INTRODUCTION
Natural products provide some of the most promising and effective known anti-cancer agents. Even if not used directly, they offer a template to derivatize and/or mimic. Non-synthetically derived small molecules account for almost 75% of all clinically used anti-cancer agents since 1940 with almost 50% being either natural products or their direct derivatives (1–3). However, natural products are often not optimized for human diseases and can in turn interact with many normal cells causing unwanted side effects. Biosynthetic and chemical modifications of natural products offer a great opportunity to improve the anti-cancer properties of natural product drugs while reducing their non-specific interactions.
Mithramycin (MTM) is an example of how such modifications can yield improved anti-cancer agents. MTM is an aureolic acid type anti-cancer agent produced by various soil bacteria of the genus Streptomyces. MTM exhibits compelling anti-cancer activity and a unique mode of action. MTM acts by cross-linking GC-rich DNA thereby shutting down the transcription of several proto-oncogenes, particularly pathways regulated by transcription factors Sp1 and Sp3 (4–7). The Sp1 transcription factors are important as they have been linked to the control of cell growth, survival, and differentiation and their over expression has been observed in several cancers (8–10). MTM does have a clinical history, e.g., applications for the treatment testicular cancer, Paget bone disease, and hypercalcemia, but has been limited by side effects such as hepatic, gastrointestinal, bone marrow, and renal toxicities (11–15). MTM has shown promise with respect to treating neurological diseases, glioblastomas, and other tumors in addition to showing the ability to inhibit the multi-drug resistance efflux pump MDR1 for which smaller, less toxic doses are required (16–21). Most recently MTM was identified as the lead compound against the Friend leukemia virus integration 1 (EWS-FLI1) transcription factor and in a combinational approach with betulinic acid to treat pancreatic cancer (19, 22). It is clear that MTM has high potential in the fight against cancer and new and improved analogues would find clinical relevance.
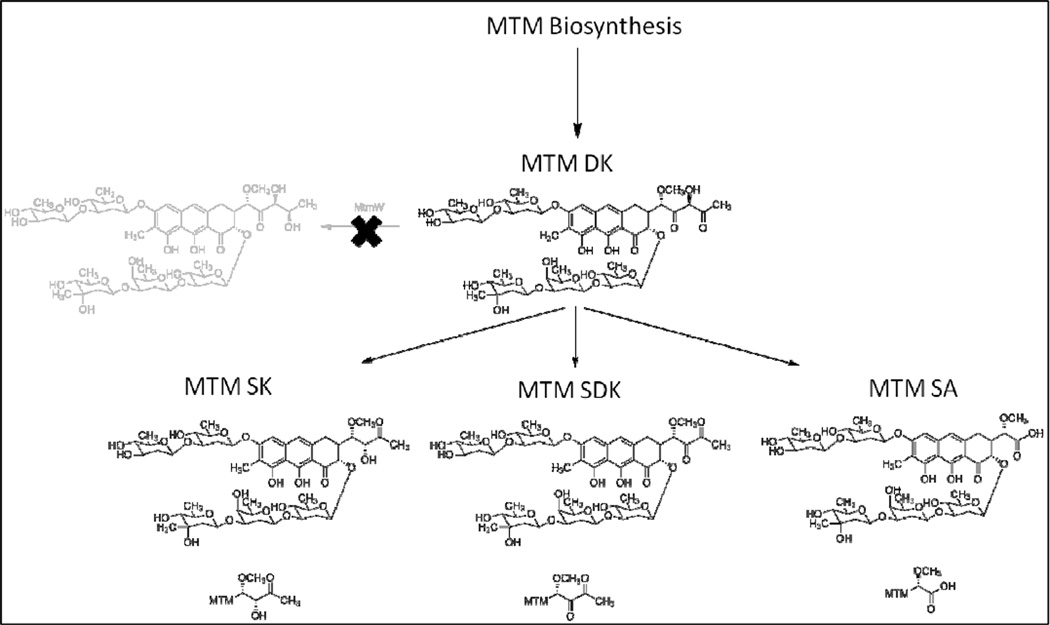
To that end, extensive combinatorial biosynthesis has been performed on the drug biosynthesis pathway to produce altered MTM analogues with hopes to improve the toxicity profiles. This has resulted in several novel compounds with intriguing characteristics. The inactivation of the mtmW gene, which is the gene encoding the last acting enzyme in the MTM biosynthetic pathway, produced the analogues MTM SK and MTM SDK (Figure 1) (23, 24). Both of these analogues possess shorter side chains at the 3-position. The 3-side chain has been identified previously as important, since it is in part responsible for MTM’s interaction with the DNA phosphate backbone (6, 25). Both MTM SK and MTM SDK showed increased activity against several cancer cell lines compared to the parent MTM (23, 24, 26). These results indicate that the 3-side chain is important for the activity of MTM and offers a base for further molecular manipulations. As an unwanted side product along with the production of the desired MTM SK and MTM SDK analogues, MTM SA also accumulated in the MtmW-minus-mutant, but showed – in contrast to MTM SK and MTM SDK – significantly decreased activity compared to MTM (26, 27). One of the discussed reasons for MTM SA’s decreased activity might be that its 3-side chain is too short and its negatively charged carboxylic acid is disadvantageous to interact with naturally negatively charged DNA. It is hypothesized that by modifying the 3-side chain of MTM SA via a semi-synthetic approach the unique carboxylic acid moiety of MTM SA can be chemically modified to introduce new functionalities into the 3-side chain. The modifications to the side chain may improve the biological activity of MTM SA resulting in new MTM analogues. Recent work by Preobrazhenskaya et al. (28–30) on olivomycin derivatizations showed that introduction of N-atoms can improve aureolic acid type anticancer drugs. Thus reaction of MTM SA with primary amines would create amides thereby elongating the 3-side chain and simultaneously incorporating one or more N-atom/s which could enhance the interaction with the DNA-phosphate backbone and result in improved MTM analogues.
Figure 1.
A schematic representation of the accumulation of MTM SK, MTM SDK, and MTM SA by the inactivation of the mtmW gene in the MTM biosynthetic pathway.
MATERIALS AND METHODS
Materials
N,N’-diisopropylcarbodiimide (DIC), N-hydroxysuccinimide (NHS), dimethylformamide (DMF), dichloromethane (DCM), O-(Benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium tetrafluoroborate (TBTU), (1-Cyano-2-ethoxy-2-oxoethylidenaminooxy)dimethylamino-morpholino-carbenium hexafluorophosphate (COMU), diphenyl phosphoryl azide (DPPA), N,N-diisopropylethylamine (DIPEA), methyl hydrazine, benzotriazol-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate (PyBOP), L-cysteine methyl ester hydrochloride, L-glycine methyl ester hydrochloride, L-alanine methyl ester hydrochloride, L-valine methyl ester hydrochloride, cystamine, DMSO (molecular biology grade, ≥99.9 %), dimethylsulfoxide-d6 (DMSO-d6), resazurin sodium salt, were purchased from Sigma-Aldrich (USA). N,N-dimethylethylenediamine was purchased from TCI America (USA). 1-amino-2-propanone was purchased from Waterstone Tech (USA). Methanol (MeOH), acetonitrile (ACN), celite, C18 RP silica gel, tryptic soy broth (TSB), LB broth, Difco agar, sucrose, potassium sulfate, magnesium chloride, glucose, casamino acids, yeast extract, MOPS, and trace elements were purchased from Fisher Scientific (USA). Steptomyces argillaceus ATCC 12956, A549 tissue culture cells, F-12K media, fetal bovine serum (FBS) were purchased from ATCC (USA).
