Abstract
Background
Medicago truncatula Gaertn. (barrel medic) is cultivated as a pasture legume for its high protein content and ability to improve soils through nitrogen fixation. Toxic concentrations of the micronutrient Boron (B) in agricultural soils hamper the production of cereal and leguminous crops. In cereals, the genetic analysis of B tolerance has led to the development of molecular selection tools to introgress and maintain the B tolerance trait in breeding lines. There is a comparable need for selection tools in legumes that grow on these toxic soils, often in rotation with cereals.
Results
Genetic variation for B tolerance in Medicago truncatula was utilised to generate two F2 populations from crosses between tolerant and intolerant parents. Phenotyping under B stress revealed a close correlation between B tolerance and biomass production and a segregation ratio explained by a single dominant locus. M. truncatula homologues of the Arabidopsis major intrinsic protein (MIP) gene AtNIP5;1 and the efflux-type transporter gene AtBOR1, both known for B transport, were identified and nearby molecular markers screened across F2 lines to verify linkage with the B-tolerant phenotype. Most (95%) of the phenotypic variation could be explained by the SSR markers h2_6e22a and h2_21b19a, which flank a cluster of five predicted MIP genes on chromosome 4. Three CAPS markers (MtBtol-1,-2,-3) were developed to dissect the region further. Expression analysis of the five predicted MIPs indicated that only MtNIP3 was expressed when leaf tissue and roots were assessed. MtNIP3 showed low and equal expression in the roots of tolerant and intolerant lines but a 4-fold higher expression level in the leaves of B-tolerant cultivars. The expression profile correlates closely with the B concentration measured in the leaves and roots of tolerant and intolerant plants. Whereas no significant difference in B concentration exists between roots of tolerant and intolerant plants, the B concentration in the leaves of tolerant plants is less than half that of intolerant plants, which further supports MtNIP3 as the best candidate for the tolerance trait-defining gene in Medicago truncatula.
Conclusion
The close linkage of the MtNIP3 locus to B toxicity tolerance provides a source of molecular selection tools to pasture breeding programs. The economical importance of the locus warrants further investigation of the individual members of the MIP gene cluster in other pasture and in grain legumes.
Keywords: Medicago truncatula, Abiotic stress, Legume, Boron toxicity tolerance, Genetic analysis, MtNIP3, Gene cluster, Differential expression
Background
Medicago truncatula Gaertn. (barrel medic; Fabaceae), native to the Mediterranean region, is a self-regenerating annual pasture species grown on over 4.5 million hectares in the dryland cereal/livestock zones in southern Australia [1]. M. truncatula is adapted to a wide range of soils from sandy loams to clays and prefers neutral to alkaline soils (pH (water) > 6.5). When grown in rotation with cereal crops their ability to fix atmospheric nitrogen contributes significantly to the nitrogen balance of following crops. This lessens the need for inorganic nitrogen fertilisers and hence increases economic returns to producers and benefits the environment [2,3]. Additionally as a non-host of many important cereal diseases, they are able to provide a valuable disease-break from pathogens such as Take-all (Gaeumannomyces graminis), crown rot (Fusarium graminearum) and cereal cyst nematode (Heterodera avenae) [4]. Together with Lotus japonicus, M. truncatula is also a model organism for legume biology with an international consortium providing researchers with numerous genetic resources including two drafts of the sequenced genome and genetic maps (http://www.medicagohapmap.org). Its relatively small genome size (ca. 500 Mbp) and high levels of synteny to major crop legumes such as pea (Pisum sativum) and alfalfa (M. sativa) [5] make it an ideal system to conduct molecular genetics analyses on biotic and abiotic stresses affecting legumes.
A major problem facing M. truncatula and other legumes in arid world regions such as southern Australia, northern Africa, and the Mediterranean is the presence of phytotoxic levels of Boron (B) in the soil which can significantly impact on seed yield and quality [6]. Because remediation of B-toxic soils is impractical [7], investigators have looked to improve plant tolerance to B. In cereal crop species such as wheat and barley, genetic variation for B tolerance has led to the identification of quantitative trait loci (QTL) linked to B toxicity tolerance [8], and more recently to the genes underlying these QTL [9-11]. This has facilitated the development of closely linked molecular markers that can be used in breeding programs to improve the tolerance of wheat and barley grown in areas where B toxicity is a problem. In M. truncatula, genetic variation for B toxicity tolerance among different cultivars had earlier been reported [6] but so far has not been exploited to identify the gene(s) or genetic regions controlling this trait.
Recent studies in barley and Arabidopsis offer clues as to what genes may control B toxicity tolerance in M. truncatula. These studies have implicated the involvement of genes encoding one of two types of proteins involved in B uptake and translocation: (1) efflux-type B transporters or (2) members of the major intrinsic protein (MIP) family. The first efflux-type B transporter gene identified was the Arabidopsis AtBOR1 gene [12]. Although AtBOR1 conferred tolerance to plants under B deficient conditions, its homologue in barley, HvBot1 (identical to HvBOR2), enabled plants to tolerate high levels of B [9,11]. The over-expression of an AtBOR1 paralog, AtBOR4, in transgenic Arabidopsis plants also increased their tolerance to high B levels [13]. AtNIP5;1 was the first B transporter gene identified that encodes a member of the MIP family [14]. MIPs, also known as aquaporins, are channels for water and/or small non-charged molecules [15] and are comprised of four subfamilies; the nodulin 26-like intrinsic proteins (NIPs), the plasma membrane intrinsic proteins (PIPs), the tonoplast intrinsic proteins (TIPs), and the small basic intrinsic proteins (SIPs). AtNIP5;1 encodes a NIP subfamily member, and like AtBOR1, was shown to be associated with B deficiency tolerance, with AtNIP5;1 functioning as a boric acid channel required for the efficient uptake of B in roots [14]. In contrast, a recently discovered NIP gene in barley, HvNIP2;1, was found to be an important determinant of B toxicity tolerance [10], with the authors proposing that B toxicity tolerance is mediated by reduced expression of HvNIP2;1 to limit B uptake as well as by increased expression of HvBot1 to remove B from roots and sensitive tissues. A MIP subfamily member, AtTIP5;1 was also recently described conferring tolerance to B toxicity when over-expressed in transgenic Arabidopsis plants [16].
In this study, a candidate gene approach combined with population genetics and phenotypic analysis was used to determine the molecular basis of B tolerance in M. truncatula. We report the identification of a homologous AtNIP5;1 gene, MtNIP3, in M. truncatula associated with B toxicity tolerance. A gene-based marker derived from MtNIP3, named MtBtol-1, and further markers co-segregated with B toxicity tolerance in two F2 populations. In addition, gene expression of MtNIP3 was up-regulated in the leaves of respective B-tolerant parental lines. This information warrants further characterisation of MtNIP3 as a candidate gene for B tolerance and provides gene-based markers as molecular tools to introgress and maintain the tolerance trait in Medicago breeding programs.
Results
Segregation analysis
The inheritance results of the phenotypic data are presented in Table 1. The segregation ratio for the B toxicity tolerance trait has a significant fit (p = 0.05) to the expected 3:1 (tolerant vs. intolerant) ratio for a single dominant locus, which is true for both F2 populations. F2 plants of the population tap × Paraggio (intolerant x tolerant) had leaf symptom scores ranging from 0 to 4, whereas in tap × Caliph the scores ranged from 0 to 5. Relative to these symptom ranges in txP and txC, plants with a score of 0 and 1 in txP and a score of 0 to 2 in txC were considered as B tolerant.
Table 1.
