Abstract
Bloom’s syndrome (BS) is an inherited disorder caused by loss of function of the recQ-like BLM helicase. It is characterized clinically by severe growth retardation and cancer predisposition. BLM localizes to PML nuclear bodies and to the nucleolus; its deficiency results in increased intra- and inter-chromosomal recombination, including hyper-recombination of rDNA repeats. Our previous work has shown that BLM facilitates RNA polymerase I-mediated rRNA transcription in the nucleolus (Grierson et al., 2012). This study uses protein co-immunoprecipitation and in vitro transcription/translation (IVTT) to identify a direct interaction of DNA topoisomerase I with the C-terminus of BLM in the nucleolus. In vitro helicase assays demonstrate that DNA topoisomerase I stimulates BLM helicase activity on a nucleolar-relevant RNA:DNA hybrid, but has an insignificant effect on BLM helicase activity on a control DNA:DNA duplex substrate. Reciprocally, BLM enhances the DNA relaxation activity of DNA topoisomerase I on supercoiled DNA substrates. Our study suggests that BLM and DNA topoisomerase I function coordinately to modulate RNA:DNA hybrid formation as well as relaxation of DNA supercoils in the context of nucleolar transcription.
Keywords: BLM helicase, DNA topoisomerase I, rRNA transcription, Bloom’s syndrome, nucleolus, RNA polymerase I
1. Introduction
Human cells in interphase contain several nucleoli, sub-nuclear structures that contain the highly repetitive ribosomal DNA (rDNA) genes mapping to the satellite regions of the acrocentric chromosomes. Nucleolar rDNA associates with the nucleolar-dedicated RNA polymerase I and numerous other proteins required for ribosome biogenesis. The predominant function of nucleoli is the transcription of ribosomal RNA (rRNA) from rDNA, occurring during S- and G2-phases of the cell cycle [1,2]. Nascent rRNAs generated during RNA polymerase I-mediated rRNA transcription have a tendency to re-associate with template rDNA and form rRNA:rDNA hybrids that can inhibit rRNA transcription and facilitate rDNA recombination (reviewed in [3]). DNA topoisomerase I, a component of the RNA polymerase I transcription complex, relaxes the negative and positive supercoiling associated with rRNA transcription and prevents the formation of inhibitory rRNA:rDNA hybrids [4-8].
Bloom’s syndrome (BS), an inherited disorder characterized by a high predisposition to cancer and severe growth retardation, is caused by loss of function of the BLM helicase [9]. BLM belongs to the conserved recQ subfamily of ATP-dependent 3′-5′ helicases [10,11]. It localizes to the nucleolus and binds rDNA [12-14]. The C-terminus of BLM is required for its nucleolar retention and rDNA binding within the 18S-coding region and the intergenic spacers (IGS) [13,14]. BLM deficiency leads to hyper-recombination within rDNA [15,16] and a reduction of overall rDNA repeat numbers in comparison to wild-type cells [13,14]. Hyper-recombination within rDNA generates extra-chromosomal rDNA circles (ERC), the accumulation of which is associated with aging in S. cerevisiae [17]. BLM-deficient cells display rDNA hyper-recombination [15,16], while some of the clinical characteristics of BS are suggestive of aging. These observations first suggested that nucleolar BLM maintains the stability of rDNA via direct binding to rDNA and implicate it in rDNA metabolism.
Our previous work demonstrated that BLM is a component of the RNA polymerase I transcription complex and unwinds RNA:DNA hybrids with 3′ overhangs of DNA [18]. It also suggested that BLM and DNA topoisomerase I may cooperatively function to limit the accumulation of rRNA:rDNA hybrids in the nucleolus. Here, we report that BLM interacts directly with DNA topoisomerase I. Protein co-immunoprecipitation from nuclear extracts and sub-fractionated nuclei from cultured cells demonstrate that this interaction occurs in nucleoli. Purified recombinant proteins co-immunoprecipitate in vitro, while in vitro transcription/translation (IVTT) coupled to immunoprecipitation demonstrates that the interaction is mediated by a domain within the C-terminus of BLM. We show using helicase assays that DNA topoisomerase I stimulates BLM helicase activity on a GC-rich rDNA-like RNA20:DNA33 duplex substrate that models a co-transcriptionally formed rRNA:rDNA hybrid, but does not do so on a DNA20:DNA33 substrate. Finally, we show that BLM stimulates the DNA relaxation activity of topoisomerase I. Our data suggest that BLM and DNA topoisomerase I interact and cooperate to promote efficient rRNA transcription by RNA polymerase I.
2. Materials and Methods
2.1 Cell lines
MCF7 and HEK 293T cells were obtained from ATCC and cultured in Dulbecco’s Modified Eagle Medium (Invitrogen) containing 10% fetal bovine serum (Hyclone). All cells were cultured at 37°C and 5% CO2.
2.2 Nucleolar isolation
Nucleoli were isolated from 293T cells according to the protocol of the Lamond Lab (www.lamondlab.com). Briefly, proliferating 293T cells were harvested by trypsinization, washed in PBS, re-suspended in buffer A (10mM HEPES, pH7.9, 10mM KCl, 1.5mM MgCl2, 0.5mM DTT) and incubated on ice for 5 min. Cell suspensions were homogenized until approximately 90% of the cells were disrupted to produce intact nuclei. Lysis was monitored by light microscopy. Homogenized suspensions were centrifuged at 218g for 5 min at 4°C, nuclear pellets re-suspended in 3ml of S1 solution (0.25M sucrose, 10mM MgCl2), layered over 3ml of S2 solution (0.38M sucrose, 0.5mM MgCl2), and centrifuged at 1430g for 5 min at 4°C. Resultant nuclear pellets were re-suspended in 3ml of S2 solution and sonicated at 4°C (Fisher Scientific Sonic Dismembrator model 500). Liberation of nucleoli was monitored by light microscopy. Resultant nucleolar suspensions were layered over 3ml of S3 solution (0.88M sucrose, 0.5mM MgCl2), centrifuged at 3000g for 10 min at 4°C and re-suspended in 500ul of S2 solution.
2.3 Protein co-immunoprecipitation
Protein co-immunoprecipitations used 293T nuclear lysates prepared according to published protocols [19] or nucleolar and nucleoplasmic lysates prepared as described above. Antibodies used in co-immunoprecipitation included α BLM (Santa Cruz Biotech, sc-7790) and α DNA topoisomerase I (Bethyl, A302-589A). Protein-antibody complexes were captured with Dynabeads Protein G (Invitrogen, 100-04D), washed, eluted and separated by 8% SDS-PAGE, and detected using standard western blotting procedures using α BLM (Bethyl Laboratories, A300-110A), α RPA194 (Santa Cruz Biotech, sc-48385) and α RNA polymerase II (Abcam, ab817).