Biosynthesis of MTM SA
MTM SA was produced by a procedure reported previously (31). S. argillaceus M7W1 was plated on R5A agar and allowed to grow for four days or until spores formed. The spores were then used to seed a culture in 100 mL of tryptic soy broth (TSB) and grown for 24 hr in an orbital shaker at 28 °C, 250 rpm. After 24 hrs 4 mL of the TSB culture was used to start a culture in R5A media in 40, 100 mL flasks. The cultures in R5A media were grown for 3 days at 28 °C, 250 rpm while the production of SA was monitored by HPLC. After 3 days the cells were collected with 50 g/L of celite and removed by filtration. The cell pellet was then re-dissolved in MeOH and sonicated for 1 hr to lyse the cells. Following the cell lysis the cellular debris was filtered off and the MeOH was evaporated from the filtrate. The dried cellular extract was then reconstituted in water and loaded onto a 5 × 12 cm C18 RP column equilibrated with 10 column volumes of water. The column was washed with 10% ACN in water, followed by a fractionation of ACN in water from 20–50%, followed by 100% ACN. The filtrate removed from the cells in step one was added to the C18 column and eluted by the same fractionation procedure. The samples were dried and re-constituted in 80% MeOH and water. SA was then completely isolated from the mixture of a few compounds by semi-preparative HPLC.
Synthetic Modification of MTM SA 3 Side Chain
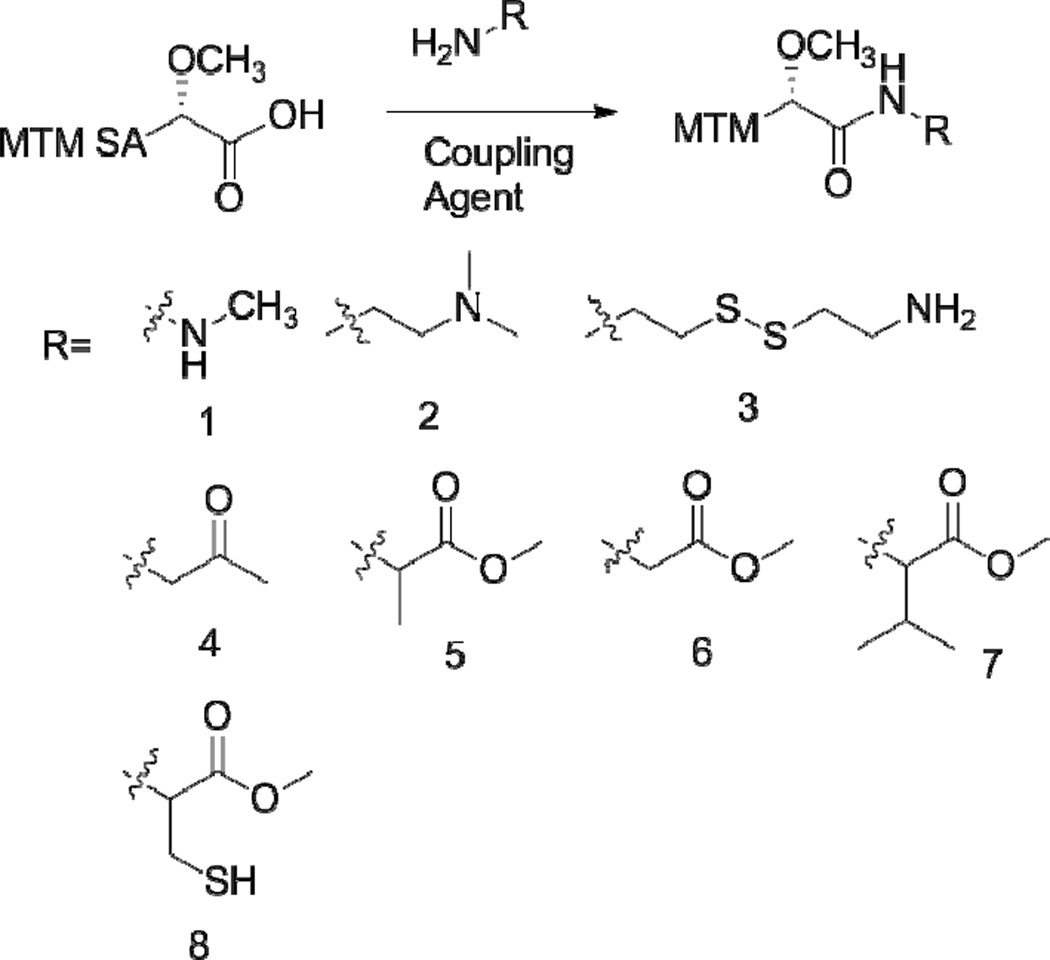
MTM SA was modified by converting the terminal carboxylic acid group with a primary amine through a coupling reaction. Several different protocols were investigated to discover the optimal reaction conditions. For all of the exploratory reactions 2 mg of MTM SA was used. The initial reaction was completed by reacting MTM SA with the desired side chain modifying molecule in a 3× molar ratio, 3× DIPEA and 1× TBTU in 500 µL DCM starting at 4 °C. The reaction was monitored at 2 h by HPLC-MS and allowed to proceed a total 24 h with the products analyzed by HPLC-MS. For HPLC-MS analysis a small aliquot was removed from the reaction mixture, the solvent dried off, and then reconstituted in methanol for analysis. The reaction was then repeated with 3× DIPEA, 10× of the side chain modification molecule, and 2× of the coupling agent DPPA in DMF, starting at 4 °C and monitored at 24 and 36 h. The protocol was modified and optimized by returning to 3× of the desired side chain modification molecule and while substituting the coupling agents COMU or PyBOP and the solvent DCM or DMF in different combinations. The reactions were checked at 24 and 48 h. After the reactions were completed the solvent was removed and the product mixture was reconstituted in methanol. The mixture was analyzed by HPLC-MS using a combination of the mass and UV absorbance to identify the elution peaks corresponding to the expected products. HPLC was then used to isolate the individual compounds. The organic solvents were removed from the samples followed by the freeze drying of the compounds. To scale up the production of the compounds showing favorable in vitro results 10 mg of SA was reacted with 3× of the side chain molecule, 3× DIPEA, 2× PyBOP in DCM, starting at 4 °C, for 24 h. The solvent was removed and the products were reconstituted in methanol and isolated by HPLC. The side chain modification molecules included methyl hydrazine (1), N,N-dimethylethylenediamine (2), cystamine (3), 1-amino-2-propanone (4), L-alanine methyl ester (5), L-glycine methyl ester (6), L-valine methyl ester (7), and L-cysteine methyl ester (8) (Figure 2). (Reaction products labeled as MTM SA(number of side chain modifier), ex MTM SA-(1) for the reaction product of MTM SA and methyl hydrazine). For the reactions that were scaled up with MTM SA and (5), (6), and (7) the reaction yields were 74 %, 66%, and 81%, respectively.
Figure 2.
The functionalization of MTM SA was achieved through a reaction with a primary amine containing compound. Representative compounds investigated are shown.
Structure Confirmation
The structures of the most active derivatives from the reactions with MTM SA and (5), (6), and (7) were confirmed through H1 and C13 NMR along with MALDI-TOF mass spectrometry. Mass spectrometry was performed by the University of Kentucky Mass Spectrometry Facility. 1H and 13C spectra were recorded using Agilent instruments (1H frequencies 500 MHz, corresponding 13C frequencies 125 MHz). Chemical shifts are quoted in parts per million (ppm) relative to TMS. J values are recorded in Hz.