Phenotypic segregation of the B toxicity tolerance trait in the F2 populations tap x Paraggio and tap x Caliph
| Population | No. of plants phenotyped | No. of B-tolerant F2 plants | No. of B-intolerant F2 plants | Segregation ratio | χ2 |
|---|---|---|---|---|---|
|
tap x Paraggio |
316 |
247 |
69 |
3 : 1 |
0.882 |
| tap x Caliph | 313 | 228 | 85 | 3 : 1 | 0.406 |
B accumulation in root and leaf tissue
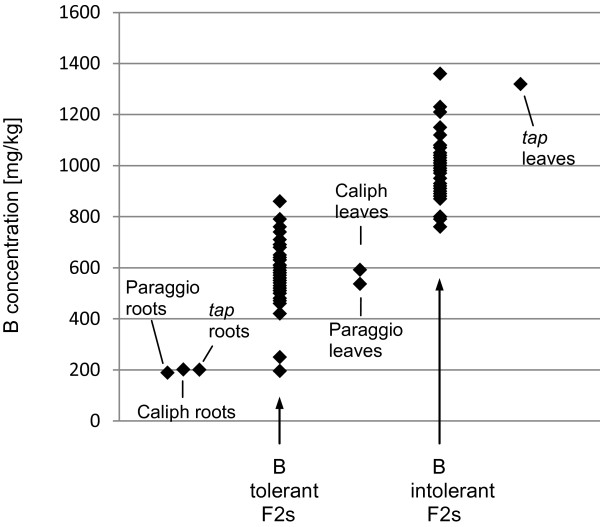
Initially, subsets of 39 and 36 F2 lines from the txP and txC populations, respectively, were selected to screen for potential molecular markers. The subsets consisted of lines that were phenotyped after three weeks of high B treatment and had clear tolerant (0 or 1) or intolerant (4 or 5) scores. ICP analysis was used to (a) quantify the amount of B accumulating in the leaf, and (b) verify the correlation between B levels and phenotypic scores. Additional ICP analysis was carried out on leaves and roots of the three parental lines to quantify the B distribution in tolerant and intolerant plants. The results are summarised in Figure 1. In the txP subset, the mean B concentration in leaves sampled from B-tolerant and -intolerant F2 lines was 559 mg/kg (range: 420–710) and 942 mg/kg (range: 760–1360), respectively. In the txC subset the mean B concentration was 605 mg/kg (range: 195–860) and 1021 mg/kg (range: 870–1210). The leaves of parental lines had mean B concentrations of 536 (Paraggio), 592 (Caliph), and 1319 (tap) mg/kg. In contrast to the mean B concentrations in the leaves, with tolerant lines accumulating less than half the amount of B compared to the intolerant lines, mean B concentrations in the roots did not differ significantly, 189 (Paraggio), 201 (Caliph) and 200 (tap) mg/kg.
Figure 1.
Distribution of B concentrations in tolerant and intolerant M.t. lines after 3 weeks of excess B. The amount of B was quantified in leaves of tolerant and intolerant F2 lines of both populations and the two tolerant parental lines Paraggio and Caliph and the intolerant parent tap. ICP data were generated on 35–40 F2 lines of the tolerant and intolerant subsets of both populations, 4–8 repeats of leaf and 4 repeats of root samples for each parent.
Biomass production and Boron tolerance
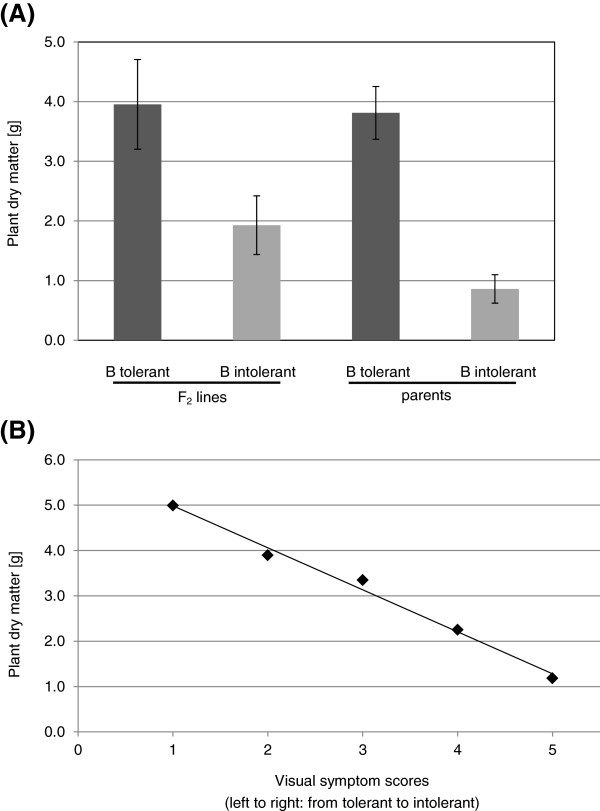
Breeding for B tolerance aims at maintaining yield under high B concentrations. As a proxy for the agronomic value of the B tolerance trait in pasture legumes, plant biomass or dry matter production (DM) was measured. The average DM production of the tolerant F2 plants and their tolerant parent, Caliph, was not significantly different, 4.0 g versus 3.8 g, respectively. In contrast, the average DM of the intolerant F2 lines was significantly higher than that of the intolerant parent tap, 1.9 g versus 0.9 g, respectively (Figure 2A). The DM and symptom scores were highly correlated with a coefficient of −0.995 (Figure 2B) indicating that the observed differences in biomass production are due to the B tolerance trait being present or absent in the individual F2 plants. With each increase in visual symptom score DM was reduced by approximately one gram.
Figure 2.
Mean plant dry matter (DM) comparison. (A) DM of tolerant and intolerant F2 lines is compared to tolerant and intolerant parental lines (Caliph, tap) of the corresponding population. Error bars represent standard deviations of the mean. (B) High negative correlation (−0.995) between DM and severity of the visual B stress symptom scores. n = 21, 42, 32, 30, 13 for plants with visual symptom score 1, 2, 3, 4, 5, respectively.
Development of molecular markers for B toxicity tolerance
The AtBOR1 and AtNIP5;1 cDNA sequences (GenBank: AB073713 and NM_117106) and their respective predicted protein sequences (GenBank: BAC20173 and NP_192776) were used to query the M. truncatula pseudomolecule database (version Mt3.5) available at the M. truncatula Hapmap project website (http://www.medicagohapmap.org). The best AtBOR1 homologue, identified on chromosome 7 (BAC AC157892; GeneID: Medtr7g110000.1) encodes a predicted protein, which shares 78% amino acid (aa) sequence identity with AtBOR1. We developed a gene based CAPS marker for this gene but found no linkage between this Boron transporter gene and B tolerance in either of the txP or txC F2 subsets.
