Abstract
Background
Human endogenous retroviruses (HERVs) are genomic sequences that resulted from ancestral germ-line infections by exogenous retroviruses and therefore are transmitted in a Mendelian fashion. Increased HERV expression and antibodies to HERV antigens have been found in various autoimmune diseases. HERV-K18 in chromosome 1 was previously associated with type one diabetes and multiple sclerosis (MS). The etiology of these complex conditions has not been completely elucidated even after the powerful genome wide association studies (GWAS) performed. Nonetheless, this approach does not scrutinize the repetitive sequences within the genome, and part of the missing heritability could lie behind these sequences. We aimed at evaluating the role of HERV-K18 in chromosome 1 on autoimmune disease susceptibility.
Methods
Two HERV-K18 SNPs (97Y/C and 154W/Stop substitutions) conforming three haplotypes were genotyped in Spanish cohorts of multiple sclerosis (n = 942), rheumatoid arthritis (n = 462) and ethnically matched controls (n = 601). Our findings were pooled in a meta-analysis including 5312 autoimmune patients and 4032 controls.
Results
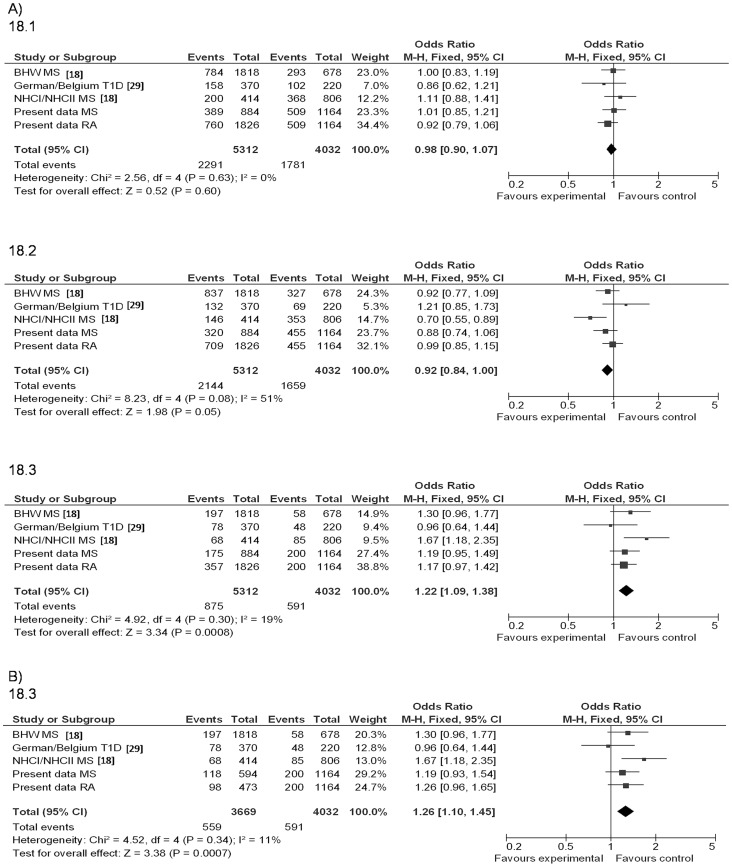
Significant associations of both HERV-K18 polymorphisms in chromosome 1 with MS patients stratified by HLA-DRB1*15∶01 were observed [97Y/C p = 0.02; OR (95% CI) = 1.5 (1.04–2.17) and 154W/Stop: p = 0.001; OR (95% CI) = 1.6 (1.19–2.16)]. Combined meta-analysis of the previously published association studies of HERV-K18 with different autoimmune diseases, together with data derived from Spanish cohorts, yielded a significant association of the HERV-K18.3 haplotype [97Y–154W: pM-H = 0.0008; ORM-H (95% CI) = 1.22 (1.09–1.38)].
Conclusion
Association of the HERV-K18.3 haplotype in chromosome 1 with autoimmune-disease susceptibility was confirmed through meta-analysis.
