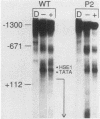
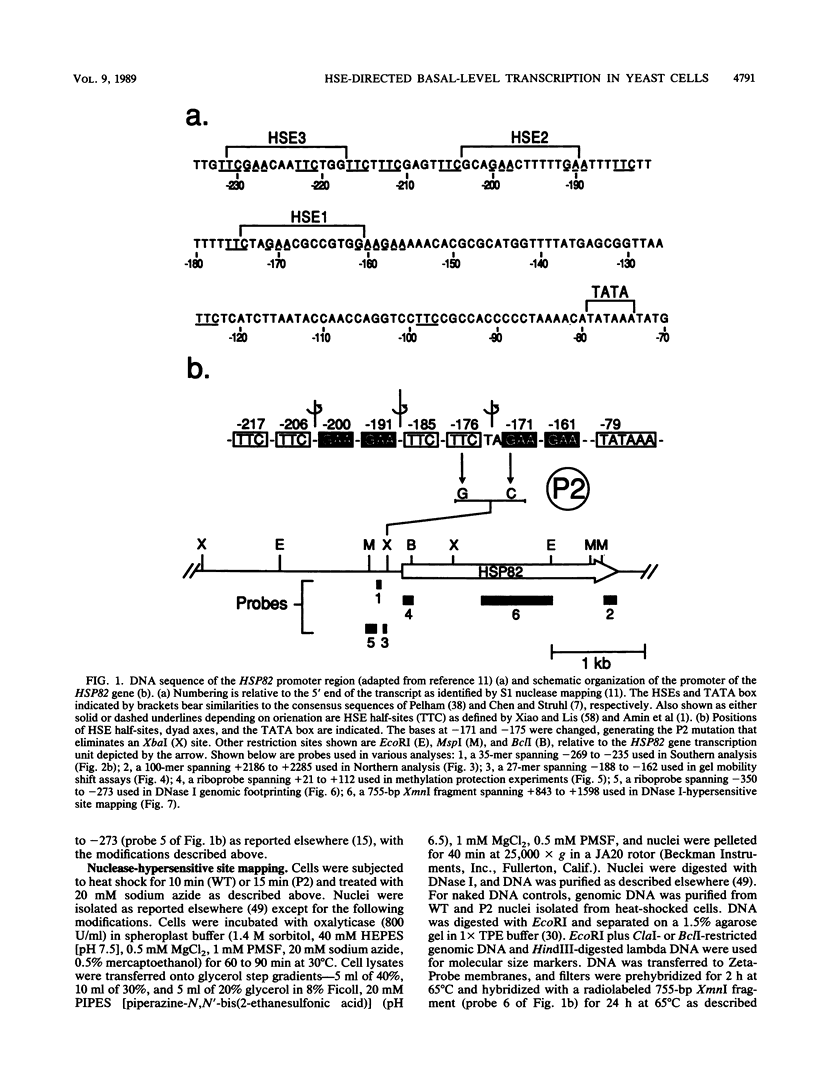
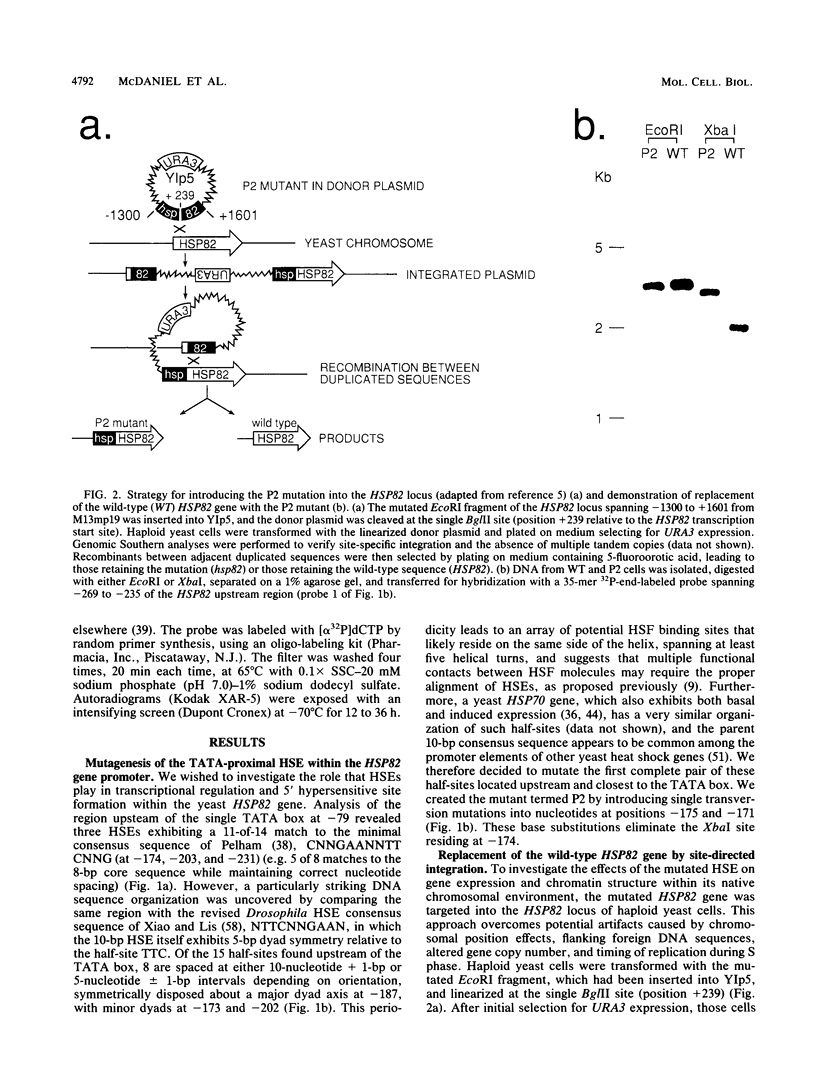
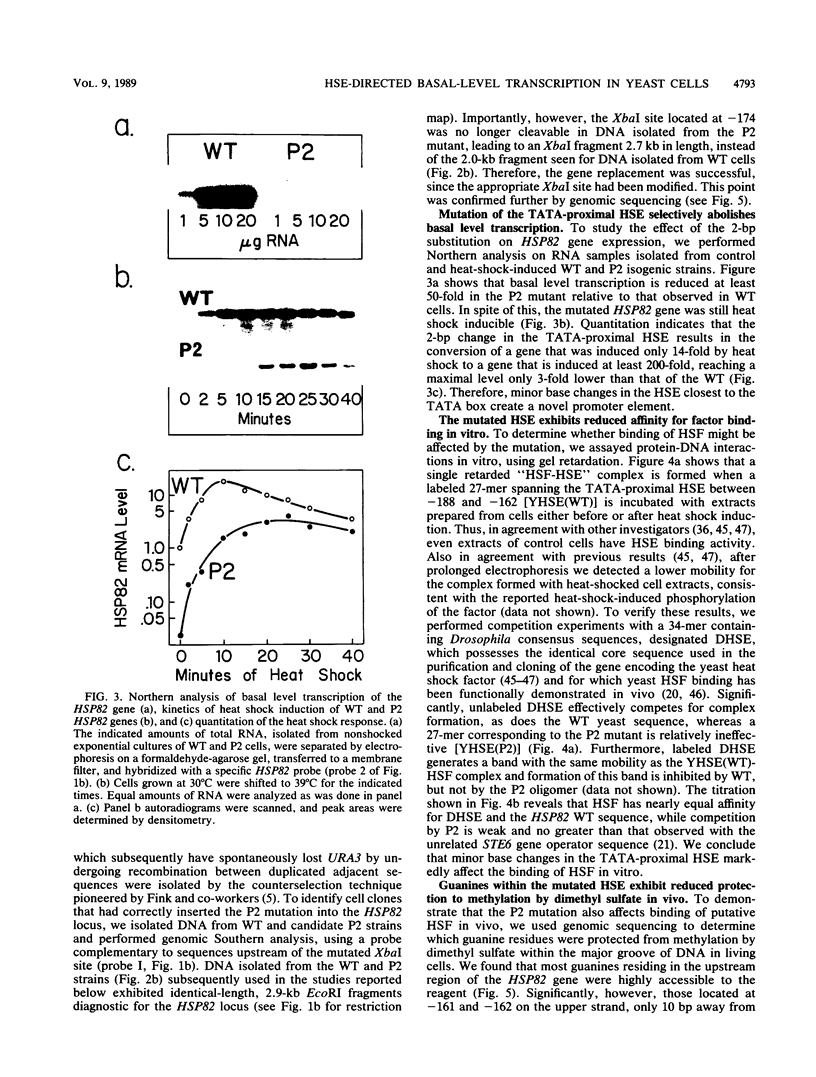
Abstract
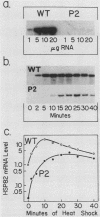
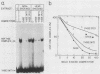
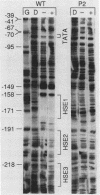
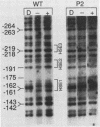
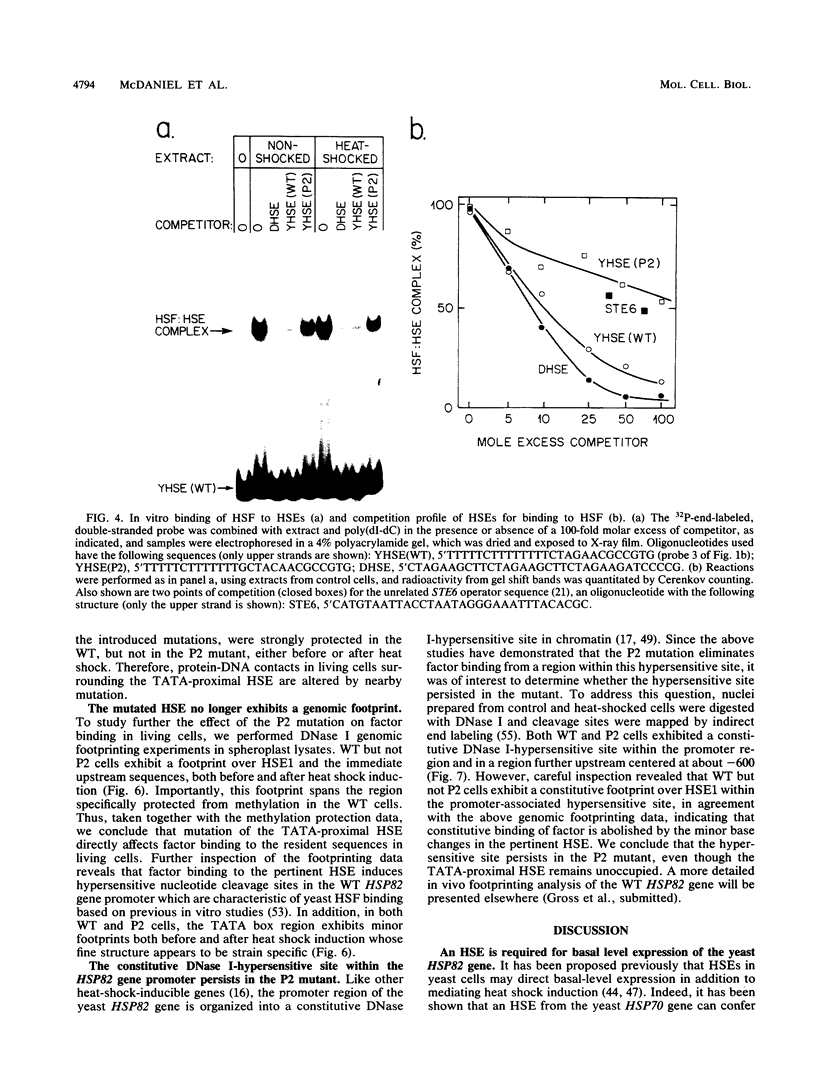
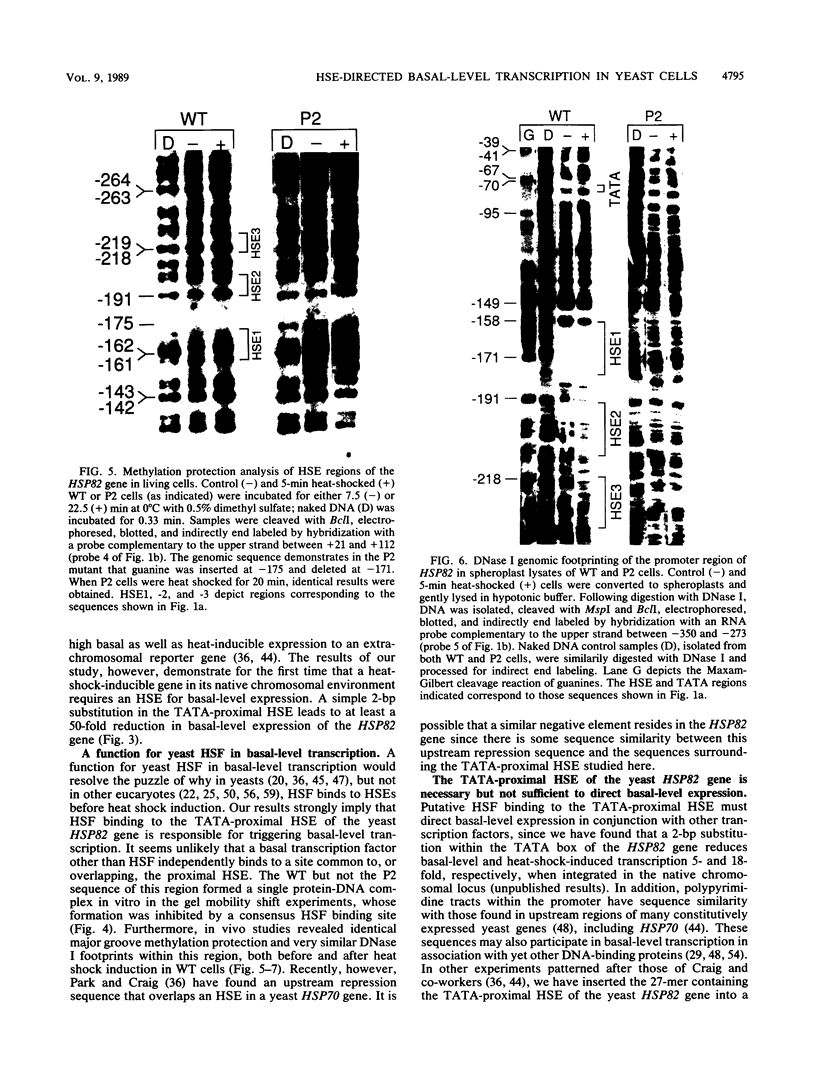
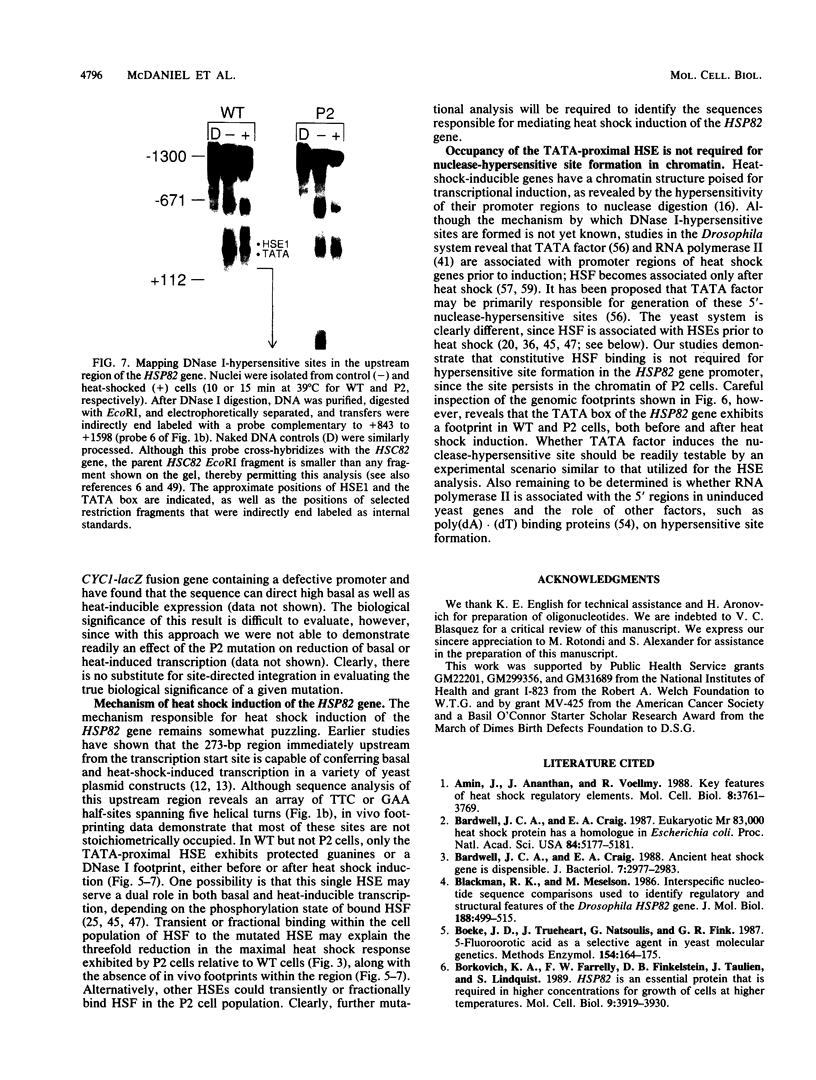
Previous studies have shown that heat shock factor is constitutively bound to heat shock elements in Saccharomyces cerevisiae. We demonstrate that mutation of the heat shock element closest to the TATA box of the yeast HSP82 promoter abolishes basal-level transcription without markedly affecting inducibility. The mutated heat shock element no longer bound putative heat shock factor, either in vitro or in vivo, but still resided within a nuclease-hypersensitive site in the chromatin. Thus, constitutive binding of heat shock factor to heat shock elements in S. cerevisiae appears to functionally direct basal-level transcription.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amin J., Ananthan J., Voellmy R. Key features of heat shock regulatory elements. Mol Cell Biol. 1988 Sep;8(9):3761–3769. doi: 10.1128/mcb.8.9.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell J. C., Craig E. A. Ancient heat shock gene is dispensable. J Bacteriol. 1988 Jul;170(7):2977–2983. doi: 10.1128/jb.170.7.2977-2983.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell J. C., Craig E. A. Eukaryotic Mr 83,000 heat shock protein has a homologue in Escherichia coli. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5177–5181. doi: 10.1073/pnas.84.15.