Abstract
Background
Natural products have been an important source of lead compounds for drug discovery. How to find and evaluate bioactive natural products is critical to the achievement of drug/lead discovery from natural products.
Methodology
We collected 19,7201 natural products structures, reported biological activities and virtual screening results. Principal component analysis was employed to explore the chemical space, and we found that there was a large portion of overlap between natural products and FDA-approved drugs in the chemical space, which indicated that natural products had large quantity of potential lead compounds. We also explored the network properties of natural product-target networks and found that polypharmacology was greatly enriched to those compounds with large degree and high betweenness centrality. In order to make up for a lack of experimental data, high throughput virtual screening was employed. All natural products were docked to 332 target proteins of FDA-approved drugs. The most potential natural products for drug discovery and their indications were predicted based on a docking score-weighted prediction model.
Conclusions
Analysis of molecular descriptors, distribution in chemical space and biological activities of natural products was conducted in this article. Natural products have vast chemical diversity, good drug-like properties and can interact with multiple cellular target proteins.
Introduction
Natural products (NPs) play an important role in drug discovery [1]–[3]. About more than 50 percent of FDA-approved drugs were NPs or natural products derivatives [4], [5]. Moreover, NPs have special selectivity to cellular targets [6]. Biologically active natural products would provide selective ligands for disease-related targets [7], and influence the disease-related pathways and eventually shift the biological network from disease status to the healthy status.
With the development of large-scale network analysis, researchers have recently begun to explore the action mechanism of bioactive compounds in the context of biological networks, e.g. drug-target network (DTN) [8]–[10], protein-protein interaction network [11], metabolic network [12], [13] and disease pathway [14]. However, most studies focused on few molecules. NPs possesses vast chemical diversity and so have enormous potential to find various different kinds of bioactive molecules [15]. Researchers have done statistics and analysis for natural products in several aspects, such as chemical diversity [15]–[18], property distribution [19], molecular scaffold [20]–[22], chemical space [23], [24] and comparison between NPs and other compound collections [22], [25], [26]. However, researchers seldom did comprehensive statistics on natural products and comparison between NPs and other types of compounds because it was difficult to obtain large quantity of data collection (both structures and annotations).
During the past decades, our laboratory has been focusing on pharmaceutically relevant natural products. In 2002, we established a 3D structure database of components from Chinese traditional medicinal herbs (CHDD) [27]. Right now, we constructed the Universal Natural Products Database (UNPD) to facilitate the high throughput virtual screening from natural products and the database comprised 197201 natural products now. To the best of our knowledge, UNPD is the largest non-commercial and freely available database for natural products (http://pkuxxj.pku.edu.cn/UNPD). UNPD comprised 197201 natural products from plants, animals and microorganisms. Based on the calculated molecular properties, we compared NPs and FDA-approved drugs in many aspects. We also explored the potential of use NPs as chemical library for drug discovery and network pharmacology by using both experimental and computational results.
Methods
1. Collection of Natural Products and Approved Drugs
The natural products were collected from Reaxys, Chinese Natural Product Database (CNPD) [28], Traditional Chinese Medicines Database (TCMD) [29] and our CHDD [27]. The number of compounds and number of duplicate structures in each databases were listed Table 1 . The 3D structures were generated by Discovery Studio. We use the absolute configuration of each natural product. For those ambiguous structures (e.g. R/S or Z/E is not clear), we create two absolute configuration and assign different number to each configuration. When one structure had two part (e.g. salts or adducts), the larger part was retained and the smaller part was deleted. The duplicates were removed according to InChIKey generated by Open Babel [30]. Therefore, each molecule in UNPD has unambiguous stereoconfiguration. All chemical structure were minimized in MMFF94 force field. The structure of approved drugs were downloaded from DrugBank.
Table 1. The number of compounds and number of duplicate structures in each databases.
| Databases | CHDD | CNPD | TCMD | Reaxys |
| Total compounds | 30564 | 57346 | 23303 | 171504 |
| Used in UNPD | 29759 | 41729 | 7528 | 118185 |
| Duplicates | 785 | 15617 | 15775 | 53319 |
2. Calculation and Statistics of Molecular Descriptors of NPs and Drugs
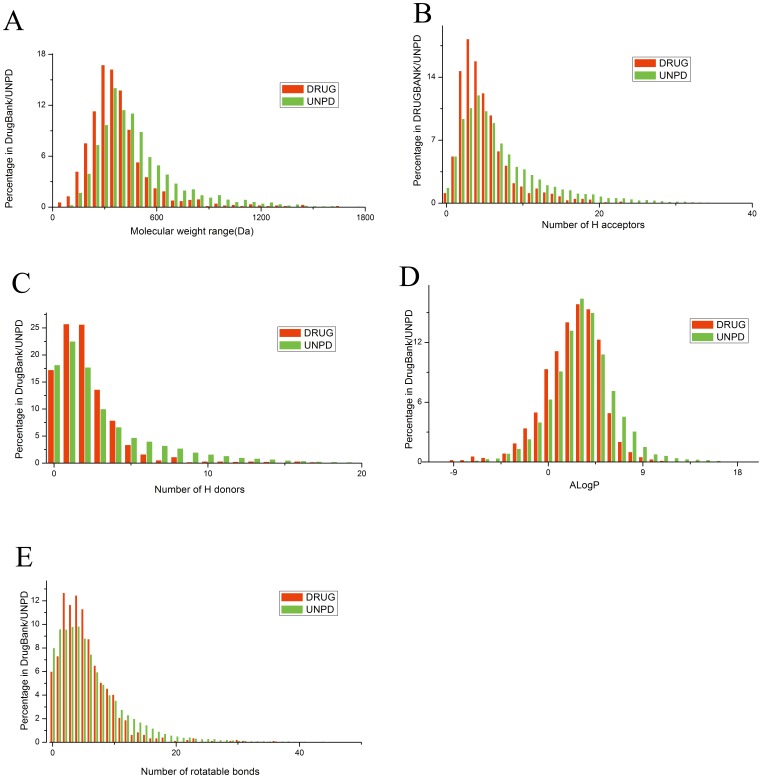
Molecular descriptors of NPs and drugs in Figure 1 and Table 2 were calculated in Discovery Studio by using default parameters. PaDEL-Descriptor [31], a free software developed by National University of Singapore, was employed to calculate substructure-related molecular descriptor and 307 substructure descriptors.