2.4 Protein purification
pYES-BLM expression vector (pJK1) was kindly provided by I. Hickson [11]. BLM was purified as previously described [19], with an additional heparin-sepharose purification step. Briefly, hexa-histidine (6X-His)-tagged BLM was over-expressed in Saccharomyces cerevisiae. Yeast were lysed at 20k psi using a French Press Cell Disrupter (Thermo) and lysates were separated by ultracentrifugation at 65,000g for 1hr at 4°C. Cleared lysates were purified by FPLC using Ni-NTA Superflow (Qiagen), followed by Heparin-Sepharose 6 Fast Flow (Amersham Biosciences) and finally Q-Sepharose (Sigma). Purity of the resultant BLM was determined by 8% SDS-PAGE, staining of gels with SYPRO Ruby Protein Gel Stain (Sigma) and analysis using ImageQuant software. The helicase-dead mutant BLMK695E was purified by batch purification [19,20].
2.5 In vitro transcription/ translation (IVTT)
IVTT reactions were performed according to Lillard-Wetherell et al., 2004 [21]. Briefly, pET24D-BLM-N, pET24D-BLM-H and pET24D-BLM-C were used in IVTT according to manufacturer’s instructions (TNT Rabbit Reticulocyte Lysate kit, Promega). IVTT products were mixed with full-length wild-type human DNA topoisomerase I (Topogen) according to published protocols [22] and incubated for 2hrs at 4°C with rotation. Subsequently, α DNA topoisomerase I (Bethyl, A302-589A) was added with an additional 2hr incubation at 4°C. Finally, Dynabeads Protein G (Invitrogen, 100-04D) were added for 2hrs at 4°C, washed 5 times with binding buffer, eluted with 1X SDS-PAGE sample buffer, separated on 10% SDS-PAGE, dried and imaged using ImageQuant software.
2.6 Helicase assays
Oligonucleotides were purchased from Invitrogen. Oligonucleotide sequences (5′-3′ orientation) are as follows: DNA20-CGCTAGCAATATTCTGCAGC, DNA33-GCTGCAGAATATTGCTAGCGGGAATTCGGCGCG and RNA20-CGCUAGCAAUAUUCUGCAGC. RNA20 and DNA were 32 20 P end-labeled using polynucleotide kinase (PNK; NEB) according to manufacturer’s instructions. Duplex substrates were generated by heating to 95°C for 5 min and slow cooling. Helicase assays were performed as previously described [18]. Helicase products were separated on 12% non-denaturing polyacrylamide gels, dried and analyzed using ImageQuant software.
2.7 Toposiomerase I assays
DNA relaxation assays mediated by human DNA topoisomerase I (Topogen) were performed as described [23] in reaction buffer supplied by the manufacturer (10 mM Tris-HCl pH 7.9, 1 mM EDTA, 150mM NaCl, 0.1% BSA, 0.1 mM spermidine, 5% glycerol). 200ng replicative form I (RF I) DNA [24] was used in 20ul reactions containing various dilutions of human DNA topoisomerase I in the presence or absence of purified BLM or helicase-dead mutant BLMK695E [20]. The only measure of activity for BLM after purification is helicase unwinding. In the case of wild-type protein a loss of unwinding activity was observed upon dilution. Since there is no direct measure of the activity of the mutant as it is helicase-dead, for the topoisomerase I stimulation assays, higher concentrations of the mutant were used to overcome any loss of activity occurring upon dilution of protein as the reason for a potential negative result (supplementary figure 3). The mutant was diluted to the same extent as wild-type BLM. Reactions were stopped at various intervals of time (10-30 minutes) in stop buffer and analyzed on a 1.3 % agarose gel in TAE buffer as described [23]. Gels were stained by ethidium bromide and the DNA bands were quantitated by ImageJ software. The relative fold-stimulation of topoisomerase I activity by BLM was estimated by dividing the ratio of products (topoisomers) to substrate (RF I) in the presence of BLM by that in its absence for each reaction.
3. Results
3.1 BLM interacts with DNA topoisomerase I from nuclear and nucleolar-enriched extracts
Our recent study revealed a novel role for BLM in facilitating RNA polymerase I-mediated rRNA transcription via interaction with RNA polymerase I and resolution of RNA-DNA hybrid structures formed during transcription [18]. This role is consistent with localization of BLM in the nucleolus [12] and suggests its participation in nucleolar DNA metabolism and subsequent effects on ribosome biogenesis and protein production. DNA topoisomerase I is a component of the RNA polymerase I transcription complex and facilitates efficient rRNA transcription in bacteria, yeast and human cells [7,8,25] by relaxation of DNA supercoils and prevention of RNA-DNA hybrids generated during transcription. Therefore, we asked whether BLM and DNA topoisomerase I interact with each other (Fig 1A, Supplementary Figure 1a). Nuclear extracts from two cell lines, MCF7 and 293T, were used to test for co-immunoprecipitation using anti-BLM (α BLM) or anti-DNA topoisomerase I (α DNA topoisomerase I) antibodies. Figure 1A shows that each antibody immunoprecipitated both BLM and DNA topoisomerase I. Control immunoprecipitation experiments using α BLM or α DNA topoisomerase I antibodies and nuclear extracts from Bloom’s syndrome cell line GM8505 did not immunoprecipitate topoisomerase I or BLM, respectively (data not shown). These results are consistent with BLM and DNA topoisomerase I interactions and their role in a common RNA polymerase I-associated complex (see Figure 1C below)[18].
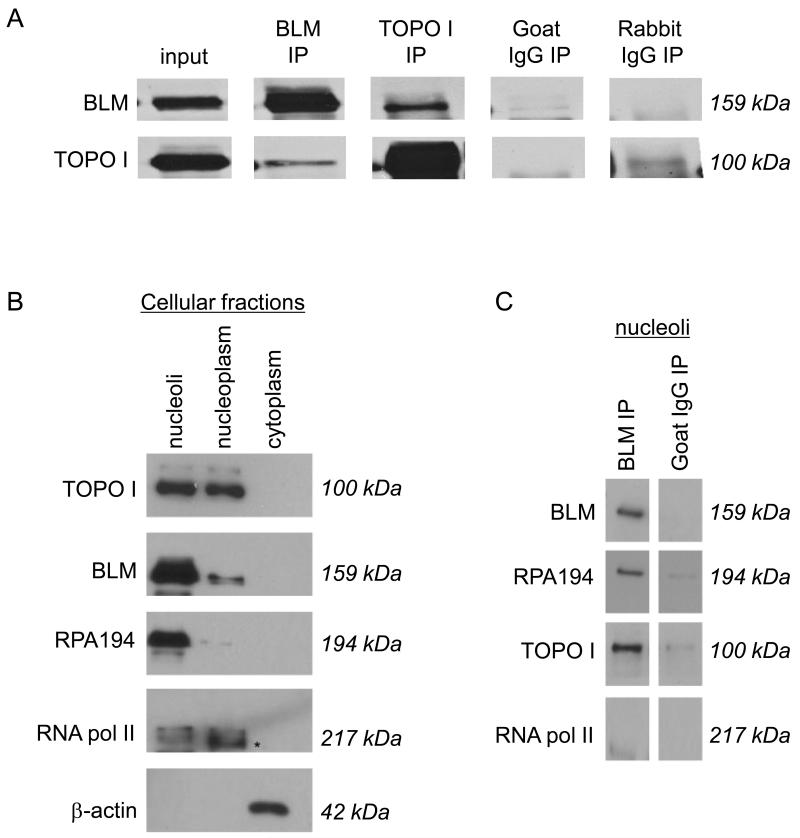
Figure 1. BLM and DNA topoisomerase I associate in mammalian cells.