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman R. K., Meselson M. Interspecific nucleotide sequence comparisons used to identify regulatory and structural features of the Drosophila hsp82 gene. J Mol Biol. 1986 Apr 20;188(4):499–515. doi: 10.1016/s0022-2836(86)80001-8. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Trueheart J., Natsoulis G., Fink G. R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Borkovich K. A., Farrelly F. W., Finkelstein D. B., Taulien J., Lindquist S. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol Cell Biol. 1989 Sep;9(9):3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Struhl K. Saturation mutagenesis of a yeast his3 "TATA element": genetic evidence for a specific TATA-binding protein. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2691–2695. doi: 10.1073/pnas.85.8.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen R. S., Meselson M. Periodic interactions of heat shock transcriptional elements. Nature. 1988 Apr 28;332(6167):856–858. doi: 10.1038/332856a0. [DOI] [PubMed] [Google Scholar]
- Dragon E. A., Sias S. R., Kato E. A., Gabe J. D. The genome of Trypanosoma cruzi contains a constitutively expressed, tandemly arranged multicopy gene homologous to a major heat shock protein. Mol Cell Biol. 1987 Mar;7(3):1271–1275. doi: 10.1128/mcb.7.3.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrelly F. W., Finkelstein D. B. Complete sequence of the heat shock-inducible HSP90 gene of Saccharomyces cerevisiae. J Biol Chem. 1984 May 10;259(9):5745–5751. [PubMed] [Google Scholar]
