IAI.73.2.722-729.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine WC, Griffin PM. Vibrio infections on the Gulf Coast: results of first year of regional surveillance. Gulf Coast Vibrio Working Group. The Journal of infectious diseases. 1993;167(2):479–483. doi: 10.1093/infdis/167.2.479. [DOI] [PubMed] [Google Scholar]
- Evans MC, Griffin PM, Tauxe RV. CDC report: Vibrio surveillance system, summary data, 1997-2000. Available from: URL: http://www.cdc.gov/ncidod/dbmd/diseaseinfo/files/CSTE_Vibrio_2000.pdf.
- Srinivasan VB, Virk RK, Kaundal A, Chakraborty R, Datta B, Ramamurthy T, Mukhopadhyay AK, Ghosh A. Mechanism of drug resistance in clonally related clinical isolates of Vibrio fluvialis isolated in Kolkata, India. Antimicrob Agents Chemother. 2006;50(7):2428–2432. doi: 10.1128/AAC.01561-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidler RJ, Allen DA, Colwell RR, Joseph SW, Daily OP. Biochemical characteristics and virulence of environmental group F bacteria isolated in the United States. Appl Environ Microbiol. 1980;40(4):715–720. doi: 10.1128/aem.40.4.715-720.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellet J, Klein B, Altieri M, Ochsenschlager D. Vibrio fluvialis, an unusual pediatric enteric pathogen. Pediatr Emerg Care. 1989;5(1):27–28. doi: 10.1097/00006565-198903000-00008. [DOI] [PubMed] [Google Scholar]
- Wu B, Ma J, Lin W. Ioslation and identif ication of causative vibrio in sea products. Journal of Da lian Institute of Light Industry. 2003;22:47–49. [Google Scholar]
- Singh R, Rajpara N, Tak J, Patel A, Mohanty P, Vinothkumar K, Chowdhury G, Ramamurthy T, Ghosh A, Bhardwaj AK. Clinical isolates of Vibrio fluvialis from Kolkata, India, obtained during 2006: plasmids, the qnr gene and a mutation in gyrase A as mechanisms of multidrug resistance. J Med Microbiol. 2012;61(3):369–374. doi: 10.1099/jmm.0.037226-0. [DOI] [PubMed] [Google Scholar]
- Igbinosa EO, Okoh AI. Vibrio fluvialis: an unusual enteric pathogen of increasing public health concern. Int J Environ Res Public Health. 2010;7(10):3628–3643. doi: 10.3390/ijerph7103628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner DJ, Hickman-Brenner FW, Lee JV, Steigerwalt AG, Fanning GR, Hollis DG, Farmer JJ 3rd, Weaver RE, Joseph SW, Seidler RJ. Vibrio furnissii (formerly aerogenic biogroup of Vibrio fluvialis), a new species isolated from human feces and the environment. J Clin Microbiol. 1983;18(4):816–824. doi: 10.1128/jcm.18.4.816-824.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty R, Sinha S, Mukhopadhyay AK, Asakura M, Yamasaki S, Bhattacharya SK, Nair GB, Ramamurthy T. Species-specific identification of Vibrio fluvialis by PCR targeted to the conserved transcriptional activation and variable membrane tether regions of the toxR gene. J Med Microbiol. 2006;55(Pt 6):805–808. doi: 10.1099/jmm.0.46395-0. [DOI] [PubMed] [Google Scholar]
- Lee SK, Wang HZ, Law SH, Wu RS, Kong RY. Analysis of the 16S-23S rDNA intergenic spacers (IGSs) of marine vibrios for species-specific signature DNA sequences. Mar Pollut Bull. 2002;44(5):412–420. doi: 10.1016/S0025-326X(01)00256-9. [DOI] [PubMed] [Google Scholar]