In Vitro Cytotoxicity Assays
The cytotoxic effects of the side chain modifications of SA were investigated in order to determine whether the modifications were beneficial to the activity of the derivatives. All cytotoxicity assays were performed with A549, human non-small cell lung cancer cells. A549 cells were cultured as specified from ATCC at 37 °C, 5% CO2. The cells were added to a 96 well plate (5,000 cells/well) and permitted to attach for 24 h. After 24 h culture media were replaced with the side chain modified MTM SA derivative containing media at differing concentrations ranging from 10 µM to 1 ×10−4 µM. The cells were incubated with the drug containing media for 72 hrs total (n=8). Cell viability was determined using a resazurin assay that signifies mitochondrial metabolic activity in living cells (32). 10 µL of a 1 mM resazurin solution in phosphate buffered saline (PBS) was added to the control and analogue-treated cells at the end of the treatment period. Cell viability was determined three hours later by reading the fluorescence at 560 nm (Ex)/ 590 nm (Em). The fluorescence signals were quantified using a Spectramax M5 plate reader (Molecular Devices) equipped with a SoftMaxPro software. Cytotoxicity was determined by calculating the half maximal inhibitory concentration (IC50) of each sample.
RESULTS
Biotechnological production of MTM SA
MTM SA was successfully produced by the S. argillaceus M7W1, and isolated through a previously developed procedure (23, 31). Spores of S. argillaceus were formed by plating of the cells of the M7W1 mutant strain on R5A agar and allowing them to grow until spores were formed. Colonies of the S. argillaceus cells began to appear after incubation for two days and after three days spores were observed. The cells were allowed to incubate for one more day to allow the majority of the cells to exist as spores and then added to TSB media. Once transferred to TSB the cells grew quickly and after 24 h the TSB culture was used to inoculate multiple flasks of R5A media to produce SA. The pH of the R5A media was adjusted to pH 6.85. HPLC was used to monitor the culture for the production of SA. After 72 h the culture production was primarily composed of the end products including MTM SK and MTM SDK in addition to MTM SA, and the culture was terminated. A large portion of the MTM SA production is excreted into the culture media so once the culture process was stopped the media and the cells were separated. Celite cell binding resin was used to bind the cells and allow the culture to more easily be separated through filtration. C18 RP silica gel was able to successfully collect MTM SA from the complex culture mixture. Other compounds were also collected by the column so initial washes of 10% and 20% ACN were used to remove many of the byproducts from the sample. Further fractionation of the culture on the C18 RP column in addition to the loading and subsequent fractionation on a new, smaller C18 RP column did not aid in the isolation of MTM SA from the other MTM analogues so all compounds were eluted to together with 100% ACN to minimize dilution. There was also still some MTM SA contained inside the cells removed in the initial filtration step so the cells were lysed by sonication in methanol to release the intracellular content including all the MTM SA. Filtration removed the cellular debris, followed by the removal of the methanol. The same procedure as with the culture media then resulted in a relatively simple mixture of MTM SA along with the other MTM analogues and a few other impurities. Since the fractionation of the sample on the C18 RP silica gel did not completely isolate MTM SA from the other analogues the final purification steps were performed by using semi-preparative HPLC. The individual peak corresponding to MTM SA was collected and analyzed for purity using HPLC-MS. MTM SA was collected at >95% purity and was stored at −20 °C until further use.
Products from the Synthetic Modification of MTM SA 3-Side Chain
Once produced, MTM SA was modified chemically by taking advantage of the terminal carboxylic acid on the 3-side chain of the molecule. Several different protocols were attempted in order to discern the most favorable reaction conditions. The reactions were monitored by HPLC and the ratio of the signal corresponding to MTM SA was compared to the appearance of any new peaks following the reaction. MTM SA eluted with a retention time of 16.5 min, showing absorption of ~410 nm and a mass to charge ratio of m/z 1026. The initial reaction with (1) and TBTU as a coupling agent in DCM only displayed SA as the major component of the mixture after 2 h with only very minor other peaks appearing. After 24 h the MTM SA peak was gone and several broad peaks with shorter retention times appeared. The initial attempt to separate the products by ethyl acetate extraction did not result in favorable results and thus was not repeated. The reaction was then repeated with methyl hydrazine with the DPPA coupling agent in DMF. The reaction was also performed with (4) under the same conditions. After 24 h a new peak at 14.0 min appeared equal in intensity to that of the MTM SA peak. The expected product had a m/z of 1054.4, the same as the 14.0 min peak of 1054.4. The absorption of the peak was shifted however to ~440 nm. The reaction was then repeated using the coupling agent COMU in DMF with the side chains (6) and N,N-dimethylethylenediamine. For the reaction with (2) a new product appeared with a retention time of 11.25 min, a UV-Vis absorption of ~410 nm and a m/z matching that of the calculated product of 1096.6 (1096.5 expected). For the reaction with (6) three new products were formed with retention times of 14.4, 15.6, and 17.0 min. The UV-Vis absorption for the peaks were ~455 nm, 455 nm, and 410 nm with a m/z of 1097.3, 1168.4, and 1097.3 m/z, respectively. The calculated m/z for the correct product of the reaction with (6) is 1097.4 m/z. There was still also a small peak corresponding to MTM SA. The reaction of MTM SA with (3) was performed in DMF with COMU and resulted in the formation of a new peak at 9.0 min with absorption of ~455 nm and m/z 1160.6. This matches the theoretical m/z of the expected product but the UV/vis absorption is shifted. The reaction solvent was then switched to DCM and (5) was used as the side chain modification molecule. This reaction resulted in the formation of one new peak with a retention time of 18 min at roughly equal intensity to that of the MTM SA peak and a UV-Vis of ~410 nm and a m/z of 1111.3. The calculated m/z of the expected molecule was 1111.4 m/z. The same conditions were used with (7) as well which resulted in a new molecule with a retention time of 20.6 min, a UV-Vis absorption of ~410 nm and a m/z of 1139.5. The peak was the major product compared to the SA peak. The calculated m/z of the expected product was 1139.5. The reactions with (6) and (2) were then repeated with DCM as the solvent and PyBOP as the coupling agent. These reactions resulted in the formation of one new product with a retention time of 11.0 min for the (2) reaction and 17.0 min for the (6) reaction. Both products had an UV-Vis absorption of ~410 nm and the m/z of the expected products identified previously. The individual peaks were collected from HPLC, the ACN removed, and freeze dried for use with the in vitro cytotoxicity assays.
Structure Confirmation by NMR
The structures of MTM SA-(6), MTM SA-(5), and MTM SA-(7) were confirmed by H1 and C13 NMR and are summarized in Table 1. The mass of the derivatives was also confirmed by mass spectrometry. For MTM SA-(6) the expected mass + Na was 1120.45 and the observed mass was 1120.45, the expected MTM SA-(5) product had a calculated mass + Na of 1134.47 and the observed mass was 1134.47, and the expected MTM SA-(7) product had a calculated mass of 1162.50 with an observed mass was 1162.50.
Table 1.