- Finkelstein D. B., Strausberg S. Heat shock-regulated production of Escherichia coli beta-galactosidase in Saccharomyces cerevisiae. Mol Cell Biol. 1983 Sep;3(9):1625–1633. doi: 10.1128/mcb.3.9.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein D. B., Strausberg S. Identification and expression of a cloned yeast heat shock gene. J Biol Chem. 1983 Feb 10;258(3):1908–1913. [PubMed] [Google Scholar]
- Finkelstein D. B., Strausberg S., McAlister L. Alterations of transcription during heat shock of Saccharomyces cerevisiae. J Biol Chem. 1982 Jul 25;257(14):8405–8411. [PubMed] [Google Scholar]
- Gross D. S., Collins K. W., Hernandez E. M., Garrard W. T. Vacuum blotting: a simple method for transferring DNA from sequencing gels to nylon membranes. Gene. 1988 Dec 30;74(2):347–356. doi: 10.1016/0378-1119(88)90168-0. [DOI] [PubMed] [Google Scholar]
- Gross D. S., Garrard W. T. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen B. K., Pelham H. R. Constitutive binding of yeast heat shock factor to DNA in vivo. Mol Cell Biol. 1988 Nov;8(11):5040–5042. doi: 10.1128/mcb.8.11.5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D., Herskowitz I. A repressor (MAT alpha 2 Product) and its operator control expression of a set of cell type specific genes in yeast. Cell. 1985 Aug;42(1):237–247. doi: 10.1016/s0092-8674(85)80119-7. [DOI] [PubMed] [Google Scholar]
- Kingston R. E., Schuetz T. J., Larin Z. Heat-inducible human factor that binds to a human hsp70 promoter. Mol Cell Biol. 1987 Apr;7(4):1530–1534. doi: 10.1128/mcb.7.4.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyasu S., Nishida E., Kadowaki T., Matsuzaki F., Iida K., Harada F., Kasuga M., Sakai H., Yahara I. Two mammalian heat shock proteins, HSP90 and HSP100, are actin-binding proteins. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8054–8058. doi: 10.1073/pnas.83.21.8054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S., Lindquist S. Changing patterns of gene expression during sporulation in yeast. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7323–7327. doi: 10.1073/pnas.81.23.7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson J. S., Schuetz T. J., Kingston R. E. Activation in vitro of sequence-specific DNA binding by a human regulatory factor. Nature. 1988 Sep 22;335(6188):372–375. doi: 10.1038/335372a0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A. The use of intensifying screens or organic scintillators for visualizing radioactive molecules resolved by gel electrophoresis. Methods Enzymol. 1980;65(1):363–371. doi: 10.1016/s0076-6879(80)65047-2. [DOI] [PubMed] [Google Scholar]
- Lindquist S., Craig E. A. The heat-shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- Lindquist S. The heat-shock response. Annu Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- Lue N. F., Buchman A. R., Kornberg R. D. Activation of yeast RNA polymerase II transcription by a thymidine-rich upstream element in vitro. Proc Natl Acad Sci U S A. 1989 Jan;86(2):486–490. doi: 10.1073/pnas.86.2.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mazzarella R. A., Green M. ERp99, an abundant, conserved glycoprotein of the endoplasmic reticulum, is homologous to the 90-kDa heat shock protein (hsp90) and the 94-kDa glucose regulated protein (GRP94). J Biol Chem. 1987 Jun 25;262(18):8875–8883. [PubMed] [Google Scholar]