Introduction
Human endogenous retroviruses (HERVs) are sequences within the genome resulting from ancestral infections by exogenous retroviruses that were incorporated into germ-line DNA and therefore are inherited in a classical Mendelian pattern. They comprise approximately an 8% of the human genome [1] and have been detected in all vertebrates studied, including most apes, indicative of their evolutionary origin at least 25 million years ago. Different mechanisms have led to inactivation of most HERVs: recombination of their long terminal repeats with the excision of the proviral genome, hypermethylation of HERV promoters or mutations preventing the expression of functional HERV proteins. These mechanisms would support the concept that HERVs constitute “junk” DNA, but mounting evidence questions this view as some of them have physiological or pathological functions. The best example of their physiological role in the host is that of the HERV-W envelope protein known as syncytin, encoded on chromosome 7. This fusogenic protein expressed in the placenta is involved in the formation of the syncytiotrophoblast and decreased placental expression of this protein has been linked with pregnancy-induced hypertension [2]. An additional physiological role in spermatogenesis has been described for HERV-K in human males [3]. Moreover, the hypothesis that HERVs may be key factors in the pathogenesis of different diseases has been proposed. Apart from their specific expression reported in several cancer types, evidence of their pathogenic potential accumulates in a number of autoimmune diseases. Increased HERV expression from different families, as HERV-W, HERV-K or HERV-H, has been reported in rheumatoid arthritis (RA) [4], psoriasis [5], systemic lupus erythematosus [6] or multiple sclerosis (MS) [7]–[10]. Circulating antibodies to various HERV antigens have also been found in autoimmune patients [11]. Additionally, many reports show genetic association of different retrovirus with autoimmune diseases (i.e. association with MS of polymorphisms located near HERV-Fc or within a gen involved in retrovirus replication, TRIM5 [12], [13]).
In this study we focused on the influence of a copy of the HERV-K family, located in transcriptional antisense within the first intron of the CD48 gene in chromosome 1q23.3. In 1997, Conrad and collaborators [14] found that the env gene of a new HERV-K endogenous retrovirus encoded a superantigen in insulin-dependent diabetes. Later, it was suggested that the tissue tropism of an exogenous virus might trigger the organ specific HERV-K18 superantigen response, leading to the expansion of autoreactive T cells [15]. Elevated HERV-K18 superantigen was also found in juvenile rheumatoid arthritis [16]. The HERV-K18 in chromosome 1 is an insert of 9235 bp with three variants and all of them encode superantigens [17]. Nonetheless, HERV-K18 expression might be a consequence rather than the cause of autoimmune conditions.
The search for the aetiology of these complex diseases has undergone a spectacular advance in recent times with the identification through genome wide association studies (GWAS) of literally hundreds of common single nucleotide polymorphisms (SNPs) involved in their susceptibility. However, although GWAS have provided valuable insights in the underlying causes of complex traits, they have explained relatively little of their heritability and the issue of where the “missing heritability” lies is a matter of debate [18]. There are several possible explanations as to the missing genetic basis. One of them points to the identified marker SNPs as imperfect proxies for the actual causal mutations that led to the association, which frequency could be low and would remain undetected. However, recent studies claim that rare variants in predisposition genes discovered by GWAS are not likely to make a large contribution to inherited risk in complex diseases [19]. Furthermore, gene-environment interactions may be relevant to fully elucidate the causes of these complex diseases. Another possible explanation is that repetitive sequences in the genome have not been adequately scrutinized through GWAS. With these considerations in mind, in the present work we aimed at studying the three haplotypes of HERV-K18 on chromosome 1 previously described associated with diabetes and MS risk [20], [21] and at testing their influence on autoimmune disease susceptibility, provided the shared genetic susceptibility evidenced among these autoimmune conditions [22]. Additionally, increased HERV-K18 transcription was reported in cultured synoviocytes derived from biopsies of RA patients compared to controls [23]. A meta-analysis has been conducted including data from MS and RA Spanish cohorts combined with previously published results. In both traits, human herpesvirus 6 (HHV-6) or Epstein–Barr virus (EBV) infections have been often times outlined as environmental triggers of the diseases [24], [25]. Moreover, HHV-6 and EBV induce transcriptional activation of the endogenous superantigen HERV-K18, independently of virus replication [26], [27].
Materials and Methods
Ethics Statement
The study was approved by the Ethics Committee of the Hospital Clínico San Carlos, Madrid, Spain. Patients and ethnically matched controls gave written informed consent.
