Abstract
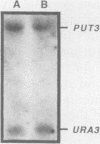
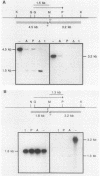
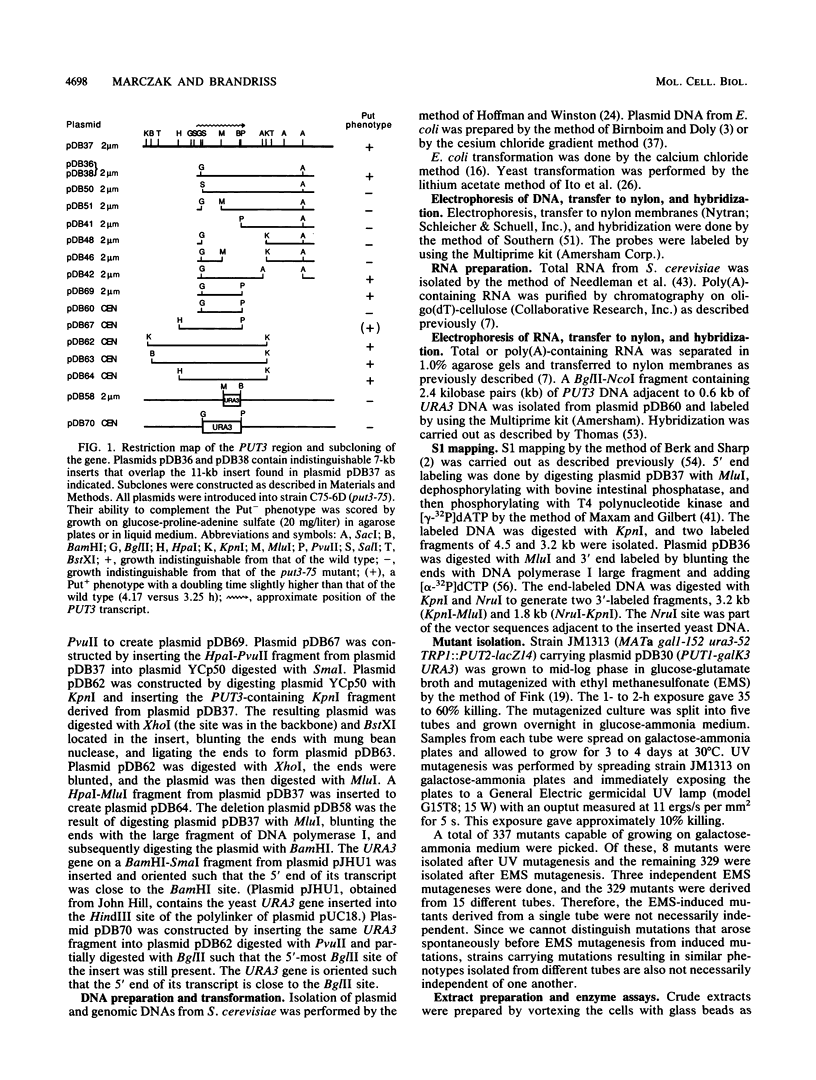
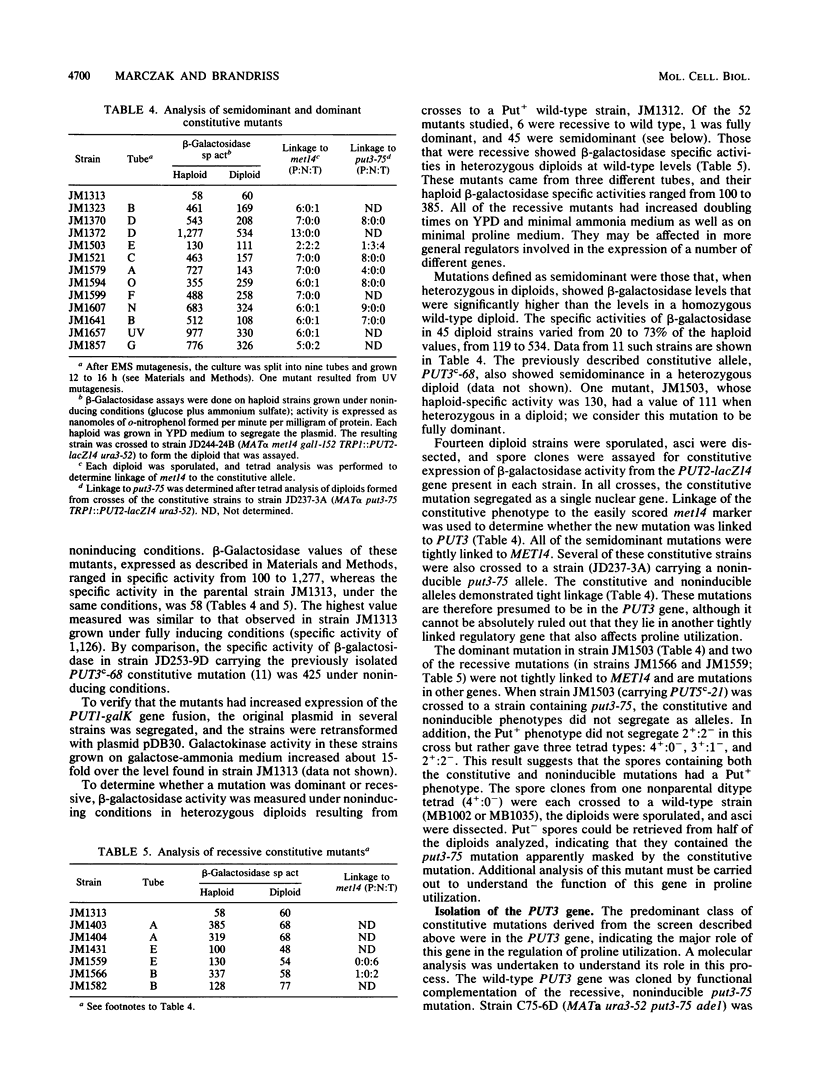
The enzymes of the proline utilization pathway (the products of the PUT1 and PUT2 genes) in Saccharomyces cerevisiae are coordinately regulated by proline and the PUT3 transcriptional activator. To learn more about the control of this pathway, constitutive mutations in PUT3 as well as in other regulators were sought. A scheme using a gene fusion between PUT1 (S. cerevisiae proline oxidase) and galK (Escherichia coli galactokinase) was developed to select directly for constitutive mutations affecting the PUT1 promoter. These mutations were secondarily screened for their effects in trans on the promoter of the PUT2 (delta 1-pyrroline-5-carboxylate dehydrogenase) gene by using a PUT2-lacZ (E. coli beta-galactosidase) gene fusion. Three different classes of mutations were isolated. The major class consisted of semidominant constitutive PUT3 mutations that caused PUT2-lacZ expression to vary from 2 to 22 times the uninduced level. A single dominant mutation in a new locus called PUT5 resulted in low-level constitutive expression of PUT2-lacZ; this mutation was epistatic to the recessive, noninducible put3-75 allele. Recessive constitutive mutations were isolated that had pleiotropic growth defects; it is possible that these mutations are not specific to the proline utilization pathway but may be in genes that control several pathways. Since the PUT3 gene appears to have a major role in the regulation of this pathway, a molecular analysis was undertaken. This gene was cloned by functional complementation of the put3-75 mutation. Strains carrying a complete deletion of this gene are viable, proline nonutilizing, and indistinguishable in phenotype from the original put3-75 allele. The PUT3 gene encodes a 2.8-kilobase-pair transcript that is not regulated by proline at the level of RNA accumulation. The presence of the gene on a high-copy-number plasmid did not alter the regulation of one of its target genes, PUT2-lacZ, suggesting that the PUT3 gene product is not limiting and that a titratable repressor is not involved in the regulation of this pathway.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bach M. L., Lacroute F., Botstein D. Evidence for transcriptional regulation of orotidine-5'-phosphate decarboxylase in yeast by hybridization of mRNA to the yeast structural gene cloned in Escherichia coli. Proc Natl Acad Sci U S A. 1979 Jan;76(1):386–390. doi: 10.1073/pnas.76.1.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brandriss M. C. Evidence for positive regulation of the proline utilization pathway in Saccharomyces cerevisiae. Genetics. 1987 Nov;117(3):429–435. doi: 10.1093/genetics/117.