- Mosser D. D., Theodorakis N. G., Morimoto R. I. Coordinate changes in heat shock element-binding activity and HSP70 gene transcription rates in human cells. Mol Cell Biol. 1988 Nov;8(11):4736–4744. doi: 10.1128/mcb.8.11.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida E., Koyasu S., Sakai H., Yahara I. Calmodulin-regulated binding of the 90-kDa heat shock protein to actin filaments. J Biol Chem. 1986 Dec 5;261(34):16033–16036. [PubMed] [Google Scholar]
- Park H. O., Craig E. A. Positive and negative regulation of basal expression of a yeast HSP70 gene. Mol Cell Biol. 1989 May;9(5):2025–2033. doi: 10.1128/mcb.9.5.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C. S., Topol J. A Drosophila RNA polymerase II transcription factor contains a promoter-region-specific DNA-binding activity. Cell. 1984 Feb;36(2):357–369. doi: 10.1016/0092-8674(84)90229-0. [DOI] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose D. W., Wettenhall R. E., Kudlicki W., Kramer G., Hardesty B. The 90-kilodalton peptide of the heme-regulated eIF-2 alpha kinase has sequence similarity with the 90-kilodalton heat shock protein. Biochemistry. 1987 Oct 20;26(21):6583–6587. doi: 10.1021/bi00395a003. [DOI] [PubMed] [Google Scholar]
- Rougvie A. E., Lis J. T. The RNA polymerase II molecule at the 5' end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988 Sep 9;54(6):795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Sanchez E. R., Meshinchi S., Tienrungroj W., Schlesinger M. J., Toft D. O., Pratt W. B. Relationship of the 90-kDa murine heat shock protein to the untransformed and transformed states of the L cell glucocorticoid receptor. J Biol Chem. 1987 May 25;262(15):6986–6991. [PubMed] [Google Scholar]
- Sanchez E. R., Redmond T., Scherrer L. C., Bresnick E. H., Welsh M. J., Pratt W. B. Evidence that the 90-kilodalton heat shock protein is associated with tubulin-containing complexes in L cell cytosol and in intact PtK cells. Mol Endocrinol. 1988 Aug;2(8):756–760. doi: 10.1210/mend-2-8-756. [DOI] [PubMed] [Google Scholar]