Summary of the 1H and 13C NMR Data
| MTM SA-Glycine methyl ester | |||
|---|---|---|---|
| Position | δH (J in Hz) | δC, mult. | HMBC |
| 1 | 216.2, C | ||
| 2 | 4.36 (bs, 1H) | 78.7, CH | |
| 3 | 2.44 – 2.58 (m, 2H) | 45.2, CH | |
| 4 | 2.44 – 2.58 (m, 2H) 2.82, d (11.0) |
28.1, CH2 | |
| 4a | 136.9, C | ||
| 5 | 6.22, s | 102.0, CH | |
| 6 | 159.8, C | ||
| 7 | 111.6, C | ||
| 7-CH3 | 2.10, bs | 8.9, CH3 | |
| 8 | 156.7, C | ||
| 8a | 108.6, C | ||
| 9 | 164.1, C | ||
| 9a | 109.1, C | ||
| 10 | 6.48, s | 118.2, CH | 102.0, 108.6, 109.1 |
| 10a | 139.5, C | ||
| 1' | 4.13 (bs, 2H) | 81.2, CH | 28.1, 45.2, 60.4, 175.2 |
| 1'-OCH3 | 3.56 (bs, 4H) | 60.4, CH3 | 81.2 |
| 2' | 175.2, C | ||
| 4' | 3.99, d (17.5) 4.13 (bs, 2H) |
41.8, CH | 175.2 |
| 5' | 171.9, C | ||
| 5'-OCH3 | 3.80 (bs, 5H) | 52.9, CH3 | 171.9 |
| 1A | 4.87 (overlap, 1H) | 97.3, CH | |
| 2A | 1.74 – 1.82 (m, 2H) 2.36, bs | 38.3, CH2 | 100.0 |
| 3A | 3.80 (bs, 5H) | 80.9, CH | |
| 4A | 3.07, t (8.5), 2H | 76.5, CH | 73.3, 80.9 |
| 5A | 3.36 – 3.48 (m, 3H) | 73.3, CH | |
| 6A | 1.32, d (6.0), 2H | 18.8, CH3 | 73.3, 76.5 |
| 1B | 4.63 – 4.80 (m, 2H) | 100.0, CH | |
| 2B | 1.53 – 1.68 (m, 3H) 2.22 (m, 1H) |
40.9, CH2 | |
| 3B | 3.56 (bs, 4H) | 72.2, CH | |
| 4B | 2.98, t (8.8) | 78.2, CH | 18.4, 72.2, 73.9 |
| 5B | 3.36 – 3.48 (m, 3H) | 73.9, CH | |
| 6B | 1.35, d (6.0) | 18.4, CH3 | 73.9, 78.2 |
| 1C | 4.97 – 5.15 (m, 2H) | 102.5, CH | |
| 2C | 1.53 – 1.68 (m, 3H) 2.62, d (7.5) |
38.4, CH2 | |
| 3C | 3.80 (bs, 5H) | 81.2, CH | |
| 4C | 3.07, t (8.5), 2H | 76.7, CH | 81.2 |
| 5C | 3.36 – 3.48 (m, 3H) | 73.6, CH | |
| 6C | 1.42, bs | 18.9, CH3 | 73.6 |
| 1D | 4.63 – 4.80 (m, 2H) | 100.3, CH | |
| 2D | 1.74 – 1.82 (m, 2H) 1.89 – 2.01 (m, 2H) |
33.3, CH2 | |
| 3D | 3.89, d (11.5) | 77.5, CH | |
| 4D | 3.71 (bs, 2H) | 70.6, CH | 77.5 |
| 5D | 3.63 – 3.68 (m, 1H) | 72.2, CH | |
| 6D | 1.32, d (6.0), 2H | 17.3, CH3 | 70.6, 72.2 |
| 1E | 4.97 – 5.15 (m, 2H) | 99.1, CH | 77.5 |
| 2E | 1.53 – 1.68 (m, 3H) 1.89 – 2.01 (m, 2H) |
45.4, CH2 | 99.1 |
| 71.9, 78.1 | |||
| 3E | 71.9, C | ||
| 3E-CH3 | 1.25, s | 27.4, CH3 | 45.4, 71.9, 72.0, 78.1 |
| 4E | 2.93, d (9.5) | 78.1, CH | 18.9, 72.0 |
| 5E | 3.71 (bs, 2H) | 72.0, CH | |
| 6E | 1.27, d (6.5) | 18.9, CH | 71.9, 72.0, 78.1 |
| MTM SA-alanine methyl ester | |||
|---|---|---|---|
| Position | δH (J in Hz) | δC, mult. | HMBC |
| 1 | 204.2, C | ||
| 2 | 4.42, d (11.5) | 78.5, CH | |
| 3 | 2.45 – 2.56 (m, 2H) | 45.0, CH | |
| 4 | 2.45 – 2.56 (m, 2H) 2.60 – 2.73 (m, 1H) |
28.3, CH2 | |
| 4a | 136.4, C | ||
| 5 | 6.34, s | 102.1, CH | 108.5, 111.8, 118.1 |
| 6 | 160.1, C | ||
| 7 | 111.8, C | ||
| 7-CH3 | 2.09, s | 8.9, CH3 | |
| 8 | 156.7, C | ||
| 8a | 108.5, C | ||
| 9 | 164.9, C | ||
| 9a | 109.0, C | ||
| 10 | 6.45, s | 118.1, CH | 28.3, 102.1, 108.5, 109.0 |
| 10a | 139.6, C | ||
| 1' | 4.13, s | 81.0, CH | 28.3, 45.0, 60.4, 78.5, 174.1 |
| 1'-OCH3 | 3.59 (bs, 4H) | 60.4, CH3 | 81.0 |
| 2' | 174.1, C | ||
| 4' | 4.56, m (7.0) | 49.8, CH | 17.8, 174.1, 174.4 |
| 4'-CH3 | 1.52, d (7.0) | 17.8, CH3 | 49.8, 174.4 |
| 5' | 174.4, C | ||
| 5'-OCH3 | 3.75, s | 53.1, CH3 | 174.4 |
| 1A | 4.94 – 5.13 (m, 3H) | 97.6, CH | |
| 2A | 1.75 – 1.87 (m, 2H) 2.36 – 2.45 (m, 1H) |
38.2, CH2 | 100.1 |
| 3A | 3.77 – 3.84 (m, 2H) | 81.0, CH | |
| 4A | 3.03 – 3.12 (m, 2H) | 76.5, CH | 18.8, 73.4, 81.0 |
| 5A | 3.44 – 3.52 (m, 1H) | 73.4, CH | |
| 6A | 1.33, d (6.5) | 18.8, CH3 | 73.4, 76.5 |
| 1B | 4.65 – 4.78 (m, 2H) | 100.1, CH | |
| 2B | 1.55 – 1.66 (m, 3H) 2.18 – 2.26 (m, 1H) |
40.9, CH2 | |
| 72.2, 78.3 | |||
| 3B | 3.59 (bs, 4H) | 72.2, CH | |
| 4B | 2.98, t (9.0) | 78.3, CH | 18.4, 72.2, 73.8 |
| 5B | 3.36 – 3.44 (m, 2H) | 73.8, CH | |
| 6B | 1.34, d (6.5) | 18.4, CH3 | 73.8, 78.3 |
| 1C | 4.94 – 5.13 (m, 3H) | 102.4, CH | |
| 2C | 1.55 – 1.66 (m, 3H) 2.60 – 2.73 (m, 1H) |
38.5, CH2 | 100.3 |
| 3C | 3.77 – 3.84 (m, 2H) | 81.2, CH | |
| 4C | 3.03 – 3.12 (m, 2H) | 76.7, CH | 19.0, 73.6, 81.2 |
| 5C | 3.36 – 3.44 (m, 2H) | 73.6, CH | |
