Abstract
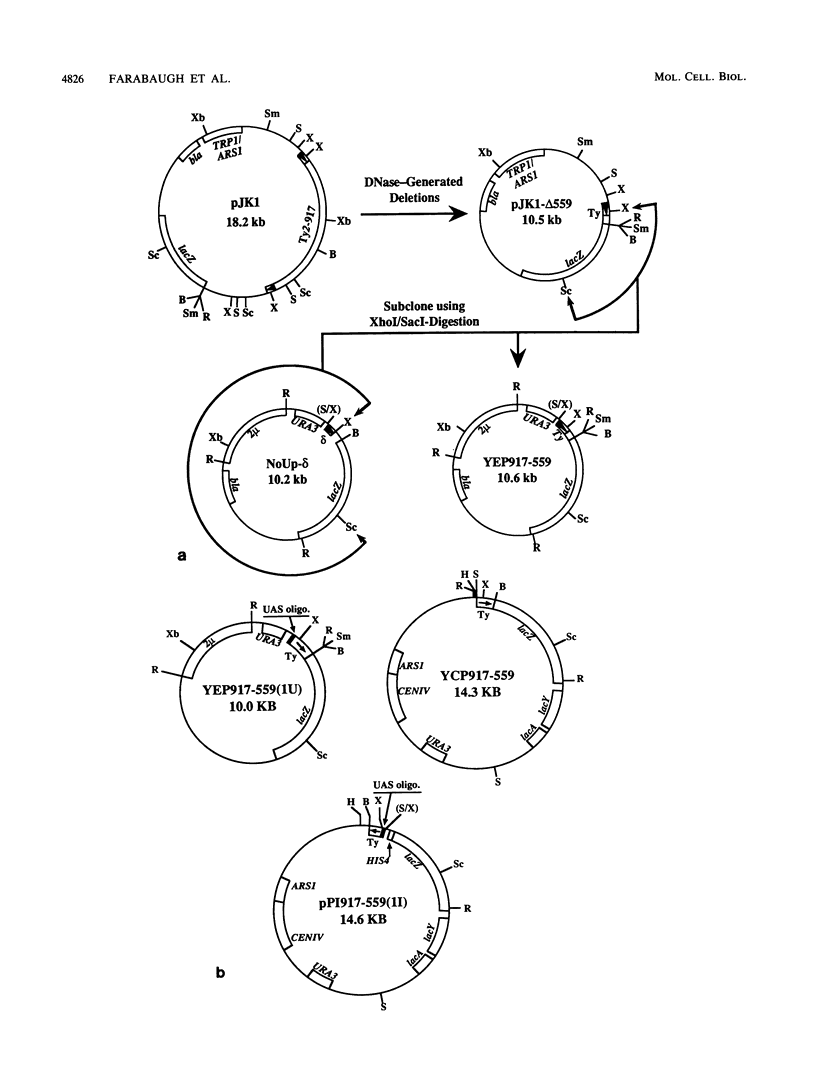
The Ty2-917 element is a member of the Ty2 class of retroviruslike transposable elements of Saccharomyces cerevisiae. We showed that regions downstream of the Ty2-917 transcription start site modulate its transcription. One region was located downstream of the transcription initiation site (position 240) and within the first 559 base pairs of the element. This region had a dramatic effect, causing an approximately 1,000-fold increase in steady-state levels of RNA. The region stimulated transcription when placed in either orientation upstream of a heterologous gene, HIS4, lacking its own upstream activation sequence (UAS). We termed this positively acting region an enhancer, by analogy to sites described in higher cells, to distinguish it from yeast UASs which do not function when placed within the transcribed portion of the gene. Though, like some higher eucaryotic enhancers, the Ty2-917 enhancer is located within the transcribed region, it is unlike them in that it occurs within a coding region rather than in an intron. The Ty2-917 enhancer and the Ty2-917 UAS had a synergistic effect on transcription, together stimulating transcription 15-fold over the predicted additive effect. We also identified a site which decreases RNA accumulation, located about 750 base pairs into the element. This site functioned in only one orientation when inserted upstream of the UAS-less heterologous gene. The site was similar to silencers, or negative enhancers, in that it acted to repress transcription from outside the transcribed region, but was distinct in that the function of a canonical silencer was independent of orientation.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams S. E., Mellor J., Gull K., Sim R. B., Tuite M. F., Kingsman S. M., Kingsman A. J. The functions and relationships of Ty-VLP proteins in yeast reflect those of mammalian retroviral proteins. Cell. 1987 Apr 10;49(1):111–119. doi: 10.1016/0092-8674(87)90761-6. [DOI] [PubMed] [Google Scholar]
- Arndt K. T., Styles C. A., Fink G. R. A suppressor of a HIS4 transcriptional defect encodes a protein with homology to the catalytic subunit of protein phosphatases. Cell. 1989 Feb 24;56(4):527–537. doi: 10.1016/0092-8674(89)90576-x. [DOI] [PubMed] [Google Scholar]
- Arndt K. T., Styles C., Fink G. R. Multiple global regulators control HIS4 transcription in yeast. Science. 1987 Aug 21;237(4817):874–880. doi: 10.1126/science.3303332. [DOI] [PubMed] [Google Scholar]
- Barnes W. M., Bevan M., Son P. H. Kilo-sequencing: creation of an ordered nest of asymmetric deletions across a large target sequence carried on phage M13. Methods Enzymol. 1983;101:98–122. doi: 10.1016/0076-6879(83)01008-3. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Garfinkel D. J., Styles C. A., Fink G. R. Ty elements transpose through an RNA intermediate. Cell. 1985 Mar;40(3):491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- Brand A. H., Breeden L., Abraham J., Sternglanz R., Nasmyth K. Characterization of a "silencer" in yeast: a DNA sequence with properties opposite to those of a transcriptional enhancer. Cell. 1985 May;41(1):41–48. doi: 10.1016/0092-8674(85)90059-5. [DOI] [PubMed] [Google Scholar]
- Cameron J. R., Loh E. Y., Davis R. W. Evidence for transposition of dispersed repetitive DNA families in yeast. Cell. 1979 Apr;16(4):739–751. doi: 10.1016/0092-8674(79)90090-4. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Martinez-Arias A., Shapira S. K., Chou J. Beta-galactosidase gene fusions for analyzing gene expression in escherichia coli and yeast. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- Ciriacy M., Williamson V. M. Analysis of mutations affecting Ty-mediated gene expression in Saccharomyces cerevisiae. Mol Gen Genet. 1981;182(1):159–163. doi: 10.1007/BF00422784. [DOI] [PubMed] [Google Scholar]