- Yu L, Zhou Y, Wang R, Lou J, Zhang L, Li J, Bi Z, Kan B. Multiple antibiotic resistance of Vibrio cholerae serogroup O139 in China from 1993 to 2009. PLoS One. 2012;7(6):e38633. doi: 10.1371/journal.pone.0038633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochhut B, Lotfi Y, Mazel D, Faruque SM, Woodgate R, Waldor MK. Molecular analysis of antibiotic resistance gene clusters in vibrio cholerae O139 and O1 SXT constins. Antimicrob Agents Chemother. 2001;45(11):2991–3000. doi: 10.1128/AAC.45.11.2991-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux TM, Lee R, Love J. Complete genome sequence of a free-living Vibrio furnissii sp. nov. strain (NCTC 11218) J Bacteriol. 2011;193(6):1487–1488. doi: 10.1128/JB.01512-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidinost C, Saka HA, Aliendro O, Sola C, Panzetta-Duttari G, Carranza P, Echenique J, Patrito E, Bocco JL. Virulence factors of non-O1 non-O139 Vibrio cholerae isolated in Cordoba, Argentina. Rev Argent Microbiol. 2004;36(4):158–163. [PubMed] [Google Scholar]
- Ramamurthy T, Pal A, Pal SC, Nair GB. Taxonomical implications of the emergence of high frequency of occurrence of 2,4-diamino-6,7-diisopropylpteridine-resistant strains of Vibrio cholerae from clinical cases of cholera in Calcutta, India. J Clin Microbiol. 1992;30(3):742–743. doi: 10.1128/jcm.30.3.742-743.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez JA, Silva AJ, Finkelstein RA. Environmental signals controlling production of hemagglutinin/protease in Vibrio cholerae. Infect Immun. 2001;69(10):6549–6553. doi: 10.1128/IAI.69.10.6549-6553.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Mekalanos JJ. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev Cell. 2003;5(4):647–656. doi: 10.1016/S1534-5807(03)00295-8. [DOI] [PubMed] [Google Scholar]
- Cooper KL, Luey CK, Bird M, Terajima J, Nair GB, Kam KM, Arakawa E, Safa A, Cheung DT, Law CP, Watanabe H, Kubota K, Swaminathan B, Ribot EM. Development and validation of a PulseNet standardized pulsed-field gel electrophoresis protocol for subtyping of Vibrio cholerae. Foodborne Pathog Dis. 2006;3(1):51–58. doi: 10.1089/fpd.2006.3.51. [DOI] [PubMed] [Google Scholar]
- Kaysner CA, Jr, DePaola A. Bacteriological Analytical Manual online, 8th edition. Washington, DC. USA; 2001. Chapter 9. Vibrio. Available from: URL: http://www.fda.gov/Food/ScienceResearch/LaboratoryMethods/BacteriologicalAnalyticalManualBAM/ucm070830.htm. [Google Scholar]
- Chen KC, Culbertson NJ, Knapp JS, Kenny GE, Holmes KK. Rapid method for simultaneous detection of the arginine dihydrolase system and amino acid decarboxylases in microorganisms. J Clin Microbiol. 1982;16(5):909–919. doi: 10.1128/jcm.16.5.909-919.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulde M, Willenborg J, de Greeff A, Benga L, Smith HE, Valentin-Weigand P, Goethe R. ArgR is an essential local transcriptional regulator of the arcABC operon in Streptococcus suis and is crucial for biological fitness in an acidic environment. Microbiology. 2011;157(Pt 2):572–582. doi: 10.1099/mic.0.043067-0. [DOI] [PubMed] [Google Scholar]
- Hase CC, Finkelstein RA. Cloning and nucleotide sequence of the Vibrio cholerae hemagglutinin/protease (HA/protease) gene and construction of an HA/protease-negative strain. J Bacteriol. 1991;173(11):3311–3317. doi: 10.1128/jb.173.11.3311-3317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JC, Shao CP, Hor LI. Cloning and nucleotide sequencing of the protease gene of Vibrio vulnificus. Gene. 1996;183(1–2):255–257. doi: 10.1016/s0378-1119(96)00488-x. [DOI] [PubMed] [Google Scholar]
- Miyoshi S, Sonoda Y, Wakiyama H, Rahman MM, Tomochika K, Shinoda S, Yamamoto S, Tobe K. An exocellular thermolysin-like metalloprotease produced by Vibrio fluvialis: purification, characterization, and gene cloning. Microb Pathog. 2002;33(3):127–134. doi: 10.1006/mpat.2002.0520. [DOI] [PubMed] [Google Scholar]