Figure 1. Distribution of five molecular descriptors of natural products and approved drugs.
Table 2. Statistics of molecular descriptors of natural products in UNPD and FDA-approved drugs in DrugBank.
| Descriptors | Natural products in UNPD | Approved drugs | ||||||
| Mean | Median | Min | Max | Mean | Median | Min | Max | |
| AlogP | 2.788±3.352 | 2.710 | −36.007 | 53.473 | 1.899±2.814 | 2.164 | −12.834 | 14.242 |
| Molecular_Weight | 472.6±265.7 | 406.5 | 16.0 | 3973.5 | 360.8±199.1 | 322.1445 | 6.9 | 1639.9 |
| Num_Rotatable_Bonds | 6.6±6.8 | 5 | 0 | 140 | 5.5±4.9 | 4 | 0 | 44 |
| Num_Rings | 3.7±2.4 | 3 | 0 | 32 | 2.7±1.6 | 3 | 0 | 10 |
| Num_Aromatic_Rings | 0.9±1.3 | 0 | 0 | 20 | 1.3±1.1 | 1 | 0 | 8 |
| Num_H_Acceptors | 7.5±6.7 | 6 | 0 | 104 | 5.2±4.2 | 4 | 0 | 51 |
| Num_H_Donors | 3.4±4.1 | 2 | 0 | 64 | 2.3±2.6 | 2 | 0 | 23 |
| Molecular_Volume | 323.8±172.5 | 278.5 | 19.2 | 2576.2 | 238.0±128.1 | 217.8 | 6.8 | 1053.3 |
| Molecular_Surface_Area | 462.4±249.4 | 400.2 | 33 | 4020.9 | 347.7±186.4 | 312.45 | 16.6 | 1586.9 |
| Molecular_Polar_Surface_Area | 122.2±110.6 | 87.0 | 0 | 1917.9 | 93.9±84.1 | 75.0 | 0 | 878.8 |
| Molecular_Fractional_PolarSurfaceArea | 0.248±0.128 | 0.233 | 0 | 1 | 0.277±0.171 | 0.242 | 0 | 1 |
| Molecular_SASA | 683.0±295.7 | 612.5 | 0 | 5106.7 | 563.7±232.6 | 523.3 | 137.9 | 2167.1 |
| Molecular_PolarSASA | 194.9±169.3 | 142.1 | 0 | 3101.9 | 152.1±131.0 | 126.0 | 0 | 1321.8 |
| Molecular_FractionalPolarSASA | 0.268±0.144 | 0.242 | 0 | 0.967 | 0.272±0.171 | 0.239 | 0 | 0.933 |
| Molecular_SAVol | 591.7±253.7 | 531.5 | 0 | 4468.8 | 495.9±202.2 | 462.8 | 124.3 | 1900.9 |
Note: the descriptors of 197201 natural products in UNPD and 1380 FDA-approved small molecule drugs in DrugBank were calculated by Discovery Studio.
3. Chemical Space Analysis
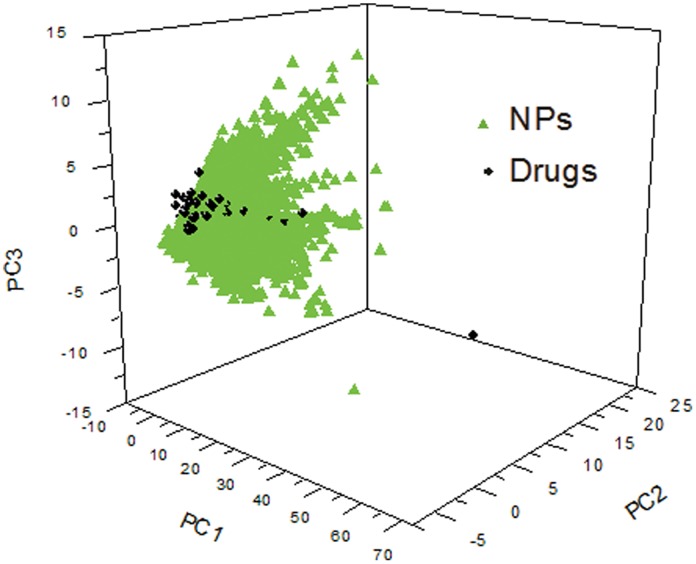
Principal component analysis (PCA) was conducted in library analysis module of Discovery Studio and the input parameters were listed in Table 2 . PCA was an orthogonal linear transformation technique which can transform the data into a new coordinate system, which is in three-dimensional system in our analysis. The variance of the data which was maximized on the first coordinate was called first principal component. The rest of variance maximized on the second coordinate, and so on. The PCA model was built with 8 descriptors: AlogP, Molecular_Weight, Num_H_Donors, Num_H_Acceptors, Num_RotatableBonds, Num_Rings, Num_AromaticRings and Molecular_FractionalPolarSurfaceArea. these descriptors were not pre-scaled. The variances of PC1, PC2 and PC3 for UNPD and drugs in Figure 2 were 0.506,0.202,0.136 and 0.427,0.315,0.099, respectively.
Figure 2. The distribution in chemical space according to principal component analysis of natural products in UNPD and FDA-approved drugs.