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandriss M. C. Isolation and preliminary characterization of Saccharomyces cerevisiae proline auxotrophs. J Bacteriol. 1979 Jun;138(3):816–822. doi: 10.1128/jb.138.3.816-822.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandriss M. C., Krzywicki K. A. Amino-terminal fragments of delta 1-pyrroline-5-carboxylate dehydrogenase direct beta-galactosidase to the mitochondrial matrix in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Oct;6(10):3502–3512. doi: 10.1128/mcb.6.10.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandriss M. C., Magasanik B. Genetics and physiology of proline utilization in Saccharomyces cerevisiae: enzyme induction by proline. J Bacteriol. 1979 Nov;140(2):498–503. doi: 10.1128/jb.140.2.498-503.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandriss M. C., Magasanik B. Genetics and physiology of proline utilization in Saccharomyces cerevisiae: mutation causing constitutive enzyme expression. J Bacteriol. 1979 Nov;140(2):504–507. doi: 10.1128/jb.140.2.504-507.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandriss M. C., Magasanik B. Proline: an essential intermediate in arginine degradation in Saccharomyces cerevisiae. J Bacteriol. 1980 Sep;143(3):1403–1410. doi: 10.1128/jb.143.3.1403-1410.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandriss M. C. Proline utilization in Saccharomyces cerevisiae: analysis of the cloned PUT2 gene. Mol Cell Biol. 1983 Oct;3(10):1846–1856. doi: 10.1128/mcb.3.10.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Cherry J. R., Johnson T. R., Dollard C., Shuster J. R., Denis C. L. Cyclic AMP-dependent protein kinase phosphorylates and inactivates the yeast transcriptional activator ADR1. Cell. 1989 Feb 10;56(3):409–419. doi: 10.1016/0092-8674(89)90244-4. [DOI] [PubMed] [Google Scholar]
- Clifton D., Weinstock S. B., Fraenkel D. G. Glycolysis mutants in Saccharomyces cerevisiae. Genetics. 1978 Jan;88(1):1–11. doi: 10.1093/genetics/88.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G., Böhni P. C., Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem. 1982 Nov 10;257(21):13028–13033. [PubMed] [Google Scholar]
- Dubois E., Messenguy F. Isolation and characterization of the yeast ARGRII gene involved in regulating both anabolism and catabolism of arginine. Mol Gen Genet. 1985;198(2):283–289. doi: 10.1007/BF00383008. [DOI] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- Hashimoto H., Kikuchi Y., Nogi Y., Fukasawa T. Regulation of expression of the galactose gene cluster in Saccharomyces cerevisiae. Isolation and characterization of the regulatory gene GAL4. Mol Gen Genet. 1983;191(1):31–38. doi: 10.1007/BF00330886. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol Rev. 1988 Dec;52(4):536–553. doi: 10.1128/mr.52.4.536-553.