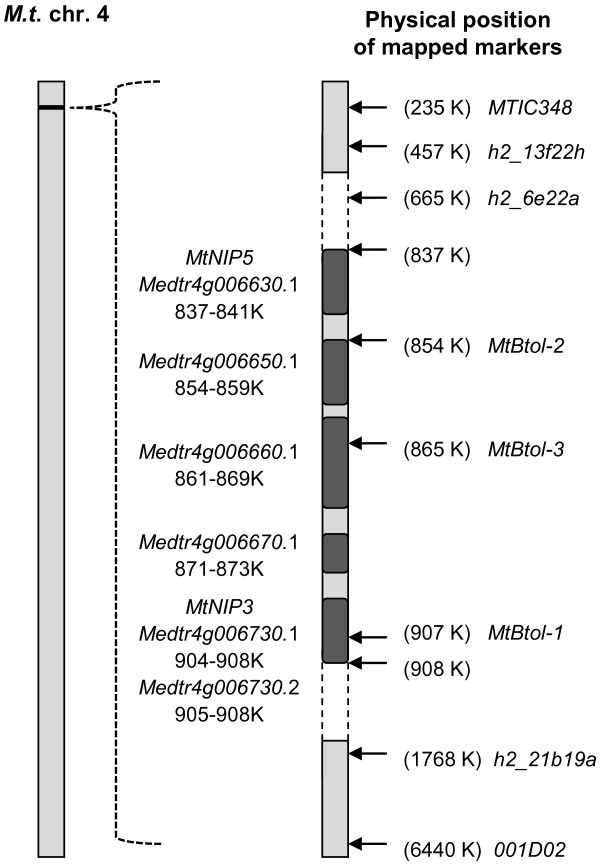
Four Medicago database matches to AtNIP5;1 were identified on chromosomes 1 (BACs AC181971 and AC225543) and 4 (BACs AC168305 and AC153003). Both chromosome 1 candidates (GeneID: Medtr1g097840.1 and Medtr1g098080.1) encode exactly the same predicted protein, which shares 69% sequence identity with AtNIP5;1, whereas the chromosome 4 candidates encode different predicted proteins with 60% (GeneID: Medtr4g006730.1) and 54% (GeneID: Medtr4g006630.1) identity to AtNIP5;1, respectively. The Medtr4g006730.1 protein product corresponds to the aquaporin MtNIP3 (GenBank: AY539749) identified [17] and its corresponding gene is referred to as MtNIP3. The two BACs on each chromosome were adjacent to one another and the physical distance between candidate genes was small – approximately 84 kb and 71 kb for chromosome 1 and 4 candidates, respectively. Two flanking SSR markers (002G02, h2_13c11a) located within 500 kb of the chromosome 1 candidate genes showed polymorphisms between tap and Paraggio but proved not to be linked to B tolerance in the subset of F2 lines tested. Thus, like the AtBOR1 homologue on chromosome 7, the chromosome 1 candidate genes were ruled out from further analysis. In contrast, SSR markers flanking the NIP candidate genes on chromosome 4 showed strong linkage with the B tolerance phenotype. The SSR markers MTIC348, h2_13f22h, h2_6e22a, h2_21b19a and 001D02, which flank a 6205 kb interval were polymorphic in either one or both F2 populations and explained 94% to 96% of the phenotypic variation (Table 2, Figure 3). Amongst these SSRs, the markers which were more distant from the 71 kb NIP gene cluster were less closely linked to the tolerance phenotype within the segregating populations (Table 2, Figure 3). Together with the segregation ratio characteristic of a single dominant locus, the strong linkage of these SSR makers to the tolerant phenotypes indicated that the trait-defining gene(s) reside at this 71 kb NIP gene locus.
Table 2.
Molecular markers and the phenotypic variation they explain in the two F2 populations studied
| Marker | Marker, Mt chromosome | Primer sequence (forward and reverse, 5’ – 3’) | Restriction enzyme |
Expected CAPS fragment size [bp] |
Phenotypic variation explained [%] |
||
|---|---|---|---|---|---|---|---|
| tap | Par/Cal | t x P* | t x C** | ||||
|
MTIC348 |
SSR, chr. 4 |
TGGAGGAGGGGTAGGATAGG ATGATGATGAGGCGGAGAAG |
- |
- |
- |
95 |
94 |
|
h2_13f22h |
SSR, chr. 4 |
GGCTTCCTGATGCTGGTTAG ACAAGCAGGTTGGACACACA |
- |
- |
- |
95 |
94 |
|
h2_6e22a |
SSR, chr. 4 |
CCATGGTGTCATTTGATCCT CCATGGTGTCATTTGATCCT |
- |
- |
- |
96 |
n.p. |
|
MtBtol-1 |
CAPS (gene), chr. 4 |
CTCTTTACCTCCCCCTCCTG TTGAACTACGATCAGGGTCGTTCCTA |
ApoI |
387 |
170/217 |
96 |
94 |
|
MtBtol-2 |
CAPS (gene), chr. 4 |
TTGAACTACGATCAGGGTCGTTCCTA TGCTCCAGCACATCCAATTA |
DraI |
102/131 |
102/256 |
96 |
94 |
| 256/280 |
411 |
||||||
|
MtBtol-3 |
CAPS (gene), chr. 4 |
TCCCACTGAAAATCGATTCAA GGCCAAAGCTCATTCCATT |
MspI |
125/450 |
575 |
96 |
94 |
|
h2_21b19a |
SSR, chr. 4 |
GGAACACCGGACACAATACC CACTTCTCAGGCACAGCAAA |
- |
- |
- |
96 |
n.p. |
|
001D02 |
SSR, chr. 4 |
TCCCAACGCTTTTTCATTTC GAACTTGAAGAAGAACGCCG |
- |
- |
- |
94 |
89 |
|
MtChr7 |
CAPS, chr. 7 |
TTGGAGCATTTATACCAGCAA TTGTATGCATCGGAGACTGC |
NlaIII |
306 |
84/222 |
n.a. |
n.a. |
|
002G04 |
SSR, chr. 1 |
AGGGTCGCCTCAACTATTA TCAACACCATTTTCTCAATG |
- |
- |
- |
n.a. |
n.a. |
|
002G02 |
SSR, chr. 1 |
TAACGTTACTCCCTCCTCCG TCTCCAACGAAGTTCAAGGG |
- |
- |
- |
n.a. |
n.a. |
| h2_13c11a | SSR, chr. 1 | TCACAACCTTCTTGCACCAG TCATAACTTGACCTGCACCA | - | - | - | n.a. | n.a. |
Most closely linked markers are in bold font. * n = 200; ** n = 36; n.a., not applicable; n.p., not polymorphic.
Figure 3.
Chromosome region linked to B tolerance. Scheme of the genome region on linkage group 4 in M. truncatula (version Mt3.5) showing the physical position of the B tolerance candidate genes and markers.
To dissect this locus, we derived CAPS markers from or closer to the NIP genes themselves. The M. truncatula Hapmap project gene annotations revealed that the candidate AtNIP5;1 homologue on BAC AC153003 has a potential splice variant, with both gene products (GeneID: Medtr4g006730.1 and Medtr4g006730.2) predicted to encode NIPs. Likewise, the second chromosome 4 candidate on BAC AC168305 (GeneID: Medtr4g006630.1) is also predicted to encode a NIP, with the 71 kb genomic region in between the two genes (Medtr4g006730.1 and Medtr4g006630.1) containing three more potential NIP genes (GeneIDs: Medtr4g006650.1/660.1/670.1). Therefore, according to the gene annotations, the locus as a whole contains up to five predicted NIP genes encoding six gene products potentially involved in controlling B toxicity tolerance. Three CAPS markers, named MtBtol-1, MtBtol-2 and MtBtol-3 (Table 2, Figure 3, Figure 4A-C) were developed that were polymorphic in both txP (200 F2 lines tested) and txC (36 F2 lines tested) populations (Figure 4D, E) within this region and explained 96% and 94% of the phenotypic variation, respectively. MtBtol-1 is specific to the first candidate AtNIP5;1 homologue, Medtr4g006730.1/730.2, MtBtol-2 is specific to the Medtr4g006650.1 gene which is situated 13 kb away from the second candidate AtNIP5;1 homologue, Medtr4g006630.1, and MtBtol-3 is specific to the Medtr4g006660.1 gene. It was not possible to design a gene specific marker for Medtr4g006630.1 due to its close similarity with the Medtr4g006730.1/730.2 gene.
Figure 4.