- Clare J., Farabaugh P. Nucleotide sequence of a yeast Ty element: evidence for an unusual mechanism of gene expression. Proc Natl Acad Sci U S A. 1985 May;82(9):2829–2833. doi: 10.1073/pnas.82.9.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Company M., Adler C., Errede B. Identification of a Ty1 regulatory sequence responsive to STE7 and STE12. Mol Cell Biol. 1988 Jun;8(6):2545–2554. doi: 10.1128/mcb.8.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Company M., Errede B. A Ty1 cell-type-specific regulatory sequence is a recognition element for a constitutive binding factor. Mol Cell Biol. 1988 Dec;8(12):5299–5309. doi: 10.1128/mcb.8.12.5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Company M., Errede B. Cell-type-dependent gene activation by yeast transposon Ty1 involves multiple regulatory determinants. Mol Cell Biol. 1987 Sep;7(9):3205–3211. doi: 10.1128/mcb.7.9.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coney L. R., Roeder G. S. Control of yeast gene expression by transposable elements: maximum expression requires a functional Ty activator sequence and a defective Ty promoter. Mol Cell Biol. 1988 Oct;8(10):4009–4017. doi: 10.1128/mcb.8.10.4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder R. T., Loh E. Y., Davis R. W. RNA from the yeast transposable element Ty1 has both ends in the direct repeats, a structure similar to retrovirus RNA. Proc Natl Acad Sci U S A. 1983 May;80(9):2432–2436. doi: 10.1073/pnas.80.9.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder R. T., St John T. P., Stinchcomb D. T., Davis R. W., Scherer S., Davis R. W. Studies on the transposable element Ty1 of yeast. I. RNA homologous to Ty1. II. Recombination and expression of Ty1 and adjacent sequences. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):581–591. doi: 10.1101/sqb.1981.045.01.075. [DOI] [PubMed] [Google Scholar]
- Emori Y., Shiba T., Kanaya S., Inouye S., Yuki S., Saigo K. The nucleotide sequences of copia and copia-related RNA in Drosophila virus-like particles. 1985 Jun 27-Jul 3Nature. 315(6022):773–776. doi: 10.1038/315773a0. [DOI] [PubMed] [Google Scholar]
- Errede B., Cardillo T. S., Wever G., Sherman F., Stiles J. I., Friedman L. R., Sherman F. Studies on transposable elements in yeast. I. ROAM mutations causing increased expression of yeast genes: their activation by signals directed toward conjugation functions and their formation by insertion of Ty1 repetitive elements. II. deletions, duplications, and transpositions of the COR segment that encompasses the structural gene of yeast iso-1-cytochrome c. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):593–607. [PubMed] [Google Scholar]
- Errede B., Company M., Ferchak J. D., Hutchison C. A., 3rd, Yarnell W. S. Activation regions in a yeast transposon have homology to mating type control sequences and to mammalian enhancers. Proc Natl Acad Sci U S A. 1985 Aug;82(16):5423–5427. doi: 10.1073/pnas.82.16.5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errede B., Company M., Hutchison C. A., 3rd Ty1 sequence with enhancer and mating-type-dependent regulatory activities. Mol Cell Biol. 1987 Jan;7(1):258–265. doi: 10.1128/mcb.7.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P. J., Fink G. R. Insertion of the eukaryotic transposable element Ty1 creates a 5-base pair duplication. Nature. 1980 Jul 24;286(5771):352–356. doi: 10.1038/286352a0. [DOI] [PubMed] [Google Scholar]
- Fulton A. M., Rathjen P. D., Kingsman S. M., Kingsman A. J. Upstream and downstream transcriptional control signals in the yeast retrotransposon, TY. Nucleic Acids Res. 1988 Jun 24;16(12):5439–5458. doi: 10.1093/nar/16.12.5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel D. J., Boeke J. D., Fink G. R. Ty element transposition: reverse transcriptase and virus-like particles. Cell. 1985 Sep;42(2):507–517. doi: 10.1016/0092-8674(85)90108-4. [DOI] [PubMed] [Google Scholar]