(A) Co-immunoprecipitations used MCF7 and 293T nuclear extracts, and either anti-BLM or anti-DNA topoisomerase I antibodies (referred as α BLM or α DNA topoisomerase I respectively). Immunoprecipitated proteins were separated using 8% SDS-PAGE and detected by western blotting using α BLM or α DNA topoisomerase I. Goat IgG was chosen as an isotype-matched negative control for α BLM; rabbit IgG was an isotype-matched negative control for α DNA topoisomerase I. (B) 293T nuclei were sub-fractionated into nucleoli and nucleoplasm. The purity of the sub-cellular fractions is shown using the RNA polymerase I-specific subunit RPA194 as a nucleolar marker; RNA polymerase II as a nucleoplasmic marker (indicated with an *); and α -actin as a cytoplasmic marker. Proteins were separated using 8% SDS-PAGE and detected by western blotting using the respective antibodies as indicated. (C) Nucleolar fractions were used in protein co-immunoprecipitation with α BLM. Immunoprecipitated proteins were separated using 8% SDS-PAGE and detected by western blotting using α BLM, α DNA topoisomerase I, α RPA194 and α RNA polymerase II. Goat IgG was used as an isotype-matched negative control for α BLM.
As BLM and DNA topoisomerase I localize to the nucleolus and the nucleoplasm, we asked whether the BLM-DNA topoisomerase I interaction occurs specifically in the nucleolus. Nuclei were fractionated into nucleoli and nucleoplasm (Figure 1B) and the resultant sub-nuclear fractions used in co-immunoprecipitation experiments with α BLM antibodies. The RNA polymerase I-specific subunit RPA194 was used as a nucleolar marker, RNA polymerase II as a nucleoplasmic marker and α -actin as a cytoplasmic marker (Figure 1B). Results from co-immunoprecipitation experiments using the sub-nuclear fractions showed specific interactions of BLM with DNA topoisomerase I and RNA polymerase I subunit RPA194 in fractions from nucleoli (Figure 1C; Supplementary Figure 1c). These results suggest that their function may be specific to nucleolar metabolism and are consistent with the presence of a majority of BLM in the nucleolar fraction (Fig 1B).
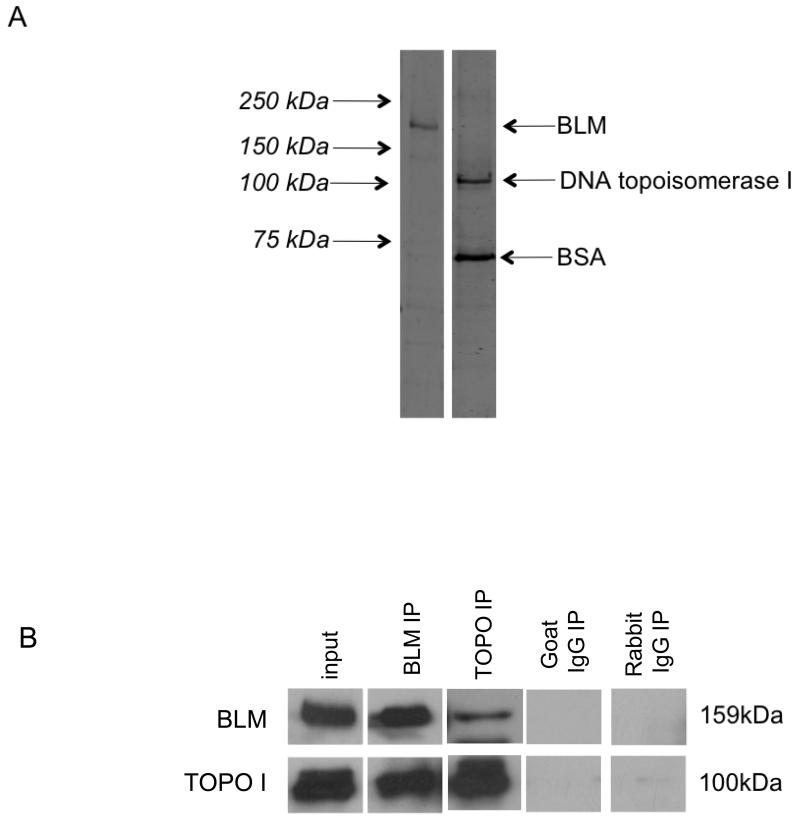
Purified full-length human proteins were used for in vitro protein co-immunoprecipitations to determine whether the interaction between BLM and DNA topoisomerase I is direct or indirect. Recombinant BLM was expressed in S. cerevisiae and FPLC-purified; DNA topoisomerase I was purchased from Topogen (cat # TG2005H-RC1). Purified proteins are shown in Figure 2A. Immunoprecipitation with either α BLM or α DNA topoisomerase I antibodies recovered both proteins demonstrating a direct protein-protein interaction (Figure 2B, Supplementary Figure 2).
Figure 2. BLM and DNA topoisomerase I directly interact in vitro.
(A) The purity of recombinant FPLC-purified BLM and purchased DNA topoisomerase I (Topogen) was determined using 8% SDS-PAGE and staining with Sypro Ruby Protein Gel Stain (Sigma). DNA topoisomerase I is supplied with 50ng/ul of bovine serum albumin (BSA), as indicated. (B) BLM and DNA topoisomerase I were used in in vitro protein co-immunoprecipitation using α BLM or α DNA topoisomerase I. Immunoprecipitated proteins were separated using 8% SDS-PAGE and detected by western blotting using α BLM and α DNA topoisomerase I. Goat IgG was used as an isotype-matched negative control for α BLM; rabbit IgG was used as an isotype-matched negative control for α DNA topoisomerase I.