Meta-analysis
We performed a comprehensive search strategy of various electronic databases (MEDLINE (1966 - December 2012), Cochrane Database of Systematic Reviews (1991- December 2012) and EMBASE (1980- December 2012) by combining the terms: “HERVk-18”, “IDDMK(1,2)22” and “Human endogenous retrovirus-k18”. Additionally, a manual search of all references was conducted among the identified studies and relevant review articles (See Figure 1 and Checklist S1). This search rendered 37 articles published to date and all the association studies involved the copy on chromosome 1. Neither date nor language restrictions were imposed. The association studies considered for further analysis were required to hold information about HERV-K18 haplotypes. We also included non published data from Hospital Clinico San Carlos (HCSC) cohort.
Figure 1. Strategy used for the selection of studies finally included in the meta-analysis.
Patients
The Spanish case-control study included 942 MS, 462 RA patients and 601 ethnically matched controls consecutively recruited from a single centre (Hospital Clínico San Carlos, Madrid, Spain).
MS Spanish patients (63% female) were diagnosed based on the McDonald criteria [28] and 36% of them carried the human leukocyte antigen HLA DRB1*1501 risk allele. Most patients suffered relapsing remitting MS (79%), 11% of them showed the secondary progressive clinical form and 9% of them were classified as primary progressive patients. Their mean age at MS onset was 29±9.
RA Spanish patients (74% female) were diagnosed according to American College of Rheumatology criteria [29]. The mean age of the RA patients at the time of disease onset was 51±15, 56% of patients were shared epitope (SE) positive and 52% were anti-cyclic citrullinated peptide (anti-CCP) antibody positive. SE identification was achieved by using Lifecodes HLA-DRB Typing Kit (Tepnel Diagnostics Ltd., Abingdon, Oxon, UK).
Genotyping
DNA was extracted from peripheral blood by a standard salting out method. A fragment of 6417 bp of HERV-K18 env (accession number AF333069, GRCh37.p10 (160666969–160673386); www.ensembl.org, October-2012) was amplified by polymerase chain reaction (PCR) [Cycle conditions: initial denaturation: 94°C/5 min, 35 cycles: 94°C/30 sec, 62°C/30 sec, 68°C/5 min, and final extension of 68°C/10 min] using a primer located in the proviral sequence and the other mapping to the CD48 sequence (5′-CCCACTCTAATGCAAGCTC-3′ and 5′-CATGGGAAATAGGGAAGCTG-3′). Within this amplicon, two SNPs, a Y/C substitution at amino acid position 97 (SNP1) and another W/Stop at position 154 (SNP2), discriminate the three haplotypes previously described (AF134984, Y18890, and AF333069) [20], [21]. They were analyzed by Taqman technology using 384 well plates in a 7900HT Fast Real-Time PCR system, under the conditions recommended by the manufacturer (Applied Biosystems, Foster City, CA, USA). Genotyping success was over 95% for all groups of patients and controls. No departure from Hardy Weinberg equilibrium was observed in the control group.
Statistical Analysis
Statistical analyses were performed with standard software (SPSS v15 and Review Manager RevMan v. 5.0.).
Haplotypic frequencies were estimated using the expectation-maximization and partition-ligation algorithms implemented in the Haploview software [30].
For the meta-analysis, Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated by using raw data for each study and for the pooled population. The Der Simonian and Laird random effects model was used according to the results of the tests of heterogeneity. The combined effect for heterogeneity was calculated by taking the estimated inverse variance. The effect of each study was weighted for its number of patients. P value <0.10 defines a significant degree of heterogeneity and we also used the I2 statistic, with a cut-off point of 25%, to assess heterogeneity between the studies. A sensitivity analysis was performed to test the relative influence of each study on the results. Studies were sequentially dropped, and the effect on the change in the overall degree of heterogeneity was determined.
Results
In order to ascertain the real impact of the HERV-K18 haplotypes in chromosome 1 on autoimmune disease risk, we performed a meta-analysis with 5312 patients and 4032 controls. After the systematic review, only 4 studies remained for further analysis [20], [21], [31], [32]. Due to remarkable differences in allele frequencies found between Japanese and European populations, data from a non-European ancestry study [32] were not included in the meta-analysis. Other Transmission Disequilibrium Test (TDT) study lacking case-control data [20] was also discarded. Finally the meta-analysis pooled data from the previously published cohorts [21], [31] and our MS and RA Spanish cohorts (Figure 2).
Figure 2. Role of HERV-K18 haplotypes A) Meta-analysis of case-control studies in different autoimmune disease cohorts; B) Spanish data stratified by HLA susceptibility alleles (DRB1*15∶01 in MS and shared epitope in RA).
