Abstract
Biological aging alters the metabolism and volume of adipose tissue depots. Recent evidence suggests that circadian mechanisms play a role in promoting adipogenesis, obesity, and lipodystrophy. The current study compared cohorts of younger (5–9 months) and older (24–28 months) C57BL/6 mice as a function of biological age and circadian time. Advanced age significantly reduced the weight of the brown, epididymal, inguinal, and retroperitoneal adipose depots but not total body weight. The older mice reduced their physical activity by >50% and delayed their activity initiation after light offset. The expressed transcriptome in brown and white adipose depots and liver of both cohorts displayed evidence of circadian rhythmicity; however, the oscillating mRNAs differed significantly between age groups and across tissues. The amplitude of Cry1, a component of the negative arm of the circadian apparatus, and downstream regulators such as Rev-erbα were elevated in the older relative to the younger cohorts as a function of circadian time. Overall, transcript levels differed significantly for 557 (inguinal adipose), 1,016 (liver), and 1,021 (brown adipose) expressed sequences between the cohorts as a function of age. These included transcripts encoding proteins within the canonical and non-canonical Wnt pathways. Since the Wnt pathway regulates adipose stem cell differentiation and shares a critical enzyme, glycogen synthase kinase 3β, with the circadian mechanism, the intersection between these two fundamental regulatory mechanisms merits further investigation with respect to biological aging of adipose tissues.
Electronic supplementary material
The online version of this article (doi:10.1007/s11357-012-9389-7) contains supplementary material, which is available to authorized users.
Keywords: Brown adipose, Circadian, Liver, Oscillation, Transcriptomics, White adipose
Introduction
Biological aging has profound effects in every major adipose tissue depot (Cartwright et al. 2007; Huffman and Barzilai 2009; Tchkonia et al. 2010). The loss of subcutaneous white adipose tissue (WAT) or lipodystrophy in the elderly leads to thinning of the skin, impairing its mechanical resilience, and increasing the risk for bed-sores or decubitus ulcers during prolonged periods of bed rest (Davies 1994). Visceral WAT accumulates with advancing age in men and women (Weisberg et al. 2003; Ortega et al. 2008; Gavi et al. 2007), and inflammatory processes within visceral WAT place the elderly at increased risk for metabolic syndrome, diabetes, and cardiovascular disease (Trayhurn and Beattie 2001; Trayhurn 2005; Cartwright et al. 2007; Huffman and Barzilai 2009). Recent evidence indicates that thermogenic brown fat decreases with age and this change contributes to obesity (Nedergaard et al. 2007; Cypess et al. 2009; van Marken Lichtenbelt et al. 2009; Virtanen et al. 2009). Furthermore, biological aging is associated with increased bone marrow adipogenesis, an etiologic factor underlying osteopenia and osteoporosis (Meunier et al. 1971; Gimble et al. 2006).
Circadian regulation is mediated by transcriptional and post-translational modification of a discrete group of genes/proteins (Hirota and Fukada 2004; Lowrey and Takahashi 2004). Transcriptional regulatory proteins belonging to the basic helix-loop-helix/Per–ARNT–single-minded domain family interact with each other and downstream target proteins to create a self-perpetuating, rhythmic pattern of gene transcription. The proteins BMAL1 and CLOCK (or its ortholog, NPAS2) form a transcriptional regulatory heterodimer that acts as a “positive arm” of the circadian pathway. The BMAL1/CLOCK heterodimer drives transcription of multiple downstream targets. These include the “negative arm” of the circadian pathway, formed by heterodimers of the PERIOD (Per1, Per2, Per3) and CRYPTOCHROME (Cry1, Cry2) family members. The PER/CRY heterodimer provides a feedback loop that suppresses the BMAL1/CLOCK complex. Additional BMAL1/CLOCK downstream targets include the leucine zipper transcriptional regulatory proteins albumin D box binding protein (DBP), E4BP4 (also known as nuclear factor IL-3), hepatocyte leukemia factor, nocturnin, and thyrotroph embryonic factor, as well as Rev-Erb α, a member of the nuclear hormone receptor family (Fonjallaz et al. 1996; Green and Besharse 1996; Mitsui et al. 2001; Yin et al. 2007). The serine/threonine kinases, casein kinase Iε and glycogen synthase kinase 3β (GSK3 β), serve as post-translational regulators of BMAL1, PER, and other proteins. Once phosphorylated, these proteins are targeted to the ubiquitin/proteasomal pathway for degradation. Together, this core circadian regulatory complex constitutes a self-contained feedback loop whose rate-limiting protein levels oscillate in a ∼24-h manner. It is noteworthy that BMAL1, nocturnin, and Rev-erb α have all been implicated as transcriptional regulators of adipogenesis in pre-adipocyte cell models (Shimba et al. 2004, 2005; Kawai et al. 2010; Kumar et al. 2010).
Additional evidence links circadian mechanisms to adipose biology, obesity, and their related metabolic co-morbidities (Green et al. 2008; Bray and Young 2009; Gimble and Floyd 2009; Marcheva et al. 2009). Outside of the central nervous system, adipose tissue physiology exhibits some of the most robust associations with seasonal, diurnal, and circadian rhythms. The serum levels of glucocorticoids and melatonin, factors with potent adipogenic properties, exhibit circadian oscillations (Hirota and Fukada 2004; Lowrey and Takahashi 2004). Eating during periods of darkness (Holmback et al. 2002, 2003; Spiegel et al. 2004) and antipsychotic drugs influencing circadian rhythms such as lithium chloride and valproic acid (Baptista et al. 1995; Elmslie et al. 2001; Atmaca et al. 2002; Chengappa et al. 2002; Fagiolini et al. 2002, 2003; Iwahana et al. 2004) have been associated with an increased incidence of obesity. The serum levels of multiple adipose-derived proteins, including adiponectin, interleukin-6, leptin, lipoprotein lipase, PAI-1, and tumor necrosis factor-α, display a diurnal profile characterized by a distinct amplitude, zenith (peak), and nadir (trough) (Kotlar and Borensztajn 1977; Goubern and Portet 1981; Saad et al. 1998; Ahmad et al. 2001; Arasaradnam et al. 2002; Mastronardi et al. 2002; Gavrila et al. 2003; Calvani et al. 2004; Ruge et al. 2004; Yildiz et al. 2004). Moreover, transcription factors regulating adipocyte gene markers (PPARα, Rev-Erbα, SREBP) display a circadian expression profile (Lemberger et al. 1996; Torra et al. 2000; Patel et al. 2001). Global transcriptomic approaches by ourselves and others have demonstrated the diurnal/circadian oscillation of up to 20% of expressed genes in adipose tissue as well as the suprachiasmatic nuclei, liver, and heart (Akhtar et al. 2002; Duffield et al. 2002; Panda et al. 2002a, b; Ueda et al. 2002; Hogenesch et al. 2003; Oishi et al. 2003; Sato et al. 2003, 2004). Oscillating genes in adipose tissue included those encoding proteins related to glycolysis, lipolysis, steroidogenesis, redox potential, and iron/heme metabolism (Ptitysn et al. 2006; Zvonic et al. 2006). While “conventional wisdom” (Monk 2005) assumes that biological age impacts circadian biology, this relationship in adipose tissue remains poorly defined. To explore the relation between circadian rhythms and adipose tissue biology, the current study has compared young and old C57BL/6 mice with respect to their physical activity profiles, body composition, and circadian gene expression in brown and white adipose tissue depots and liver. The results confirm the effects of biological aging on the adipose tissue depots; moreover, the data demonstrate that biological aging results in pronounced alteration of the circadian mechanisms in metabolically active organs.
Methods
Materials
All reagents were purchased from Fisher Scientific (Dallas, TX, USA) or Sigma-Aldrich (St. Louis, MO, USA) unless otherwise noted.
Animal and wheel-running experiments
All studies were approved by the Pennington Biomedical Research Center’s Institutional Animal Care and Use Committee. Male 5 (n = 9) or 24 months (n = 9) old C57BL/6J mice purchased from the National Institute of Aging rodent colony run by Charles River Laboratory were maintained on a 12-h LD period (lights on 0600–1800 hours or circadian time CT0 and CT12). Wheel-running activity data (MiniMitter, Bend, OR, USA) was recorded as previously described (Sutton et al. 2008). Mice were allowed 14 days of acclimation in the wheel cages, and data were collected over the following 14 day period. For subsequent experiments under constant dark conditions, lights were turned off at 0800 hours on day 1, and mice were allowed to free run for 3 weeks before data were collected and analyzed (over a total of two additional weeks). By the conclusion of all aspects of the activity/behavioral study including quarantine, acclimatization, and study analyses, the ages of the mice in the younger and older cohorts had increased to ∼8–9 and ∼28–29 months, respectively.
Body composition
Fat mass and fat free mass were measured using nuclear magnetic resonance (NMR; Bruker Mice Minispec NMR Analyzer, Bruker Optics Inc., Billerica, MA, USA).