The green triangles and black dots represent natural products and FDA-approved drugs, respectively.
4. Constructing of DTNe
We downloaded the experimental binding data of natural products from BindingDB [32] on Oct. 21, 2011. Molecular structures were compared according to InChIKey to identify natural products in BindingDB. Those binding data which target had definite UniProt entry were retained. NPs and experimental targets were connected in Cytoscape [33] to construct the drug-target network based on experimental data (DTNe). The network properties and node centralities were calculated by network plugin and CentiBin [34].
5. Constructing of DTNd
The target proteins of approved Drugs in DrugBank were marked out with “Targets”. There were 4152 target proteins and we used the crystal or NMR structures in RCSB Protein Data Bank (http://www.rcsb.org/pdb/home/home.do) to screen potential lead compounds. The protein-ligand complex structures of target proteins of approved drugs in DrugBank were download and hetero atoms were removed and then hydrogen atoms were added by using Discovery Studio. The original ligands in the complex structures were used as reference compounds to judge the affinity of NPs to corresponding targets. For each target protein, the binding site was defined as a 40×40×40 Å cube centered on the occupied space of the original ligand with a spacing of 0.375 Å between the grid points. Docking was performed by autodock4.01 in DOVIS 2.0 [35] and parameters were listed in File S1. The procedure of constructing of drug-target network based on docking data (DTNd) was the same with that of constructing of DTNe.
Results and Discussion
1. Statistics of Molecular Properties of Natural Products and Comparison between Natural Products and FDA-approved Drugs
Some important molecular descriptors of natural products in UNPD and FDA-approved drugs in DrugBank [36] were listed in Table 2 . Typically, statistical means and standard deviations of natural products were larger than those of FDA-approved drugs. Consequently, these complex and diverse chemical structures of natural products would provide more polypharmacology by interacting with multiple target proteins [6].
Lipinski’s “rule of five” [37] which was derived from the statistics of oral drugs was often used in first screening. Although wemi-empirical rules are not necessarily valid [38], Lipinski’s “rule of five” can be used to help find drug-like molecules from large componds library. The drug-like properties basically contain four aspects which have their own limits: molecular weight should be less than 500 Da, hydrogen bond acceptors (HBA) should be less than 10, hydrogen bond donors (HBD) should be less than 5, partition coefficient AlogP should be less than five. Recently, Leeson emphasized a point that Lipinski's rule of five would mislead drug discovery because some effective drugs did not meet all four cut-off criteria [39]. We checked the satisfied conditions for “rule of five” of all natural products in UNPD and found that only 102605 (52.0%) out of 197201 natural products met “rule of five” ( Table 3 ). However, 141628 (71.8%) natural products met at least three cut-off criteria. Meanwhile, 1065 drugs, 77.17% of the total (1380), obey the “rule of five”. Table 3 shows the count of the molecules obeying all the four limitations or three of them which shows a small fluctuation between different cut-off criteria. This is reasonable for that if molecular weight is bigger, the hydrogen bond acceptors or donors may become more at the same time. And AlogP has definitely the same relationship with these properties.
Table 3. Statistics of satisfied conditions for “rule of five” of natural products in UNPD and approved small drugs in DrugBank.
| Rule of five | UNPD (total 197201) | DrugBank (total 1380) |
| all satisfied | 102605 | 1065 |
| except mw | 113008 | 1074 |
| except acceptors | 103701 | 1074 |
| except donors | 106105 | 1081 |
| except AlogP | 126629 | 1150 |
UNPD contained a fair number of molecules only published in Chinese publications or even some of them have not be published till now. We compared UNPD molecules with FDA-approved drugs in several properties which have been mentioned before in “rule of five”. The histograms ( Figure 1 ) of each descriptor of molecules in UNPD (197201 molecules) and Drugs (1380 molecules) showed that a vast majority of properties in two groups had a very similar distribution (both are non-normal distributions), which indicated that natural products can be a drug-like molecule resource for drug development. Considering our huge size of UNPD, this result will be more persuasive. From the histogram of molecule weight, drugs tended to be smaller than natural products. Most drugs were in the [250,300] interval while natural products were in the [300,350] interval. And natural products had less chiral centers. In the interval of less than 5 in histogram of ALogP, the distributions of NPs and drugs were quite similar. However, we still found that NPs had large ALogP which indicated that they would not dissolve in water easily. Provided that the solubility has large impact to therapeutic effectiveness, the distribution of ALogP may provide useful information.
2. Drug-like Space and Lead Compounds Discovery from Natural Products
The widely used concept of drug-like chemical space was important for drug discovery [23], [39]–[44]. Rosen and colleagues analyzed the chemical space occupancy of natural products and found that natural products exhibited similar activity to drugs with their neighborhood [24]. By using FDA-approved drugs as a reference in chemical space, we can screen potential lead compounds from large chemical libraries [41]. Drugs tended to have more aromatic or heterocyclic and less chiral centers, which was in agreement with the data in a recent study [45]. The median and mean of F-Chirality (number of chiral carbon atoms divided by total carbon count) in DrugBank and Natural products are 0.44, 0.38 and 0.45, 0.41, respectively. It shows that drugs had larger proportion of chiral centers than that of natural products. However, natural products had more carbons and so the total counts of chiral carbon are larger than that of drugs. Other properties were smaller than those of natural products, respectively. To get a better understanding of two groups of molecules, principal component analysis was employed to give visual illustration in chemical space. The 3D plot in Figure 2 offered us an opportunity to compare the distribution between the NPs and drugs easily. The wide distribution in chemical space indicated that there would be vast property diversity in NPs. The large overlap in chemical space showed that natural products could be a large source for drug discovery.