In the Spanish samples, a 6417 bp fragment of chromosome 1q22.3 was amplified by PCR using a primer located in the HERV-K18 env proviral sequence and the other mapping to the CD48 sequence (as described in Methods). Case-control studies from the pre-amplified fragment of the Spanish samples were conducted with the two SNPs covering the haplotypic diversity of the HERV-K18 chromosomal region: a change Y/C at amino acid position 97 and another change W/Stop at 154. As shown in Fig. 2A, no association was observed for haplotype 18.1 (97Y-154Stop; SNP1*A-SNP2*A: p = 0.60, OR = 0.98) or for haplotype 18.2 after performing sensitivity analyses and eliminating the source of heterogeneity (97C-154W; SNP1*G-SNP2*G: p = 0.31, OR = 0.95). Haplotype 18.3 evidenced an overall effect [97Y-154W; SNP1*A-SNP2*G: p = 0.0008; OR (95% CI) = 1.22 (1.09–1.38)] with low heterogeneity among data of the reported cohorts and ours (I2 = 19%). When the RA and MS Spanish cohorts were stratified by the HLA risk factor (Fig. 2B), the shared epitope alleles and HLA-DRB1*15∶01 respectively, even higher homogeneity among cohorts was found (I2 = 11%).
The genotypic frequencies for SNP1 and SNP2 in the Spanish patients and controls are summarized in Table 1. Significant differences were observed for both SNPs when MS patients were stratified by the well-known susceptibility factor HLA-DRB1*15∶01 (MS 1501+ vs. Controls: GG vs. AG+AA: p = 0.02, OR = 1.50 and p = 0.001, OR = 1.60, for SNP1 and SNP2 respectively). No significant association of the tested SNPs with RA risk was reached in the Spanish cohort, most probably due to statistical power restrictions.
Table 1. Genotypic frequencies of polymorphisms within the HERV-K18 sequence in autoimmune disease Spanish patients and controls.
| SNP1 | SNP2 | ||||||||
| AA | AG | GG | GG | AG | AA | ||||
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | ||||
| Controls | 225 (37) | 286 (48) | 90 (15) | 188 (31) | 298 (50) | 112 (19) | |||
| MS | Overall | 372 (39) | 417 (44) | 153 (16) | 334 (36) | 422 (45) | 175 (19) | ||
| 1501 | 1501+ | 114 (37) | 128 (42) | 64 (21)1 | 128 (42)2 | 121 (40) | 53 (18) | ||
| 1501− | 196 (39) | 241 (48) | 70 (14) | 155 (31) | 246 (49) | 102 (20) | |||
| RA | Overall | 197 (43) | 192 (42) | 65 (14) | 132 (30) | 231 (52) | 79 (18) | ||
| SE | SE+ | 107 (44) | 105 (43) | 31 (13) | 65 (28) | 133 (56) | 38 (16) | ||
| SE− | 64 (40) | 69 (43) | 27 (17) | 51 (32) | 77 (49) | 29 (18) | |||
GG vs AG+AA: MS 1501+ vs 1501−: p = 0.01; MS 1501+ vs. Controls: p = 0.02, OR (95%CI) = 1.50 (1.04–2.17).
GG vs AG+AA: MS 1501+ vs 1501−: p = 0.0009; MS 1501+ vs. Controls: p = 0.001, OR (95%CI) = 1.60 (1.19–2.16).
Discussion
Autoimmune diseases are multifactorial conditions determined by the interplay of environmental and genetic components. Particularly, viral infections have been proposed to trigger these diseases in the background of genetic predisposition. In both, multiple sclerosis and rheumatoid arthritis, the main genetic contribution was long ago attributed to specific human leukocyte antigen (HLA) class II alleles on the short arm of chromosome 6p21. Other genes increasing risk to both diseases have been recently ascertained through GWAS [33], [34], and a shared genetic background among them evidenced. However, the genetic component has been incompletely disentangled maybe due to partial coverage of the genome. In fact, associated genes which initially passed undetected by GWAS were sometimes identified through other experimental approaches. One limitation of whole genome association studies corresponds to the difficulty to unambiguously identify repetitive elements by the standard hybridization-based methods used, and consequently polymorphisms in HERVs would remain undetected in GWAS platforms. Prior amplification of genomic target regions, as the one performed in the present work, is required to successfully characterize endogenous retroviral sequences.