Circadian analysis
A total of 18 young (5 months) and old (24 months) C57BL/6 mice were acclimated to a set 12:12-h light/dark cycle and fed ad lib on standard lab chow for 2 weeks in the PBRC Comparative Biology Core Facility. Five days prior to the study, the mice were converted to constant darkness (red light) regimen. On the day of the study, mice were euthanized by CO2 asphyxiation after ad lib feeding overnight in groups of three animals per young or old cohort at 4-h intervals beginning at 0700 hours and ending at 0300 hours on a single day (July 2009). The body weight of each animal was recorded prior to dissection of the following tissues: brown adipose tissue (BAT), epididymal white adipose tissue (eWAT), inguinal WAT (iWAT), and liver. Individual tissues were weighed prior to freezing in liquid nitrogen. Samples were stored at −80°C until use.
RNA isolation and qRT-PCR
Total RNA was isolated from the BAT, eWAT, iWAT, and liver tissues using TriReagent (MRC, Cincinnati, OH, USA) in accordance with the manufacturer’s recommendations as previously published (Ptitysn et al. 2006; Zvonic et al. 2006, 2007). A Nanodrop ND-1000 Spectrophotometer (Wilmington, DE, USA) was used to determine total RNA concentrations, and 2 μg of total RNA was reverse-transcribed using Moloney Murine Leukemia Virus Reverse Transcriptase (MMLV-RT; Promega, Madison, WI, USA), with Oligo dT at 42°C for 1 h in a 20-μL reaction. Primers for genes of interest were identified using Primer Express software (Applied Biosystems, Foster City, CA, USA). A complete list of primers used in these studies is listed in Supplemental Table 1. qRT-PCR was performed on diluted cDNA samples with SYBR® Green PCR Master Mix (Applied Biosystems) using the 7900 Real Time PCR system (Applied Biosystems) under universal cycling conditions (95°C for 10 min; 40 cycles of 95°C for 15 s; then 60°C for 1 min). All results were normalized relative to a Cyclophilin B expression control.
Illumina microarray
The Illumina TotalPrep RNA Amplification Kit (Applied Biosystems Inc., Foster City, CA, USA; Catalog #AMIL1791) was used to create labeled cRNA from 750 ng of input total RNA according to the manufacturer’s protocol. The labeled cRNA samples were then assessed for quality and quantity using a NanoDrop and an Agilent Bioanalyzer. The MouseWG-6 v2 Beadchip (Illumina Sentrix Beadchip Array #11278593) contains 45,200 transcripts and allows six samples to be interrogated in parallel; 1.5 μg of each labeled cRNA was hybridized to each array according to the manufacturer’s protocol. Experimental group samples were distributed randomly across all beadchips. After an 18-h hybridization at 58°C, the beadchips were processed according to manufacturer’s protocol and scanned using an Illumina BeadArray Reader (Illumina, Inc., San Diego, CA, USA). The data discussed in this publication were deposited in NCBI’s Gene Expression Omnibus (GEO). Data are accessible through GEO Series accession number GSE25325 (linked to subseries GSE25323 and GSE25324).
Statistics and data analysis strategy for Illumina beadchips
Metrics files from the bead scanner were checked to ensure that all samples fluoresced at comparable levels before importing samples into BeadStudio (Framework version 3.1.1.0) Gene Expression module v.3.2. Reference, hybridization control, stringency, and negative control genes were checked for proper chip detection. Two datasets were created and exported for downstream analysis. Each contained the average signal for each transcript and the detection p value. Both data sets had background subtracted from the transcript signals. One data set was quantile-normalized and the other was not normalized.
Inference of differentially expressed genes
Values are presented as the mean ± SD. Pair-wise Student t test was used to evaluate significance (p < 0.05) of mean expression levels between young and old mice. The difference in amplitude between the young and old cohorts was estimated by Fisher’s test.
Analysis of periodicity
Expression profiles were smoothed using a third degree polynomial procedure and median-subtracted using the seven-point Savitzky–Golay algorithm (Savitzky 1964). To take advantage of all points in the time series, a single-pass smoothing was applied in a circular manner, with the last points contributing to smoothing of the starting points. The same smoothing and median subtraction procedure was applied to all data sets. The significance of the observed periodicity can be estimated by Fisher’s g statistic, as recommended by Wichert et al. (2004). This algorithm closely follows the guidelines recommended for analysis of periodicities in time-series microarray data except that we applied C++ code (written by and available from AP) instead of R scripts. Fisher’s g test has low power on short time series under 50 samples (Liew et al. 2009). The problem can be mitigated by application of g test in phase continuum setting with digital filters (Ptitsyn et al. 2007). We have also applied alternative tests for periodicity such as autocorrelation (as described in Zvonic et al. 2006) and Pt test (Ptitsyn et al. 2006a).
False discovery rate adjustment
We have applied false discovery rate (FDR) selectively at different levels of the analysis. Tests for periodicity in microarray expression time series have been deliberately left without adjustment for the lack of multiple hypothesis testing settings and violation of basic assumptions required to FDR analysis. These reasons are explained in greater in previous publications (Ptitsyn 2008; Ptitsyn et al. 2008). We have applied Benjamini–Hochberg procedure (Benjamini and Hochberg 1995) (p < 0.05) for FDR control in analysis of differential expression between age cohorts. Since we interpret and discuss differential genes in the context of their molecular function and gene interaction neighborhood, we apply FDR to adjust the discovery of statistically significant biological pathways.
Biological pathway analysis
We have conducted two types of pathway analysis: the analysis of over-represented canonical pathway maps and analysis of groups (pathways) interlinked by gene interaction within the list of identified genes of interest. The analysis of biological pathways as well as graphic representation of gene interaction maps have been produced using GeneGo Metacore software (GeneGo Inc., A Thomson Reuters Business St. Josef, MI, USA). Significance of pathway enrichment was estimated by Metacore software with p = 0.05 cutoff and FDR adjustment.
Statistical methods
Values are presented as the mean ± SD. Student t test was used to evaluate significance (p < 0.05).
Results
Biological aging impacts activity profiles
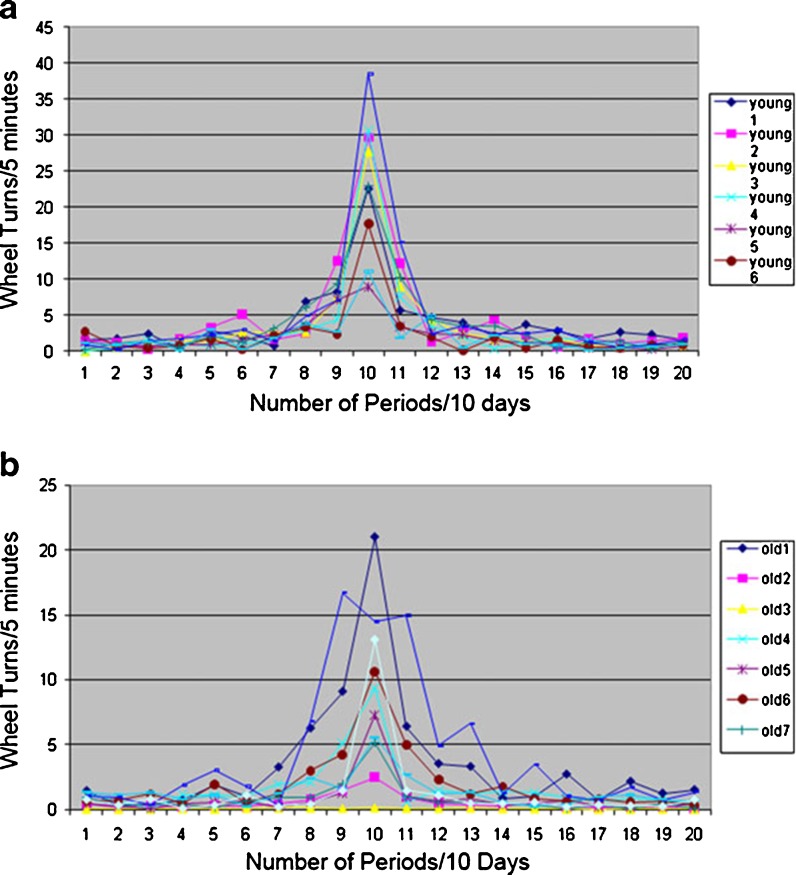
Physical activity is a recognized marker of circadian biological rhythms. As an initial evaluation of the circadian behavior of the younger and older mice, their wheel-running activity was compared under conditions of a constant 12:12-h light/dark cycle (Fig. 1). These studies extended over several months such that the younger and older cohorts had reached the ages of 8–9 and 28–29 months, respectively, upon completion of the evaluation. Consistent with published observations (Valentinuzzi et al. 1997), the nocturnally active older animals initiated wheel-running activity significantly later than their younger controls following the onset of the lights out period. Furthermore, in contrast to the younger cohort, the older animals continued their wheel running after the lights were turned on. Additional studies evaluated the same cohorts under an extended period of constant darkness (free running condition) (Fig. 2). While the older animals maintained an oscillatory physical activity pattern under these conditions (Fig. 2b), the timing of their peak level of activity drifted as compared to the younger cohort (Fig. 2a) based on periodogram analysis. Additionally, while the younger cohort maintained a period length of ∼24 h under constant darkness, period length in the older cohort changes of drifted by ±2.4 to 4.8 h (Fig. 2).
Fig. 1.
