Abstract
The severe acute respiratory syndrome (SARS) epidemic started in late 2002 and swiftly spread across 5 continents with a mortality rate of around 10%. Although the epidemic was eventually controlled through the implementation of strict quarantine measures, there continues a need to investigate the SARS coronavirus (SARS-CoV) and develop interventions should it re-emerge. Numerous studies have shown that neutralizing antibodies against the virus can be found in patients infected with SARS-CoV within days upon the onset of illness and lasting up to several months. In contrast, there is little data on the kinetics of T cell responses during SARS-CoV infection and little is known about their role in the recovery process. However, recent studies in mice suggest the importance of T cells in viral clearance during SARS-CoV infection. Moreover, a growing number of studies have investigated the memory T cell responses in recovered SARS patients. This review covers the available literature on the emerging importance of T cell responses in SARS-CoV infection, particularly on the mapping of cytotoxic T lymphocyte (CTL) epitopes, longevity, polyfunctionality and human leukocyte antigen (HLA) association as well as their potential implications on treatment and vaccine development.
Keywords: SARS, epitope, T-cell immunity, CD4+, CD8+, virus clearance, coronavirus
Introduction
Severe acute respiratory syndrome (SARS) first emerged in Guangdong, China in late 20021 and infected more than 8000 people in 29 countries across 5 continents.2 According to the World Health Organization (WHO), the fatality rate of the SARS outbreak was estimated to be 9.6%. Of those infected, healthcare workers and care-takers accounted for the majority. The SARS epidemic was officially controlled by July 2003 after the implementation of strict isolation of patients. Sometime into the epidemic, a novel coronavirus, the SARS coronavirus (SARS-CoV), was identified as the causative agent.3,4,5 Molecular epidemiology showed that at least two strains of SARS-CoV infected the patients in Hong Kong,6 suggesting that the virus had jumped from animal sources to humans on two separate occasions. Later in 2005, reports from two laboratories identified a virus resident in Chinese horseshoe bats that is genetically similar to the human SARS-CoV, pinpointing the horseshoe bat to be a likely natural reservoir of the SARS-CoV.7,8 If this is indeed the case, a re-emergence of SARS-CoV cannot be ruled out.
Coronaviruses are a diverse group of large, enveloped positive-stranded RNA viruses in the order Nilovirales, family Coronaviridae, and genus Coronavirus. Typically, they cause respiratory and enteric diseases in humans and animals. Using the open reading frame (ORF) 1a sequences, SARS-CoV was categorized as a subgroup of the group II coronaviruses.9 The 30 kb poly-adenylated positive-stranded RNA genome10,11 has an genomic organization typical of a coronavirus where the first two ORFs (1a and 1b) encode the viral replicase that requires processing by the viral cysteine proteinases to yield the functional membrane-bound replicase complex and a group of 16 non-structural proteins (NSP).12 Although some functions of these NSPs have been investigated and known, there are many others that still require further characterization (reviewed by Cheng et al.13).
The SARS-CoV genome encodes four structural proteins: spike (S), envelope (E), membrane (M) and nucleocaspid (N). In addition, a set of unique accessory proteins (namely ORF 3a, 3b, 6, 7a, 7b, 8a, 8b and 9b) is also found. Functionally, the N protein packs the RNA into a helical nucleocapsid; while the S protein forms the characteristic projections on the virion surface for the attachment and entry into the host cells; and together, N, M and E control the assembly of the virion. At present, no significant homology has been found for the accessory proteins to the viral proteins of other coronaviruses; in fact, they were found to be dispensable for virus replication in cell culture despite contributing to viral pathogenesis.14,15
Neutralizing antibodies against SARS-CoV found in patients and animals infected with SARS-CoV block viral entry by binding to the S glycoprotein.16 Besides the humoral response, the role of T cells in viral infections has been known to be just as important. Whilst neutralizing antibodies can prevent viral entry, the body also requires SARS-CoV specific CD4+ T helper cells for the development of these specific antibodies. Similarly, CD8+ cytotoxic T cells are important for the recognition and killing of infected cells, particularly in the lungs of infected individuals. Despite the increasing number of reports that investigated memory CD4+ and CD8+ T cell responses in recovered SARS patients, there is a lack of data that describes the kinetics of the T cell response during a SARS-CoV infection. This review will focus on the memory T cell studies and its possible implications on treatment and vaccine development. For easy reference, all the T cell epitopes identified are summarized in Table 1.