| 6C | 1.39, d (5.5) | 19.0, CH3 | 73.6, 76.7 |
| 1D | 4.65 – 4.78 (m, 2H) | 100.3, CH | 81.2 |
| 2D | 1.75 – 1.87 (m, 2H) 1.96 – 2.02 (m, 1H) |
33.3, CH2 | |
| 3D | 3.86 – 3.93 (m, 1H) | 77.4, CH | |
| 4D | 3.68 – 3.72 (bs, 2H) | 70.6, CH | 17.3, 33.3, 77.4 |
| 5D | 3.65 (m, 1H) | 72.2, CH | 70.6 |
| 6D | 1.31, d (6.5) | 17.3, CH3 | 70.6 |
| 1E | 4.94 – 5.13 (m, 3H) | 99.1, CH | 77.4 |
| 2E | 1.55 – 1.66 (m, 3H) 1.93, d (13.0) |
45.4, CH2 | 99.1 |
| 71.9, 78.1, 99.1 | |||
| 3E | 71.9, C | ||
| 3E-CH3 | 1.25, s | 27.4, CH3 | 45.4, 71.9, 78.1, 99.1 |
| 4E | 2.93, d (10.0) | 78.1, CH | 18.9, 27.4, 71.9, 72.0 |
| 5E | 3.68 – 3.72 (bs, 2H) | 72.0, CH | |
| 6E | 1.27, d (6.5) | 18.9, CH | 72.0, 78.1 |
| MTM SA-valine methyl ester | |||
|---|---|---|---|
| Position | δH (J in Hz) | δC, mult. | HMBC |
| 1 | 202.8, C | ||
| 2 | 4.53, d (10.0) | 78.2, CH | |
| 3 | 2.37 – 2.58 (m, 3H) | 45.0, CH | |
| 4 | 2.37 – 2.58 (m, 3H) 2.81 – 2.90 (m, 1H) |
28.5, CH2 | |
| 4a | 136.3 C | ||
| 5 | 6.42 (bs, 2H) | 102.0, CH | |
| 6 | 160.2, C | ||
| 7 | 111.9, C | ||
| 7-CH3 | 2.08 (bs, 3H) | 8.9, CH3 | |
| 8 | 156.8, C | ||
| 8a | 108.7, C | ||
| 9 | 150.5, C | ||
| 9a | 109.0, C | ||
| 10 | 6.42 (bs, 2H) | 118.0, CH | |
| 10a | 139.6, C | ||
| 1' | 4.17, s | 81.3, CH | 28.5, 45.0, 60.4 |
| 1'-OCH3 | 3.60 (bs, 5H) | 60.4, CH3 | 81.3 |
| 2' | 174.3, C | ||
| 4' | 4.44, d (3.5) | 59.1, CH | 173.4 |
| 5' | 173.4, C | ||
| 5'-OCH3 | 3.77 (bs, 5H) | 52.9, CH3 | 173.4 |
| 6’ | 2.32 (m, 1H) | 31.8, CH | 19.2, 20.1, 59.1 |
| 7’ | 1.05, d (6.5) | 20.1, CH3 | 19.2, 31.8, 59.1 |
| 8’ | 1.05, d (6.5) | 19.2, CH3 | 20.1, 31.8, 59.1 |
| 1A | 5.17, d (7.0) | 97.6, CH | |
| 2A | 1.74 – 1.88 (m, 2H) 2.37 – 2.58 (m, 3H) |
38.2, CH2 | |
| 3A | 3.77 (bs, 5H) | 81.0, CH | |
| 4A | 3.03 – 3.13 (m, 2H) | 76.5, CH | 18.8 |
| 5A | 3.47 – 3.53 (m, 1H) | 73.4, CH | |
| 6A | 1.26 – 1.44 (m, 5H) | 18.8, CH3 | 73.4, 76.5 |
| 1B | 4.65 – 4.75 (m, 2H) | 100.2, CH | |
| 2B | 1.52 – 1.65 (m, 3H) 2.17 – 2.24 (m, 1H) |
40.9, CH2 | |
| 3B | 3.60 (bs, 5H) | 72.2, CH | |
| 4B | 2.97, t (9.3) | 78.3, CH | 18.3, 72.2, 73.8 |
| 5B | 3.34 – 3.42 (m, 2H) | 73.8, CH | |
| 6B | 1.26 – 1.44 (m, 5H) | 18.3, CH3 | 73.8, 78.3 |
| 1C | 5.06 (bs, 1H) | 102.2, CH | |
| 2C | 1.52 – 1.65 (m, 3H) 2.58 – 2.67 (m, 1H) |
38.4, CH2 | |
| 3C | 3.77 (bs, 5H) | 81.2, CH | |
| 4C | 3.03 – 3.13 (m, 2H) | 76.7, CH | 18.8, 73.6 |
| 5C | 3.34 – 3.42 (m, 2H) | 73.6, CH | |
| 6C | 1.26 – 1.44 (m, 5H) | 18.8, CH3 | |
| 1D | 4.65 – 4.75 (m, 2H) | 100.2, CH | |
| 2D | 1.74 – 1.88 (m, 2H) 1.95 – 2.02 (m, 1H) |
33.3, CH2 | |
| 3D | 3.85 – 3.93 (m, 1H) | 77.5, CH | |
| 4D | 3.68 – 3.73 (bs, 2H) | 70.6, CH | |
| 5D | 3.60 (bs, 5H) | 72.2, CH | |
| 6D | 1.26 – 1.44 (m, 5H) | 17.3, CH3 | 70.6 |
| 1E | 4.99, d (9.0) | 99.1, CH | |
| 2E | 1.52 – 1.65 (m, 3H) 1.92, d (13.0) |
45.4, CH2 | 99.1 |
| 71.9, 78.1 | |||
| 3E | 71.9, C | ||
| 3E-CH3 | 1.25, s | 27.4, CH3 | 45.4, 78.1, 99.1 |
| 4E | 2.93, d (9.5) | 78.1, CH | 18.9, 72.0 |
| 5E | 3.68 – 3.73 (bs, 2H) | 72.0, CH | |
| 6E | 1.26 – 1.44 (m, 5H) | 18.9, CH | 72.0, 78.1 |
In Vitro Cell Toxicity Assays
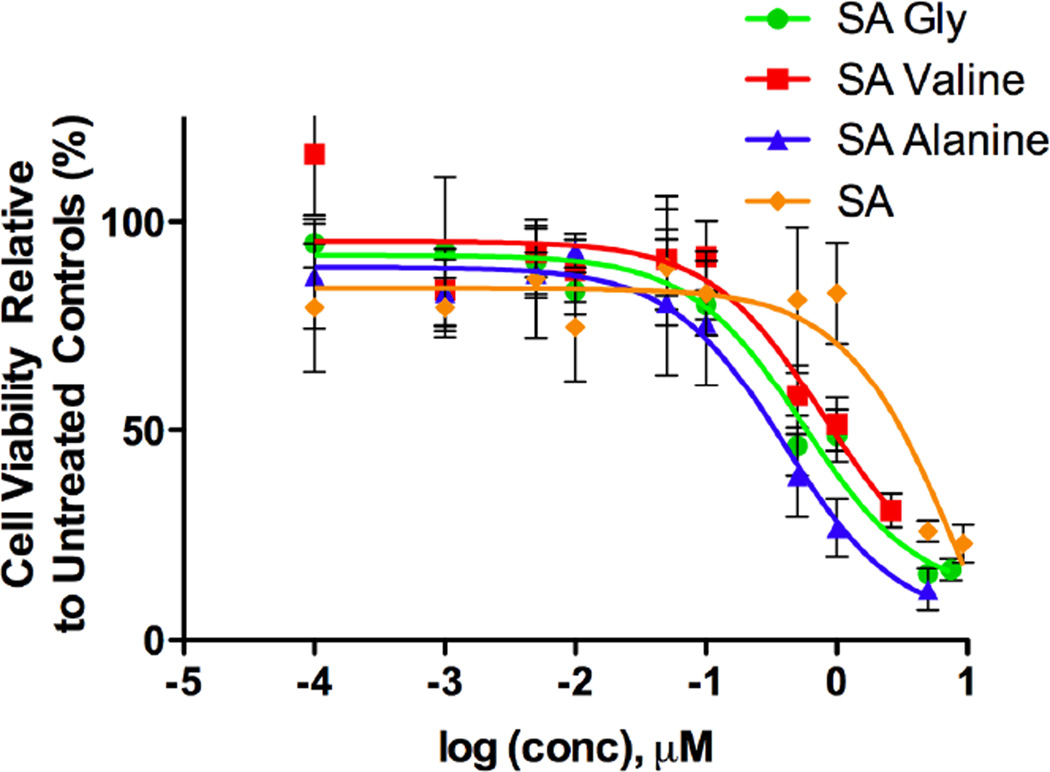
The cytotoxicity of the MTM SA analogues was tested against the A549 non-small cell lung cancer cell line (Figure 3). Each analogue was checked individually with a range of concentrations to determine the IC50 of the molecule. For comparison sake the cytotoxicity of MTM SA, and MTM SK, an analogue discovered previously and found to be more active than the regular MTM were also investigated (23). MTM SA showed an IC50 of 8.7 ± 0.37 µM, and MTM SK an IC50 of 0.28 µM. The IC50 of the synthetically modified MTM SA analogues are as follows: MTM SA-(1): 8.8 ± 1.69 µM; MTM SA-(2): 2.7 ± 0.22 µM; MTM SA-(3): N/A; MTM-SA-(4): N/A; MTM SA-(8): 1.0 ± 0.19 µM; MTM SA-(6): 0.55 ± 0.11 µM; MTM SA-(5): 0.36 ± 0.12 µM; MTM SA-(7): 0.80 ± 0.20 µM (Table 2). MTM-SA-(3) and MTM-SA-(4) did not show significant cytotoxicity.