3. Biological Activity of Natural Products
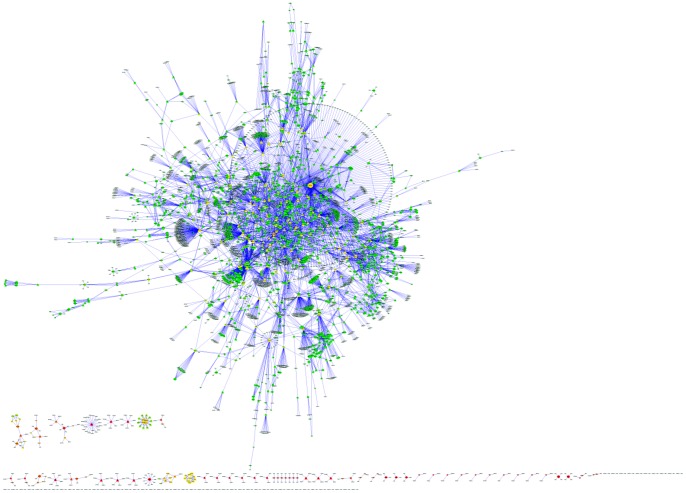
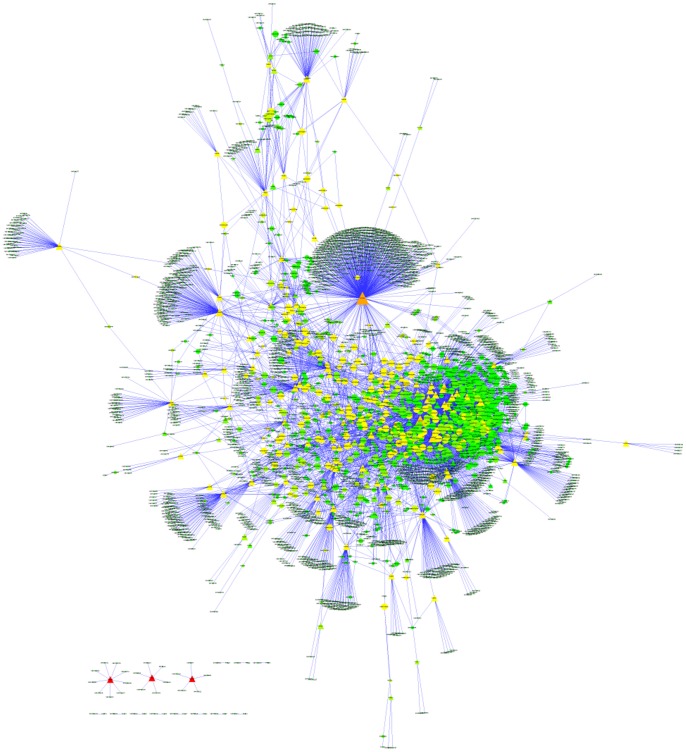
Natural products have many biological activities and they can interact with multiple cellular targets since they are created by nature [6]. Presently, more than 17,000 records of such interactions have been reported according to BindingDB [32] and ChEMBL [46]. We extracted these interaction information (Tables S1) and constructed a drug-target network (DTNe) by connecting the natural products and their experimental targets ( Figure 3 ).
Figure 3. Drug-target network of natural products and their experimental targets (DTNe).
The size of each node is proportional to its degree. The nodes are colored according to their shortest-path betweenness in the network. Circles and triangles correspond to small compounds (natural products or drugs) and target proteins, respectively.
Degree and betweenness centrality were two primary parameters to evaluate the importance of nodes in a network. Degree was defined as the number of neighbors of a node in a undirected graph. Betweenness reflected the important role nodes would play in information transmission in the network. Nodes with the highest local connectivity and the highest global centrality measured by degree and betweenness centrality were defined as hubs and bottlenecks, respectively [47]. Such nodes would be highly influential in the whole network.
DTNe was a typical scale-free network (degree distribution P(x) = 180.77*×∧(−1.125), r = 0.84), like most biological networks. This would be very important for network robustness and information transmission. Most natural products had only one or two experimental targets, and the average was 2.66. However, there were several natural compounds who had many targets, such as UNPD68000 (298 targets) and UNPD49205 (82 targets). UNPD68000 (staurosporine, STS) was a natural product isolated from the bacterium Streptomyces [48]. The main biological activity of STS was the inhibition of protein kinases by occupying the ATP-binding site of the target, with a high affinity and low selectivity. Staurosporine was was also the precursor of midostaurin which was a novel potent kinase inhibitor [49]. Right now, several staurosporine cognates are in advanced clinical trials for anticancer [50].
UNPD49205 (quercetin) was a flavonoid widely distributed in plants. As an antioxidant, it was similar to many other phenolic heterocyclic compounds. Quercetin has been effective against a wide variety of diseases, such as viral disease [51], [52], inflammations [53], and even cancer [54]. Moreover, several cellular models as well as animal models showed that the quercetin can also exert a direct effect in blocking the growth of tumor cells in different phases [55].
STS and quercetin had not only large degree but also high betweenness centrality. However, some natural products had low degree but high betweenness centrality in DTNe. UNPD152676 (genistein) was a well-known isoflavone in several plants. There were many biological functions of genistein reported to date, such as antioxidation and inhibition of epidermal growth factor receptor [55]. It was also reported that it can be potentially used to inhibit the growth of tumor cells [56].
Natural products have extensive biological activities and so can be used as a chemical library for drug discovery. However, there was lack of adequate information of the interactions between natural products and cellular targets. Fortunately, with the increasing development of computer technology, high throughput virtual screening gives us such ability to generate sufficient data. As a result, molecular docking by AutoDock4 [57] was adopted to simulate the interactions between natural products and cellular targets.