Table 1. Summary of T cell epitopes found in the SARS-CoV.
| Protein | Amino acid position | HLA restriction | Identification | References |
|---|---|---|---|---|
| Spike | 1203 to 1211 | HLA-A*02:01 | Human PBMCs | 19 |
| Spike | 978 to 986 | HLA-A*02:01 | Human PBMCs | 19 |
| Spike | 1167 to 1175 | HLA-A*02:01 | Transgenic mouse and verified in human PBMCs | 20 |
| Spike | 787 to 795 | HLA-A*02:01 | Human PBMCs | 22 |
| Spike | 1042 to 1050 | HLA-A*02:01 | Human PBMCs | 22 |
| Spike | 411 to 420 | HLA-A*02:01 | Human PBMCs | 23 |
| Spike | 958 to 966 | HLA-A*02:01 | Transgenic mouse and verified in human PBMCs | 24 |
| Nucleocapsid | 223 to 231 | HLA-A*02:01 | Transgenic mouse | 22 |
| Nucleocapsid | 227 to 235 | HLA-A*02:01 | Transgenic mouse | 22 |
| Nucleocapsid | 317 to 325 | HLA-A*02:01 | Transgenic mouse and verified in human PBMCs | 22 |
| Nucleocapsid | 331 to 347 | HLA-A*02:01 | Human PBMCs | 28 |
| Nucleocapsid | 346 to 362 | HLA-A*02:01 | Human PBMCs | 28 |
| Nucleocapsid | 211 to 235 | HLA-A*02:01 | Human PBMCs | 32 |
| Nucleocapsid | 330 to 354 | HLA-A*02:01 | Human PBMCs | 32 |
| Nucleocapsid | 216 to 225 | HLA-B*40:01 | Human PBMCs | 33, 34 |
| Membrane | 21 to 44 | ND | Human PBMCs | 41 |
| Membrane | 65 to 91 | ND | Human PBMCs | 41 |
| Membrane | 117 to 140 | ND | Human PBMCs | 41 |
| Membrane | 200 to 220 | ND | Human PBMCs | 41 |
| Membrane | 146 to 160 | ND | Human PBMCs | 34 |
| 3a | 36 to 50 | ND | Human PBMCs | 34 |
| 3a | 6 to 20 | HLA-B*58:01 | Human PBMCs | Unpublished |
*ND indicates not determined.
Characterization of T cell epitopes in the spike (S) glycoprotein
Amongst the SARS-CoV structural proteins, the S protein has been found to elicit neutralizing antibodies17,18 with its major immunodominant epitope found between residues 441 to 700. Using an online database and with verification from T2 binding assays, the first two HLA-A*02:01-restricted T cell epitopes (S1203–1211 and S978–986) were identified in the S protein of SARS-CoV.19 They were immunogenic and elicited high IFNγ-specific T cell response in patients who have recovered from SARS. In comparison, the homologous peptide from HCoV229e did not elicit a significant response. A third CTL epitope, S1167–1175 (also known as SSp-1) was reported shortly after by another group.20 This peptide was able to induce CTL response in HLA-A*02:01 transgenic mice immunized with peptide loaded dendritic cells (DCs). At the same time, they were able to generate peptide-specific CD8+ CTL in peripheral blood mononuclear cells (PBMCs) from healthy human donor. The same group also showed that heat inactivated SARS-CoV particles elicited CTL response to all three S epitopes (SSp-1, S978 and S1202) in patients' PBMCs one year post-infection.21 Interestingly, 5 healthy individuals without contact history with SARS-CoV also exhibited SSp-1-specific responses in their PBMCs but exhibited lower cytotoxic activity and cytokine release when compared to the recovered SARS patients.
Other T cell epitopes identified include S787–795, S1042–1050 (found in the S2 domain) and S411–420 (P15) (found in the S1 domain) .22,23 These epitopes were found to be immunogenic and able to induce strong IFNγ production from PBMCs of recovered SARS patients. At the same time, HLA-A*02:01 transgenic mice immunized with DNA vaccines encoding the S protein were able to induce significant peptide-specific response.24 In this study, the HLA-A*02:01 restricted epitope S958–966 (also known as Sp8), first identified based on HLA-A*02:01 binding peptide and proteosomal cleavage prediction systems, was found capable of inducing specific CTLs in the PBMCs of healthy individuals as well as in transgenic mice immunized with S DNA vaccine.24 This suggests that there may already be SARS-CoV-specific CTL precursor cells within the T-cell repertoire of healthy individuals.
Animal studies using mice primed intramuscularly with S DNA vaccine and boosted with subcutaneous HLA-A*02:01 restricted peptides25 or with the S DNA vaccine alone26 elicited antigen-specific CD8+ T cell responses. In fact, one recent study showed that prime-boost immunization of transgenic mice with 5 of the HLA-A*02:01 S peptides together with CpG oligodeoxynucleotide (ODN) could significantly enhance the frequency of peptide-specific CD8+ T cells.27
Taken together, the S protein is not only capable of inducing neutralizing antibodies but also contains several immunogenic T cell epitopes. Some of these epitopes found in either the S1 or S2 domain of the protein should therefore be considered during SARS-CoV vaccine development.