Figure 3.
The in vitro cytotoxicity assays are shown of the three most active MTM SA derivatives compared to MTM SA itself.
Table 2.
The calculated IC50 values of the derivatives in vitro against the A549 cell line.
| Side Chain Modifying Agent + MTM SA | IC50 (µM) |
|---|---|
| MTM SK | 0.28 ± 0.11 |
| MTM SA | 8.7 ± 0.37 |
| Methyl hydrazine (1) | 8.8 ± 1.69 |
| N,N-dimethylethylenediamine (2) | 2.7 ± 0.22 |
| Cystamine (3) | N/A |
| 1-amino-2-propanone (4) | N/A |
| L-alanine methyl ester (5) | 0.36 ± 0.12 |
| L-glycine methyl ester (6) | 0.55 ± 0.11 |
| L-valine methyl ester (7) | 0.80 ± 0.20 |
| L-cysteine methyl ester (8) | 1.0 ± 0.19 |
DISCUSSION
The biosynthesis of MTM SK and MTM SDK is accomplished through a genetically engineered S. argillaceus strain, M7W1, which contains an inactivated mtmW gene coding for the MtmW enzyme. Both the MTM SK and MTM SDK analogues have been shown to be improved analogues compared to the parent MTM compound, thus it would be optimal if these were the only two compounds produced by the M7W1 strain (23, 24). However, this is not the case, and two other major compounds are produced alongside of MTM SK and MTM SDK (24). One of these compounds, MTM SA, has previously been regarded as not useful due to the lack of biological activity compared to the parent compound (26, 27). This is unfortunate as MTM SA is produced in many fermentations in higher amounts than MTM SK or MTM SDK, and the production yield can be shifted even further in favor of the production of MTM SA by altering the pH of the culture media (33). Since MTM SK and MTM SDK are separated chromatographically MTM SA is easily collected and isolated alongside MTM SK and MTM SDK during the normal isolation procedure.
In an effort to develop a useful molecule from the previously unfortunate side product, the 3-side chain of the molecule was targeted (23). It is known that the 3-side chain of the MTM structure is responsible for an interaction with the DNA-phosphate backbone (23). Thus by altering the functionality of the 3-side chain we hoped to enhance and/or increase the specificity for the DNA of diseased cells. The 3-side chain of MTM SA is terminated by a carboxyl acid functional group which is likely ionized at a physiological pH, repulsing from the negative charge of the DNA-phosphate backbone thereby weakening MTM SA’s ability to bind to DNA. Side chain functionalizations were rationally selected to contain amine residues capable of positively charged ionization in order to enhance the interaction with DNA-phosphate backbone. Both MTM SK and MTM SDK have shorter side chains than MTM. It was unclear whether modifications adding an N-atom but also chain length would have an overall positive effect. To test this several different side chain modification were investigated with differing lengths and additional functionalities (Figure 2).
Various reaction conditions were examined before discovering the optimal protocol. The mass, UV absorptions and retention times of the products separated by HPLC were used to identify the products. Initial reactions were performed in DMF with the different coupling agents. These reactions tended to result in multiple product peaks. The products typically had the expected mass or the expected mass plus one additional side chain. MTM, MTM SK, and MTM SDK have a chromophore which absorbs light at roughly 410 nm. The UV-absorption maxima of the different products was typically either ~410 nm as expected, or near 450 nm range, which was not expected. The products that had a mass corresponding to two side chains being attached also showed the ~450 nm absorption. This led us to believe that a second side chain was being attached somewhere on or in the proximity of the chromophore, although it was not clear as to exactly where or how this was happening. It could be possible that DMF and excess side chain promoted conditions capable of allowing the carbonyl group of the A-ring to be attacked by the amine side chain, however this was not confirmed. These reactions also typically had one product that did match the expected mass and UV absorption profile although it was not always the major product. Once DCM was substituted as the reaction solvent the number of reaction products was typically reduced to one with the correct mass and UV. The retention times also matched with the product from the DMF reaction also having the correct mass and UV absorption. PyBOP outperformed the other coupling agents (COMU, DPPA, and TBTU) in terms of product yield and reaction time. With COMU, DPPA, and TBTU there was unreacted MTM SA left in the product mixture even with excess side chain molecules and coupling reagents. However when PyBOP was used in combination with DCM only minute traces of MTM SA were observed in the production chromatograph. Initially reactions were allowed to proceed for up to 48 h before stopping the reaction, which still contained unreacted MTM SA at the time of reaction termination. However when PyBOP was used reactions were complete within 12 h. All initial reactions were done on a small scale of only 2 mg MTM SA in order to optimize the reaction conditions without wasting precious reagents. For these reasons DCM and PyBOP were chosen as the optimal solvent and coupling reagent for the subsequent reactions and scaled up production.
To test the effectiveness of the side chain modifications cytotoxicity assays were performed against the human non-small cell lung cancer cell line (A549). The least effective side chain modification was with (3) as it eliminated all cytotoxicity effects against the cell line. The modification of the side chain with (1) did not improve the cytotoxicity of the molecule compared to MTM SA, but it should be noted that it also did not decrease the activity either. A modest 3–4 fold increase in cytotoxicity activity of was observed with the (2) functionalization. The increase in activity was still well below the value of MTM SK. The modification of the side chain with O-methyl protected amino acids residues yielded much better results. L-Ala showed the greatest activity with >23-fold improvement over MTM SA and an IC50 value very near to that of MTM SK. L-Gly and L-Val also showed promising results with a 15- and 10-fold improvement, respectively. L-Cys did also show an 8-fold improvement over MTM SA.