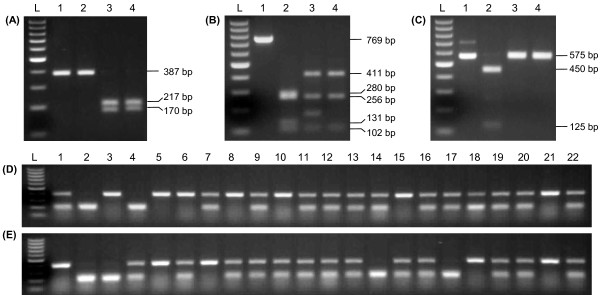
CAPS markers linked to B toxicity tolerance. (A) MtBtol-1; (B) MtBtol-2; (C) MtBtol-3; Lane L - DNA ladder, Lane 1 – undigested amplicon from tap, Lanes 2 to 4 – digested amplicons from tap, Caliph, and Paraggio, respectively. (D) and (E), Application of MtBtol-1 on genomic DNA from 22 randomly selected F2 plants derived from the tap x Paraggio and tap x Caliph crosses, respectively, distinguishing homozygous and heterozygous F2 plants.
NIP gene expression
We used Q-PCR to investigate the expression profiles of the NIP genes in roots and shoots of the three parental lines over a 48 h period following excess B treatment. Due to the high level of coding sequence similarity between the Medtr4g006630.1/730.1/730.2 gene products, it was only possible to design gene specific primers for Medtr4g006630.1 and the splice variant Medtr4g006730.2. Design of gene specific primers was also hindered by the absence of information related to the untranslated regions of these gene sequences. Hence, a third primer pair, specific to all three genes, was designed to generate a cumulative expression profile of B transporter candidates. Each of the primer pairs amplified the expected sized fragment when genomic DNA was used as template, however, only the primer pair specific for all three gene products amplified clear fragments of expected size from the cDNAs of the time course experiment. Two Medtr4g006630.1 specific primer pairs with different annealing positions in the gene amplified no product at all whereas the Medtr4g006730.2 specific primers yielded a small amount of expected product corresponding to <10 copies/μL in the leaf and root tissue examined, which is at the very limit of detection for Q-PCR. Therefore, by exclusion, the cumulative expression profile that was generated for all three genes was in fact presumed to represent one, Medtr4g006730.1.
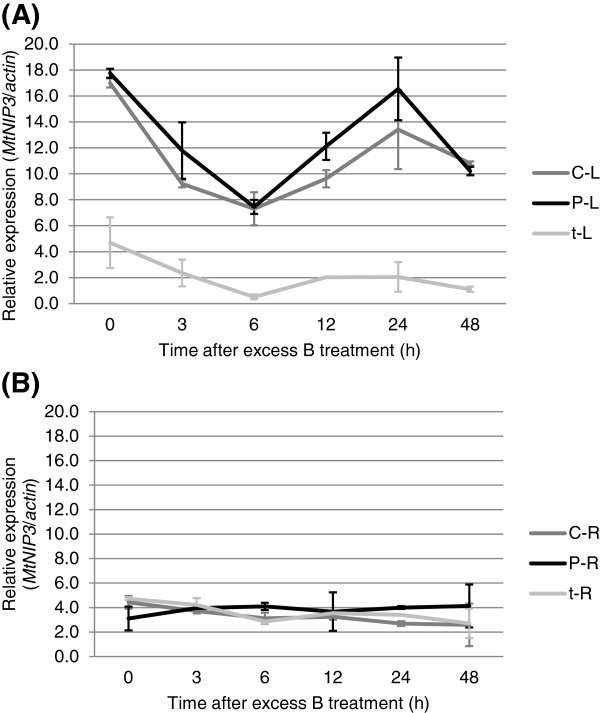
The expression profile for Medtr40006730.1 showed that the gene is constitutively expressed in the leaves, with approximately 4 times more transcripts present in both B-tolerant lines compared to the intolerant line (Figure 5A). In roots, the expression was equally low and non-differential in all three genotypes irrespective of their B tolerance phenotype. The expression level in the roots was comparable with the leaf expression of the intolerant line tap (Figure 5B). The stronger gene expression of Medtr40006730.1 in the leaves of the tolerant lines correlated with the B toxicity tolerance phenotype.
Figure 5.
Quantitative RT-PCR of MtNIP3. (A) MtNIP3 expression in Paraggio (P), Caliph (C), and tap (t) is regulated in the leaves (B) but not in roots following excess B treatment. Error bars represent standard error of the means, n = 2.
Q-PCR assays were also designed to investigate the possible involvement of the other three predicted MIP genes at this locus, Medtr4g006650.1/660.1/670.1. Again, the specificity of the primers was confirmed with the correct sized PCR fragment being amplified from DNA template. However, none of the primer pairs amplified any product on the cDNAs of the time course experiment from leaf and root tissue, suggesting no detectable expression before or after B treatment.
Analysis of MtNIP3 (Medtr4g006730.1) gene copy number
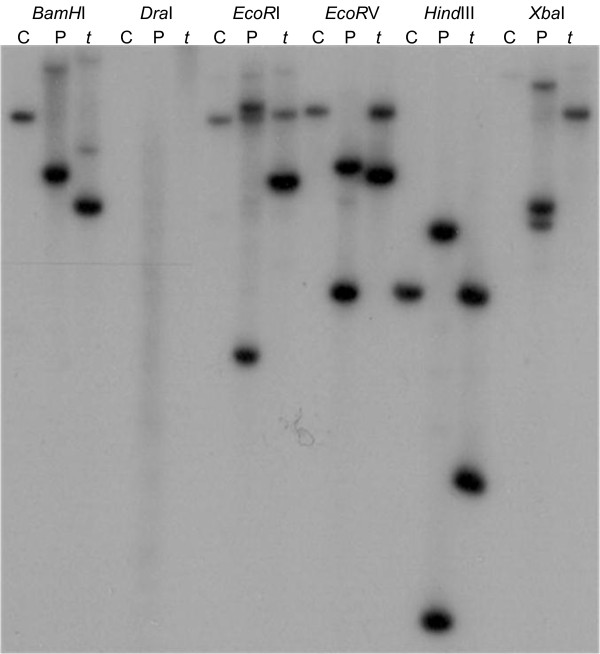
We used Southern hybridisation to determine if the presence of additional MtNIP3 gene copies in the two B-tolerant lines could account for the increased mRNA transcript levels observed for this gene in the leaves. As it was not possible to design a specific hybridisation probe for Medtr4g006730.1 (MtNIP3) due to the high level of homology it shares with Medtr4g006630.1 (MtNIP5), the designed probe would detect both genes. The resulting hybridisation revealed the presence of two gene copies in tap (as expected) and Paraggio, and one copy in Caliph (Figure 6). Thus, the observed increase in MtNIP3 transcript levels in the leaves of the two B-tolerant lines was not associated with a higher gene copy number.
Figure 6.
Southern hybridisation with an MtNIP3 (Medtr4g006730.1)-specific probe in Caliph (C), Paraggio (P) and tap (t).
MtNIP3 coding sequence and promoter region
The MtNIP3 coding region was sequenced from Caliph, Paraggio, and tap to identify potential differences in the predicted amino acid composition of the corresponding MIP protein. A 918 bp cDNA fragment was amplified from all three genotypes. None of the seven single nucleotide polymorphisms (SNPs) identified were specific to Caliph/Paraggio or tap (Additional file 1). The seven SNPs translated into three changes among the 305 amino acid residues of the NIP protein in the three genotypes. Again, the changes were not specific to the B-tolerant genotypes (Additional file 2).