Characterization of T cell epitopes in the nucleocapsid (N) protein
Besides the S glycoprotein, persistently high levels of anti-N protein antibodies and T cell responses were also found in the SARS-recovered individuals 2 years post-infection.28,29 For other coronaviruses, some protective effects were found to be conferred through N-specific CD8+ T cells.30,31 Using a similar approach of HLA peptide binding prediction algorithm with validation from T2-cell binding assay, Tsao et al. identified several HLA-A*02:01 restricted epitopes in the N protein (peptide N223–231, N227–235 and N317–325) and showed that they could induce specific CTL responses in transgenic mice immunized with N proteins or peptides with CpG ODN.22 In addition, peptide N317–325 was able to stimulate the recall of CD8+ T cell response in PBMCs of recovered SARS patients.
There had been numerous attempts to screen for CTL epitopes in the N protein through the use of overlapping peptides spanning the entire N protein. One such study that used PBMCs from recovered SARS patients 2 years post-infection has revealed that the major dominant antigenic site of the N protein lies in the C-terminal region (amino acids 331 to 362). At least 2 different T cell epitopes (N331–347 and N346–362) have been found in this region when the PBMCs were stimulated with a pool of 57 overlapping N peptides in vitro, followed by IFNγ Enzyme-linked immunosorbent spot (ELISPOT) assay.28 Using the same approach, another group identified 2 potential CTL epitopes at positions N211–235 and N330–354 in the N protein.32 More recently, we also identified the same dominant response (N216–230) in SARS-recovered patients 6 years post-infection.33 This response was observed in 19% [3/16] of our cohort of recovered SARS patients. Similarly, a comprehensive study of T cell responses against all the SARS-CoV proteins conducted by Li et al. showed that 11% of their SARS subjects gave positive T cell responses against peptide N211–225, and it was identified as the most recognized epitope in the N protein.34 Exact epitope mapping by our group further indicated that the CTL epitope was a 10 mer (N216–225) restricted by HLA-B*40:01 and that PBMCs from healthy individuals can be transduced to become N peptide-specific T cells.33
In one of the first animal studies conducted in monkeys, adenoviral-based vectors were used to test the efficacy of the S, M and N proteins.35 The monkeys were injected intramuscularly with adenoviral-based vectors that expressed codon-optimized S1 domain, M and N proteins. The S1 domain of the S protein was found to induce strong humoral response, while the N protein elicited high frequency of IFNγ-producing T cells as determined using N peptides as the antigen in the ELISPOT assay. This was the first indication that the N protein could be a good vaccine candidate for cell-mediated T cell response. This phenomenon was also found in mice where DNA vaccines encoding the N protein elicited good T cell responses.36,37,38,39 C3H/He mice intramuscularly immunized with N protein pcDNA-fn vector showed both high antibody titre and CTL activity after 3 injections;36 and using Balb/c mice, two other groups showed that DNA vaccines encoding N protein alone could elicit T cell proliferation, IFNγ release, delayed-type hypersensitivity (DTH) and in vivo cytotoxic T cell activity.37,38 Further experiments reported enhanced T cell response when calreticulin (CRT)-linked DNA vaccine was used39 or DNA vaccination was performed with the addition of a chemical adjuvant levamisole.38 Synthetic N peptides coupled to the surface of liposomes were also reported to enhance T cell response.40 These synthetic N peptides not only induced CTL response, but the mice were also able to clear vaccinia virus-expressing SARS-CoV epitopes when challenged.40
In summary, several different studies have identified immunogenic regions in amino acids 211 to 362 of the N protein to contain T cell epitopes. However, to date, the only epitope characterized in detail is the 10-mer epitope (N216–225) which is restricted by HLA-B*40:01.33
Characterization of T cell epitopes in other SARS-CoV proteins
There are very few studies of T cell response to other SARS-CoV proteins. Nonetheless, animal studies using DNA vaccines suggest that the M protein may induce T cell response, albeit to a lesser degree than the S and N proteins.38 Yang et al. demonstrated that it was possible to induce recall T cell response from the PBMCs of SARS patients who have recovered for more than 1 year by using overlapping peptides spanning the entire M protein.41 In this study, four human T cell immunodominant peptides, M21–44, M65–91, M117–140 and M200–220, were subsequently identified. Similarly, Li et al. also reported that 9% of their SARS subjects had T cell response against the M peptide region, M146–160.34 The largest accessory protein of SARS-CoV is the 3a protein of 274 amino acids. However, other than Li et al.'s report, there had been no demonstration of T cell responses against this protein. The 3a protein peptide 3a36–50 was one of the three most frequently recognized T cell epitopes identified in their study.34 Similar to the results reported by Li et al. our data showed that the 3a protein peptide 3a6-20 was able to elicit both CD8+ and CD4+ responses.33 Interestingly, mice immunized with 3a DNA vaccine were shown to have high levels of humoral response as well as Th1 response.42 These observations indicated that the accessory 3a protein is immunogenic and able to induce T cell response.