4. Network Pharmacology
Network pharmacology was proposed by Hopkins [58], [59] in 2007 and it could take advantage of network analysis methods to explore the pharmaceutical action of molecules in the context of biological networks. By analyzing the network properties or exploring the influence of compounds to the biological networks, it help us to understand the action mechanism and to evaluate the drug efficacy [14], [60]. Now network pharmacology is regarded as the next paradigm in drug discovery [59].
Because there were only 1.8% natural products which biological activities have been reported, we have an urgent need to obtain a large quantity of binding data between natural products and target proteins. By using Autodock4, all natural products were docked to 332 target proteins (all have protein-ligand complex structures in RCSB protein data bank) of FDA-approved drugs and screened according to docking score.
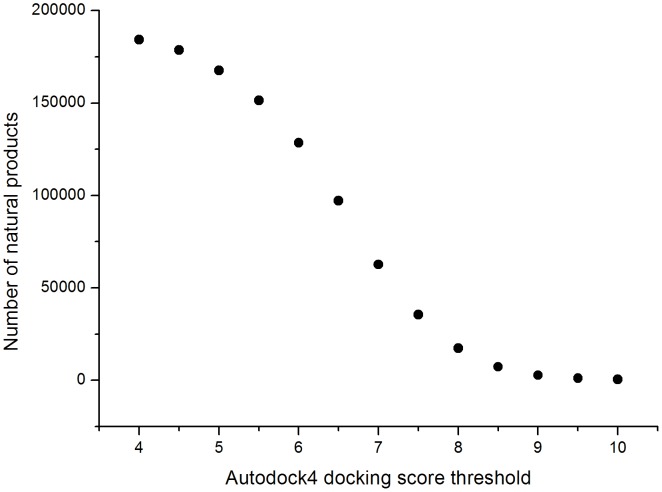
UNPD contained more than 65 millions of docked conformations of natural products and FDA-approved drugs. Although the potential binding of natural products in cavities that may be different from the binding site of drugs, most proteins had limited binding sites. In most cases, the binding sites of natural products and drugs were essentially the same.Generally, the hit rate of virtual screening is about 35% [61]. In this work, the number of natural products which docking score was higher than 7 and higher than the score of original ligand of complex structure of the target protein was 62918, accounting for 32% of total compound ( Figure 4 ). Consequently, it would be an criterion to predict whether a natural product has certain kind of biological activity. In order to promote the accuracy of predicted results and lower the complexity of data handling, we set the threshold as that the docking score was higher than 9 and higher than the score of original ligand of complex structure of the target protein. Then we constructed drug-target network (DTNd, Figure 5 ). Typically, a natural product was linked to a target protein if the docking score exceeded the threshold (Table S2).
Figure 4. Distribution of docking score of natural products.
Figure 5. Drug-target network of natural products and their computational targets.
Representations of the symbols are the same to Figure 3.
Natural products targeted at an average of 2.14 target proteins in DTNd and each target protein contained an average of 25 hits (natural products). Meanwhile, the two values of DTNe were 2.66 and 5.35 ( Table 4 ), respectively. It would mean that most natural products have not conducted experimental test of biological activity. DTNd was comprised of 15 subgraphs. The giant component (the largest connected subnetwork) contained 2810 natural products and 228 target proteins, that is, accounting for 98.6% of all nodes. However, DTNe was comprised of 110 subgraphs and the giant component accounted for 90.1% of total nodes. Therefore, present studies on biological activities of natural products were far from systematic and molecular docking in a large-scale would be an effective supplement.
Table 4. General characteristics of three drug-target networks.
| DTN | No. of compounds | No. of targets | <node degree> | <shortest path> | network density |
| DTNd | 2884 | 243 | 3.96 | 4.30 | 0.0013 |
| DTNe | 2840 | 1413 | 3.56 | 5.95 | 0.0008 |
| DTN* | 1279 | 1328 | 3.68 | 7.16 | 0.0014 |
Drug-target network of FDA-approved drugs and their pharmacological targets in DrugBank.
Most nodes in DTNd had high degree centrality. Especially, UNPD43323, UNPD194973, UNPD107682 and UNPD141622 ( Table 5 ) had more than forty targets. These natural products would be noteworthy because polypharmacology is greatly enriched for high-degree compounds. UNPD43323, UNPD194973, UNPD129237, UNPD162694 and UNPD10433 had highest betweenness centrality, and the first two were also those compounds with largest degree.
Table 5. Most potential natural products for lead discovery.
| UNPD ID | chemical name | CAS | Degree | Betweenness |
| UNPD43323 | Ormojine | 14710-67-9 | 90 | 0.072 |
| UNPD194973 | Ormosinin | NOT Available | 63 | 0.035 |
| UNPD107682 | vatamidine | 129741-48-6 | 46 | 0.014 |
| UNPD141622 | Vatamine | 129741-49-7 | 40 | 0.020 |
| UNPD61603 | strychnohexamine | 442123-70-8 | 35 | 0.017 |
| UNPD38223 | caledonine | 235099-24-8 | 31 | 0.009 |
| UNPD21224 | Lycopodium Base B | 54352-31-7 | 28 | 0.012 |
| UNPD5255 | Vatine | 129741-50-0 | 28 | 0.005 |
| UNPD41999 | Lycopodium Base A | 54352-30-6 | 26 | 0.005 |
| UNPD2675 | Seldomycin 5 | 56276-26-7 | 25 | 0.004 |
5. Predicted Diseases for Natural Products
Natural products have been used to treat diseases for thousands of years. However, the molecular mechanism was rarely elucidated clearly. Here, we predicted the potential indications for natural products based on DTNd. Typically, natural products, especially high-degree compounds, would interact with several target proteins and target protein would concern a lot of diseases. After extracting the target-related diseases from Therapeutic Targets Database [62], we constructed a docking score-weighted prediction model ( Figure 6 ) to predict the possibility of a natural product to treat some diseases ( Table 6 and Table S3). Typically, UNPD194973 and UNPD43323 would have very large latent capacity as drugs for bacterial infections and several cancers.