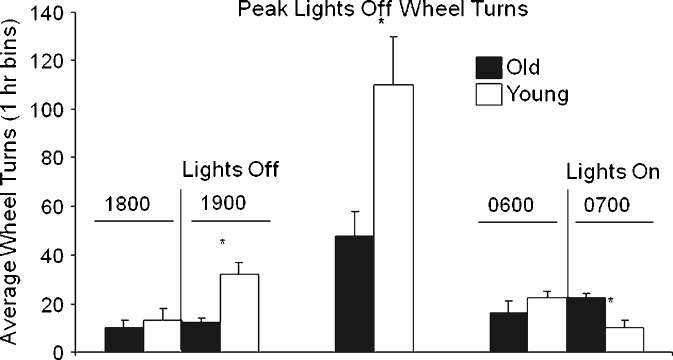
Younger and older mice display differential wheel revolutions and peak wheel-running activity at light offset and onset. Younger (open columns) and older (black columns) male mice (n = 9/10 per group) were housed in wheel cages individually under a constant 12:12-h light/darkness cycle and 24-h wheel-running activity monitored. The average wheel turn activity (± SD) is displayed for the following periods: left panel the final hour of “lights on” (1800 hours) and the initial hour of “lights off/darkness” (1900 hours); middle panel the hour of peak activity level during the “lights off” period (1900 to 0700 hours); right panel the final hour of “lights off/darkness” (0600 hours) and “lights on” (0700 hours). *p < 0.01 indicates significant differences between younger and older cohorts at the indicated time points
Fig. 2.
Biological aging and regularity of peak physical activity. Fourier analysis was performed on the wheel-running activity of cohorts (n = 6–7) young adult (a) and old (b) mice collected over 4 weeks under constant darkness conditions. The resulting periodogram shows discrete harmonic components (number of complete cycles over the period of observation) on the X-axis vs. intensity of exercise on the Y-axis. Integral values on the X-axis correspond to the number of periods completed over a 10-day cycle. Thus, an X value of 1 corresponds to a single period of 10 days length, a value of 10:10 periods of 24-h length, and a value of 20:20 periods of 12-h length. Integral values on the Y-axis correspond to wheel turns per 5 min bins. While the cohort of younger animals retained a tightly regulated peak activity period (resulting in consistent 10 peaks over 10 days and thus dominating peak in the center of periodograms a), there was significantly greater drift in the older cohort (less regular exercise pattern resulting in “erosion” of a single dominating peak of periodograms b)
Biological aging impacts body composition
Biological aging has been correlated with alterations in adipose depots in rodent models (Kirkland et al. 1990). Consistent with these findings, the size of the BAT, subcutaneous inguinal, and visceral epididymal and retroperitoneal WAT depots were reduced significantly in the older relative to the younger cohorts of male mice (Table 1). Nevertheless, total body weights were nearly identical between the two cohorts (Table 1), and consistent with this, their relative percentage body fat and lean muscle mass were equivalent based on NMR-derived body composition (Table 2). It should be noted that body composition data were collected on a subpopulation of each age cohort at the completion of the activity/behavioral analyses. Consequently, the animal ages were 8–9 and 28–29 months, respectively, in the younger and older cohorts. Thus, the animals in the younger cohort had approached a more mature status by the time this information was obtained.
Table 1.
Effect of age on murine adipose depot, liver, and total body weights
| Age group | iWAT | eWAT | rWAT | BAT | Liver | Body Wt |
|---|---|---|---|---|---|---|
| 8–9 months | 0.35 ± 0.14 | 0.55 ± 0.16 | 0.19 ± 0.10 | 0.19 ± 0.04 | 2.08 ± 0.39 | 32.24 ± 2.63 |
| 28–29 months | 0.21 ± 0.08 | 0.34 ± 0.14 | 0.09 ± 0.05 | 0.11 ± 0.02 | 3.01 ± 1.62 | 32.21 ± 3.21 |
| p value | 0.0265 | 0.0126 | 0.0141 | 0.0003 | 0.1553 | 0.9860 |
Body composition data were collected after the completion of all activity/behavioral analyses in a subset of the murine cohorts. Values reflect the mean weight in grams ± SD of organs or adipose depots collected from n = 8–9 mice per cohort. A single mouse in the older age group displayed a liver tumor (removed from mean liver weight). p values were determined by two-tailed Student’s t test
BAT brown adipose tissue, eWAT epididymal white adipose tissue, iWAT inguinal WAT, rWAT retroperitoneal WAT
Table 2.
Effect of age on murine body composition as determined by NMR
| Age group | % Fat mass | % Lean mass | % Free fluid | Body Wt (g) |
|---|---|---|---|---|
| 8–9 months | 13.5 ± 2.8 | 83.7 ± 2.3 | 2.8 ± 0.6 | 30.7 ± 1.9 |
| 28–29 months | 12.0 ± 2.0 | 84.2 ± 2.4 | 3.8 ± 0.7 | 31.4 ± 3.8 |
| p value | 0.329 | 0.752 | 0.037 | 0.730 |
Total body NMR was conducted on n = 5 or 6 animals per age cohort at the completion of the activity/behavioral aspects of the study immediately prior to euthanasia. Values reflect the mean ± SD as either percentile of total body weight or body weight in grams. p values were determined by two-tailed Student’s t test
Biological aging alters circadian gene expression
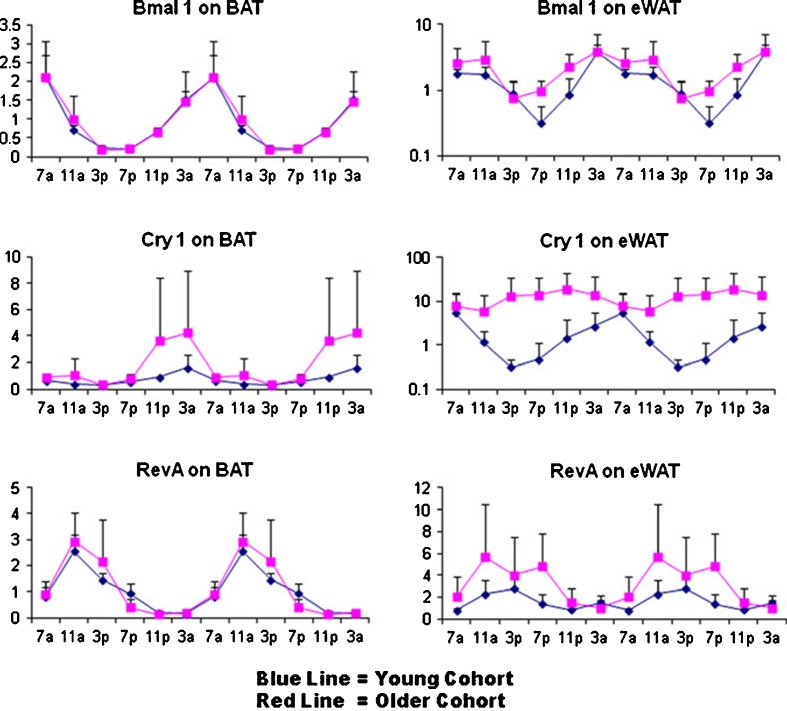
Expression of circadian regulatory genes has been found to oscillate in both the adipose tissue and liver of young mice (Ando et al. 2005; Zvonic et al. 2006). Using qRT-PCR, the impact of biological aging on the expression of circadian rhythm associated genes was evaluated (Fig. 3 and Supplement Figures 1–4). Young (5 months) and old (24 months) male C57BL/6 mice were purchased from the NIA aging colony. Upon arrival, the animals were acclimated for 2 weeks to a constant 12:12-h light/dark cycle with ad libitum access to food and then converted to constant darkness (red light) for 5 days to remove any photic stimuli. Groups of mice (n = 3) from each age group were then euthanized at 4 h intervals over a 24-h period. Isolated total RNA from BAT, eWAT, and liver were used in RT-PCR assays to determine circadian regulatory gene mRNA levels normalized to cyclophilin B (Fig. 3). In both adipose depots, the Bmal1 profile, reflecting the “positive arm” of the CCRP feedback pathway, showed a similar acrophase (i.e., point of peak expression or zenith) and amplitude in both the young (blue) and old (red) cohorts. In contrast, the Cry1 profile, reflecting the “negative” CCRP arm, displayed greater amplitude and, in the case of eWAT, a phase advanced acrophase, in the older relative to the younger cohort. Likewise, the related “negative arm” mRNA Per1 showed a similar age-dependent profile. The Rev-erbα profile, reflecting an immediate “downstream” circadian target, displayed a similar acrophase in both cohorts and tissues; however, the aged cohort amplitude was greater in the eWAT. Other downstream targets, including DBP and E4BP4, showed similar age-dependent expression patterns to Rev-erbα; the temporal-dependent expression profile of these and related adipogenic mRNAs in BAT, epididymal and inguinal WAT, and liver are shown in Supplement Figures 1–4.
Fig. 3.