Although T cell response could be observed for the M protein, current studies seem to suggest that the 3a protein is more immunogenic in comparison, and T cell epitopes identified in it may play an important role in recovery from a primary SARS-CoV infection and in vaccine development.
Longevity and phenotype of CD4+ and CD8+ T cell responses
To date, there is only one study that investigated T cell response against whole SARS-CoV in humans.34 In this study, PBMCs from 1-year post-infected patients showed T cell response to eight (replicase, S, N, E, M, 3a, 3b, and 9b) out of the fourteen SARS-CoV proteins when tested using overlapping peptides spanning the entire SARS-CoV genome. Of the 70% of T cell responses elicited against the structural proteins, the S protein induced the most dominant responses (41%). In fact, the three most commonly recognized T cell epitope were that of one found in 3a, and the other two in the S protein (S435–451 and S633–650). The latter were not reported in the other studies. Although both CD4+ and CD8+ T cell responses were observed in the study, the frequency and magnitude of the CD8+ T cell responses were greater than the CD4+ T cell responses. However, the CD4+ and CD8+ T cells were found to have similar central memory phenotypes (CD27+ and CD45RO+). A separate study by Peng et al. showed that the N-specific CD4+ T cells had central memory (CD45RA− CCR7+ CD62L−), whereas most of the CD8+ T cells had effector memory (CD45RA+ CCR7− CD62L−) phenotype.28 Similar observations were made with the M-specific CD4+ and CD8+ T cells,41 whilst the S-specific CD8+ T cells were reported to have effector memory (CD45RA+ CCR7− CD62L−) phenotype.21
Using peptides in the four SARS-CoV structural proteins, an analysis of the memory T cell response in recovered SARS patients four years post-infection revealed that both CD4+ and CD8+ T cells produced IFNγ.43 In support of Li et al.'s study,34 Fan et al. also found that CD4+ memory T cells produce IL-2, TNFα and IFNγ, with the exception of one patient.43 It was also observed that S peptides induced the highest percentages of IFNγ producing cells. Interestingly, the frequency of these polyfunctional CD4+ T cells (T cells producing multiple cytokines) was higher in the individuals with severe SARS infection than in moderately severe patients.34 On the other hand, this difference between moderate severe and severe patients was not observed with the CD8+ T cells which produced mainly IFNγ. Nonetheless, a proportion of the CD8+ T cells were found to produce TNFα and degranulate (with the detection of CD107a).
Since polyfunctional T cells were associated with better control of human immunodeficiency virus (HIV) infection and vaccination efficacy,44,45 we further characterized the cytokine profile of the SARS-specific T cells in our recent study.33 A summary of our findings is shown in Figure 1. We have observed that the majority of CD4+ T cells produced IFNγ, IL-2, and TNFα, with a small percentage of the cells also simultaneously producing inflammatory cytokines such as macrophage inflammatory protein (MIP) 1α, MIP 1β and granulocyte-macrophage colony-stimulating factor (GM-CSF). Of these, the majority of the CD8+ T cells produced IFNγ, TNFα, MIP 1α or MIP 1β alone or in combination. Only a small percentage produced IFNγ, IL-2, and TNFα. Moving forward, we cloned the α and β T cell receptor (TCR) chains of one immunodominant CTL epitope in the N protein (amino acid 216 to 225) from the SARS-CoV specific CD8+ T cells and used them to redirect the specificity of lymphocytes of healthy subjects lacking SARS-CoV specific memory T cells. These TCR-redirected T cells were found to possess a cytokine production profile similar to SARS-CoV specific memory CD8+ T cells in recovered SARS patients (as mentioned above). Thus we proposed that these T cells may be potential therapeutic treatments for this life threatening infection.
Figure 1.
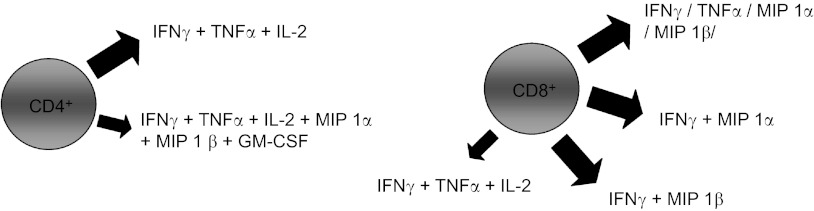
Diagram showing the cytokine profiles of the CD4+ and CD8+ T cells from SARS recovered patients. The big and small arrows indicate the major and minor populations of CD4+ and CD8+ T cells producing the cytokines indicated respectively.