Figure 6. Prediction model of indications for natural products.
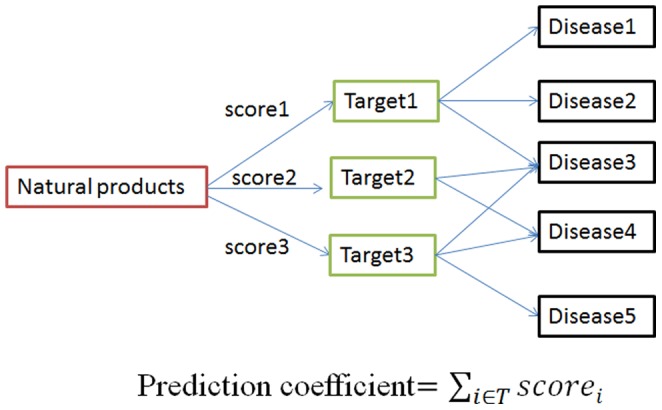
Table 6. Predicted indications for natural products.
| Naturalproducts | Prediction coefficient | Indications |
| UNPD194973 | 58.12 | Bacterial infections |
| UNPD43323 | 55.26 | Prostate cancer |
| UNPD43323 | 51.60 | Asthma |
| UNPD43323 | 50.08 | Cancer, unspecific |
| UNPD107682 | 49.74 | Bacterial infections |
| UNPD194973 | 47.89 | Prostate cancer |
| UNPD112143 | 47.62 | Prostate cancer |
| UNPD194973 | 44.34 | Asthma |
| UNPD43323 | 43.37 | Bacterial infections |
| UNPD107682 | 41.47 | Prostate cancer |
| UNPD141622 | 39.76 | Bacterial infections |
| UNPD141622 | 39.62 | Prostate cancer |
| UNPD141622 | 39.00 | Lung Cancer |
| UNPD141622 | 39.00 | Osteoarthritis |
| UNPD194973 | 38.99 | Cancer, unspecific |
| UNPD107682 | 38.24 | Asthma |
| UNPD112143 | 37.97 | Non-small Cell Lung Cancer |
| UNPD43323 | 35.82 | Diabetes mellitus |
| UNPD43323 | 34.29 | Non-small Cell Lung Cancer |
| UNPD43323 | 33.89 | Brain Cancer |
Conclusions
Natural products have vast chemical diversity, not only structural diversity but also various biological activity, so as to guarantee the opportunities to find different kinds of lead compounds for different diseases. We find that NPs and FDA-approved drugs share a lot of space in chemical space. Moreover, NPs have a large quantity of lead-like molecules, which could be used as scaffolds to expand the chemical library.
Notwithstanding the recent advances in omics, the data collection of NPs is largely incomplete. First of all, the inventory of NPs remains incomplete and new chemical structures are being discovered [7]. Secondly, researchers explored only a small part of biological functions of NPs. Thirdly, there were mistakes and errors in existing data. Many chemical structures of NPs are questionable. Data of biological activity obtained from different laboratories for one compounds would vary greatly. While no adequate data is available, a good and useful complement is virtual screening results. Last but not least, more research methods both experimental and computational to afford more overall and more accurate data are needed urgently. We are extending the computational targets to all proteins if it has protein-ligand complex structure.
Presently, most studies on network pharmacology are based on static networks. However, biological networks is always changing. Recently, Hoeng and colleagues proposed that using of network analysis to prediction the efficacy or toxicity for chronic diseases by estimating the perturbation of biological networks would be particularly useful [60].
Supporting Information
Lists experimental interaction between natural products and target proteins.
(XLSX)
Lists computational interaction between natural products and target proteins.
(XLSX)
Lists the prediction of indications for natural products.
(XLSX)
Lists the parameters used in the virtual screening by autodock4.0.
(DOCX)
Funding Statement
The present work was supported by the Major Scientific and Technological Special Project for ‘Significant New Drugs Formulation’ number 2012ZX09501001; 012ZX09301002-001 (http://www.most.gov.cn). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Newman DJ, Cragg GM (2012) Natural Products As Sources of New Drugs over the 30 Years from 1981 to 2010. Journal of Natural Products 75: 311–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ji HF, Li XJ, Zhang HY (2009) Natural products and drug discovery Can thousands of years of ancient medical knowledge lead us to new and powerful drug combinations in the fight against cancer and dementia? Embo Reports 10: 194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Harvey A (2008) Natural products in drug discovery. Drug Discovery Today 13: 894–901. [DOI] [PubMed] [Google Scholar]
- 4. Kingston DGI (2011) Modern Natural Products Drug Discovery and Its Relevance to Biodiversity Conservation. Journal of Natural Products 74: 496–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chin YW, Balunas MJ, Chai HB, Kinghorn AD (2006) Drug discovery from natural sources. The AAPS Journal 8: E239–E253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lagunin A, Filimonov D, Poroikov V (2010) Multi-Targeted Natural Products Evaluation Based on Biological Activity Prediction with PASS. Current Pharmaceutical Design 16: 1703–1717. [DOI] [PubMed] [Google Scholar]
- 7. Clardy J, Walsh C (2004) Lessons from natural molecules. Nature 432: 829–837. [DOI] [PubMed] [Google Scholar]
- 8. Gu J, Zhang H, Chen L, Xu S, Yuan G, et al. (2011) Drug-target network and polypharmacology studies of a Traditional Chinese Medicine for type II diabetes mellitus. Computational Biology and Chemistry 35: 293–297. [DOI] [PubMed] [Google Scholar]
- 9. Vogt I, Mestres J (2010) Drug-Target Networks. Molecular Informatics 29: 10–14. [DOI] [PubMed] [Google Scholar]
- 10. Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M (2007) Drug-target network. Nature Biotechnology 25: 1119–1126. [DOI] [PubMed] [Google Scholar]