Analysis of circadian gene oscillation by qRT-PCR. Total RNA isolated from brown adipose tissue (BAT) and epididymal white adipose tissue (eWAT) of younger (blue) and older (red) mice (n = 3) harvested at serial 4-h time points over a 24-h period were analyzed by qRT-PCR for expression of representative “positive arm” (BMAL1), “negative arm” (CRY1), and a downstream target (Rev-erb α). The RT-PCR data is expressed on the “Y”-axis normalized relative to cyclophilin B as a control and double-plotted relative to time (“X”-axis) over 24-h periods. Additional qRT-PCR data from multiple tissues and genes are presented in Supplement Figures 1–4
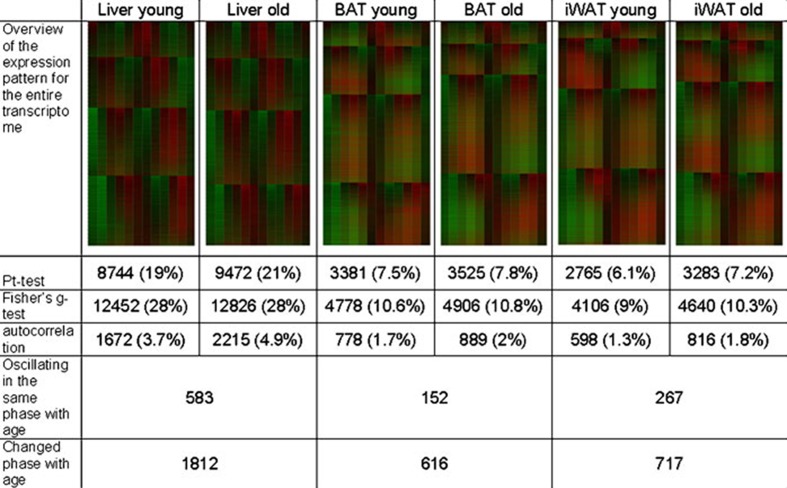
Analyses of the RNA samples collected from the younger and older cohorts were extended using duplicate transcriptomic microarrays covering the 24-h period (Fig. 4). The oscillatory gene expression profile determined using published algorithms (Ptitsyn et al. 2006b) was compared between the younger and older cohorts and determined that a total of 616, 717, and 1,812 expressed transcripts differed significantly in BAT, WAT, and liver, respectively. Table 3 summarizes the major biochemical pathways most significantly impacted in each tissue site. While cell signaling related pathways account for >25% of the top 50 pathways detected in all tissues, those related to the adrenergic receptor are detected readily in BAT and WAT but not liver (Table 3). This is consistent with the large body of evidence supporting a role for adrenergic signaling in regulating lipolysis and/or differentiation in adipose depots (Lafontan et al. 1985; Houstek et al. 1990; Rehnmark et al. 1990; Elabd et al. 2009).
Fig. 4.
Biological aging and the circadian oscillation of the transcriptome in BAT, iWAT, and Liver. The heat map of genes in BAT, iWAT, and liver is organized into genes oscillating in phase with peak (zenith) expression levels at circadian time CT 0 (light onset), CT 4 (4 h after light onset), CT 8 (8 h after light onset), and CT 16 (4 h after light offset). The number of expressed oscillating genes detected by the three algorithms (permutation (Pt), Fishers g test, or autocorrelation) are indicated along with the percentage and number of genes maintaining the same phase or different phase between age groups
Table 3.
Top 50 GeneGo canonic pathways significantly impacted by biological aging in BAT, WAT, and liver of young (5 months) vs. old (24 months) C57BL/6 mice
| Pathway family | BAT | WAT | Liver |
|---|---|---|---|
| Cell adhesion/cytoskeleton | 13 | 3 | 7 |
| Immune | 4 | 7 | 18 |
| Signaling/development (adrenergic/G protein) | 19 (8) | 13 (9) | 11 (0) |
| Cell cycle/proliferation | 3 | 2 | 7 |
| Metabolism | 2 | 7 | 2 |
| Apoptosis | 4 | 8 | 0 |
| Other | 5 | 10 | 5 |
The number of biochemical pathways specific to a particular general category is reported for each tissue type (numbers reflected in each column). These were detected by Gene Ontology Functional Ontology Enrichment analyses (using Metacore software, Tompson Reuters) of the transcriptome data from all circadian times as a function of cohort age. Under the signaling/development pathways, the parenthetical number refers to signaling pathways that involve adrenergic receptors and/or G coupled protein signal transduction
Biological aging differentially modulates a subset of expressed genes in BAT, iWAT, and liver
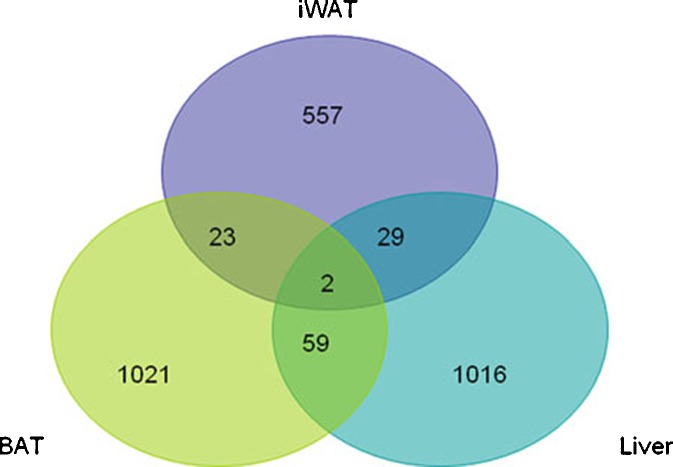
Biological aging has been reported to modulate adipose and cardiac gene expression between young and old rats fed ad libitum (Linford et al. 2007). While the majority of genes were induced 2-fold in young vs. older cardiac tissue (82%), only a minority of genes displayed this pattern in adipose tissue (47%) (Linford et al. 2007). Of these expressed transcripts, only 10% (WAT) or 21% (cardiac) of the total were common to both tissues (Linford et al. 2007). Using a similar analytical method, the relative expression was compared between microarrays collected at multiple time points from BAT, iWAT, and liver of younger and older mice. This information is summarized in a Venn diagram (Fig. 5). Within these subsets, which ranged from 557 (iWAT) to 1,021 (BAT) expressed sequences, the majority were induced in the younger relative to the older cohort in BAT (94%) and liver (88%). In contrast, only a minority of the transcripts were increased in the younger relative to the older cohort in iWAT (39%). Furthermore, circadian expressed transcripts shared in common by two tissues ranged from 23 to 59 (2% to 6%), and only two transcripts encoding a solute carrier anion exchanger and a Golgi protein were modulated consistently in all three tissues as a function of age (Supplemental Table 2). It should be noted that current study monitored all changes in gene expression, not just those at or above a 2-fold threshold.
Fig. 5.
Biological age-dependent mRNA expression levels in the transcriptome of BAT, iWAT, and liver. A Venn diagram depicts the number of independent transcripts differentially expressed between young and old cohorts within each tissue; the areas of overlap depict the number of genes with a shared differential expression profile between two or more tissues
Discussion
The current study has extended the literature linking circadian biology, metabolism, and obesity (Gimble et al. 2009) by evaluating circadian mechanisms in adipose tissue and liver of younger and older murine cohorts. At the morphological level, all adipose depots decreased significantly in total weight as a function of advanced age. These alterations are consistent with patterns reported in rat models (Kirkland et al. 1990, 1993, 1994; Tchkonia et al. 2010). The body composition of the younger and older cohorts was similar with respect to total fat and lean masses, suggesting that lipid content re-distributed to other depots such as the liver or skeletal muscle; while liver weights were increased with age, this did not achieve statistical significance. Physical activity, as monitored by wheel running, decreased overall and displayed an altered profile with advanced age. While younger mice became more active at light offset (beginning of darkness), the older mice exhibited a delayed onset in activity. Likewise, the peak activity of the younger mice was ∼2-fold greater than that of the older cohort. In contrast, the older mice showed increased activity following lights on relative to the younger cohort. This activity pattern is consistent with that reported in the literature where older mice displayed up to a 50% reduction in total wheel revolutions relative to younger controls (Valentinuzzi et al. 1997; Weinert and Waterhouse 1999).
The amplitude of the mRNAs encoding the “negative arm” circadian apparatus was greater in the older mice relative to the younger cohort (Fig. 3), and similar features were displayed in downstream targets such as DBP and E4BP4 (Supplement Figures 1–4). These features are similar to those displayed by the alpha MUPA transgenic mice which overexpress a serine protease involved in brain development (Froy et al. 2006). These mice have increased the amplitude of Cry1, Per1, and Per2, but not Bmal1, mRNAs in the liver relative to wild-type controls (Froy et al. 2006). Furthermore, the alpha MUPA transgenic mice eat less than their wild-type controls and exhibit an increased life span in a manner similar to that displayed by caloric restricted mice (Froy and Miskin 2007; Froy et al. 2008). The dynamic amplification of the expression of Cry, Per, and repressive transcription factors such as DBP may be a protective adaptation to biological aging. In the Bmal1−/− strain, where maintenance of a homeostatic balance between the negative elements Cry and Per and the positive arm of the circadian apparatus is disrupted, the mice display evidence of accelerated aging (Kondratov et al. 2006; Kondratov 2007). The Bmal1−/− die at an earlier age, have accelerated loss of fat and muscle mass during their lifetime, and show evidence of elevated levels of reactive oxygen species (Kondratov et al. 2006). Thus, the circadian transcriptional mechanisms can directly impact biological aging, and further studies investigating this relationship are warranted.