Our aim in the present work was the analysis of the role of the HERV-K18 haplotypes in chromosome 1 on autoimmune disease susceptibility. A search with the HERV-K18 env sequence (accession number AF333069, GRCh37.p10 (160666969–160673386); www.ensembl.org, October-2012) evidenced 332 positions in the genome showing total or partial homology, and 27 out of them presented over 85% homology and over 80% of the total length of HERV-K18 env. Among those 27 copies, 21 have open reading frames (ORFs) with one of both of the studied SNPs. No SNP previously found associated with any autoimmune disease through GWAS was observed in the 200 Kb- sequence surrounding those 27 copies. As mentioned above, we focused on the association previously observed within chromosome 1. Former studies regarding the association of HERV-K18 with other autoimmune disease, type 1 diabetes, reported apparently inconsistent results [20], [31], [32]. The genetic evidence presented here corroborates the HERV-K18 influence on autoimmune disease predisposition. Our data is fully concordant with a published report showing the involvement of HERV-K18 on the genetic risk to MS [21]. Provided that HERV-K18 superantigens are strictly dependent on Major Histocompatibility Complex class II for T cell activation, genetic epistasis between both loci is plausible and it seems from our data that a stronger association is found in the subgroup of MS patients carrying the HLA-DRB1*15∶01 risk allele (Table 1).
Given the location of HERV-K18 within the intron of the CD48 gene, the etiology of a putative CD48 variant in linkage disequilibrium with the 18.3 haplotype found associated could not be disregarded. In terms of epigenetic regulation, the ENCODE project data show the signal of the H3K4ME1 histone in the promoter region of CD48 in an immortalized cell line of B lymphocytes, GM12873, indicative of a DNAse hypersensitive site [35]. The impact of CD48 on T cell activation has been recognized with the description of severe T cell signaling defects in the CD48-knockout mouse [36]. T cells reorganize their surface molecules to form a well-structured contact zone with antigen presenting cells, known as immunological synapse, and CD48 in particular was firmly demonstrated to amplify T-cell receptor signaling [37]. Moreover, it has been recently reported that retrotransposable elements, including endogenous retroviruses, can act as an evolutionary mechanism for coordinately boosting the expression of many genes, suggesting that they become functional promoters [38].
The present results reside at the crossroads of environmental and genetic factors influencing autoimmune disease predisposition. Recently, mechanistic data about the transactivation of HERV-K18 by EBV have been released [39], [40], and our work supports the role of the HERV-K18 haplotype on chromosome 1 in autoimmune pathogenesis. In summary, we report the association of the HERV-K18 18.3 haplotype on chromosome 1 with autoimmune disease susceptibility and further studies are warranted to fully elucidate the specific mechanism underlying this association.
Supporting Information
Reported items for meta-analysis.
(DOCX)
Acknowledgments
We thank patients and controls for making this study feasible. Angel García-Martínez and Carmen Martínez provided expert technical assistance.
Funding Statement
Belén de la Hera is recipient of a PhD scholarship from ‘Fondo de Investigaciones Sanitarias’ (FI11/00560), Jezabel Varadé is recipient of a contract from “Ministerio de Economía y Competitividad” (PTA2011-6137-1) and Elena Urcelay works for the Fundación para la Investigación Biomédica-Hospital Clínico San Carlos. Financial support for the study was provided by: Fondo de Investigaciones Sanitarias FEDER-FIS (grant numbers: PI08/1636 and PI10/1985) and Fundación Alicia Koplowitz. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- 2. Kudaka W, Oda T, Jinno Y, Yoshimi N, Aoki Y (2008) Cellular localization of placenta-specific human endogenous retrovirus (HERV) transcripts and their possible implication in pregnancy-induced hypertension. Placenta 29: 282–289. [DOI] [PubMed] [Google Scholar]