3.2 BLM and DNA topoisomerase I interact directly in vitro via the BLM C-terminus
BLM is a modular protein with distinct domains dictating specific functions and localization to sub-nuclear regions. The C-terminus of BLM (amino acids 1116-1331) is required for localization to nucleoli and binding to rDNA [13,14]. BLM binds nucleolar rDNA both in the rDNA transcription unit as well as in the intergenic spacer region (IGS) [14]. The N-terminus of BLM is required for its accumulation in PML bodies and is also primarily used for interaction with DNA topoisomerases IIα and IIIα [13,19,26].
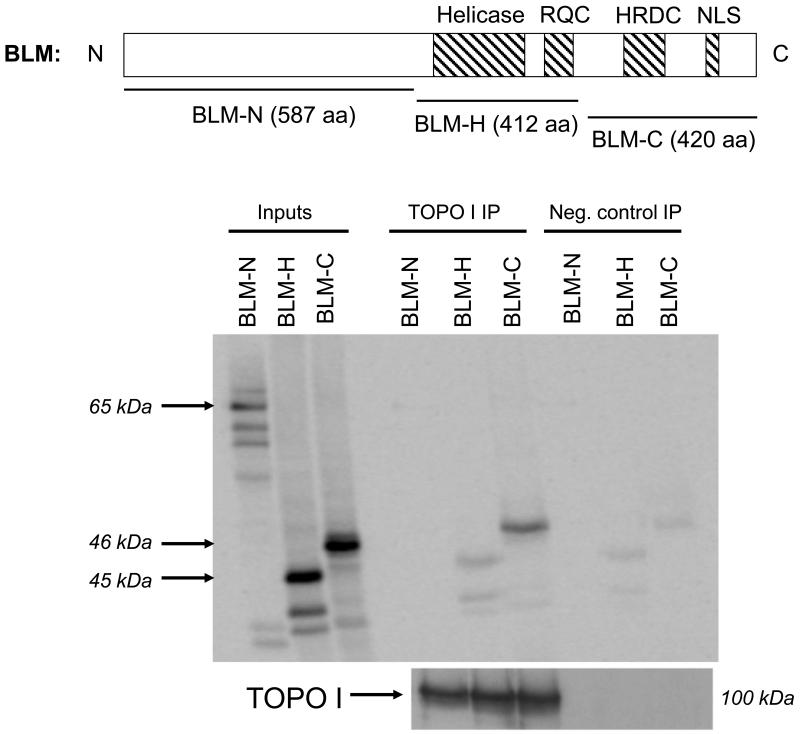
In vitro transcription/translation (IVTT) of BLM using 35S-methionine was used to generate the N-terminal (N: amino acids 25-612), helicase (H: amino acids 613-1025) and C-terminal (C: amino acids 1021-1417) segments of BLM. Co-immunoprecipitation of these segments with full-length human DNA topoisomerase I showed that the C-terminal region of BLM interacts with DNA topoisomerase I (Figure 3). These data suggest that this region of BLM may not only mediate BLM nucleolar localization and rDNA binding [13,14], but also direct protein-protein interactions with other nucleolar proteins.
Figure 3. DNA topoisomerase I interacts in vitro with the C-terminus of BLM.
In vitro transcription/translation (IVTT) was performed using the TNT Rabbit Reticulocyte Lysate kit (Promega) using pET24D-BLM-N, pET24D-BLM-H and pET24D-BLM-C. The resultant 35S-methionine-labeled input BLM fragments are shown on the left. N-, H- and C-fragments were used in co-immunoprecipitation with purified DNA topoisomerase I protein (Topogen) and α DNA topoisomerase I. Following capture with Protein G Dynabeads (Invitrogen), immunoprecipitated proteins were separated using 8% SDS-PAGE; western blotting identified DNA topoisomerase I; autoradiography identified BLM fragments. Negative control immunoprecipitations were carried out by omitting DNA topoisomerase I to control for non-specific binding of BLM fragments to α DNA topoisomerase I or to the beads.
3.3 DNA topoisomerase I stimulates BLM helicase activity on GC-rich RNA20:DNA33 substrate designed to model rRNA/rDNA hybrids, but not on DNA20:DNA33
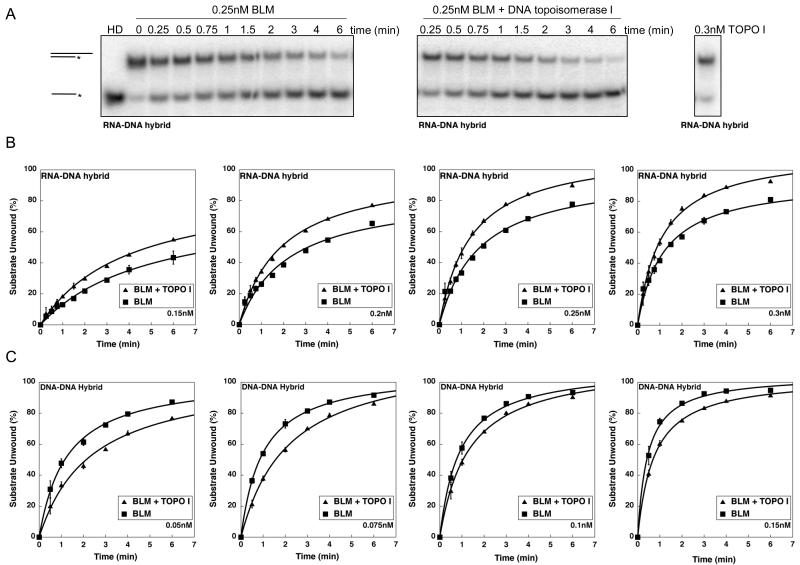
The direct interaction of DNA topoisomerase I and BLM and their functions as components of an RNA polymerase I-associated transcription complex [7,18] suggested that BLM and DNA topoisomerase I may cooperate to unwind hybrid secondary structures presumed to form in the nucleolus during rRNA transcription (RNA-DNA hybrids) and rDNA replication (DNA-DNA hybrids). In vitro helicase assays used FPLC-purified BLM and equimolar human DNA topoisomerase I (Topogen) with the GC-rich RNA20:DNA33 duplex substrate (modeling co-transcriptionally formed rRNA:rDNA hybrids) or a DNA20:DNA33 duplex substrate (modeling rDNA:rDNA hybrids formed during replication). Helicase assays evaluated different BLM concentrations ranging from 0.15nM-0.3nM at time points from 15 sec-6 min in the presence or absence of equimolar amounts of DNA topoisomerase I. Reactions were stopped at the indicated time point and analyzed on native PAGE; bands were quantified to calculate the amount of substrate unwound for each time point.