Despite the numerous reports describing the elevation of inflammatory cytokines in primary infected patients (reviewed by Zhu et al.46), it is not known if these cytokines are beneficial or contribute to the pathogenicity of the infection. Moreover, there is currently no report confirming the protective effect of T cells during a primary SARS-CoV infection in humans. In fact, research in this area is hampered by the lack of systematic sample collection during the 2003 SARS outbreak which lasted for a relatively short period of ∼ 16 weeks. Since there is no second major outbreak of SARS, the protective effect of memory T cell response in recovered SARS patients is not known. Nevertheless, the phenotype and cytokine profile of the T cells in these recovered individuals indicate the possible protective effect of T cell response in the initial infection or during any subsequent infections.
HLA association
The association of certain HLA genotypes with increased resistance or the ability to clear viral infections have been reported in hepatitis C virus (HCV) and human papillomavirus studies.47,48,49,50 Although earlier studies done on SARS patients from Taiwan and Hong Kong suggested that the HLA-B, HLA-Cw and HLA-DR alleles were highly associated with SARS infection and disease development,51,52,53 further investigation is required. Of these literature, SARS individuals from Hong Kong showed that HLA-B*07:03 and HLA-DR*03:01 conferred factors for susceptibility and resistance to SARS infection, respectively.53 In agreement with this, a study on a Taiwanese cohort of SARS patients found that both HLA-Cw*15:02 and HLA-DR*03:01 were associated with resistance to SARS infection.54 These observations suggested the important role of HLA-DR*03:01 in viral disease progression through enhancing the function of CD4+ T helper cells. Similarly, we observed that the CD8+ T cell responses against both the N and 3a proteins were all restricted by HLA-B subtype (unpublished data), thus pointing to the possible role of HLA-B subtypes in viral immunity. Among the HLA class I genes, HLA-B is known to be the most polymorphic,55 and was associated in protective roles against the HIV,56,57,58 HCV59 and acute influenza infections.60
Conclusion
Currently, no antiviral therapy has yet been proven useful for SARS. Attempts to test potential anti-SARS agents using antiviral antibodies, entry inhibitors, proteinase inhibitors, calpain inhibitors, ribavirin (nucleoside analogues), interferons, and short interfering RNAs were riddled with contradictory reports from different laboratories. The lack of clinical trials also prevented the reaching of a conclusive agreement for effective anti-SARS strategies (reviewed by Weiss et al.61). Nevertheless, human convalescent-phase plasma seemed to shorten hospitalization without adverse effects if it is administered as an immunotherapy to SARS patients early in the course of infection.62 With the finding that recovered SARS patients have higher and more sustainable levels of neutralizing antibodies when compared to those who had succumbed to the disease,63 monoclonal antibodies for passive immunization were also obtained using phage-display antibody libraries and immortalization of B cells from convalescent SARS patients.64,65
Although it is still not known whether naturally acquired immune responses can confer protection from re-infection of SARS-CoV, vaccines are likely to be the most effective way to provide protection against a future re-emergence of SARS-CoV. Several strategies for vaccine development included DNA vaccines, inactivated whole virus vaccines,66,67 virus-like particles,68,69 recombinant virus vector vaccines,70 and recombinant protein vaccine.71 Most SARS-CoV vaccines that elicited neutralizing antibodies are believed to be protective, but as described, T cells may also play an important role in viral clearance in a primary SARS-CoV infection.72,73 Zhao et al. suggested that inefficient immune activation and a poor virus-specific T cell response underlay severe disease in SARS-CoV infected mice.74 In their recent report, they showed that virus-specific T cells were necessary and sufficient for virus clearance and protection from clinical disease in mouse-adapted SARS-CoV (MA15) virus-infected mice.73 In addition, CD4+ T cells in a senescent mouse model were found to play an important role in viral clearance in a primary infection with SARS-CoV.72 In humans, SARS-CoV-specific memory T cells were found to persist in the peripheral blood of SARS patients up to 6 years post-infection despite a lack of specific memory B cell response in these patients.75 This seems to suggest that SARS- CoV-specific T cell response could persist longer and thus indicating that cell-mediated immune response is important for protecting against re-infection. As all these studies suggest that T cell may play a crucial role in the clearance of SARS-CoV, there is therefore a need for detailed characterization of the T cell response to SARS-CoV for the development of future vaccine candidates.
Finally, it is important to note that T cells can play a protective and/or pathological role during an infection. In the case of mouse-hepatitis virus (MHV), an increase of morbidity and mortality in infected mice has been associated with memory T cells.76 Although no direct evidence have shown that SARS-CoV-specific T cell responses contribute to immunopathology in SARS, it is a question that needs to be further addressed.
Acknowledgments
Research work on the T cell immune response in SARS coronavirus infection in the corresponding author's laboratory is supported by a grant from the Biomedical Research Council, Agency of Science Technology and Research (A*Star), Singapore No: 10/1/21/16/652.