A comparative Medtr4g006730.1 promoter analysis between Caliph, Paraggio, and tap was also performed to search for differences which could explain the variation in transcript levels observed between the B-tolerant and -intolerant lines. Sequencing of the 1.1 kb region directly upstream of the transcription start site revealed a 99% basepair identity between the three genotypes. The differences were not specific to the intolerant or the tolerant genotypes and further work would be required to address the impact of these single and dinucleotide differences on gene expression or search for potential differences further upstream.
Discussion
In this study, a candidate gene approach was used to identify the underlying genetic loci for tolerance to B toxicity in M. truncatula. Similar to the recently reported identification of the genetic basis for herbicide tolerance in M. truncatula[18], we utilised the molecular and physiological information on B homeostasis first reported in Arabidopsis [12,14]. Although the corresponding proteins of the Arabidopsis B transporter /channel genes AtBOR1 and AtNIP5;1 are associated with the efficient transport of B across the plasma membrane under B deficiency [12,14], similar proteins in barley were shown to aid the plant in conditions of B excess [9-11]. Therefore, it was hypothesised that homologous genes in M. truncatula of AtNIP5;1 and AtBOR1 could also play a role in supporting plants to tolerate high levels of B in the soil.
Genetic variation for B toxicity tolerance was identified in the M. truncatula lines Paraggio and Caliph (tolerant, [6]) and tap (intolerant). Paraggio and Caliph were each crossed with tap to generate two F2 populations segregating for B tolerance. The phenotypic segregation of the B tolerance trait was measured in over 300 individuals for each of the F2 populations and agreed with a single dominant locus that might host a single or several closely linked tolerance genes. A very close correlation between the visual symptom scores on individual plants and their biomass production was also evident suggesting B uptake as the direct and major cause of reduced plant growth and validating the symptom scores as a valid tool to estimate plant performance. All tolerant plants analysed showed a reduced (on average ca. 50%) level of B in their leaf tissue compared to intolerant lines implying an exclusion mechanism being the underlying B tolerance mechanism in this pasture legume. An exclusion mechanism was also proposed in barley where B-tolerant cultivars showed decreased B concentrations in their leaves compared to intolerant cultivars [11,19].
With the aid of the 2nd draft (version Mt3.5) of the sequenced M. truncatula genome, candidate AtNIP5;1 and AtBOR1 homologues were identified on chromosomes 1, 4, and 7. A subset of highly B-tolerant and -intolerant lines from the two segregating F2 populations was used initially for genotyping to establish linkage between the B-tolerant phenotype and SSR markers located close to the homologous Bor and NIP genes on all three chromosomes. Only SSR markers located near two candidate AtNIP5;1 homologues, separated by only 71 kb, on chromosome 4 showed linkages of approximately 95% to the tolerance trait in both F2 populations. The strong linkage was then upheld across 200 individual F2 lines from the tap x Paraggio cross which further supported that the corresponding genetic region was associated with B toxicity tolerance. We referred to one of these candidate genes as MtNIP3 and the predicted protein as MtNIP3, which was one of six aquaporins first identified in M. truncatula[17]. The other predicted gene was referred to as MtNIP5.
Although both corresponding proteins encoded by MtNIP3 and MtNIP5 confirmed good amino acid sequence similarity to AtNIP5;1, they both showed a greater level of amino acid sequence identity to AtNIP6;1 – another MIP family member and B channel identified in Arabidopsis [20]. MtNIP3 has 75% sequence identity to AtNIP6;1 whereas MtNIP5 shares only 57% identity. Within the 71 kb interval, between MtNIP3 and MtNIP5, there are up to three additional predicted MIP genes and a splice variant of MtNIP3. Fine mapping this locus with three CAPS markers derived from the actual MIP genes themselves or sequences nearby did not improve the linkage obtained with the two most closely linked SSR markers. However, the slight decrease in linkage for flanking SSR markers either side of this locus gave a strong indication that one or a combination of the MIP genes in the 71 kb interval were involved in regulating the transport of B in the plant. Therefore, as an output of the genetic analysis we identified eight molecular markers that were closely linked to the B tolerance trait, the best of which were CAPS markers MtBtol-1, MtBtol-2, and MtBtol-3 and SSR markers h2_6e22a and h2_21b19a, which are all 95% linked. The observation that these markers explain 95% and not 100% of the phenotypic variation is expected considering that B-intolerant F2 plants still show a greater level of B tolerance, when quantified as DM production, than the intolerant parent (Figure 2A). Plant characteristics independent of B exclusion such as morphological differences e.g. a stronger vigour of Caliph and Paraggio over the intolerant tap mutant, are likely to account for the remaining 5% of phenotypic variation in these two populations. Aside from DM production, the ICP analysis clearly showed differences in the range of B levels in the leaves of the F2 lines from the txC compared to the txP population. These differences can again be explained by genotype-specific traits (e.g. maturity, morphology) that are inherited independently of the B exclusion locus.
As the chromosome 4 locus associated with MtBtol-1 contains up to five predicted MIP genes, we tried to narrow down the association with B toxicity tolerance to individual genes by monitoring their expression in Caliph, Paraggio, and tap prior to and after treatment with excess B. Although a specific Q-PCR assay could not be designed for MtNIP3, by excluding the low or non-expression of its predicted splice variant and the similar gene MtNIP5, respectively, we deduced that MtNIP3 was the only MIP gene expressed at this locus as no expression signals were detected for the remaining three MIP candidates when using alternative sets of primers. This meant that they were either not expressed, expressed at extremely low levels, or that the gene predictions were not correct and these genes were simply not there. Ultimately, the presence and function of in silico predicted genes has to be confirmed through the analysis of their gene product [21]. Most interesting was the observation that the constitutive expression of MtNIP3 in the leaves was almost four-fold higher in both B-tolerant lines compared to the intolerant line. By contrast, there was no difference in expression levels in the roots of tolerant and intolerant lines. A similar pattern was observed with the distribution of B in the leaves and roots of the tolerant and intolerant parental lines. There was no significant difference of B in the roots but twice the B concentration in the leaves of the intolerant parent compared to that in both tolerant lines. Several mechanisms of alleviating B toxicity are being discussed, including efflux from the roots, redistribution of B in the leaves and leaching of B by rain or removing B from the plant through guttation [22]. Based on the observed B distribution in the roots and leaves we can exclude that efflux from the roots is responsible for the different levels of B tolerance in these genotypes. Nevertheless, other B tolerant medic genotypes [6] might well employ such a mechanism. Possible mechanisms could be either the redistribution of B in the leaves from sensitive symplastic to apoplastic spaces in the leaves and subsequent leaching through rain [22] and/or the removal of B via guttation [9,22,23]. After B treatment, MtNIP3 expression was regulated in the leaves only, with the level of differential expression being 4 times higher in B-tolerant compared to intolerant lines. The fact that the two B-tolerant lines also have substantially less B in their leaves suggests that MtNIP3 itself or homologs at the MtNIP3 locus may function in removing B from the leaf.
Southern hybridisation with an MtNIP3/MtNIP5 specific probe ruled out the possibility that additional copies of MtNIP3 were responsible for the increased expression of this gene in the leaves of B-tolerant lines. In fact, Caliph only had one gene copy as opposed to the two detected in Paraggio and tap. In barley, it was shown that a B-tolerant genotype had approximately four times as many copies of the B transporter gene Bot1 compared to a number of intolerant genotypes [9], however, in Medicago this does not appear to be the case for MtNIP3.