The cytotoxicity improvement with L-Cys is not as impressive as some of the other analogues, however this and similar modifications have the potential to be of value for a different reason. Producing a potent anti-cancer molecule is only half of the battle in the fight of cancer. The other half is being able to get the drug to the area of interest in high enough concentrations to be effective without damaging the rest of the body and healthy tissues. We have reported previously the development of a nanoparticle delivery system compatible with MTM SK and MTM SDK to complement the potency of these molecules and enhance the cytotoxicity (31). The unique thiol group introduced with the L-Cys side chain offers the potential to load this molecule through a disulfide bond into a compatible delivery system. This bond would be stable throughout the blood stream with low glutathione concentrations, however once the delivery system was internalized by the cancer cells and the system was exposed to the increased intracellular glutathione concentrations (up to the millimolar concentration) which would reduce the disulfide bond and release the L-Cys functionalized MTM analogue. Several drug delivery systems have previously shown the ability to take advantage of the increased intracellular glutathione concentration to modulate drug release (34, 35). The fact that less drug would be circulating systemically and interacting with normal tissue may allow a higher dose of drug to be administered without affecting the normal tissue and overcome the lower cytotoxicity of the molecule compared to MTM SK. The L-Cys modified MTM SA is still a very active compound compared to other anti-cancer agents.
While working on the modifications of MTM SA, Preobrazhenskaya et al. published the side chain derivatives of another aureolic acid antibiotic, olivomycin A (36). Here the side chain of olivomycin A was chemically shortened, to achieve olivomycin SA, and additionally functionalized as amides with selected amines. The authors observed favorable results when the modifications of the side chain were achieved using (2) and O-methyl alanine. As described above, the latter modification also improved MTM SA. However, the reported methods are not compatible with MTM, because an oxidative sodium periodate cleavage was used to cut the side chain to olivomycin SA. Applying that periodate cleavage to MTM would also destroy the two terminal sugar residues (sugars B and E), thereby interfering with two structural elements important for the MTM bioactivity. We circumvented the problem by using biotechnologically produced MTM SA followed by synthetic modifications similar to those reported by Preobrazhenskaya et al (29).
CONCLUSIONS
Natural products are not always optimized for human purposes. Combining biosynthetic derivatization with chemical synthesis offers to produce unique molecules unattainable by either method individually. Here we combined these methods to modify the previously useless (since biologically inactive) MTM SA that is accumulated alongside the biologically improved MTM analogues MTM SK and MTM SDK. The latter two molecules, which are both considerably more active and significantly less toxic than the natural product MTM itself, pointed the way that 3-side chain modifications can be advantageous. The modification of the 3-side chain of MTM SA with amino acid derivatives yielded several active compounds with the O -methyl-alanine showing the most potent activity. Although none of the so far synthesized MTM SA derivatives are as active as MTM SK or MTM SDK, the O-methyl-alanine and the O-methyl-glycine derivatives show activities comparable to MTM SK, and are clearly improved derivatives compared to the rather inactive MTM SA. Furthermore, this type of modification also allows the incorporation of important drug loading moieties for combination with specialized drug delivery systems. It is envisioned that the modified MTM SA analogues will give us more insight into the MTM mode of action and allow us to better tailor future generations of drug molecules towards more effective anti-cancer drugs.
ACKNOWLEDGEMENTS
This research was supported in part by the National Institues of Health (grant CA 091901 to JR) and the Kentucky Lung Cancer Research Program (to YB). DS acknowledges the financial support from a NCI-CNTC postdoctoral traineeship and the project described was supported by Grant Number 5R25CA153954 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Contributor Information
Daniel Scott, Email: Daniel.scott@uky.edu.
Jhong-Min Chen, Email: jch249@g.uky.edu.
Younsoo Bae, Email: younsoo.bae@uky.edu.
Jürgen Rohr, Email: jrohr2@email.uky.edu.
REFERENCES
- 1.Cragg GM, Newman DJ, Snader KM. Natural products in drug discovery and development. J Nat Prod. 1997;60:52–60. doi: 10.1021/np9604893. [DOI] [PubMed] [Google Scholar]
- 2.Newman DJ, Cragg GM, Snader KM. Natural products as sources of new drugs over the period 1981–2002. J Nat Prod. 2003;66:1022–1037. doi: 10.1021/np030096l. [DOI] [PubMed] [Google Scholar]
- 3.Newman DJ, Cragg GM. Natural Products As Sources of New Drugs over the 30 Years from 1981 to 2010. Journal of Natural Products. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller DM, Polansky DA, Thomas SD, Ray R, Campbell VW, Sanchez J, et al. Mithramycin selectively inhibits transcription of G-C containing DNA. Am J Med Sci. 1987;294:388–394. doi: 10.1097/00000441-198711000-00015. [DOI] [PubMed] [Google Scholar]
- 5.Sastry M, Fiala R, Patel DJ. Solution structure of mithramycin dimers bound to partially overlapping sites on DNA. J Mol Biol. 1995;251:674–689. doi: 10.1006/jmbi.1995.0464. [DOI] [PubMed] [Google Scholar]
- 6.Sastry M, Patel DJ. Solution structure of the mithramycin dimer-DNA complex. Biochemistry. 1993;32:6588–6604. doi: 10.1021/bi00077a012. [DOI] [PubMed] [Google Scholar]
- 7.Blume SW, Snyder RC, Ray R, Thomas S, Koller CA, Miller DM. Mithramycin inhibits SP1 binding and selectively inhibits transcriptional activity of the dihydrofolate reductase gene in vitro and in vivo. J Clin Invest. 1991;88:1613–1621. doi: 10.1172/JCI115474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Safe S, Abdelrahim M. Sp transcription factor family and its role in cancer. Eur J Cancer. 2005;41:2438–2448. doi: 10.1016/j.ejca.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 9.Abdelrahim M, Smith R, 3rd, Burghardt R, Safe S. Role of Sp proteins in regulation of vascular endothelial growth factor expression and proliferation of pancreatic cancer cells. Cancer Res. 2004;64:6740–6749. doi: 10.1158/0008-5472.CAN-04-0713. [DOI] [PubMed] [Google Scholar]
- 10.Black AR, Black JD, Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 11.Kennedy BJ. Metabolic and toxic effects of mithramycin during tumor therapy. The American Journal of Medicine. 1970;49:494–503. doi: 10.1016/s0002-9343(70)80044-4. [DOI] [PubMed] [Google Scholar]