- Goel A., Pearlman R. E. Transposable element-mediated enhancement of gene expression in Saccharomyces cerevisiae involves sequence-specific binding of a trans-acting factor. Mol Cell Biol. 1988 Jun;8(6):2572–2580. doi: 10.1128/mcb.8.6.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Hoar E. Upstream activation sites of the CYC1 gene of Saccharomyces cerevisiae are active when inverted but not when placed downstream of the "TATA box". Proc Natl Acad Sci U S A. 1984 Dec;81(24):7860–7864. doi: 10.1073/pnas.81.24.7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983 Apr;32(4):1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L., Holmgren-König M., Khoury G. Transcriptional "silencer" element in rat repetitive sequences associated with the rat insulin 1 gene locus. Proc Natl Acad Sci U S A. 1986 May;83(10):3151–3155. doi: 10.1073/pnas.83.10.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X. B., Clare J. J., Farabaugh P. J. The upstream activation site of a Ty2 element of yeast is necessary but not sufficient to promote maximal transcription of the element. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8520–8524. doi: 10.1073/pnas.84.23.8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb D. D., Padgett R. W., Hardies S. C., Shehee W. R., Comer M. B., Edgell M. H., Hutchison C. A., 3rd The sequence of a large L1Md element reveals a tandemly repeated 5' end and several features found in retrotransposons. Mol Cell Biol. 1986 Jan;6(1):168–182. doi: 10.1128/mcb.6.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J., Malim M. H., Gull K., Tuite M. F., McCready S., Dibbayawan T., Kingsman S. M., Kingsman A. J. Reverse transcriptase activity and Ty RNA are associated with virus-like particles in yeast. Nature. 1985 Dec 12;318(6046):583–586. doi: 10.1038/318583a0. [DOI] [PubMed] [Google Scholar]
- Mount S. M., Rubin G. M. Complete nucleotide sequence of the Drosophila transposable element copia: homology between copia and retroviral proteins. Mol Cell Biol. 1985 Jul;5(7):1630–1638. doi: 10.1128/mcb.5.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagawa F., Fink G. R. The relationship between the "TATA" sequence and transcription initiation sites at the HIS4 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8557–8561. doi: 10.1073/pnas.82.24.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Rathjen P. D., Kingsman A. J., Kingsman S. M. The yeast ROAM mutation--identification of the sequences mediating host gene activation and cell-type control in the yeast retrotransposon, Ty. Nucleic Acids Res. 1987 Sep 25;15(18):7309–7324. doi: 10.1093/nar/15.18.7309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder G. S., Farabaugh P. J., Chaleff D. T., Fink G. R. The origins of gene instability in yeast. Science. 1980 Sep 19;209(4463):1375–1380. doi: 10.1126/science.6251544. [DOI] [PubMed] [Google Scholar]
- Roeder G. S., Rose A. B., Pearlman R. E. Transposable element sequences involved in the enhancement of yeast gene expression. Proc Natl Acad Sci U S A. 1985 Aug;82(16):5428–5432. doi: 10.1073/pnas.82.16.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Genetic properties and chromatin structure of the yeast gal regulatory element: an enhancer-like sequence. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7865–7869. doi: 10.1073/pnas.81.24.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teem J. L., Rosbash M. Expression of a beta-galactosidase gene containing the ribosomal protein 51 intron is sensitive to the rna2 mutation of yeast. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4403–4407. doi: 10.1073/pnas.80.14.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmington J. R., Waring R. B., Newlon C. S., Indge K. J., Oliver S. G. Nucleotide sequence characterization of Ty 1-17, a class II transposon from yeast. Nucleic Acids Res. 1985 Sep 25;13(18):6679–6693. doi: 10.1093/nar/13.18.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson V. M., Young E. T., Ciriacy M. Transposable elements associated with constitutive expression of yeast alcohol dehydrogenase II. Cell. 1981 Feb;23(2):605–614. doi: 10.1016/0092-8674(81)90156-2. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]