With MtNIP5 not showing measurable transcript levels in roots and youngest leaves, our findings suggest that MtNIP3 is the most promising candidate to be involved in the mechanism controlling B toxicity tolerance in M. truncatula as its expression is strongly linked with B tolerance and distribution in the plant. Given that B is essential for cell wall elongation, where it is required for the cross-linking of the pectic polysaccharide rhamnogalacturonan II [24], the constitutive expression of MtNIP3 in the roots and leaves is not surprising as MtNIP3 may always be needed to give B access to sink tissues for normal plant growth. The lower constitutive and then non-differential MtNIP3 expression observed in the roots following B treatment is consistent with what was reported for AtNIP6;1, the Arabidopsis gene most similar to MtNIP3[20]. It would be interesting to know if this closest Medicago match to AtNIP6;1 does share the water impermeability of the AtNIP6;1 protein [20,25]. Considering that M. truncatula grows in low rainfall environments of northern Africa, Mediterranean Europe and southern Australia, a water conserving mechanism to remove excess B would be advantageous to the plant. It is likely that additional MtNIP3 alleles, homologous or orthologous genes to the efflux transporter genes AtBOR1[12] or HvBot1[9,11] control B toxicity tolerance in other M. truncatula lines than the ones investigated here. The information gained in M. truncatula could also be used to facilitate the identification of genes in other pasture legumes (e.g. other Medicago species [6]) and also grain legumes (lentils [26], peas [6]) as recently demonstrated for alfalfa and the resistance to the fungal disease anthracnose [27].
The generated molecular markers are currently being used in SARDI’s pasture breeding program to confirm that B tolerance is conserved in advanced lines that also contain tolerance to the sulfonylurea family of herbicides [28], which is being selected for by using the gene-specific marker CAng [18]. These breeding aims support a crop/pasture rotation system where self-regenerating annual medics are grown in rotation with wheat and barley to fix atmospheric nitrogen to lower input costs, provide a cereal pathogen disease-break and serve as a high protein feed for livestock. At an estimated rate of approximately 25 kg nitrogen per tonne DM and a production of 4 tonnes DM per hectare, a medic crop can fix approximately 100 kg nitrogen per hectare [29], reducing the need for expensive inorganic fertilisers and at the same time avoiding CO2 emissions that would arise from the methane-driven fertiliser production.
Conclusions
In the present study, a candidate gene approach was applied to establish a genetic linkage to the B toxicity tolerance trait observed in different cultivars of Medicago truncatula. Whereas no genetic linkage between other Boron exclusion and efflux transporter genes was detected, strong linkages were evident to genetic markers flanking a cluster comprising five predicted NIP genes explaining most (95%) of the phenotypic variation observed in the segregating Medicago populations. The derived markers are valuable tools for selection of this trait in a pasture breeding program and the close correlation of biomass and B toxicity qualifies B toxicity tolerance as a priority trait. The close correlation of MtNIP3 expression and B concentration in roots and leaves of tolerant and intolerant lines suggests that this member of the gene cluster is the most promising candidate for a B tolerance gene in this species. It further suggests that the molecular basis of B detoxification in Medicago differs from that observed in the monocot barley where differential expression of HvNIP2;1 in the roots [10] and a greater Boron-efflux transporter gene activity in the leaves [9] has previously been identified as the molecular basis of B toxicity alleviation. Further physiological studies on guttation and B leaching through rain, as recently described for barley [22] are required to quantify the possible role of these two mechanisms in mediating B toxicity tolerance.
Methods
Genetic material
Two F2 populations of M. truncatula were developed by crossing two B toxicity tolerant cultivars, Caliph and Paraggio (Caliph tested as Z-602 [6]), with the B toxicity intolerant line, tap. Paraggio and Caliph are cultivars developed by the South Australian Research and Development Institute (SARDI) and were registered as cultivars in 1982 and 1993, respectively, whereas tap (mtapetala) is a male sterile mutant derived from EMS treatment of A17 (Jemalong A17), the genotype used for M. truncatula genome sequencing [30]. For determination of the B tolerance trait inheritance, 316 and 313 F2 plants were assessed from the tap x Paraggio (txP) and tap x Caliph (txC) crosses, respectively, whereas 200 and 36 F2 plants from the txP and txC crosses, respectively, were used for molecular marker analysis.
Plant growth conditions
Seeds from the three parental lines and the F2 progeny were surface sterilised, scarified with a small nick to the seedcoat with a scalpel blade, and allowed to germinate on moist filter paper in an inverted position. When the radicals had reached approximately 10 mm in length seeds were transferred onto a wire mesh and suspended over a 32 x 45 cm tub containing 10 L of a modified version of Hoagland’s nutrient solution (2 mM KNO3, 2 mM Ca(NO3)2.4H2O, 0.8 mM MgSO4.7H2O, 0.4 mM KH2PO4, and 0.036 mM NaFe(iii).EDTA of macronutrients, and 18.50 μM H3BO3, 3.66 μM MnCl2.4H2O, 0.31 μM ZnS04.7H2O, 0.41 μM Na2MoO4.2H2O, and 0.13 μM CuSO4.5H2O of micronutrients, at pH 6.0). One 32 x 45 cm tub was sufficient to grow up to 200 plants. The nutrient solution was continually mixed with an aquarium pump and replaced weekly. The plants were grown in a glasshouse under natural light (12 hr day, 12 hr night) for 3 weeks and then treated with excess B by supplementing the aforementioned nutrient solution with 1 mM H3BO3.
Quantification of boron tolerance
Visual phenotyping was performed 3 weeks after B treatment when the three oldest trifoliate leaves (of about 10–15 leaves) of each plant were scored using the following 0–5 scale: 0 = green leaf, 1 = slight chlorosis, 2 ≥ 10% leaf chlorosis, 3 = slight necrosis, 4 = 10-20% necrosis, 5 > 20% necrosis. The three leaves per plant were then harvested, pooled, and dried in a 65°C oven overnight.
The dried leaf samples from parental lines and a subset of approximately 40 highly B toxicity tolerant and intolerant F2 progeny from each cross were subjected to ICP-OES analysis [31] to measure their B concentrations. In an independent hydroponics experiment, root and leaf samples from the two tolerant and the intolerant parents, four plants each, were separately harvested in the manner as described above for ICP analysis. The wet roots were not rinsed but only blotted between paper towels to avoid B leaching as previously described [11]. The youngest two leaves were harvested to extract high quality genomic DNA for genotypic analysis.
Quantification of plant biomass
The weight of total plant dry matter [mg plant-1] was measured from plants under high B stress in the txC F2 population using 137 F2 lines and 5–9 repeats of their corresponding parents. Visual leaf symptom scores were taken before harvest, eight weeks after B treatment when the plants were a total of 11 weeks old.
Development and detection of DNA markers
DNA was extracted from freeze-dried leaf tissue using a previously documented protocol [32] with modifications as described [33]. To identify M. truncatula homologues of AtBOR1 and AtNIP5;1, their corresponding cDNA (GenBank: AB073713 and NM_117106) and protein (GenBank: BAC20173 and NP_192776) sequences were used to query the M. truncatula pseudomolecule database, version Mt3.5 (http://www.medicagohapmap.org). Once identified, nearby SSR markers were screened by PCR on a subset of lines from both txP and txC F2 populations to establish linkage. PCRs with the SSR primers were carried out in 12.5 μL reactions containing 1X reaction buffer (QIAGEN, Hilden, Germany), 1X Q solution (QIAGEN), 0.16 mM MgCl2, 0.32 μM of each primer, 0.4 U of Taq DNA polymerase (QIAGEN) and approximately 50 ng of DNA as template. The PCRs were effected using the following cycling conditions: 90 s at 95°C, then 35 cycles of 20 s at 95°C, 30 s at 58°C, and 30 s at 72°C, and a final extension step of 5 min at 72°C. All PCR products were electrophoresed on 8% polyacrylamide gels which were stained with ethidium bromide and visualised under UV light.