References
- Zhong NS, Zheng BJ, Li YM, Xie ZH, Chan KH, et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003. 2003. Available at http://www.who.int/csr/sars/country/table2003_09_23/en/.
- Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki, Peret T, Emery S, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Guan Y, Peiris JS, Zheng B, Poon LL, Chan KH, Zeng FY, et al. Molecular epidemiology of the novel coronavirus that causes severe acute respiratory syndrome. Lancet. 2004;363:99–104. doi: 10.1016/S0140-6736(03)15259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, Wong BH, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LL, et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Marra MA, Jones SJ, Astell CR, Holt RA, Brooks-Wilson A, Butterfield YS, et al. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Ziebuhr J, Snijder EJ, Gorbalenya AE. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- Cheng VC, Lau SK, Woo PC, Yuen KY. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20:660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YJ, Lim SG, Hong W. Understanding the accessory viral proteins unique to the severe acute respiratory syndrome (SARS) coronavirus. Antiviral Res. 2006;72:78–88. doi: 10.1016/j.antiviral.2006.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K, Huang C, Makino S. SARS coronavirus accessory proteins. Virus Res. 2008;133:113–121. doi: 10.1016/j.virusres.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh PR, Huang LM, Chen PJ, Kao CL, Yang PC. Chronological evolution of IgM, IgA, IgG and neutralisation antibodies after infection with SARS-associated coronavirus. Clin Microbiol Infect. 2004;10:1062–1066. doi: 10.1111/j.1469-0691.2004.01009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz UJ, Bukreyev A, Yang L, Lamirande EW, Murphy BR, Subbarao K, et al. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc Natl Acad Sci USA. 2004;101:9804–9809. doi: 10.1073/pnas.0403492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Manopo I, Leung BP, Chng HH, Ling AE, Chee LL, et al. Immunological characterization of the spike protein of the severe acute respiratory syndrome coronavirus. J Clin Microbiol; 42:1570–1576. doi: 10.1128/JCM.42.4.1570-1576.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YD, Sin WY, Xu GB, Yang HH, Wong TY, Pang XW, et al. T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus spike protein elicit a specific T-cell immune response in patients who recover from SARS. J Virol. 2004;78:5612–5618. doi: 10.1128/JVI.78.11.5612-5618.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Chen H, Jiang X, Zhang M, Wan T, Li N, et al. Identification of an HLA-A*0201-restricted CD8+ T-cell epitope SSp-1 of SARS-CoV spike protein. Blood. 2004;104:200–206. doi: 10.1182/blood-2003-11-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Hou J, Jiang X, Ma S, Meng M, Wang B, et al. Response of memory CD8+ T cells to severe acute respiratory syndrome (SARS) coronavirus in recovered SARS patients and healthy individuals. J Immunol. 2005;175:591–598. doi: 10.4049/jimmunol.175.1.591. [DOI] [PubMed] [Google Scholar]
- Tsao YP, Lin JY, Jan JT, Leng CH, Chu CC, Yang YC, et al. HLA-A*0201 T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus nucleocapsid and spike proteins. Biochem Biophys Res Commun. 2006;344:63–71. doi: 10.1016/j.bbrc.2006.03.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Xu D, Li X, Li H, Shan M, Tang J, et al. Screening and identification of severe acute respiratory syndrome-associated coronavirus-specific CTL epitopes. J Immunol. 2006;177:2138–2145. doi: 10.4049/jimmunol.177.4.2138. [DOI] [PubMed] [Google Scholar]
- Lv Y, Ruan Z, Wang L, Ni B, Wu Y. Identification of a novel conserved HLA-A*0201-restricted epitope from the spike protein of SARS-CoV. BMC Immunol. 2009;10:61. doi: 10.1186/1471-2172-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Yang B, Xu Y, Wu C. CD8+ T cell response in HLA-A*0201 transgenic mice is elicited by epitopes from SARS-CoV S protein. Vaccine. 2010;28:6666–6674. doi: 10.1016/j.vaccine.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Ma R, Wu CY. Immunization with SARS-CoV S DNA vaccine generates memory CD4+ and CD8+ T cell immune responses. Vaccine. 2006;24:4905–4913. doi: 10.1016/j.vaccine.2006.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Wang H, Wu C. The immune responses of HLA-A*0201 restricted SARS-CoV S peptide-specific CD8 T cells are augmented in varying degrees by CpG ODN, PolyI:C and R848. Vaccine. 2011;29:6670–6678. doi: 10.1016/j.vaccine.2011.06.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Yang LT, Wang LY, Li J, Huang J, Lu ZQ, et al. Long-lived memory T lymphocyte responses against SARS coronavirus nucleocapsid protein in SARS-recovered patients. Virology. 2006;351:466–475. doi: 10.1016/j.virol.2006.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DT, Tam FC, Ma CH, Chan PK, Cheung JL, Niu H, et al. Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. J Infect Dis. 2004;190:379–386. doi: 10.1086/422040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collisson EW, Pei J, Dzielawa J, Seo SH. Cytotoxic T lymphocytes are critical in the control of infectious bronchitis virus in poultry. Dev Comp Immunol. 2000;24:187–200. doi: 10.1016/s0145-305x(99)00072-5. [DOI] [PubMed] [Google Scholar]