- Slater M. R., Craig E. A. Transcriptional regulation of an hsp70 heat shock gene in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987 May;7(5):1906–1916. doi: 10.1128/mcb.7.5.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger P. K., Lewis M. J., Pelham H. R. Heat shock factor is regulated differently in yeast and HeLa cells. Nature. 1987 Sep 3;329(6134):81–84. doi: 10.1038/329081a0. [DOI] [PubMed] [Google Scholar]
- Sorger P. K., Pelham H. R. Purification and characterization of a heat-shock element binding protein from yeast. EMBO J. 1987 Oct;6(10):3035–3041. doi: 10.1002/j.1460-2075.1987.tb02609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger P. K., Pelham H. R. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell. 1988 Sep 9;54(6):855–864. doi: 10.1016/s0092-8674(88)91219-6. [DOI] [PubMed] [Google Scholar]
- Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8419–8423. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szent-Györgyi C., Finkelstein D. B., Garrard W. T. Sharp boundaries demarcate the chromatin structure of a yeast heat-shock gene. J Mol Biol. 1987 Jan 5;193(1):71–80. doi: 10.1016/0022-2836(87)90628-0. [DOI] [PubMed] [Google Scholar]
- Thomas G. H., Elgin S. C. Protein/DNA architecture of the DNase I hypersensitive region of the Drosophila hsp26 promoter. EMBO J. 1988 Jul;7(7):2191–2201. doi: 10.1002/j.1460-2075.1988.tb03058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuite M. F., Bossier P., Fitch I. T. A highly conserved sequence in yeast heat shock gene promoters. Nucleic