For CAPS (cleaved amplified polymorphic sequence) marker development, PCR primers were designed on the sequence of the A17 genotype. Genomic regions were amplified by PCR in 12.5 μL reactions as described earlier but with a 72°C extension step ranging from 30–90 s depending on the expected size of the amplicon. PCR products were resolved on 1.2% agarose gels and sequenced using the Big Dye Terminator Version 3.1 labelling kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s protocol but using one-eighth of the recommended reaction mix. DNA sequencing was performed on an ABI 3730 DNA Sequencer (Applied Biosystems) and DNA sequences were analysed using Vector NTI software (Invitrogen). The SNP2CAPS program [34] was then used to develop CAPS markers from the sequence alignments. PCR products were digested with the relevant restriction enzymes according to the manufacturer’s protocol and separated on 1.5% agarose gels.
Southern hybridization of parental DNA was carried out according to standard protocols [35] using 10 μg of digested DNA per lane and the restriction enzymes BamHI, DraI, EcoRI, EcoRV, HindIII and XbaI (5 U/μg).
Analysis of candidate B transporter gene expression
To monitor the expression of candidate B transporter genes, plants were grown in a controlled-environment room for 3 weeks using the supported hydroponics system previously described [36] but with the same nutrient solution as described earlier. Whole roots and the three youngest trifoliates from each plant were harvested and frozen in liquid nitrogen immediately before supplementing the nutrient solution with 1 mM H3BO3 (+B) and then at 3, 6, 12, 24, and 48 hr after B treatment. A control set of plants was allowed to grow for 3 weeks after B treatment to ensure that adequate symptoms developed.
Total RNA was extracted from frozen root and young leaf tissue using TRI Reagent (Sigma-Aldrich, St. Louis, MO). For each time point, RNA was extracted from three independent biological replicates which were pooled to average out biological variability. To prepare cDNA template for quantitative real-time RT-PCR (Q-PCR), RNA was first treated with 2 U RNase-free DNase I (Ambion, Austin, TX) to remove residual DNA. cDNA was then synthesised from 3 μg of RNA using 200 U of SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer’s instructions with quality being confirmed via PCR detection of transcripts from the gene actin using earlier described primers [37]. Q-PCR was performed in an RG 3000 Rotor-Gene Real Time Thermal Cycler (Corbett Research, Sydney, Australia). The final reaction volume of 8 μL comprised a mixture of 1X Sensimix SYBR green PCR reagent (Bioline), 0.4 μM of each forward and reverse gene-specific primer, and 3 μL of a 1:20 dilution of cDNA. The PCR was effected using the following cycling conditions, 10 min at 95°C, followed by 35 cycles of 20 s at 95°C, 30 s at 58°C and 30 s at 72°C. All Q-PCR reactions were carried out in duplicate and melt curve analysis was performed at the end of each run to confirm that there was no signal from non-specific binding products. No-template controls were also included in each run to test for possible contamination of assay reagents. The sequences of all target gene primers and the reference gene actin primers are shown in additional file 3.
To quantify gene expression levels, DNA standards were generated by cloning each target and reference gene PCR product into the pDrive cloning vector (QIAGEN) according to the manufacturer’s instructions. After spectrophotometric determination of plasmid DNA concentration, the vector was linearised with the restriction enzyme NruI, and the copy number of standard DNA molecules calculated using the formula (x g/μL DNA / (plasmid length in base pairs × 660)) × 6.022 × 1023 = y molecules/μL. Serial ten-fold dilutions of a given DNA standard (107-101 copies/ μL) were then incorporated into each run to quantify the number of target gene transcripts. These values were then normalised against the quantities of transcripts detected from the reference gene actin.
Abbreviations
B: Boron; Mt: Medicago truncatula; T: tap; C: Caliph; P: Paraggio; ICP spectrometry: Inductively coupled plasma spectrometry; MIP: Major intrinsic protein; SSR marker: Single sequence repeat marker; CAPS marker: Cleaved amplified polymorphic sequence marker.
Competing interests
The authors declare that they have no competing interest.
Authors’ contribution
PB performed most of the experiments, was involved in experimental design, and drafted the manuscript, DMP prepared the crosses of the segregating populations and was involved in the setup of the hydroponics system, RMN was involved in the conceptualisation of the project, JH helped with the selection of the population parents, KHO conceptualised the idea, was involved in experimental design and coordination, and drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
MtNIP3 cDNA sequence alignments for parental lines Caliph, Paraggio, and the intolerant tap. Single nucleotide polymorphisms between sequences are highlighted.
MtNIP3 protein sequence alignments for parental lines Caliph, Paraggio, and the intolerant tap. Amino acid differences between sequences are highlighted.
Q-PCR primers, primers for Southern probe and promoter analysis.
Contributor Information
Paul Bogacki, Email: paul.bogacki@sa.gov.au.
David M Peck, Email: david.peck@sa.gov.au.
Ramakrishnan M Nair, Email: ramakrishnan.nair@worldveg.org.
Jake Howie, Email: jake.howie@sa.gov.au.
Klaus H Oldach, Email: klaus.oldach@sa.gov.au.
Acknowledgements
This work was supported by the Rural Industries Research and Development Corporation of Australia (RIRDC project PRJ-003521 to KHO). We acknowledge Waite Analytical Services (WAS), University of Adelaide, for ICP-OES analysis of the plant samples.