- Seo SH, Wang L, Smith R, Collisson EW. The carboxyl-terminal 120-residue polypeptide of infectious bronchitis virus nucleocapsid induces cytotoxic T lymphocytes and protects chickens from acute infection. J Virol. 1997;71:7889–7894. doi: 10.1128/jvi.71.10.7889-7894.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Xie J, He Y, Fan H, Baril L, Qiu Z, et al. Long-term persistence of robust antibody and cytotoxic T cell responses in recovered patients infected with SARS coronavirus. PLoS One. 2006;1:e24. doi: 10.1371/journal.pone.0000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh HL, Chia A, Chang CX, Leong HN, Ling KL, Grotenbreg GM, et al. Engineering T cells specific for a dominant severe acute respiratory syndrome coronavirus CD8 T cell epitope. J Virol. 2011;85:10464–10471. doi: 10.1128/JVI.05039-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CK, Wu H, Yan H, Ma S, Wang L, Zhang M, et al. T cell responses to whole SARS coronavirus in humans. J Immunol. 2008;181:5490–5500. doi: 10.4049/jimmunol.181.8.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Tamin A, Soloff A, D'Aiuto L, Nwanegbo E, Robbins PD, et al. Effects of a SARS-associated coronavirus vaccine in monkeys. Lancet. 2003;362:1895–1896. doi: 10.1016/S0140-6736(03)14962-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu MS, Pan Y, Chen HQ, Shen Y, Wang XC, Sun YJ, et al. Induction of SARS-nucleoprotein-specific immune response by use of DNA vaccine. Immunol Lett. 2004;92:237–243. doi: 10.1016/j.imlet.2004.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Cao J, Zhao LJ, Qin ZL, Ke JS, Pan W, et al. Immune responses against SARS-coronavirus nucleocapsid protein induced by DNA vaccine. Virology. 2005;331:128–135. doi: 10.1016/j.virol.2004.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Xiao C, Chen Z, Kang Y, Ma Y, Zhu K, et al. Induction of Th1 type response by DNA vaccinations with N, M, and E genes against SARS-CoV in mice. Biochem Biophys Res Commun. 2005;328:979–986. doi: 10.1016/j.bbrc.2005.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Lee JH, Hung CF, Peng S, Roden R, Wang MC, et al. Generation and characterization of DNA vaccines targeting the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J Virol. 2004;78:4638–4645. doi: 10.1128/JVI.78.9.4638-4645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S, Kohyama S, Taneichi M, Moriya O, Hayashi H, Oda H, et al. Synthetic peptides coupled to the surface of liposomes effectively induce SARS coronavirus-specific cytotoxic T lymphocytes and viral clearance in HLA-A*0201 transgenic mice. Vaccine. 2009;27:3912–3920. doi: 10.1016/j.vaccine.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Peng H, Zhu Z, Li G, Huang Z, Zhao Z, et al. Persistent memory CD4+ and CD8+ T-cell responses in recovered severe acute respiratory syndrome (SARS) patients to SARS coronavirus M antigen. J Gen Virol. 2007;88:2740–2748. doi: 10.1099/vir.0.82839-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Tao L, Wang T, Zheng Z, Li B, Chen Z, et al. Humoral and cellular immune responses induced by 3a DNA vaccines against severe acute respiratory syndrome (SARS) or SARS-like coronavirus in mice Clin Vaccine Immunol2009;1673–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan YY, Huang ZT, Li L, Wu MH, Yu T, Koup RA, et al. Characterization of SARS-CoV-specific memory T cells from recovered individuals 4 years after infection. Arch Virol. 2009;154:1093–1099. doi: 10.1007/s00705-009-0409-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Precopio ML, Betts MR, Parrino J, Price DA, Gostick E, Ambrozak DR, et al. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8(+) T cell responses. J Exp Med. 2007;204:1405–1416. doi: 10.1084/jem.20062363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M. SARS Immunity and Vaccination. Cell Mol Immunol. 2004;1:193–198. [PubMed] [Google Scholar]