Acids Res. 1988 Dec 23;16(24):11845–11845. doi: 10.1093/nar/16.24.11845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederrecht G., Seto D., Parker C. S. Isolation of the gene encoding the S. cerevisiae heat shock transcription factor. Cell. 1988 Sep 9;54(6):841–853. doi: 10.1016/s0092-8674(88)91197-x. [DOI] [PubMed] [Google Scholar]
- Wiederrecht G., Shuey D. J., Kibbe W. A., Parker C. S. The Saccharomyces and Drosophila heat shock transcription factors are identical in size and DNA binding properties. Cell. 1987 Feb 13;48(3):507–515. doi: 10.1016/0092-8674(87)90201-7. [DOI] [PubMed] [Google Scholar]
- Winter E., Varshavsky A. A DNA binding protein that recognizes oligo(dA).oligo(dT) tracts. EMBO J. 1989 Jun;8(6):1867–1877. doi: 10.1002/j.1460-2075.1989.tb03583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Wu C. Two protein-binding sites in chromatin implicated in the activation of heat-shock genes. Nature. 1984 May 17;309(5965):229–234. doi: 10.1038/309229a0. [DOI] [PubMed] [Google Scholar]
- Wu C., Wilson S., Walker B., Dawid I., Paisley T., Zimarino V., Ueda H. Purification and properties of Drosophila heat shock activator protein. Science. 1987 Nov 27;238(4831):1247–1253. doi: 10.1126/science.3685975. [DOI] [PubMed] [Google Scholar]
- Xiao H., Lis J. T. Germline transformation used to define key features of heat-shock response elements. Science. 1988 Mar 4;239(4844):1139–1142. doi: 10.1126/science.3125608. [DOI] [PubMed] [Google Scholar]
- Zimarino V., Wu C. Induction of sequence-specific binding of Drosophila heat shock activator protein without protein synthesis. 1987 Jun 25-Jul 1Nature. 327(6124):727–730. doi: 10.1038/327727a0. [DOI] [PubMed] [Google Scholar]