References
- Vickery PJ, Hill MJ, Donald GE. Satellite derived maps of pasture growth status, Association of classification with botanical composition. Aust J Exp Agric. 1997;37:547–562. [Google Scholar]
- Jensen ES, Hauggaard-Nielsen H. How can increased use of biological N2 fixation in agriculture benefit the environment? Plant Soil. 2003;252:177–186. [Google Scholar]
- Russelle MP, Birr AS. Large-scale assessment of symbiotic dinitrogen fixation by crops, soybean and alfalfa in the Mississippi River Basin. Agron J. 2004;96:1754–1760. [Google Scholar]
- Rovira AD. The impact of soil and crop management practices on soil-borne root diseases and wheat yields. Soil Use Manage. 1990;6:195–200. [Google Scholar]
- Choi HK, Mun JH, Kim DJ, Zhu H, Baek JM, Mudge J, Roe B, Ellis N, Doyle J, Kiss GB, Young ND, Cook DR. Estimating genome conservation between crop and model legume species. Proc Natl Acad Sci USA. 2004;101:15289–15294. doi: 10.1073/pnas.0402251101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull JG, Nable RO, Lake AWH, Materne MA, Rathjen AJ. Response of annual medics (Medicago spp.) and field peas (Pisum sativum) to high concentration of Boron, genetic variation and the mechanism of tolerance. Aust J Agric Res. 1992;43:203–213. [Google Scholar]
- Yau SK, Ryan J. Boron toxicity tolerance in crops, a viable alternative to soil amelioration. Crop Sci. 2008;48:854–865. [Google Scholar]
- Jefferies SP, Barr AR, Karakousis A, Kretschmer JM, Manning S, Chalmers KJ, Nelson JC, Islam AKMR, Langridge P. Mapping of chromosome regions conferring boron toxicity tolerance in barley (Hordeum vulgare L.) Theor Appl Genet. 1999;98:1293–1303. [Google Scholar]
- Sutton T, Baumann U, Hayes J, Collins NC, Shi B-J, Schnurbusch T, Hay A, Mayo G, Pallotta M, Tester M, Langridge P. Boron-toxicity tolerance in barley arising from efflux transporter amplification. Science. 2007;318:1446–1449. doi: 10.1126/science.1146853. [DOI] [PubMed] [Google Scholar]
- Schnurbusch T, Hayes J, Hrmova M, Baumann U, Ramesh SA, Tyerman SD, Langridge P, Sutton T. Boron toxicity tolerance in barley through reduced expression of the multifunctional aquaporin HvNIP2;1. Plant Physiol. 2010;153:1706–1715. doi: 10.1104/pp.110.158832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid R. Identification of Boron transporter genes likely to be responsible for tolerance to Boron toxicity in wheat and barley. Plant Cell Physiol. 2007;48:1673–1678. doi: 10.1093/pcp/pcm159. [DOI] [PubMed] [Google Scholar]
- Takano J, Noguchi K, Yasumori M, Kobayashi M, Gajdos Z, Miwa K, Hayashi H, Yoneyama T, Fujiwara T. Arabidopsis boron transporter for xylem loading. Nature. 2002;420:337–340. doi: 10.1038/nature01139. [DOI] [PubMed] [Google Scholar]
- Miwa K, Takano J, Omori H, Seki M, Shinozaki K, Fujiwara T. Plants tolerant of high Boron levels. Science. 2007;318:1417. doi: 10.1126/science.1146634. [DOI] [PubMed] [Google Scholar]
- Takano J, Wada M, Ludewig U, Schaaf G, von Wirén N, Fujiwara T. The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell. 2006;18:1498–1509. doi: 10.1105/tpc.106.041640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyerman SD, Niemietz CM, Bramley H. Plant aquaporins, multifunctional water and solute channels with expanding roles. Plant Cell Environ. 2002;25:173–194. doi: 10.1046/j.0016-8025.2001.00791.x. [DOI] [PubMed] [Google Scholar]
- Pang Y, Li L, Ren F, Lu P, Wei P, Cai J, Xin L, Zhang J, Chen J, Wang X. Overexpression of the tonoplast aquaporin AtTIP5;1 conferred tolerance to boron toxicity in Arabidopsis. J Genet Genomics. 2010;37:389–397. doi: 10.1016/S1673-8527(09)60057-6. [DOI] [PubMed] [Google Scholar]
- Uehlein N, Fileschi K, Eckert M, Bienert GP, Bertl A, Kaldenhoff R. Arbuscular mycorrhizal symbiosis and plant aquaporin expression. Phytochemistry. 2007;68:122–129. doi: 10.1016/j.phytochem.2006.09.033. [DOI] [PubMed] [Google Scholar]
- Oldach KH, Peck DM, Cheong J, Williams KJ, Nair RM. Identification of a chemically induced point mutation mediating herbicide tolerance in annual medics (Medicago spp.) Ann Bot. 2008;101:997–1005. doi: 10.1093/aob/mcn028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nable RO, Lance RCM, Cartwright B. Uptake of boron and silicon by barley genotypes with differing susceptibilities to boron toxicity. Ann Bot. 1990;66:83–90. [Google Scholar]
- Tanaka M, Wallace IS, Takano J, Roberts DM, Fujiwara T. NIP6;1 is a boric acid channel for preferential transport of boron to growing shoot tissues in Arabidopsis. Plant Cell. 2008;20:2860–2875. doi: 10.1105/tpc.108.058628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleator RD. An overview of the current status of eukaryote gene prediction strategies. Gene. 2010;461:1–4. doi: 10.1016/j.gene.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Reid R, Fitzpatrick K. Influence of leaf tolerance mechanisms and rain on Boron toxicity in barley and wheat. Plant Physiol. 2009;151:413–420. doi: 10.1104/pp.109.141069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl HC, Oertli JJ. Distribution of Boron in leaves. Plant Physiol. 1961;36:420–424. doi: 10.1104/pp.36.4.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill MA, Eberhard S, Albersheim P, Darvill AG. Requirement of borate cross-linking of cell wall rhamnogalacturonan II for Arabidopsis growth. Science. 2001;294:846–849. doi: 10.1126/science.1062319. [DOI] [PubMed] [Google Scholar]
- Wallace IS, Roberts DM. Homology modeling of representative subfamilies of Arabidopsis major intrinsic proteins, classification based on the aromatic/arginine selectivity filter. Plant Physiol. 2004;135:1059–1068. doi: 10.1104/pp.103.033415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson K, Armstrong R, Nicolas M, Connor D, Materne M. Response of lentil (Lens culinaris) germplasm to high concentrations of soil boron. Euphytica. 2006;151:371–382. [Google Scholar]
- Yang S, Gao M, Xu C, Gao J, Deshpande S, Lin S, Roe BA, Zhu H. Alfalfa benefits from Medicago truncatula: The RCT1 gene from M. truncatula confers broad-spectrum resistance to anthracnose in alfalfa. Proc Natl Acad Sci USA. 2008;105:12164–12169. doi: 10.1073/pnas.0802518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck DM, Howie JH. Development of an early season barrel medic (Medicago truncatula Gaertn.) with tolerance to sulfonylurea herbicide residues. Crop & Past Sci. in press.
- Peoples MB, Bowman AM, Gault RR, Herridge DF, McCallum MH, McCormick KM, Norton RM, Rochester IJ, Scammell GJ, Schwenke GD. Factors regulating the contributions of fixed nitrogen by pasture and crop legumes to different farming systems of eastern Australia. Plant Soil. 2001;228:29–41. [Google Scholar]
- Penmetsa RV, Cook DR. Production and characterization of diverse developmental mutants of Medicago truncatula. Plant Physiol. 2000;123:1387–1397. doi: 10.1104/pp.123.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheal MS, Fowles TO, Palmer LT. A cost-effective acid digestion method using closed polypropylene tubes for inductively coupled plasma optical emission spectrometry (ICP-OES) analysis of plant essential elements. Anal Methods. 2011;3:2854–2863. [Google Scholar]
- Rogowsky PM, Guider FLY, Langridge P, Shepherd KW, Koebner RMD. Isolation and characterization of wheat-rye recombinants involving chromosome arm 1DS of wheat. Theor Appl Genet. 1991;82:537–544. doi: 10.1007/BF00226788. [DOI] [PubMed] [Google Scholar]
- Williams KJ, Platz GJ, Barr AR, Cheong J, Willsmore K, Cakir M, Wallwork H. A comparison of the genetics of seedling and adult plant resistance to the spot form of net blotch (Pyrenophora teres f. maculata) Aust J Agric Res. 2003;54:1387–1394. [Google Scholar]
- Thiel T, Kota R, Grosse I, Stein N, Graner A. SNP2CAPS, a SNP and INDEL analysis tool for CAPS marker development. Nucleic Acids Res. 2004;32:e5. doi: 10.1093/nar/gnh006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, a Laboratory Manual. 2. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Genc Y, McDonald GK, Tester M. Reassessment of tissue Na+ concentration as a criterion for salinity tolerance in bread wheat. Plant Cell Environ. 2007;30:1486–1498. doi: 10.1111/j.1365-3040.2007.01726.x. [DOI] [PubMed] [Google Scholar]
- Kuppusamy KT, Endre G, Prabhu R, Penmetsa RV, Veereshlingam H, Cook DR, Dickstein R, VandenBosch KA. LIN, a Medicago truncatula gene required for nodule differentiation and persistence of rhizobial infections. Plant Physiol. 2004;136:3682–3691. doi: 10.1104/pp.104.045575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MtNIP3 cDNA sequence alignments for parental lines Caliph, Paraggio, and the intolerant tap. Single nucleotide polymorphisms between sequences are highlighted.
MtNIP3 protein sequence alignments for parental lines Caliph, Paraggio, and the intolerant tap. Amino acid differences between sequences are highlighted.
Q-PCR primers, primers for Southern probe and promoter analysis.