- Booth BA, Boesman-Finkelstein M, Finkelstein RA. Vibrio cholerae hemagglutinin/protease nicks cholera enterotoxin. Infect Immun. 1984;45(3):558–560. doi: 10.1128/iai.45.3.558-560.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamune K, Yamamoto K, Naka A, Matsuyama J, Miwatani T, Honda T. In vitro proteolytic processing and activation of the recombinant precursor of El Tor cytolysin/hemolysin (pro-HlyA) of Vibrio cholerae by soluble hemagglutinin/protease of V. cholerae, trypsin, and other proteases. Infect Immun. 1996;64(11):4655–4658. doi: 10.1128/iai.64.11.4655-4658.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RA, Boesman-Finkelstein M, Holt P. Vibrio cholerae hemagglutinin/lectin/protease hydrolyzes fibronectin and ovomucin: F.M. Burnet revisited. Proc Natl Acad Sci USA. 1983;80(4):1092–1095. doi: 10.1073/pnas.80.4.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AJ, Pham K, Benitez JA. Haemagglutinin/protease expression and mucin gel penetration in El Tor biotype Vibrio cholerae. Microbiology. 2003;149(Pt 7):1883–1891. doi: 10.1099/mic.0.26086-0. [DOI] [PubMed] [Google Scholar]
- Igbinosa EO, Obi CL, Okoh AI. Seasonal abundance and distribution of Vibrio species in the treated effluent of wastewater treatment facilities in suburban and urban communities of Eastern Cape Province, South Africa. J Microbiol. 2011;49(2):224–232. doi: 10.1007/s12275-011-0227-x. [DOI] [PubMed] [Google Scholar]
- Amel BK, Amine B, Amina B. Survival of Vibrio fluvialis in seawater under starvation conditions. Microbiol Res. 2008;163(3):323–328. doi: 10.1016/j.micres.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Baker-Austin C, McArthur JV, Tuckfield RC, Najarro M, Lindell AH, Gooch J, Stepanauskas R. Antibiotic resistance in the shellfish pathogen Vibrio parahaemolyticus isolated from the coastal water and sediment of Georgia and South Carolina, USA. J Food Prot. 2008;71(12):2552–2558. doi: 10.4315/0362-028x-71.12.2552. [DOI] [PubMed] [Google Scholar]
- Wang R, Lou J, Liu J, Zhang L, Li J, Kan B. Antibiotic resistance of Vibrio cholerae O1 El Tor strains from the seventh pandemic in China, 1961–2010. Int J Antimicrob Agents. 2012;40(4):361–364. doi: 10.1016/j.ijantimicag.2012.06.010. [DOI] [PubMed] [Google Scholar]
- Ahmed AM, Shinoda S, Shimamoto T. A variant type of Vibrio cholerae SXT element in a multidrug-resistant strain of Vibrio fluvialis. FEMS Microbiol Lett. 2005;242(2):241–247. doi: 10.1016/j.femsle.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Ahmed AM, Nakagawa T, Arakawa E, Ramamurthy T, Shinoda S, Shimamoto T. New aminoglycoside acetyltransferase gene, aac(3)-Id, in a class 1 integron from a multiresistant strain of Vibrio fluvialis isolated from an infant aged 6 months. J Antimicrob Chemother. 2004;53(6):947–951. doi: 10.1093/jac/dkh221. [DOI] [PubMed] [Google Scholar]
- Rajpara N, Patel A, Tiwari N, Bahuguna J, Antony A, Choudhury I, Ghosh A, Jain R, Ghosh A, Bhardwaj AK. Mechanism of drug resistance in a clinical isolate of Vibrio fluvialis: involvement of multiple plasmids and integrons. Int J Antimicrob Agents. 2009;34(3):220–225. doi: 10.1016/j.ijantimicag.2009.03.020. [DOI] [PubMed] [Google Scholar]
- Okoh A, Igbinosa E. Antibiotic susceptibility profiles of some Vibrio strains isolated from wastewater final effluents in a rural community of the Eastern Cape Province of South Africa. BMC Microbiol. 2010;10(1):143. doi: 10.1186/1471-2180-10-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldor MK, Tschape H, Mekalanos JJ. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J Bacteriol. 1996;178(14):4157–4165. doi: 10.1128/jb.178.14.4157-4165.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33(9):2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Information and biological features of V. fluvialis strains used in this study.