Transcriptomic analyses of pre-adipocytes isolated from young (3 months) and old (30 months) rats have revealed similar outcomes (Cartwright et al. 2010). Pre-adipocytes obtained from the epididymal and retroperitoneal fat pads exhibited an 8.4% depot-dependent difference in gene expression profiles; however, aging accounted for only a 0.02% difference in gene expression within each depot (Cartwright et al. 2010). It is possible that the collagenase isolation and subsequent culture expansion of the pre-adipocytes from the young and old rats masked differences in their gene expression profiles. Alternatively, other cell components within the intact adipose tissue could be responsible for the age-dependent difference in gene expression profiles.
Similar studies have evaluated the transcriptome in adipose tissue as a function of aging and/or caloric restriction in rodent models (Higami et al. 2004; Fu et al. 2006; Linford et al. 2007; Swindell 2008). Studies have examined 10–11-month-old male C57BL/6 mice maintained under control or long-term (9-month) caloric restriction (Higami et al. 2004). Caloric restriction altered expression of 345 expressed genes (5.5%) by 1.5-fold relative to ad libitum fed controls (Higami et al. 2004). The modulated pathways included those relating to amino acid, carbohydrate, and lipid metabolism, mitochondrial function, insulin signaling, and inflammation (Higami et al. 2004). Independent studies compared the effect of aging on gene expression in heart, liver, and hypothalamus from young (4–6 months) and old (26–28 months) C57BL/6 mice (Fu et al. 2006). Age-dependent significant differences were determined in 309 (heart), 1,819 (liver), and 1,085 (hypothalamus) gene expression profiles (Fu et al. 2006). Furthermore, similar to the current study, only 32% (100, heart), 13% (240, liver), and 20% (215, hypothalamus) of these genes were coordinately expressed as a function of aging in two tissues and only nine genes total in all three tissues (Fu et al. 2006). Comparisons between visceral adipose tissue transcriptomes of ad libitum fed 4-month and 28-month rats determined that a total of 910 genes displayed a 2-fold age-dependent variation (Linford et al. 2007). Parallel analyses of the heart transcriptome identified 415 genes with age-dependent 2-fold variation and a subset of 88 genes showed equivalent variation in both tissues (Linford et al. 2007). This subset included genes related to immune, mitochondrial, proteasomal, and ribosomal function (Linford et al. 2007). Caloric restriction returned many of the alterations in the aged rat WAT transcriptome to younger levels (Linford et al. 2007), and caloric restriction led to consistent changes in Per1 and Per2 expression in heart, liver, and other organs (Swindell 2008); however, analysis of multiple experiments indicates that caloric restriction does not reverse biological aging associated expression profiles universally (Swindell 2009).
While the general patterns of age-dependent gene modulation in the current study was similar to that reported in the literature (Linford et al. 2007; Swindell 2009), the absolute number of transcripts increased by 2-fold or reduced by 1.66-fold in younger vs. older cohorts was limited to 57 (BAT), 29 (iWAT), and 65 (liver), respectively. This lower value may reflect the fact that circadian oscillation was taken into account due to the use of multiple microarray time points. Alternatively, but not exclusively, differences in tissue depots and specie may be contributory factors. Nevertheless, several transcripts displaying the most dynamic modulation in younger vs. older cohorts merit further consideration as potential biomarkers in the context of circadian biology. For example, in the liver, genes within the circadian pathway were reduced by 2–3-fold (RORγ) or induced by 10.5-fold (NR1D1, Rev-erbα). Furthermore, the Wnt pathway gene, Wnt5B, was reduced by 1.7-fold. The Wnt pathway is of significance since its mechanism, like that of the circadian regulatory apparatus, is dependent on GSK3β post-translational phosphorylation events. In BAT, the hydroquinone (NADH) oxidase ENOX1 was reduced by 1.7-fold in younger vs. older cohorts. The ENOX1 protein has been found to display a robust time-dependent enzymatic oscillation (Klein and Melton 1996; Jiang et al. 2008; Kromkowski et al. 2008). While the oscillations of ENOX1 are ultradian with a period length of 24 min, unlike the circadian clock, its periodicity can be modulated by lithium chloride, an inhibitor of GSK3β and known modifier of circadian activity (Klein and Melton 1996; Padiath et al. 2004; Kromkowski et al. 2008). In iWAT, the homeobox transcripts, Hoxd4, was increased 2-fold in younger vs. older cohorts. Overexpression of Hoxd4 in the cartilage of transgenic mice has been found to reduce expression of Wnt3a, a canonical Wnt pathway ligand, suggesting a functional role for this homeobox transcription factor in multipotential stem cells capable of undergoing adipogenesis or chondrogenesis and a potential relationship to Wnt-related mechanisms (Kruger and Kappen 2010). This interpretation would be consistent with earlier studies determining that Wnt10b, another canonical Wnt pathway ligand, suppressed adipogenesis and promoted osteogenesis in multipotential stem cell models (Ross et al. 2000; Bennett et al. 2002, 2005).
Studies of human subjects have determined that insulin sensitivity modulates canonical Wnt pathway mRNAs in adipose tissue and cells (Yang et al. 2003, 2004). In adipose tissues biopsied from non-diabetic, non-obese males related to diabetic subjects, the expression levels of mRNAs encoding β-catenin, disheveled 1, frizzled 1, GSK3β, and Wnt-1 correlated significantly with that of FoxC2, a transcription factor responsible for brown adipogenesis (Yang et al. 2003). With increased insulin resistance, expression of each of these mRNAs was reduced (Yang et al. 2003). Consistent with this, further analyses determined that Wnt mRNA expression levels declined in correlation with adipose cell size (Yang et al. 2004). Since adipocyte hypertrophy is an indicator of insulin resistance, it can be used to predict risk of type 2 diabetes mellitus (Yang et al. 2004). Recent studies have determined that the nuclear membrane protein, lamin, mediates the canonical Wnt signal transduction pathway via β-catenin and the Tcf/Lef transcription factor (Hernandez et al. 2010). This work used a lamin mutant murine model which displays age-dependent subcutaneous adipose fat loss as well as the cranial and skeletal pathologies and the reduced longevity characteristic of Hutchinson–Gifford progeria syndrome (Hernandez et al. 2010). It is noteworthy that lamin gene mutations have been identified as causative in some patients with familial partial lipodystrophy (Garg and Agarwal 2009). Together, these observations highlight the importance of the Wnt pathway in adipose development and biological aging. Since GSK3β stands out as a critical intersection between the circadian and Wnt pathways, our future studies will focus on the impact of biological aging and circadian mechanisms on Wnt pathway elements in adipose depots.
Electronic supplementary material
Analysis of Circadian Gene Oscillation by qRT-PCR. Total RNA isolated from brown adipose tissue (Fig. 1), epididymal WAT (Fig. 2), inguinal WAT (Fig. 3), and liver (Fig. 4) of younger and older mice was harvested at serial 4-h time points beginning at 0700 hours and continuing until 0300 hours during a single 24-h period. The level of expression of the following mRNAs representative of the circadian apparatus and adipogenesis was determined and normalized relative to cyclophilin B: Bmal1, Cry1, Cry2, DBP, E4BP4, Leptin, LPL, Npas2, Per1, PGC-1, PPARγ, Rev-Erbα, and Rev-Erbβ. Values presented are the mean ± SD, and these have been double-plotted. (JPEG 94 kb)
(JPEG 75 kb)
(JPEG 87 kb)
(JPEG 61 kb)
(JPEG 80 kb)
(JPEG 44 kb)
(JPEG 71 kb)
(JPEG 56 kb)
The forward and reverse primers as well as the PubMed accession numbers used for qRT-PCR are listed in Table 8. (DOC 34 kb)
The transcripts that were expressed differentially between the young and old cohorts as a function of biological age in two or more tissues are summarized in Table 7. Shared transcripts in BAT/iWAT, BAT/Liver, and iWAT/Liver as well as all three tissues are displayed. (DOC 113 kb)
Acknowledgments
The investigators express their appreciation to Barry Robert DVM, Ph.D., Cindy Kloster, MS, and the staff of the Comparative Biology Core Facility for providing outstanding care for the animals used in this study; the Genomics Core Facility staff for assistance in performing the microarrays with support in part from the NORC Center Grant # 1P30 DK072476 entitled “Nutritional Programming: Environmental and Molecular Interactions” sponsored by NIDDK and COBRE grant NIH-P20-RR021945; Laura Dallam for administrative assistant supporting this work; and Jeffrey Keller, Ph.D. for logistical assistance. The study was supported in part through funding from the American Diabetes Association (GMS, AC, ZEF), Pennington Biomedical Research Foundation (YG, XW, JMG), COBRE grant NIH-P20-RR021945 (GMS, ZEF), the NIH/NCRR P50RR00164 (BAB), and the Louisiana Gene Therapy Research Consortium (BAB).