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heusterspreute M., Ha Thi V., Davison J. Expression of galactokinase as a fusion protein in Escherichia coli and Saccharomyces cerevisiae. DNA. 1984 Oct;3(5):377–386. doi: 10.1089/dna.1984.3.377. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev. 1988 Jun;52(2):248–273. doi: 10.1128/mr.52.2.248-273.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987 Dec;51(4):458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., Davis R. W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S. A., Hopper J. E. Isolation of the yeast regulatory gene GAL4 and analysis of its dosage effects on the galactose/melibiose regulon. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6971–6975. doi: 10.1073/pnas.79.22.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S. A., Salmeron J. M., Jr, Dincher S. S. Interaction of positive and negative regulatory proteins in the galactose regulon of yeast. Cell. 1987 Jul 3;50(1):143–146. doi: 10.1016/0092-8674(87)90671-4. [DOI] [PubMed] [Google Scholar]
- Kaput J., Brandriss M. C., Prussak-Wieckowska T. In vitro import of cytochrome c peroxidase into the intermembrane space: release of the processed form by intact mitochondria. J Cell Biol. 1989 Jul;109(1):101–112. doi: 10.1083/jcb.109.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywicki K. A., Brandriss M. C. Primary structure of the nuclear PUT2 gene involved in the mitochondrial pathway for proline utilization in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Dec;4(12):2837–2842. doi: 10.1128/mcb.4.12.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughon A., Gesteland R. F. Isolation and preliminary characterization of the GAL4 gene, a positive regulator of transcription in yeast. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6827–6831. doi: 10.1073/pnas.79.22.6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljelund P., Losson R., Kammerer B., Lacroute F. Yeast regulatory gene PPR1. II. Chromosomal localization, meiotic map, suppressibility, dominance/recessivity and dosage effect. J Mol Biol. 1984 Dec 5;180(2):251–265. doi: 10.1016/s0022-2836(84)80003-0. [DOI] [PubMed] [Google Scholar]
- Losson R., Fuchs R. P., Lacroute F. In vivo transcription of a eukaryotic regulatory gene. EMBO J. 1983;2(12):2179–2184. doi: 10.1002/j.1460-2075.1983.tb01720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Ptashne M. The carboxy-terminal 30 amino acids of GAL4 are recognized by GAL80. Cell. 1987 Jul 3;50(1):137–142. doi: 10.1016/0092-8674(87)90670-2. [DOI] [PubMed] [Google Scholar]
- Mason T. L., Poyton R. O., Wharton D. C., Schatz G. Cytochrome c oxidase from bakers' yeast. I. Isolation and properties. J Biol Chem. 1973 Feb 25;248(4):1346–1354. [PubMed] [Google Scholar]