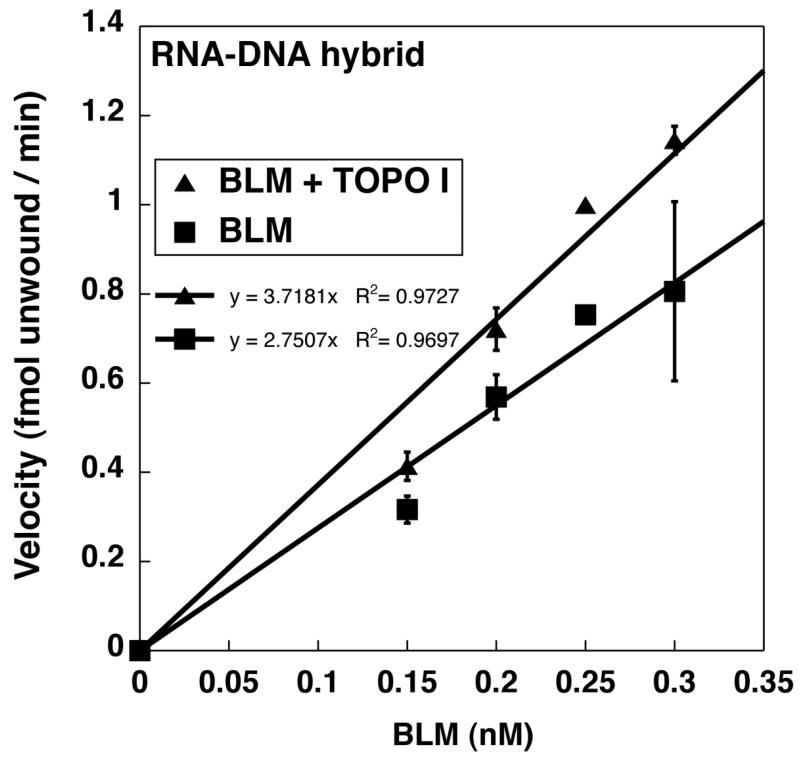
Figure 4A shows a representative experiment performed with 0.25nM BLM on a RNA-DNA hybrid duplex substrate with a 3′ DNA overhang. DNA topoisomerase I did not display unwinding activity by itself on the RNA-DNA hybrid substrate (Figure 4A) or the DNA-DNA substrate (data not shown). BLM by itself or BLM with equimolar DNA topoisomerase I unwound a GC-rich RNA20:DNA33 hybrid duplex following Michaelis-Menten kinetics (Figure 4B). Equimolar DNA topoisomerase I stimulated BLM unwinding activity at all protein concentrations studied for the RNA:DNA hybrid duplex (Figure 4B). Such stimulation was not observed on the corresponding GC-rich DNA20:DNA33 hybrid duplex (Figure 4C), suggesting substrate specificity for the stimulation observed with RNA20:DNA33 hybrid duplex. The kinetics of BLM unwinding activity using RNA-DNA or DNA-DNA hybrids were used to calculate the change in specific activity of BLM in the presence or absence of equimolar DNA topoisomerase I. BLM was pre-complexed with equimolar DNA topoisomerase I in these experiments. With a DNA20:DNA33 hybrid duplex, equimolar DNA topoisomerase I had a modest inhibitory effect on BLM activity at lower BLM concentrations that was insignificant with increased BLM concentrations (Figure 4C, data not shown). In contrast, with a RNA-DNA hybrid duplex, DNA topoisomerase I increased the specific activity of BLM unwinding from 2.75 fmol unwound/min/nM BLM (BLM alone) to 3.72 fmol unwound/min/nM BLM (in the presence of DNA topoisomerase I) (Figure 5). Our data are analogous to those of Hu et al. [26], Bhattacharyya et al., [27] and Russell et al. [19], which demonstrate that DNA topoisomerase IIIα and DNA topoisomerase IIα , respectively, modulate BLM helicase activity. These results suggest a productive interaction of the two proteins specifically in the context of rRNA transcription.
Figure 4. Equimolar DNA topoisomerase I stimulates the helicase activity of BLM using RNA20:DNA33 hybrid duplex substrates.
A. Representative gels demonstrate the unwinding activity of BLM. RNA20:DNA33 hybrid duplex substrate was 0.25nM was incubated with BLM in the absence or presence of equimolar DNA topoisomerase I for 15 sec, 30 sec, 45 sec, 1 min, 1.5 min, 2 min, 3 min, 4 min and 6 min as described in Materials and Methods. Reaction products were separated using 12% non-denaturing acrylamide gels, dried and imaged using ImageQuant software. Unwinding is evaluated by conversion of the double-stranded substrate to single-stranded product. Heat-denatured substrate (HD) is generated by heating to 95°C for 5 min followed by rapid cooling. DNA topoisomerase I (topo I) does not display unwinding activity by itself. B and C. Kinetics of BLM-catalyzed unwinding of RNA20:DNA33 (panel B) or DNA20:DNA33 (panel C) hybrid duplex substrates in the presence or absence of equimolar DNA topoisomerase I. Reactions were performed as in A, with different amounts of proteins as indicated in each panel. Unwinding was calculated by quantifying the amount of single-stranded product relative to the total amount of substrate in the reaction. The percent substrate unwound was plotted against time. All unwinding reactions followed Michaelis-Menten kinetics. Kinetics of the BLM-catalyzed reactions in the absence of DNA topoisomerase I is represented by closed squares and that in the presence of DNA topoisomerase I is represented by closed triangles.
Figure 5. DNA topoisomerase I stimulates BLM unwinding activity using RNA20:DNA33hybrid duplex substrates.
The specific activity of BLM on RNA20:DNA33 hybrid duplex substrates in the presence or absence of DNA topoisomerase I was calculated from curves in Fig 4B by plotting the initial velocity of the reaction at 1 minute against protein concentration used. The slope of the line indicates the specific activity and is indicated in the graph (BLM alone: 2.75 fmol unwound/ min/ nM BLM); BLM with DNA topoisomerase I: 3.72 fmol unwound/ min/ nM BLM).