References
- Ahmad AM, Guzder R, Wallace AM, Thomas J, Fraser WD, Vora JP. Circadian and ultradian rhythm and leptin pulsatility in adult GH deficiency: effects of GH replacement. J Clin Endocrinol Metab. 2001;86:3499–3506. doi: 10.1210/jcem.86.8.7716. [DOI] [PubMed] [Google Scholar]
- Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12:540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- Ando H, Yanagihara H, Hayashi Y, Obi Y, Tsuruoka S, Takamura T, Kaneko S, Fujimura A. Rhythmic mRNA expression of clock genes and adipocytokines in mouse visceral adipose tissue. Endocrinology. 2005;146:5631–5636. doi: 10.1210/en.2005-0771. [DOI] [PubMed] [Google Scholar]
- Arasaradnam MP, Morgan L, Wright J, Gama R. Diurnal variation in lipoprotein lipase activity. Ann Clin Biochem. 2002;39:136–139. doi: 10.1258/0004563021901883. [DOI] [PubMed] [Google Scholar]
- Atmaca M, Kuloglu M, Tezcan E, Ustundag B. Weight gain and serum leptin levels in patients on lithium treatment. Neuropsychobiology. 2002;46:67–69. doi: 10.1159/000065414. [DOI] [PubMed] [Google Scholar]
- Baptista T, Teneud L, Contreras Q, Alastre T, Burguera JL, de Burguera M, de Baptista E, Weiss S, Hernandez L. Lithium and body weight gain. Pharmacopsychiatry. 1995;28:35–44. doi: 10.1055/s-2007-979586. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Stat Soc B. 1995;57:289–300. [Google Scholar]
- Bennett CN, Ross SE, Longo KA, Bajnok L, Hemati N, Johnson KW, Harrison SD, MacDougald OA. Regulation of Wnt signaling during adipogenesis. J Biol Chem. 2002;277:30998–31004. doi: 10.1074/jbc.M204527200. [DOI] [PubMed] [Google Scholar]
- Bennett CN, Longo KA, Wright WS, Suva LJ, Lane TF, Hankenson KD, MacDougald OA. Regulation of osteoblastogenesis and bone mass by Wnt10b. Proc Natl Acad Sci U S A. 2005;102:3324–3329. doi: 10.1073/pnas.0408742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray MS, Young ME. The role of cell-specific circadian clocks in metabolism and disease. Obes Rev. 2009;10(Suppl 2):6–13. doi: 10.1111/j.1467-789X.2009.00684.x. [DOI] [PubMed] [Google Scholar]
- Calvani M, Scarfone A, Granato L, Mora EV, Nanni G, Castagneto M, Greco AV, Manco M, Mingrone G. Restoration of adiponectin pulsatility in severely obese subjects after weight loss. Diabetes. 2004;53:939–947. doi: 10.2337/diabetes.53.4.939. [DOI] [PubMed] [Google Scholar]
- Cartwright MJ, Tchkonia T, Kirkland JL. Aging in adipocytes: potential impact of inherent, depot-specific mechanisms. Exp Gerontol. 2007;42:463–471. doi: 10.1016/j.exger.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright MJ, Schlauch K, Lenburg ME, Tchkonia T, Pirtskhalava T, Cartwright A, Thomou T, Kirkland JL. Aging, depot origin, and preadipocyte gene expression. J Gerontol A Biol Sci Med Sci. 2010;65:242–251. doi: 10.1093/gerona/glp213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chengappa KN, Chalasani L, Brar JS, Parepally H, Houck P, Levine J. Changes in body weight and body mass index among psychiatric patients receiving lithium, valproate, or topiramate: an open-label, nonrandomized chart review. Clin Ther. 2002;24:1576–1584. doi: 10.1016/s0149-2918(02)80061-3. [DOI] [PubMed] [Google Scholar]
- Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, Kolodny GM, Kahn CR. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies K. Pressure sores: aetiology, risk factors and assessment scales. Br J Nurs. 1994;3:256–262. doi: 10.12968/bjon.1994.3.6.256. [DOI] [PubMed] [Google Scholar]
- Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, Dunlap JC. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol. 2002;12:551–557. doi: 10.1016/s0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- Elabd C, Chiellini C, Carmona M, Galitzky J, Cochet O, Petersen R, Penicaud L, Kristiansen K, Bouloumie A, Casteilla L, Dani C, Ailhaud G, Amri EZ. Human multipotent adipose-derived stem cells differentiate into functional brown adipocytes. Stem Cells. 2009;27:2753–2760. doi: 10.1002/stem.200. [DOI] [PubMed] [Google Scholar]
- Elmslie JL, Mann JI, Silverstone JT, Williams SM, Romans SE. Determinants of overweight and obesity in patients with bipolar disorder. J Clin Psychiatry. 2001;62:486–491. doi: 10.4088/jcp.v62n0614. [DOI] [PubMed] [Google Scholar]
- Fagiolini A, Frank E, Houck PR, Mallinger AG, Swartz HA, Buysse DJ, Ombao H, Kupfer DJ. Prevalence of obesity and weight change during treatment in patients with bipolar I disorder. J Clin Psychiatry. 2002;63:528–533. doi: 10.4088/jcp.v63n0611. [DOI] [PubMed] [Google Scholar]
- Fagiolini A, Kupfer DJ, Houck PR, Novick DM, Frank E. Obesity as a correlate of outcome in patients with bipolar I disorder. Am J Psychiatry. 2003;160:112–117. doi: 10.1176/appi.ajp.160.1.112. [DOI] [PubMed] [Google Scholar]
- Fonjallaz P, Ossipow V, Wanner G, Schibler U. The two PAR leucine zipper proteins, TEF and DBP, display similar circadian and tissue-specific expression, but have different target promoter preferences. EMBO J. 1996;15:351–362. [PMC free article] [PubMed] [Google Scholar]
- Froy O, Miskin R. The interrelations among feeding, circadian rhythms and ageing. Prog Neurobiol. 2007;82:142–150. doi: 10.1016/j.pneurobio.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Froy O, Chapnik N, Miskin R. Long-lived alphaMUPA transgenic mice exhibit pronounced circadian rhythms. Am J Physiol Endocrinol Metab. 2006;291:E1017–E1024. doi: 10.1152/ajpendo.00140.2006. [DOI] [PubMed] [Google Scholar]
- Froy O, Chapnik N, Miskin R. Relationship between calorie restriction and the biological clock: lessons from long-lived transgenic mice. Rejuvenation Res. 2008;11:467–471. doi: 10.1089/rej.2008.0669. [DOI] [PubMed] [Google Scholar]
- Fu C, Hickey M, Morrison M, McCarter R, Han ES. Tissue specific and non-specific changes in gene expression by aging and by early stage CR. Mech Ageing Dev. 2006;127:905–916. doi: 10.1016/j.mad.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A, Agarwal AK. Lipodystrophies: disorders of adipose tissue biology. Biochim Biophys Acta. 2009;1791:507–513. doi: 10.1016/j.bbalip.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavi SFJ, Melendez MM, Mynarcik DC, Gelato MC, McNurlan MA. Limb fat to trunk fat ratio in elderly persons is a strong determinant of insulin resistance and adiponectin levels. J Gerontol A Biol Sci Med Sci. 2007;62:997–1001. doi: 10.1093/gerona/62.9.997. [DOI] [PubMed] [Google Scholar]
- Gavrila A, Peng CK, Chan JL, Mietus JE, Goldberger AL, Mantzoros CS. Diurnal and ultradian dynamics of serum adiponectin in healthy men: comparison with leptin, circulating soluble leptin receptor, and cortisol patterns. J Clin Endocrinol Metab. 2003;88:2838–2843. doi: 10.1210/jc.2002-021721. [DOI] [PubMed] [Google Scholar]
- Gimble JM, Floyd ZE. Fat circadian biology. J Appl Physiol. 2009;107:1629–1637. doi: 10.1152/japplphysiol.00090.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimble JM, Zvonic S, Floyd ZE, Kassem M, Nuttall ME. Playing with bone and fat. J Cell Biochem. 2006;98:251–266. doi: 10.1002/jcb.20777. [DOI] [PubMed] [Google Scholar]
- Gimble JM, Bray MS, Young A. Circadian biology and sleep: missing links in obesity and metabolism? Obes Rev. 2009;10(Suppl 2):1–5. doi: 10.1111/j.1467-789X.2009.00672.x. [DOI] [PubMed] [Google Scholar]
- Goubern M, Portet R. Circadian rhythm and hormonal sensitivity of lipoprotein lipase activity in cold acclimated rats. Horm Metab Res. 1981;13:73–77. doi: 10.1055/s-2007-1019177. [DOI] [PubMed] [Google Scholar]
- Green CB, Besharse JC. Identification of a novel vertebrate circadian clock-regulated gene encoding the protein nocturnin. Proc Natl Acad Sci U S A. 1996;93:14884–14888. doi: 10.1073/pnas.93.25.14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez L, Roux KJ, Wong ES, Mounkes LC, Mutalif R, Navasankari R, Rai B, Cool S, Jeong JW, Wang H, Lee HS, Kozlov S, Grunert M, Keeble T, Jones CM, Meta MD, Young SG, Daar IO, Burke B, Perantoni AO, Stewart CL. Functional coupling between the extracellular matrix and nuclear lamina by Wnt signaling in progeria. Dev Cell. 2010;19:413–425. doi: 10.1016/j.devcel.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higami Y, Pugh TD, Page GP, Allison DB, Prolla TA, Weindruch R. Adipose tissue energy metabolism: altered gene expression profile of mice subjected to long-term caloric restriction. FASEB J. 2004;18:415–417. doi: 10.1096/fj.03-0678fje. [DOI] [PubMed] [Google Scholar]