- 3. Prudhomme S, Bonnaud B, Mallet F (2005) Endogenous retroviruses and animal reproduction. Cytogenet Genome Res 110: 353–364. [DOI] [PubMed] [Google Scholar]
- 4. Nakagawa K, Brusic V, McColl G, Harrison LC (1997) Direct evidence for the expression of multiple endogenous retroviruses in the synovial compartment in rheumatoid arthritis. Arthritis Rheum 40: 627–638. [DOI] [PubMed] [Google Scholar]
- 5. Moles JP, Tesniere A, Guilhou JJ (2005) A new endogenous retroviral sequence is expressed in skin of patients with psoriasis. Br J Dermatol 153: 83–89. [DOI] [PubMed] [Google Scholar]
- 6. Ogasawara H, Kageyama M, Yamaji K, Takasaki Y (2001) The possibility that autoimmune disease can be induced by a molecular mimicry mechanism between autoantigen and human endogenous retrovirus. Lupus 19: 111–113. [DOI] [PubMed] [Google Scholar]
- 7. Johnston JB, Silva C, Holden J, Warren KG, Clark AW, et al. (2001) Monocyte activation and differentiation augment human endogenous retrovirus expression: implications for inflammatory brain diseases. Ann Neurol 50: 434–442. [DOI] [PubMed] [Google Scholar]
- 8. Perron H, Lazarini F, Ruprecht K, Pechoux-Longin C, Seilhean D, et al. (2005) Human endogenous retrovirus (HERV)-W ENV and GAG proteins: physiological expression in human brain and pathophysiological modulation in multiple sclerosis lesions. J Neurovirol 11: 23–33. [DOI] [PubMed] [Google Scholar]
- 9. Brudek T, Christensen T, Aagaard L, Petersen T, Hansen HJ, et al. (2009) B cells and monocytes from patients with active multiple sclerosis exhibit increased surface expression of both HERV-H Env and HERV-W Env, accompanied by increased seroreactivity. Retrovirology 6: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Laska MJ, Brudek T, Nissen KK, Christensen T, Moller-Larsen A, et al. (2012) Expression of HERV-Fc1, a human endogenous retrovirus, is increased in patients with active multiple sclerosis. J Virol 86: 3713–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bengtsson A, Blomberg J, Nived O, Pipkorn R, Toth L, et al. (1996) Selective antibody reactivity with peptides from human endogenous retroviruses and nonviral poly(amino acids) in patients with systemic lupus erythematosus. Arthritis Rheum 39: 1654–1663. [DOI] [PubMed] [Google Scholar]
- 12. Nexo BA, Christensen T, Frederiksen J, Moller-Larsen A, Oturai AB, et al. (2011) The etiology of multiple sclerosis: genetic evidence for the involvement of the human endogenous retrovirus HERV-Fc1. PLoS One 6: e16652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hansen B, Oturai AB, Harbo HF, Celius EG, Nissen KK, et al. (2011) Genetic association of multiple sclerosis with the marker rs391745 near the endogenous retroviral locus HERV-Fc1: analysis of disease subtypes. PLoS One 6: e26438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Conrad B, Weissmahr RN, Boni J, Arcari R, Schupbach J, et al. (1997) A human endogenous retroviral superantigen as candidate autoimmune gene in type I diabetes. Cell 90: 303–313. [DOI] [PubMed] [Google Scholar]
- 15. Portis JL (2002) Perspectives on the role of endogenous human retroviruses in autoimmune diseases. Virology 296: 1–5. [DOI] [PubMed] [Google Scholar]
- 16. Sicat J, Sutkowski N, Huber BT (2005) Expression of human endogenous retrovirus HERV-K18 superantigen is elevated in juvenile rheumatoid arthritis. J Rheumatol 32: 1821–1831. [PubMed] [Google Scholar]
- 17. Stauffer Y, Marguerat S, Meylan F, Ucla C, Sutkowski N, et al. (2001) Interferon-alpha-induced endogenous superantigen. a model linking environment and autoimmunity. Immunity 15: 591–601. [DOI] [PubMed] [Google Scholar]
- 18. Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, et al. (2009) Finding the missing heritability of complex diseases. Nature 461: 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Momozawa Y, Mni M, Nakamura K, Coppieters W, Almer S, et al. (2011) Resequencing of positional candidates identifies low frequency IL23R coding variants protecting against inflammatory bowel disease. Nat Genet 43: 43–47. [DOI] [PubMed] [Google Scholar]
- 20. Marguerat S, Wang WY, Todd JA, Conrad B (2004) Association of human endogenous retrovirus K-18 polymorphisms with type 1 diabetes. Diabetes 53: 852–854. [DOI] [PubMed] [Google Scholar]