3.4 BLM stimulates DNA topoisomerase I activity on supercoiled DNA
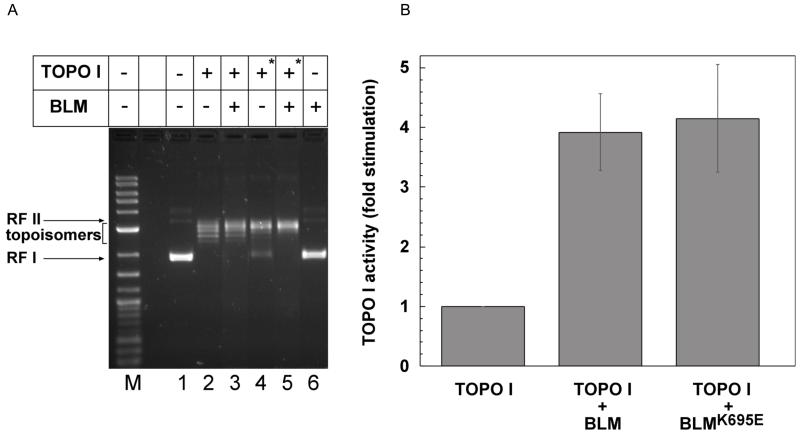
DNA topoisomerase I plays crucial roles in replication and transcription by relieving torsional stress generated during these processes [5,28-31]. It is intricately involved in rDNA transcription by RNA polymerase I and facilitates its movement by relaxation of supercoils generated on both sides of the RNA polymerase complex [4,31,32]. Our earlier studies revealed a role of BLM in facilitating RNA polymerase I-mediated rDNA transcription via its interaction with RNA pol I [18]. In addition to the resolution of RNA-DNA hybrids presented in the previous section, we tested the effect of BLM on relaxation of supercoiled DNA by DNA topoisomerase I. A 2.9kbp supercoiled plasmid was used to assess the relaxation activity of purified recombinant topoisomerase I in the presence or absence of purified BLM. DNA topoisomers were separated on agarose gels and quantified. A representative gel is shown in Figure 6A. Topoisomerase I activity was judged by the conversion of the negatively supercoiled substrate (RFI) to its topoisomers; more relaxed topoisomers migrate slower than the substrate. BLM alone did not alter the supercoiled state of the plasmid (Figure 6A, lane 6). In contrast, the activity of topoisomerase I at two concentrations (Figure 6A: 0.5 Units in lane 2 and 3, or 0.05 Units in lane 4 and 5) was enhanced in the presence of BLM (Figure 6A: compare lanes 2 and 3 or lanes 4 and 5). In order to estimate the fold-stimulation of topoisomerase activity by BLM, the amount of topoisomerase in the reaction was adjusted to get a maximum of 20% conversion of substrate to topoisomers in 30 minutes. Reactions were performed in the presence or absence of BLM or its helicase-dead mutant, BLMK695E, and analyzed. The fold stimulation of topoisomerase activity by BLM or its mutant was calculated by comparing the ratio of products (topoisomers) to substrate (RF I) in the presence of BLM to that in its absence. The results indicated that BLM stimulated topoisomerase activity nearly 4-fold (Figure 6B). Similar stimulation of DNA topoisomerase I was observed using wild-type BLM or the helicase-dead mutant, BLMK695E, suggesting that the stimulation may be independent of the BLM helicase activity (Figure 6B, Supplementary Figure 3). Our results are consistent with the conclusion that the interaction of BLM and topoisomerase I is reciprocally productive and suggests an active participation of BLM in rRNA transcription.
Figure 6. BLM stimulates DNA topoisomerase I activity.
(A). Human DNA topoisomerase I (Topogen) was incubated with a 2.9 kb RF I DNA in the presence or absence of 0.65nM BLM, as indicated, for 30 minutes. Reactions were stopped with stop buffer containing SDS and analyzed on a 1.3% agarose gel as described in Material and Methods. Two concentrations of topoisomerase I were used: 0.5 units (lanes 2 and 3) and 0.05 units (lanes 4 and 5, indicated by asterisk); Lane 1: no protein; Lane 6: no topoisomerase. (B). Graph representing the relative fold-stimulation of topoisomerase activity in the presence of BLM. Reactions were performed as described in A except that the DNA topoisomerase I concentration was adjusted in order to get a maximum of 20% conversion of RF I DNA to relaxed DNA in 30 minutes. Reactions were performed for 10-30 minutes and the ratio of product (topoisomers) to substrate (RF I) for each reaction measured as described in Materials and Methods. The fold stimulation was obtained by dividing the ratio obtained in the presence of BLM or its helicase-dead mutant, BLMK695E by that in the absence of BLM. The graph is a representation of at least 5 independent experiments. The only measure of activity for BLM after purification is helicase unwinding. In the case of wild-type protein a loss of unwinding activity was observed upon dilution. Since there is no direct measure of the activity of the mutant as it is helicase-dead, for the topoisomerase I stimulation assays, higher concentrations of the mutant (9nM, supplementary figure 3) were used to overcome any loss of activity occurring upon dilution of protein as the reason for a potential negative result. The mutant was diluted to the same extent as wild-type BLM.
4. Discussion
Transcription of rDNA repeats by RNA polymerase I is crucial for ribosome biogenesis and cellular growth. Our previous study identifying a role for BLM in facilitating rRNA transcription suggests limited rRNA biogenesis as a potential mechanism underlying the ubiquitous growth retardation observed in Bloom’s syndrome [18]. Regulation of rRNA transcription occurs at both transcription initiation and elongation. Due to the highly repetitive nature of rDNA loci, nascent rRNA generated during on-going transcription have the propensity to form stable rRNA:rDNA hybrids that cause RNA polymerase I stalling, inhibit transcription elongation and slow the overall rRNA transcriptional output. In bacteria, yeast and human cells, DNA topoisomerase I functions as a component of the RNA polymerase I transcription complex to relax rDNA template supercoiling to disfavor the formation of rRNA:rDNA hybrids and facilitate rRNA transcription [7,8,33]. The role of BLM in facilitating rRNA transcription and its ability to unwind RNA:DNA hybrids [18] suggested that BLM and DNA topoisomerase I may interact directly and cooperate to unwind these rRNA transcription hybrids.
These studies demonstrate that BLM and DNA topoisomerase I interact in nucleolar-enriched extracts of MCF7 and 293T cells, consistent with both proteins being components of the RNA polymerase I transcription complex. The C-terminal domain of BLM, the same region required for BLM nucleolar localization, mediates this direct interaction. Previously characterized interactions of BLM with other DNA topoisomerases, for example IIIα and IIα , occur primarily via the N-terminal region of BLM [19,26]. Our studies show that more than one region of BLM mediates interaction with all topoisomerases, suggesting that specific interactions modulate specific functions of BLM and that these interactions may be mediated independently.