- 12.Brown JH, Kennedy BJ. Mithramycin in the Treatment of Disseminated Testicular Neoplasms. N Engl J Med. 1965;272:111–118. doi: 10.1056/NEJM196501212720301. [DOI] [PubMed] [Google Scholar]
- 13.Koller CA, Miller DM. Preliminary observations on the therapy of the myeloid blast phase of chronic granulocytic leukemia with plicamycin and hydroxyurea. N Engl J Med. 1986;315:1433–1438. doi: 10.1056/NEJM198612043152301. [DOI] [PubMed] [Google Scholar]
- 14.Ryan WG. Mithramycin for Paget's disease of bone. N Engl J Med. 1970;283:1171. doi: 10.1056/nejm197011192832120. [DOI] [PubMed] [Google Scholar]
- 15.Ryan WG, Schwartz TB, Northrop G. Experiences in the treatment of Paget's disease of bone with mithramycin. JAMA. 1970;213:1153–1157. [PubMed] [Google Scholar]
- 16.Jin H-J, Kanthasamy A, Anantharam V, Rana A, Kanthasamy AG. Transcriptional Regulation of Pro-apoptotic Protein Kinase Cδ: Implications for Oxidative Stress-induced Neuronal Cell Death. J Biol Chem. 2011;286:19840–19859. doi: 10.1074/jbc.M110.203687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seznec J, Silkenstedt B, Naumann U. Therapeutic effects of the Sp1 inhibitor mithramycin A in glioblastoma. J Neuro-Oncol. 2011;101:365–377. doi: 10.1007/s11060-010-0266-x. [DOI] [PubMed] [Google Scholar]
- 18.Sleiman SF, Langley BC, Basso M, Berlin J, Xia L, Payappilly JB, et al. Mithramycin is a gene-selective Sp1 inhibitor that identifies a biological intersection between cancer and neurodegeneration. J Neurosci. 2011;31:6858–6870. doi: 10.1523/JNEUROSCI.0710-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao Y, Jia Z, Kong X, Li Q, Chang DZ, Wei D, et al. Combining Betulinic Acid and Mithramycin A Effectively Suppresses Pancreatic Cancer by Inhibiting Proliferation, Invasion, and Angiogenesis. Cancer Res. 2011;71:5182–5193. doi: 10.1158/0008-5472.CAN-10-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tagashira M, Kitagawa T, Isonishi S, Okamoto A, Ochiai K, Ohtake Y. Mithramycin represses MDR1 gene expression in vitro, modulating multidrug resistance. Biol Pharm Bull. 2000;23:926–929. doi: 10.1248/bpb.23.926. [DOI] [PubMed] [Google Scholar]
- 21.Tagashira M, Kitagawa T, Nozato N, Isonishi S, Okamoto A, Ochiai K, et al. Two novel C-glycosides of aureolic acid repress transcription of the MDR1 gene. Chem Pharm Bull (Tokyo) 2000;48:575–578. doi: 10.1248/cpb.48.575. [DOI] [PubMed] [Google Scholar]
- 22.Grohar PJ, Woldemichael GM, Griffin LB, Mendoza A, Chen Q-R, Yeung C, et al. Identification of an Inhibitor of the EWS-FLI1 Oncogenic Transcription Factor by High-Throughput Screening. J Natl Cancer Inst. 2011;103:962–978. doi: 10.1093/jnci/djr156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albertini V, Jain A, Vignati S, Napoli S, Rinaldi A, Kwee I, et al. Novel GC-rich DNA-binding compound produced by a genetically engineered mutant of the mithramycin producer Streptomyces argillaceus exhibits improved transcriptional repressor activity: implications for cancer therapy. Nucleic Acids Res. 2006;34:1721–1734. doi: 10.1093/nar/gkl063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Remsing LL, Gonzalez AM, Nur-e-Alam M, Fernandez-Lozano MJ, Brana AF, Rix U, et al. Mithramycin SK a novel antitumor drug with improved therapeutic index, mithramycin SA demycarosyl-mithramycin SK: three new products generated in the mithramycin producer Streptomyces argillaceus through combinatorial biosynthesis. J Am Chem Soc. 2003;125:5745–5753. doi: 10.1021/ja034162h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sastry M, Fiala R, Patel DJ. Solution structure of mithramycin dimers bound to partially overlapping sites on DNA. J Mol Biol. 1995;251:674–689. doi: 10.1006/jmbi.1995.0464. [DOI] [PubMed] [Google Scholar]
- 26.Remsing LL, Gonzalez AM, Nur-e-Alam M, Fernandez-Lozano MJ, Brana AF, Rix U, et al. Mithramycin SK a novel antitumor drug with improved therapeutic index, mithramycin SA demycarosyl-mithramycin SK: three new products generated in the mithramycin producer Streptomyces argillaceus through combinatorial biosynthesis. J Am Chem Soc. 2003;125:5745–5753. doi: 10.1021/ja034162h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Remsing LL, Bahadori HR, Carbone GM, McGuffie EM, Catapano CV, Rohr J. Inhibition of c-src transcription by mithramycin: structure-activity relationships of biosynthetically produced mithramycin analogues using the c-src promoter as target. Biochemistry. 2003;42:8313–8324. doi: 10.1021/bi034091z. [DOI] [PubMed] [Google Scholar]
- 28.Tevyashova AN, Olsufyeva EN, Turchin KF, Balzarini J, Bykov EE, Dezhenkova LG, et al. Reaction of the antitumor antibiotic olivomycin I with aryl diazonium salts. Synthesis, cytotoxic and antiretroviral potency of 5-aryldiazenyl-6-O-deglycosyl derivatives of olivomycin I. Bioorg Med Chem. 2009;17:4961–4967. doi: 10.1016/j.bmc.2009.05.076. [DOI] [PubMed] [Google Scholar]
- 29.Tevyashova AN, Shtil AA, Olsufyeva EN, Luzikov YN, Reznikova MI, Dezhenkova LG, et al. Modification of olivomycin A at the side chain of the aglycon yields the derivative with perspective antitumor characteristics. Bioorg Med Chem. 2011;19:7387–7393. doi: 10.1016/j.bmc.2011.10.055. [DOI] [PubMed] [Google Scholar]
- 30.Tevyashova AN, Zbarsky EN, Balzarini J, Shtil AA, Dezhenkova LG, Bukhman VM, et al. Modification of the antibiotic olivomycin I at the 2'-keto group of the side chain. Novel derivatives, antitumor and topoisomerase I-poisoning activity. J Antibiot. 2009;62:117. doi: 10.1038/ja.2008.7. [DOI] [PubMed] [Google Scholar]
- 31.Scott D, Rohr J, Bae Y. Nanoparticulate formulations of mithramycin analogs for enhanced cytotoxicity. Int J Nanomedicine. 2011;6:2757–2767. doi: 10.2147/IJN.S25427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mann CM, Markham JL. A new method for determining the minimum inhibitory concentration of essential oils. Journal of Applied Microbiology. 1998;84:538–544. doi: 10.1046/j.1365-2672.1998.00379.x. [DOI] [PubMed] [Google Scholar]
- 33.Gibson M, Nur-e-Alam M, Lipata F, Oliveira MA, Rohr J. Characterization of Kinetics and Products of the Baeyer-Villiger Oxygenase MtmOIV, The Key Enzyme of the Biosynthetic Pathway toward the Natural Product Anticancer Drug Mithramycin from Streptomyces argillaceus. J Am Chem Soc. 2005;127:17594–17595. doi: 10.1021/ja055750t. [DOI] [PubMed] [Google Scholar]
- 34.Li Y, Lokitz BS, Armes SP, McCormick CL. Synthesis of Reversible Shell Cross-Linked Micelles for Controlled Release of Bioactive Agents†. Macromolecules. 2006;39:2726–2728. [Google Scholar]
- 35.Koo AN, Lee HJ, Kim SE, Chang JH, Park C, Kim C, et al. Disulfide-cross-linked PEG-poly(amino acid)s copolymer micelles for glutathione-mediated intracellular drug delivery. Chemical Communications. 2008:6570–6572. doi: 10.1039/b815918a. [DOI] [PubMed] [Google Scholar]
- 36.Tevyashova AN, Shtil AA, Olsufyeva EN, Luzikov YN, Reznikova MI, Dezhenkova LG, et al. Modification of olivomycin A at the side chain of the aglycon yields the derivative with perspective antitumor characteristics. Bioorg Med Chem. 2011;19:7387–7393. doi: 10.1016/j.bmc.2011.10.055. [DOI] [PubMed] [Google Scholar]