- 11. Wang XJ, Wei XM, Thijssen B, Das J, Lipkin SM, et al. (2012) Three-dimensional reconstruction of protein networks provides insight into human genetic disease. Nature Biotechnology 30: 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang K, Ma WZ, Liang HH, Qi OY, Tang C, et al. (2007) Dynamic simulations on the arachidonic acid metabolic network. Plos Computational Biology 3: 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang Y, Thiele I, Weekes D, Li ZW, Jaroszewski L, et al. (2009) Three-Dimensional Structural View of the Central Metabolic Network of Thermotoga maritima. Science 325: 1544–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li QA, Li XD, Li CH, Chen LR, Song J, et al.. (2011) A Network-Based Multi-Target Computational Estimation Scheme for Anticoagulant Activities of Compounds. Plos One 6. [DOI] [PMC free article] [PubMed]
- 15. Hong JY (2011) Role of natural product diversity in chemical biology. Current Opinion in Chemical Biology 15: 350–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Firn RD, Jones CG (2003) Natural products - a simple model to explain chemical diversity. Natural Product Reports 20: 382–391. [DOI] [PubMed] [Google Scholar]
- 17. Basso LA, da Silva LHP, Fett-Neto AG, Junior WFD, Moreira ID, et al. (2005) The use of biodiversity as source of new chemical entities against defined molecular targets for treatment of malaria, tuberculosis, and T-cell mediated diseases - A Review. Memorias Do Instituto Oswaldo Cruz 100: 575–606. [DOI] [PubMed] [Google Scholar]
- 18. Quinn RJ, Carroll AR, Pham NB, Baron P, Palframan ME, et al. (2008) Developing a drug-like natural product library. Journal of Natural Products 71: 464–468. [DOI] [PubMed] [Google Scholar]
- 19. Feher M, Schmidt JM (2003) Property distributions: Differences between drugs, natural products, and molecules from combinatorial chemistry. Journal of Chemical Information and Computer Sciences 43: 218–227. [DOI] [PubMed] [Google Scholar]
- 20. Yongye AB, Waddell J, Medina-Franco JL (2012) Molecular scaffold analysis of natural products databases in the public domain. Chemical Biology & Drug Design 80: 717–724. [DOI] [PubMed] [Google Scholar]
- 21. Grabowski K, Baringhaus KH, Schneider G (2008) Scaffold diversity of natural products: inspiration for combinatorial library design. Natural Product Reports 25: 892–904. [DOI] [PubMed] [Google Scholar]
- 22. Lee ML, Schneider G (2001) Scaffold architecture and pharmacophoric properties of natural products and trade drugs: Application in the design of natural product-based combinatorial libraries. Journal of Combinatorial Chemistry 3: 284–289. [DOI] [PubMed] [Google Scholar]
- 23. Dobson CM (2004) Chemical space and biology. Nature 432: 824–828. [DOI] [PubMed] [Google Scholar]
- 24. Rosen J, Gottfries J, Muresan S, Backlund A, Oprea TI (2009) Novel Chemical Space Exploration via Natural Products. Journal of Medicinal Chemistry 52: 1953–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grabowski K, Schneider G (2007) Properties and Architecture of Drugs and Natural Products Revisited. Current Chemical Biology 1: 115–127. [Google Scholar]
- 26. Henkel T, Brunne RM, Muller H, Reichel F (1999) Statistical investigation into the structural complementarity of natural products and synthetic compounds. Angewandte Chemie-International Edition 38: 643–647. [DOI] [PubMed] [Google Scholar]
- 27. Qiao XB, Hou TJ, Zhang W, Guo SL, Xu XJ (2002) A 3D structure database of components from Chinese traditional medicinal herbs. Journal of Chemical Information and Computer Sciences 42: 481–489. [DOI] [PubMed] [Google Scholar]
- 28. Shen JH, Xu XY, Cheng F, Liu H, Luo XM, et al. (2003) Virtual screening on natural products for discovering active compounds and target information. Current Medicinal Chemistry 10: 2327–2342. [DOI] [PubMed] [Google Scholar]
- 29. He M, Yan XJ, Zhou JJ, Xie GR (2001) Traditional Chinese medicine database and application on the Web. Journal of Chemical Information and Computer Sciences 41: 273–277. [DOI] [PubMed] [Google Scholar]
- 30.O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, et al.. (2011) Open Babel: An open chemical toolbox. Journal of Cheminformatics 3. [DOI] [PMC free article] [PubMed]
- 31. Yap CW (2011) PaDEL-Descriptor: An Open Source Software to Calculate Molecular Descriptors and Fingerprints. Journal of Computational Chemistry 32: 1466–1474. [DOI] [PubMed] [Google Scholar]
- 32. Liu TQ, Lin YM, Wen X, Jorissen RN, Gilson MK (2007) BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Research 35: D198–D201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27: 431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Junker BH, Koschutzki D, Schreiber F (2006) Exploration of biological network centralities with CentiBiN. Bmc Bioinformatics 7: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang XH, Kumar K, Hu X, Wallqvist A, Reifman J (2008) DOVIS 2.0: an efficient and easy to use parallel virtual screening tool based on AutoDock 4.0. Chemistry Central Journal 2. [DOI] [PMC free article] [PubMed]
- 36. Knox C, Law V, Jewison T, Liu P, Ly S, et al. (2011) DrugBank 3.0: a comprehensive resource for 'Omics' research on drugs. Nucleic Acids Research 39: D1035–D1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Reviews 23: 3–25. [DOI] [PubMed] [Google Scholar]