BLM interactions with DNA topoisomerase IIα and DNA topoisomerase IIIα modify BLM helicase activity on unique nucleic acid substrates [19,26,27]. We asked whether the direct interaction between BLM and DNA topoisomerase I similarly influences BLM as well as DNA topoisomerase I activity. Using in vitro helicase assays, we were unable to demonstrate a significant functional effect of DNA topoisomerase I on BLM-mediated unwinding of DNA20:DNA33 duplex substrate. However, DNA topoisomerase I stimulated BLM unwinding activity on RNA20:DNA33 duplex substrate, which models a co-transcriptionally formed rRNA:rDNA hybrid. The role of BLM in facilitating rRNA transcription is also supported by its stimulatory effect on DNA topoisomerase I function in relaxation of supercoiled DNA independent of helicase activity. Since DNA topoisomerase I activity is independent of ATP, the results suggest a direct modulation of topoisomerase I activity by BLM through protein-protein interactions. Such a role is consistent with the increased sensitivity of BLM-deficient cells to topoisomerase I inhibitors like camptothecin as well as increased formation of topoisomerase I-DNA cleavage complexes in treated cells [34,35]. Topoisomerase I also plays a significant role in preventing replication stress from the collapse between transcription and replication forks by reducing the formation of RNA-DNA hybrids [36]. Our data suggest a coordinated role for BLM and DNA topoisomerase I in removing or preventing these hybrids at the rDNA loci. Whether BLM and topoisomerase I play redundant or cooperative roles in removal of RNA-DNA hybrids outside of rRNA transcription, as well as relaxing torsional stress during replication, is not clear. However, our results pose an attractive possibility for such functions in preventing increased replication stress in both BLM- and DNA topoisomerase I-deficient cells.
In the context of nucleolar rRNA transcription, data presented in this study and our earlier study [18] suggest a model in which BLM and DNA topoisomerase I are components of the RNA polymerase I transcription complex and serve to exclude rRNA:rDNA hybrids from the rDNA as well as relieve the torsional stress generated during transcription. Prevention of RNA:DNA hybrid formation by DNA topoisomerase I is imperfect, as some RNA:DNA hybrids form in DNA topoisomerase I-proficient cells [8]. The presence of BLM within the RNA polymerase I transcription complex may allow those RNA:DNA hybrids that form to be unwound, thereby preventing interference with transcription and replication. The ability of DNA topoisomerase I to stimulate this ability of BLM may further ensure exclusion of RNA:DNA hybrids from the rDNA. Our studies have also uncovered a unique role for BLM in facilitating the relaxation of supercoiled DNA by topoisomerase I. This observation is consistent with the decreased overall rRNA transcription rate seen in BLM-deficient cells [18] and suggests active participation of BLM in RNA polymerase I transcriptional efficiency. Our results are consistent with the inhibition of RNA polymerase I transcription in yeast deficient for the BLM homologue SGS1 and another 3′-5′ helicase, SRS2, suggesting a conserved role for RecQ-like helicases in nucleolar rRNA transcription [37].
The collaborative functioning of BLM and DNA topoisomerase I to promote efficient rRNA transcription and thus cell growth has implications for cancer therapeutics. Cells deficient in either BLM or DNA topoisomerase I display slower growth rates [7,38]. Drugs targeting DNA topoisomerase I are used chemotherapeutically, such as topotecan to treat small cell lung cancer and irinotecan to treat colon cancer (www.uptodate.com). Understanding the nucleolar mechanisms by which BLM functions would provide the opportunity to inhibit selectively its growth-promoting activity in nucleolar metabolism while preserving its role in maintaining extra-nucleolar genomic stability. Thus, chemotherapeutic dual targeting of DNA topoisomerase I and BLM nucleolar function may synergistically inhibit tumor cell growth.
5. Conclusion
The BLM helicase actively participates in nucleolar metabolism by facilitating rRNA transcription by preventing rRNA-rDNA hybrid formation and relieving torsional stress through its interactions with DNA topoisomerase I. Such a nucleolar role implicates BLM in promoting ribosome biogenesis and cell growth, and provides a basis for understanding the growth defects and camptothecin sensitivity associated with BLM-deficient cells and in those affected by Bloom’s syndrome.
Supplementary Material
Acknowledgements
The authors thank Dr. April Sandy Gocha for help with experiments associated with the Bloom’s syndrome cell line, GM8505; Jeremy Kiersey for help in purification of BLMK695E. This work was supported by the National Institutes of Health [CA117898 to J.G.]; The Ohio State University Pelotonia Fellowship Program to [P.G.]; and The Ohio State University Medical Scientist Training Program (MSTP) to [P.G.]. The project described was supported by Award Number 8UL1TR000090-05, 8KL2TR000112-05, and 8TL1TR000091-05 from the National Center For Advancing Translational Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center For Advancing Translational Sciences or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement: The authors declare that there are no conflicts of interest.
References
- [1].Schwarzacher HG, Wachtler F. The nucleolus. Anat Embryol. 1993;188:515–536. doi: 10.1007/BF00187008. [DOI] [PubMed] [Google Scholar]
- [2].Ayrault O, Andrique L, Fauvin D, Eymin B, Gazzeri S, Seite P. Human tumor suppressor p14ARF negatively regulates rRNA transcription and inhibits UBF1 transcription factor phosphorylation. Oncogene. 2006;25:7577–7586. doi: 10.1038/sj.onc.1209743. [DOI] [PubMed] [Google Scholar]
- [3].Aguilera A. The connection between transcription and genomic instability. EMBO J. 2002;21:195–201. doi: 10.1093/emboj/21.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wang JC. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- [5].Champoux JJ. DNA topisomerases: structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- [6].Gentry AC, Juul S, Veigaard C, Knudsen BR, Osheroff N. The geometry of DNA supercoils modulates the DNA cleavage activity of human topoisomerase I. Nucl Acids Res. 2011;39:1014–1022. doi: 10.1093/nar/gkq822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hraiky C, Raymond MA, Drolet M. RNase H overproduction corrects a defect at the level of transcription elongation during rRNA synthesis in the absence of DNA topoisomerase I in Escherichia coli. J Biol Chem. 2000;275:11257–11263. doi: 10.1074/jbc.275.15.11257. [DOI] [PubMed] [Google Scholar]