- Hirota T, Fukada Y. Resetting mechanism of central and peripheral circadian clocks in mammals. Zool Sci. 2004;21:359–368. doi: 10.2108/zsj.21.359. [DOI] [PubMed] [Google Scholar]
- Hogenesch JB, Panda S, Kay S, Takahashi JS. Circadian transcriptional output in the SCN and liver of the mouse. Novartis Found Symp. 2003;253:171–180. [PubMed] [Google Scholar]
- Holmback U, Forslund A, Forslund J, Hambraeus L, Lennernas M, Lowden A, Stridsberg M, Akerstedt T. Metabolic responses to nocturnal eating in men are affected by sources of dietary energy. J Nutr. 2002;132:1892–1899. doi: 10.1093/jn/132.7.1892. [DOI] [PubMed] [Google Scholar]
- Holmback U, Forslund A, Lowden A, Forslund J, Akerstedt T, Lennernas M, Hambraeus L, Stridsberg M. Endocrine responses to nocturnal eating—possible implications for night work. Eur J Nutr. 2003;42:75–83. doi: 10.1007/s00394-003-0386-6. [DOI] [PubMed] [Google Scholar]
- Houstek J, Kopecky J, Baudysova M, Janikova D, Pavelka S, Klement P. Differentiation of brown adipose tissue and biogenesis of thermogenic mitochondria in situ and in cell culture. Biochim Biophys Acta. 1990;1018:243–247. doi: 10.1016/0005-2728(90)90258-6. [DOI] [PubMed] [Google Scholar]
- Huffman DM, Barzilai N. Role of visceral adipose tissue in aging. Biochim Biophys Acta. 2009;1790:1117–1123. doi: 10.1016/j.bbagen.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahana E, Akiyama M, Miyakawa K, Uchida A, Kasahara J, Fukunaga K, Hamada T, Shibata S. Effect of lithium on the circadian rhythms of locomotor activity and glycogen synthase kinase-3 protein expression in the mouse suprachiasmatic nuclei. Eur J Neurosci. 2004;19:2281–2287. doi: 10.1111/j.0953-816X.2004.03322.x. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Gorenstein NM, Morre DM, Morre DJ. Molecular cloning and characterization of a candidate human growth-related and time-keeping constitutive cell surface hydroquinone (NADH) oxidase. Biochemistry. 2008;47:14028–14038. doi: 10.1021/bi801073p. [DOI] [PubMed] [Google Scholar]
- Kawai M, Green CB, Lecka-Czernik B, Douris N, Gilbert MR, Kojima S, Ackert-Bicknell C, Garg N, Horowitz MC, Adamo ML, Clemmons DR, Rosen CJ. A circadian-regulated gene, Nocturnin, promotes adipogenesis by stimulating PPAR-gamma nuclear translocation. Proc Natl Acad Sci U S A. 2010;107:10508–10513. doi: 10.1073/pnas.1000788107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland JL, Hollenberg CH, Gillon WS. Age, anatomic site, and the replication and differentiation of adipocyte precursors. Am J Physiol. 1990;258:C206–C210. doi: 10.1152/ajpcell.1990.258.2.C206. [DOI] [PubMed] [Google Scholar]
- Kirkland JL, Hollenberg CH, Gillon WS. Ageing, differentiation, and gene expression in rat epididymal preadipocytes. Biochem Cell Biol. 1993;71:556–561. doi: 10.1139/o93-079. [DOI] [PubMed] [Google Scholar]
- Kirkland JL, Hollenberg CH, Kindler S, Gillon WS. Effects of age and anatomic site on preadipocyte number in rat fat depots. J Gerontol. 1994;49:B31–B35. doi: 10.1093/geronj/49.1.b31. [DOI] [PubMed] [Google Scholar]
- Klein PS, Melton DA. A molecular mechanism for the effect of lithium on development. Proc Natl Acad Sci U S A. 1996;93:8455–8459. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV. A role of the circadian system and circadian proteins in aging. Ageing Res Rev. 2007;6:12–27. doi: 10.1016/j.arr.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlar TJ, Borensztajn J. Oscillatory changes in muscle lipoprotein lipase activity of fed and starved rats. Am J Physiol. 1977;233:E316–E319. doi: 10.1152/ajpendo.1977.233.4.E316. [DOI] [PubMed] [Google Scholar]
- Kromkowski J, Hignite H, Morre DM, Morre DJ. Response to lithium of a cell surface ECTO-NOX protein with time-keeping characteristics. Neurosci Lett. 2008;438:121–125. doi: 10.1016/j.neulet.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Kruger C, Kappen C. Expression of cartilage developmental genes in Hoxc8- and Hoxd4-transgenic mice. PLoS One. 2010;5:e8978. doi: 10.1371/journal.pone.0008978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, Solt LA, Wang Y, Rogers PM, Bhattacharyya G, Kamenecka TM, Stayrook KR, Crumbley C, Floyd ZE, Gimble JM, Griffin PR, Burris TP. Regulation of adipogenesis by natural and synthetic REV-ERB ligands. Endocrinology. 2010;151:3015–3025. doi: 10.1210/en.2009-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontan M, Berlan M, Carpene C. Fat cell adrenoceptors: inter- and intraspecific differences and hormone regulation. Int J Obes. 1985;9(Suppl 1):117–127. [PubMed] [Google Scholar]
- Lemberger T, Saladin R, Vazquez M, Assimacopoulos F, Staels B, Desvergne B, Wahli W, Auwerx J. Expression of the peroxisome proliferator-activated receptor alpha gene is stimulated by stress and follows a diurnal rhythm. J Biol Chem. 1996;271:1764–1769. doi: 10.1074/jbc.271.3.1764. [DOI] [PubMed] [Google Scholar]
- Liew AW-C, Law N-F, Caoc X-Q, Yan H. Statistical power of Fisher test for the detection of short periodic gene expression profiles. Pattern Recogn. 2009;42:549–556. [Google Scholar]
- Linford NJ, Beyer RP, Gollahon K, Krajcik RA, Malloy VL, Demas V, Burmer GC, Rabinovitch PS. Transcriptional response to aging and caloric restriction in heart and adipose tissue. Aging Cell. 2007;6:673–688. doi: 10.1111/j.1474-9726.2007.00319.x. [DOI] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcheva B, Ramsey KM, Affinati A, Bass J. Clock genes and metabolic disease. J Appl Physiol. 2009;107:1638–1646. doi: 10.1152/japplphysiol.00698.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronardi CA, Yu WH, McCann SM. Resting and circadian release of nitric oxide is controlled by leptin in male rats. Proc Natl Acad Sci U S A. 2002;99:5721–5726. doi: 10.1073/pnas.082098499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier P, Aaron J, Edouard C, Vignon G. Osteoporosis and the replacement of cell populations of the marrow by adipose tissue. A quantitative study of 84 iliac bone biopsies. Clin Orthop Relat Res. 1971;80:147–154. doi: 10.1097/00003086-197110000-00021. [DOI] [PubMed] [Google Scholar]
- Mitsui S, Yamaguchi S, Matsuo T, Ishida Y, Okamura H. Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev. 2001;15:995–1006. doi: 10.1101/gad.873501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk TH. Aging human circadian rhythms: conventional wisdom may not always be right. J Biol Rhythms. 2005;20:366–374. doi: 10.1177/0748730405277378. [DOI] [PubMed] [Google Scholar]
- Nedergaard J, Bengtsson T, Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. Am J Physiol Endocrinol Metab. 2007;293:E444–E452. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- Oishi K, Miyazaki K, Kadota K, Kikuno R, Nagase T, Atsumi G, Ohkura N, Azama T, Mesaki M, Yukimasa S, Kobayashi H, Iitaka C, Umehara T, Horikoshi M, Kudo T, Shimizu Y, Yano M, Monden M, Machida K, Matsuda J, Horie S, Todo T, Ishida N. Genome-wide expression analysis of mouse liver reveals CLOCK-regulated circadian output genes. J Biol Chem. 2003;278:41519–41527. doi: 10.1074/jbc.M304564200. [DOI] [PubMed] [Google Scholar]
- Ortega E, Xu X, Koska J, Marie Francisco A, Scalise M, Ferrante AW, Jr, Krakoff J. Macrophage content in subcutaneous adipose tissue: associations with adiposity, age, inflammatory markers, and whole-body insulin action in healthy Pima Indians. Diabetes. 2008;58:385–393. doi: 10.2337/db08-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padiath QS, Paranjpe D, Jain S, Sharma VK. Glycogen synthase kinase 3beta as a likely target for the action of lithium on circadian clocks. Chronobiol Int. 2004;21:43–55. doi: 10.1081/cbi-120027981. [DOI] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nature. 2002;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- Patel DD, Knight BL, Wiggins D, Humphreys SM, Gibbons GF. Disturbances in the normal regulation of SREBP-sensitive genes in PPAR alpha-deficient mice. J Lipid Res. 2001;42:328–337. [PubMed] [Google Scholar]