- 38. Faller B, Ottaviani G, Ertl P, Berellini G, Collis A (2011) Evolution of the physicochemical properties of marketed drugs: can history foretell the future? Drug Discovery Today 16: 976–984. [DOI] [PubMed] [Google Scholar]
- 39. Leeson P (2012) Drug discovery: Chemical beauty contest. Nature 481: 455–456. [DOI] [PubMed] [Google Scholar]
- 40. Leeson PD, Springthorpe B (2007) The influence of drug-like concepts on decision-making in medicinal chemistry. Nature Reviews Drug Discovery 6: 881–890. [DOI] [PubMed] [Google Scholar]
- 41. Bade R, Chan H-F, Reynisson J (2010) Characteristics of known drug space. Natural products, their derivatives and synthetic drugs. European Journal of Medicinal Chemistry 45: 5646–5652. [DOI] [PubMed] [Google Scholar]
- 42. Mirza A, Desai R, Reynisson J (2009) Known drug space as a metric in exploring the boundaries of drug-like chemical space. European Journal of Medicinal Chemistry 44: 5006–5011. [DOI] [PubMed] [Google Scholar]
- 43. Bickerton GR, Paolini GV, Besnard J, Muresan S, Hopkins AL (2012) Quantifying the chemical beauty of drugs. Nature Chemistry 4: 90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lipinski C, Hopkins A (2004) Navigating chemical space for biology and medicine. Nature 432: 855–861. [DOI] [PubMed] [Google Scholar]
- 45. Lopez-Vallejo F, Giulianotti MA, Houghten RA, Medina-Franco JL (2012) Expanding the medicinally relevant chemical space with compound libraries. Drug Discovery Today 17: 718–726. [DOI] [PubMed] [Google Scholar]
- 46. Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, et al. (2012) ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Research 40: D1100–D1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang M, Deng J, Fang CV, Zhang X, Lu LJ (2010) Molecular Network Analysis and Applications. Knowledge-Based Bioinformatics: John Wiley & Sons, Ltd. 251–287.
- 48. Omura S, Iwai Y, Hirano A, Nakagawa A, Awaya J, et al. (1977) New Alkaloid Am-2282 of Streptomyces Origin Taxonomy, Fermentation, Isolation and Preliminary Characterization. Journal of Antibiotics 30: 275–282. [DOI] [PubMed] [Google Scholar]
- 49. Wang YF, Yin OQR, Graf P, Kisicki JC, Schran H (2008) Dose- and time-dependent pharmacokinetics of midostaurin in patients with diabetes mellitus. Journal of Clinical Pharmacology 48: 763–775. [DOI] [PubMed] [Google Scholar]
- 50. Gani OABSM, Engh RA (2010) Protein kinase inhibition of clinically important staurosporine analogues. Natural Product Reports 27: 489–498. [DOI] [PubMed] [Google Scholar]
- 51. Wu LL, Yang XB, Huang ZM, Liu HZ, Wu GX (2007) In vivo and in vitro antiviral activity of hyperoside extracted from Abelmoschus manihot (L) medik. Acta Pharmacologica Sinica 28: 404–409. [DOI] [PubMed] [Google Scholar]
- 52. Yu YB, Miyashiro H, Nakamura N, Hattori M, Park JC (2007) Effects of triterpenoids and flavonoids isolated from Alnus firma on HIV-1 viral enzymes. Archives of Pharmacal Research 30: 820–826. [DOI] [PubMed] [Google Scholar]
- 53. Davis JM, Murphy EA, Carmichael MD, Davis B (2009) Quercetin increases brain and muscle mitochondrial biogenesis and exercise tolerance. American Journal of Physiology-Regulatory Integrative and Comparative Physiology 296: R1071–R1077. [DOI] [PubMed] [Google Scholar]
- 54. Verschoyle RD, Steward WP, Gescher AJ (2007) Putative cancer chemopreventive agents of dietary origin - How safe are they? Nutrition and Cancer-an International Journal 59: 152–162. [DOI] [PubMed] [Google Scholar]
- 55.Gibellini L, Pinti M, Nasi M, Montagna JP, De Biasi S, et al.. (2011) Quercetin and Cancer Chemoprevention. Evidence-Based Complementary and Alternative Medicine: 1–15.
- 56. Das A, Banik NL, Ray SK (2010) Flavonoids Activated Caspases for Apoptosis in Human Glioblastoma T98G and U87MG Cells But Not in Human Normal Astrocytes. Cancer 116: 164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, et al. (2009) AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. Journal of Computational Chemistry 30: 2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hopkins AL (2007) Network pharmacology. Nature Biotechnology 25: 1110–1111. [DOI] [PubMed] [Google Scholar]
- 59. Hopkins AL (2008) Network pharmacology: the next paradigm in drug discovery. Nature Chemical Biology 4: 682–690. [DOI] [PubMed] [Google Scholar]
- 60. Hoeng J, Deehan R, Pratt D, Martin F, Sewer A, et al. (2012) A network-based approach to quantifying the impact of biologically active substances. Drug Discovery Today 17: 413–418. [DOI] [PubMed] [Google Scholar]
- 61. Doman TN, McGovern SL, Witherbee BJ, Kasten TP, Kurumbail R, et al. (2002) Molecular docking and high-throughput screening for novel inhibitors of protein tyrosine phosphatase-1B. Journal of Medicinal Chemistry 45: 2213–2221. [DOI] [PubMed] [Google Scholar]
- 62. Zhu F, Shi Z, Qin C, Tao L, Liu X, et al. (2012) Therapeutic target database update 2012: a resource for facilitating target-oriented drug discovery. Nucleic Acids Research 40: D1128–D1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lists experimental interaction between natural products and target proteins.
(XLSX)
Lists computational interaction between natural products and target proteins.
(XLSX)
Lists the prediction of indications for natural products.
(XLSX)
Lists the parameters used in the virtual screening by autodock4.0.
(DOCX)