- [8].Hage AE, French SL, Beyer AL, Tollervey D. Loss of topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010;24:1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].German J. Bloom’s syndrome I. Genetical and clinical observations in the first twenty-seven patients. Am J Hum Genet. 1969;21:196–227. [PMC free article] [PubMed] [Google Scholar]
- [10].Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- [11].Karow JK, Chakraverty RK, Hickson ID. The Bloom’s syndrome gene product is a 3′-5′ DNA helicase. J Biol Chem. 1997;272:30611–30614. doi: 10.1074/jbc.272.49.30611. [DOI] [PubMed] [Google Scholar]
- [12].Sanz MM, Proytcheva M, Ellis NA, Holloman WK, German J. BLM, the Bloom’s syndrome protein, varies during the cell cycle in its amount, distribution, and co-localization with other nuclear proteins. Cytogenet Cell Genet. 2000;91:217–223. doi: 10.1159/000056848. [DOI] [PubMed] [Google Scholar]
- [13].Yankiwski V, Noonan JP, Neff NF. The C-terminal domain of the Bloom syndrome DNA helicase is essential for genomic stability. BMC Cell Biol. 2001;2:11. doi: 10.1186/1471-2121-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Schawalder J, Paric E, Neff NF. Telomere and ribosomal DNA repeats are chromosomal targets of the bloom syndrome DNA helicase. BMC Cell Biol. 2003;4:15. doi: 10.1186/1471-2121-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Therman E, Otto PG, Shahidi NT. Mitotic recombination and segregation of satellites in Bloom’s syndrome. Chromosoma. 1981;82:627–636. doi: 10.1007/BF00285772. [DOI] [PubMed] [Google Scholar]
- [16].Killen MW, Stults DM, Adachi N, Hanakahi L, Pierce AJ. Loss of Bloom syndrome protein destabilizes human gene cluster architecture. Hum Mol Genet. 2009;18:3417–3428. doi: 10.1093/hmg/ddp282. [DOI] [PubMed] [Google Scholar]
- [17].Sinclair DA, Guarente L. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- [18].Grierson PM, Lillard K, Behbehani GK, Combs KA, Bhattacharyya S, Acharya S, Groden J. BLM helicase facilitates RNA polymerase I-mediated ribosomal RNA transcription. Hum Mol Genet. 2012;21:1172–1183. doi: 10.1093/hmg/ddr545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Russell B, Bhattacharyya S, Keirsey J, Sandy A, Grierson P, Perchiniak E, Kavecansky J, Acharya S, Groden J. Chromosome breakage is regulated by the interaction of the BLM helicase and topoisomerase IIalpha. Cancer Res. 2011;71:561–571. doi: 10.1158/0008-5472.CAN-10-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lillard-Wetherall K, Combs KA, Groden J. BLM Helicase Complements Disrupted Type II Telomere Lengthening in Telomerase-Negative sgs1 Yeast. Cancer Res. 2005;65:5520–5522. doi: 10.1158/0008-5472.CAN-05-0632. [DOI] [PubMed] [Google Scholar]
- [21].Lillard-Wetherell K, Machwe A, Langland GT, Combs KA, Behbehani GK, Schonberg SA, German J, Turchi JJ, Orren DK, Groden J. Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum Mol Genet. 2004;13:1919–1932. doi: 10.1093/hmg/ddh193. [DOI] [PubMed] [Google Scholar]
- [22].Langland G, Kordich J, Creaney J, Goss KH, Lillard-Wetherell K, Behbehani K, Kunkel TA, Groden J. The Bloom’s syndrome protein (BLM) interacts with MLH1 but is not required for DNA mismatch repair. J Biol Chem. 2001;276:30031–30035. doi: 10.1074/jbc.M009664200. [DOI] [PubMed] [Google Scholar]
- [23].Stewart L, Champoux JJ. Assaying DNA Topoisomerase I Relaxation Activity. Methods Mol Biol. 2001;95:1–11. doi: 10.1385/1-59259-057-8:1. [DOI] [PubMed] [Google Scholar]
- [24].Acharya S, Foster PL, Brooks P, Fishel R. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol Cell. 2003;12:233–246. doi: 10.1016/s1097-2765(03)00219-3. [DOI] [PubMed] [Google Scholar]
- [25].Zhang H, Wang JC, Liu LF. Involvement of DNA topoisomerase I in transcription of human ribosomal RNA genes. Proc Natl Acad Sci U S A. 1988;85:1060–1064. doi: 10.1073/pnas.85.4.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hu P, Beresten SF, van Brabant AJ, Pandolfi PP, Johnson FB, Guarente L, Ellis NA. Evidence for BLM and topoisomerase IIIalpha interaction in genomic stability. Hum Mol Genet. 2001;10:1287–1298. doi: 10.1093/hmg/10.12.1287. [DOI] [PubMed] [Google Scholar]
- [27].Bhattacharyya S, Keirsey J, Russell B, Kavecansky J, Lillard-Wetherall K, Tahmaseb K, Turchi JJ, Groden J. Telomerase-associated protein 1, HSP90, and topoisomerase IIalpha associate directly with the BLM helicase in immortalized cells using ALT and modulate its helicase activity using telomeric DNA substrates. J Biol Chem. 2009;284:14966–14977. doi: 10.1074/jbc.M900195200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Parvin JD, Sharp PA. DNA Topology and a Minimal Set of Basal Factors for Transcription by RNA Polymerase II. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- [29].Madden KR, Stewart L, Champoux JJ. Preferential binding of human topoisomerase I to superhelical DNA. EMBO J. 1995;14:5399–5409. doi: 10.1002/j.1460-2075.1995.tb00224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Shykind BM, Kim J, Stewart L, J CJ, Sharp PA. Topoisomerase I enhances TFIID-TFIIA complex assembly during activation of transcription. Genes Dev. 1997;11:397–407. doi: 10.1101/gad.11.3.397. [DOI] [PubMed] [Google Scholar]
- [31].Stewart L, Redinbo MR, Qiu X, Hol WG, Champoux JJ. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- [32].Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- [33].Drolet M. Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol Microbiol. 2006;59:723–730. doi: 10.1111/j.1365-2958.2005.05006.x. [DOI] [PubMed] [Google Scholar]
- [34].Rao VA, Fan AM, Meng L, Doe CF, North PS, Hickson ID, Pommier Y. Phosphorylation of BLM, dissociation from topoisomerase IIIalpha, and colocalization with gamma-H2AX after topoisomerase I-induced replication damage. Mol Cell Biol. 2005;25:8925–8937. doi: 10.1128/MCB.25.20.8925-8937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Mao FJ, Sidorova JM, Lauper JM, Emond MJ, Monnat RJ. The human WRN and BLM RecQ helicases differentially regulate cell proliferation and survival after chemotherapeutic DNA damage. Cancer Res. 2010;70:6548–6555. doi: 10.1158/0008-5472.CAN-10-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Tuduri S, Crabbé L, Conti C, Tourrière H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C, Pommier Y, Tazi J, Coquelle A, Pasero P. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol. 2009;11:1315–1324. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lee SK, Johnson RE, Yu SL, Prakash L, Prakash S. Requirement of yeast SGS1 and SRS2 genes for replication and transcription. Science. 1999;286:2339–2342. doi: 10.1126/science.286.5448.2339. [DOI] [PubMed] [Google Scholar]
- [38].Lechner JF, Kaighn ME, Jetten AM, Groden J, German J. Bloom’s syndrome cells have an abnormal serum growth response. Exp Cell Res. 1983;145:381–388. doi: 10.1016/0014-4827(83)90016-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.