- Ptitsyn A. Stochastic resonance reveals “pilot light” expression in mammalian genes. PLoS One. 2008;3:e1842. doi: 10.1371/journal.pone.0001842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptitsyn AA, Zvonic S, Conrad SA, Scott LK, Mynatt RL, Gimble JM. Circadian clocks are resounding in peripheral tissues. PLoS Comput Biol. 2006;2:e16. doi: 10.1371/journal.pcbi.0020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptitsyn AA, Zvonic S, Gimble JM. Permutation test for periodicity in short time series data. BMC Bioinforma. 2006;7(2):S10. doi: 10.1186/1471-2105-7-S2-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptitsyn AA, Zvonic S, Gimble JM. Digital signal processing reveals circadian baseline oscillation in majority of mammalian genes. PLoS Comput Biol. 2007;3:e120. doi: 10.1371/journal.pcbi.0030120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptitsyn AA, Weil MM, Thamm DH. Systems biology approach to identification of biomarkers for metastatic progression in cancer. BMC Bioinforma. 2008;9(9):S8. doi: 10.1186/1471-2105-9-S9-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptitysn AAZS, Conrad SA, Scott LK, Mynatt ML, Gimble JM. Circadian clocks are resounding in peripheral tissues. PLoS Comput Biol. 2006;2:e16. doi: 10.1371/journal.pcbi.0020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehnmark S, Nechad M, Herron D, Cannon B, Nedergaard J. Alpha- and beta-adrenergic induction of the expression of the uncoupling protein thermogenin in brown adipocytes differentiated in culture. J Biol Chem. 1990;265:16464–16471. [PubMed] [Google Scholar]
- Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, MacDougald OA. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289:950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- Ruge T, Wu G, Olivecrona T, Olivecrona G. Nutritional regulation of lipoprotein lipase in mice. Int J Biochem Cell Biol. 2004;36:320–329. doi: 10.1016/s1357-2725(03)00256-5. [DOI] [PubMed] [Google Scholar]
- Saad MF, Riad-Gabriel MG, Khan A, Sharma A, Michael R, Jinagouda SD, Boyadjian R, Steil GM. Diurnal and ultradian rhythmicity of plasma leptin: effects of gender and adiposity. J Clin Endocrinol Metab. 1998;83:453–459. doi: 10.1210/jcem.83.2.4532. [DOI] [PubMed] [Google Scholar]
- Sato TK, Panda S, Kay SA, Hogenesch JB. DNA arrays: applications and implications for circadian biology. J Biol Rhythms. 2003;18:96–105. doi: 10.1177/0748730403252245. [DOI] [PubMed] [Google Scholar]
- Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Savitzky AGM. Smoothing and differentiation of data by simplified least squares procedures. Anal Chem. 1964;36:1627–1639. doi: 10.1021/ac60319a045. [DOI] [PubMed] [Google Scholar]
- Shimba S, Wada T, Hara S, Tezuka M. EPAS1 promotes adipose differentiation in 3T3-L1 cells. J Biol Chem. 2004;279:40946–40953. doi: 10.1074/jbc.M400840200. [DOI] [PubMed] [Google Scholar]
- Shimba S, Ishii N, Ohta Y, Ohno T, Watabe Y, Hayashi M, Wada T, Aoyagi T, Tezuka M. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc Natl Acad Sci U S A. 2005;102:12071–12076. doi: 10.1073/pnas.0502383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel K, Tasali E, Penev P, Van Cauter E. Brief communication: sleep curtailment in healthy young men is associated with decreased leptin levels, elevated ghrelin levels, and increased hunger and appetite. Ann Intern Med. 2004;141:846–850. doi: 10.7326/0003-4819-141-11-200412070-00008. [DOI] [PubMed] [Google Scholar]
- Sutton GM, Perez-Tilve D, Nogueiras R, Fang J, Kim JK, Cone RD, Gimble JM, Tschop MH, Butler AA. The melanocortin-3 receptor is required for entrainment to meal intake. J Neurosci. 2008;28:12946–12955. doi: 10.1523/JNEUROSCI.3615-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR. Comparative analysis of microarray data identifies common responses to caloric restriction among mouse tissues. Mech Ageing Dev. 2008;129:138–153. doi: 10.1016/j.mad.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR. Genes and gene expression modules associated with caloric restriction and aging in the laboratory mouse. BMC Genomics. 2009;10:585. doi: 10.1186/1471-2164-10-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchkonia T, Morbeck DE, Von Zglinicki T, Van Deursen J, Lustgarten J, Scrable H, Khosla S, Jensen MD, Kirkland JL. Fat tissue, aging, and cellular senescence. Aging Cell. 2010;9:667–684. doi: 10.1111/j.1474-9726.2010.00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torra IP, Tsibulsky V, Delaunay F, Saladin R, Laudet V, Fruchart JC, Kosykh V, Staels B. Circadian and glucocorticoid regulation of Rev-erbalpha expression in liver. Endocrinology. 2000;141:3799–3806. doi: 10.1210/endo.141.10.7708. [DOI] [PubMed] [Google Scholar]
- Trayhurn P. Endocrine and signalling role of adipose tissue: new perspectives on fat. Acta Physiol Scand. 2005;184:285–293. doi: 10.1111/j.1365-201X.2005.01468.x. [DOI] [PubMed] [Google Scholar]
- Trayhurn P, Beattie JH. Physiological role of adipose tissue: white adipose tissue as an endocrine and secretory organ. Proc Nutr Soc. 2001;60:329–339. doi: 10.1079/pns200194. [DOI] [PubMed] [Google Scholar]
- Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, Iino M, Shigeyoshi Y, Hashimoto S. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- Valentinuzzi VS, Scarbrough K, Takahashi JS, Turek FW. Effects of aging on the circadian rhythm of wheel-running activity in C57BL/6 mice. Am J Physiol. 1997;273:R1957–R1964. doi: 10.1152/ajpregu.1997.273.6.R1957. [DOI] [PubMed] [Google Scholar]
- van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJ. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, Taittonen M, Laine J, Savisto NJ, Enerback S, Nuutila P. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- Weinert D, Waterhouse J. Daily activity and body temperature rhythms do not change simultaneously with age in laboratory mice. Physiol Behav. 1999;66:605–612. doi: 10.1016/s0031-9384(98)00342-4. [DOI] [PubMed] [Google Scholar]
- Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichert S, Fokianos K, Strimmer K. Identifying periodically expressed transcripts in microarray time series data. Bioinformatics. 2004;20:5–20. doi: 10.1093/bioinformatics/btg364. [DOI] [PubMed] [Google Scholar]
- Yang X, Enerback S, Smith U. Reduced expression of FOXC2 and brown adipogenic genes in human subjects with insulin resistance. Obes Res. 2003;11:1182–1191. doi: 10.1038/oby.2003.163. [DOI] [PubMed] [Google Scholar]
- Yang X, Jansson PA, Nagaev I, Jack MM, Carvalho E, Sunnerhagen KS, Cam MC, Cushman SW, Smith U. Evidence of impaired adipogenesis in insulin resistance. Biochem Biophys Res Commun. 2004;317:1045–1051. doi: 10.1016/j.bbrc.2004.03.152. [DOI] [PubMed] [Google Scholar]
- Yildiz BO, Suchard MA, Wong ML, McCann SM, Licinio J. Alterations in the dynamics of circulating ghrelin, adiponectin, and leptin in human obesity. Proc Natl Acad Sci U S A. 2004;101:10434–10439. doi: 10.1073/pnas.0403465101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, Lazar MA. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- Zvonic S, Ptitsyn AA, Conrad SA, Scott LK, Floyd ZE, Kilroy G, Wu X, Goh BC, Mynatt RL, Gimble JM. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006;55:962–970. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]
- Zvonic S, Ptitsyn AA, Kilroy G, Wu X, Conrad SA, Scott LK, Guilak F, Pelled G, Gazit D, Gimble JM. Circadian oscillation of gene expression in murine calvarial bone. J Bone Miner Res. 2007;22:357–365. doi: 10.1359/jbmr.061114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of Circadian Gene Oscillation by qRT-PCR. Total RNA isolated from brown adipose tissue (Fig. 1), epididymal WAT (Fig. 2), inguinal WAT (Fig. 3), and liver (Fig. 4) of younger and older mice was harvested at serial 4-h time points beginning at 0700 hours and continuing until 0300 hours during a single 24-h period. The level of expression of the following mRNAs representative of the circadian apparatus and adipogenesis was determined and normalized relative to cyclophilin B: Bmal1, Cry1, Cry2, DBP, E4BP4, Leptin, LPL, Npas2, Per1, PGC-1, PPARγ, Rev-Erbα, and Rev-Erbβ. Values presented are the mean ± SD, and these have been double-plotted. (JPEG 94 kb)
(JPEG 75 kb)
(JPEG 87 kb)
(JPEG 61 kb)
(JPEG 80 kb)
(JPEG 44 kb)
(JPEG 71 kb)
(JPEG 56 kb)
The forward and reverse primers as well as the PubMed accession numbers used for qRT-PCR are listed in Table 8. (DOC 34 kb)
The transcripts that were expressed differentially between the young and old cohorts as a function of biological age in two or more tissues are summarized in Table 7. Shared transcripts in BAT/iWAT, BAT/Liver, and iWAT/Liver as well as all three tissues are displayed. (DOC 113 kb)