- 21. Tai AK, O’Reilly EJ, Alroy KA, Simon KC, Munger KL, et al. (2008) Human endogenous retrovirus-K18 Env as a risk factor in multiple sclerosis. Mult Scler 14: 1175–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhernakova A, van Diemen CC, Wijmenga C (2009) Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat Rev Genet 10: 43–55. [DOI] [PubMed] [Google Scholar]
- 23. Freimanis G, Hooley P, Ejtehadi HD, Ali HA, Veitch A, et al. (2010) A role for human endogenous retrovirus-K (HML-2) in rheumatoid arthritis: investigating mechanisms of pathogenesis. Clin Exp Immunol 160: 340–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alvarez-Lafuente R, Fernandez-Gutierrez B, de Miguel S, Jover JA, Rollin R, et al. (2005) Potential relationship between herpes viruses and rheumatoid arthritis: analysis with quantitative real time polymerase chain reaction. Ann Rheum Dis 64: 1357–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lunemann JD, Kamradt T, Martin R, Munz C (2007) Epstein-barr virus: environmental trigger of multiple sclerosis? J Virol 81: 6777–6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sutkowski N, Conrad B, Thorley-Lawson DA, Huber BT (2001) Epstein-Barr virus transactivates the human endogenous retrovirus HERV-K18 that encodes a superantigen. Immunity 15: 579–589. [DOI] [PubMed] [Google Scholar]
- 27. Turcanova VL, Bundgaard B, Hollsberg P (2009) Human herpesvirus-6B induces expression of the human endogenous retrovirus K18-encoded superantigen. J Clin Virol 46: 15–19. [DOI] [PubMed] [Google Scholar]
- 28. McDonald WI, Compston A, Edan G, Goodkin D, Hartung HP, et al. (2001) Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the diagnosis of multiple sclerosis. Ann Neurol 50: 121–127. [DOI] [PubMed] [Google Scholar]
- 29. Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, et al. (2010) 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum 62: 2569–2581. [DOI] [PubMed] [Google Scholar]
- 30. Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265. [DOI] [PubMed] [Google Scholar]
- 31. Ramos-Lopez E, Ghebru S, Van Autreve J, Aminkeng F, Herwig J, et al. (2006) Neither an intronic CA repeat within the CD48 gene nor the HERV-K18 polymorphisms are associated with type 1 diabetes. Tissue Antigens 68: 147–152. [DOI] [PubMed] [Google Scholar]
- 32. Kinjo Y, Matsuura N, Yokota Y, Ohtsu S, Nomoto K, et al. (2001) Identification of nonsynonymous polymorphisms in the superantigen-coding region of IDDMK1,2 22 and a pilot study on the association between IDDMK1,2 22 and type 1 diabetes. J Hum Genet 46: 712–716. [DOI] [PubMed] [Google Scholar]
- 33. Sawcer S, Hellenthal G, Pirinen M, Spencer CC, Patsopoulos NA, et al. (2011) Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476: 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, et al. (2010) Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet 42: 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rosenbloom KR, Dreszer TR, Long JC, Malladi VS, Sloan CA, et al. (2012) ENCODE whole-genome data in the UCSC Genome Browser: update 2012. Nucleic Acids Res 40: D912–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gonzalez-Cabrero J, Wise CJ, Latchman Y, Freeman GJ, Sharpe AH, et al. (1999) CD48-deficient mice have a pronounced defect in CD4(+) T cell activation. Proc Natl Acad Sci U S A 96: 1019–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Muhammad A, Schiller HB, Forster F, Eckerstorfer P, Geyeregger R, et al. (2009) Sequential cooperation of CD2 and CD48 in the buildup of the early TCR signalosome. J Immunol 182: 7672–7680. [DOI] [PubMed] [Google Scholar]
- 38. Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, et al. (2012) Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hsiao FC, Lin M, Tai A, Chen G, Huber BT (2006) Cutting edge: Epstein-Barr virus transactivates the HERV-K18 superantigen by docking to the human complement receptor 2 (CD21) on primary B cells. J Immunol 177: 2056–2060. [DOI] [PubMed] [Google Scholar]
- 40. Hsiao FC, Tai AK, Deglon A, Sutkowski N, Longnecker R, et al. (2009) EBV LMP-2A employs a novel mechanism to transactivate the HERV-K18 superantigen through its ITAM. Virology 385: 261–266. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Reported items for meta-analysis.
(DOCX)