- Matsumoto K., Adachi Y., Toh-e A., Oshima Y. Function of positive regulatory gene gal4 in the synthesis of galactose pathway enzymes in Saccharomyces cerevisiae: evidence that the GAL81 region codes for part of the gal4 protein. J Bacteriol. 1980 Feb;141(2):508–527. doi: 10.1128/jb.141.2.508-527.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Toh-e A., Oshima Y. Isolation and characterization of dominant mutations resistant to carbon catabolite repression of galactokinase synthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1981 Feb;1(2):83–93. doi: 10.1128/mcb.1.2.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Needleman R. B., Kaback D. B., Dubin R. A., Perkins E. L., Rosenberg N. G., Sutherland K. A., Forrest D. B., Michels C. A. MAL6 of Saccharomyces: a complex genetic locus containing three genes required for maltose fermentation. Proc Natl Acad Sci U S A. 1984 May;81(9):2811–2815. doi: 10.1073/pnas.81.9.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman D., Hopper J. E. Constitutive synthesis of the GAL4 protein, a galactose pathway regulator in Saccharomyces cerevisiae. Cell. 1979 Jan;16(1):89–95. doi: 10.1016/0092-8674(79)90190-9. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Rymond B. C., Zitomer R. S., Schümperli D., Rosenberg M. The expression in yeast of the Escherichia coli galK gene on CYC1::galK fusion plasmids. Gene. 1983 Nov;25(2-3):249–262. doi: 10.1016/0378-1119(83)90229-9. [DOI] [PubMed] [Google Scholar]
- Siddiqui A. H., Brandriss M. C. A regulatory region responsible for proline-specific induction of the yeast PUT2 gene is adjacent to its TATA box. Mol Cell Biol. 1988 Nov;8(11):4634–4641. doi: 10.1128/mcb.8.11.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui A. H., Brandriss M. C. The Saccharomyces cerevisiae PUT3 activator protein associates with proline-specific upstream activation sequences. Mol Cell Biol. 1989 Nov;9(11):4706–4712. doi: 10.1128/mcb.9.11.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stinchcomb D. T., Mann C., Davis R. W. Centromeric DNA from Saccharomyces cerevisiae. J Mol Biol. 1982 Jun 25;158(2):157–190. doi: 10.1016/0022-2836(82)90427-2. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. S., Brandriss M. C. Proline utilization in Saccharomyces cerevisiae: analysis of the cloned PUT1 gene. Mol Cell Biol. 1986 Jul;6(7):2638–2645. doi: 10.1128/mcb.6.7.2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. S., Brandriss M. C. Proline utilization in Saccharomyces cerevisiae: sequence, regulation, and mitochondrial localization of the PUT1 gene product. Mol Cell Biol. 1987 Dec;7(12):4431–4440. doi: 10.1128/mcb.7.12.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wartell R. M., Reznikoff W. S. Cloning DNA restriction endonuclease fragments with protruding single-stranded ends. Gene. 1980 May;9(3-4):307–319. doi: 10.1016/0378-1119(90)90329-p. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]