- Breitburd F, Ramoz N, Salmon J, Orth G. HLA control in the progression of human papillomavirus infections. Semin Cancer Biol. 1996;7:359–371. doi: 10.1006/scbi.1996.0045. [DOI] [PubMed] [Google Scholar]
- Thio CL, Gao X, Goedert JJ, Vlahov D, Nelson KE, Hilgartner MW, et al. HLA-Cw*04 and hepatitis C virus persistence. J Virol. 2002;76:4792–4797. doi: 10.1128/JVI.76.10.4792-4797.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thio CL, Thomas DL, Goedert JJ, Vlahov D, Nelson KE, Hilgartner MW, et al. Racial differences in HLA class II associations with hepatitis C virus outcomes. J Infect Dis. 2001;184:16–21. doi: 10.1086/321005. [DOI] [PubMed] [Google Scholar]
- Harcourt G, Hellier S, Bunce M, Satsangi J, Collier J, Chapman R, et al. Effect of HLA class II genotype on T helper lymphocyte responses and viral control in hepatitis C virus infection. J Viral Hepat. 2001;8:174–179. doi: 10.1046/j.1365-2893.2001.00289.x. [DOI] [PubMed] [Google Scholar]
- Lin M, Tseng HK, Trejaut JA, Lee HL, Loo JH, Chu CC, et al. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med Genet. 2003;4:9. doi: 10.1186/1471-2350-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YM, Liang SY, Shih YP, Chen CY, Lee YM, Chang L, et al. Epidemiological and genetic correlates of severe acute respiratory syndrome coronavirus infection in the hospital with the highest nosocomial infection rate in Taiwan in 2003. J Clin Microbiol. 2006;44:359–365. doi: 10.1128/JCM.44.2.359-365.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng MH, Lau KM, Li L, Cheng SH, Chan WY, Hui PK, et al. Association of human-leukocyte-antigen class I (B*0703) and class II (DRB1*0301) genotypes with susceptibility and resistance to the development of severe acute respiratory syndrome. J Infect Dis. 2004;190:515–518. doi: 10.1086/421523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SF, Chen KH, Chen M, Li WY, Chen YJ, Tsao CH, et al. Human-Leukocyte Antigen Class I Cw 1502 and Class II DR 0301 Genotypes Are Associated with Resistance to Severe Acute Respiratory Syndrome (SARS) Infection. Viral Immunol. 2011;24:421–426. doi: 10.1089/vim.2011.0024. [DOI] [PubMed] [Google Scholar]
- Robinson J, Malik A, Parham P, Bodmer JG, Marsh SG. IMGT/HLA database—a sequence database for the human major histocompatibility complex. Tissue Antigens. 2000;55:280–287. doi: 10.1034/j.1399-0039.2000.550314.x. [DOI] [PubMed] [Google Scholar]
- Carrington M, O'Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- Bihl F, Frahm N, Di Giammarino L, Sidney J, John M, Yusim K, et al. Impact of HLA-B alleles, epitope binding affinity, functional avidity, and viral coinfection on the immunodominance of virus-specific CTL responses. J Immunol. 2006;176:4094–4101. doi: 10.4049/jimmunol.176.7.4094. [DOI] [PubMed] [Google Scholar]
- Neumann-Haefelin C, McKiernan S, Ward S, Viazov S, Spangenberg HC, Killinger T, et al. Dominant influence of an HLA-B27 restricted CD8+ T cell response in mediating HCV clearance and evolution. Hepatology. 2006;43:563–572. doi: 10.1002/hep.21049. [DOI] [PubMed] [Google Scholar]
- Boon AC, De Mutsert G, Fouchier RA, Sintnicolaas K, Osterhaus AD, Rimmelzwaan GF. Preferential HLA usage in the influenza virus-specific CTL response. J Immunol. 2004;172:4435–4443. doi: 10.4049/jimmunol.172.7.4435. [DOI] [PubMed] [Google Scholar]
- Weiss SR, Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol Mol Biol Rev. 2005;69:635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo YO, Cheng Y, Wong R, Hui DS, Lee CK, Tsang KK, et al. Retrospective comparison of convalescent plasma with continuing high-dose methylprednisolone treatment in SARS patients. Clin Microbiol Infect. 2004;10:676–678. doi: 10.1111/j.1469-0691.2004.00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhang F, Yu W, He T, Yu J, Yi CE, et al. Antibody responses against SARS coronavirus are correlated with disease outcome of infected individuals. J Med Virol. 2006;78:1–8. doi: 10.1002/jmv.20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traggiai E, Becker S, Subbarao K, Kolesnikova L, Uematsu Y, Gismondo MR, et al. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat Med. 2004;10:871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J, Li W, Murakami A, Tamin A, Matthews LJ, Wong SK, et al. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc Natl Acad Sci USA. 2004;101:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang W, Zhong Q, Hou W, Yang Z, Xiao SY, et al. Immunogenicity, safety, and protective efficacy of an inactivated SARS-associated coronavirus vaccine in rhesus monkeys. Vaccine. 2005;23:3202–3209. doi: 10.1016/j.vaccine.2004.11.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruth M, Kistner O, Savidis-Dacho H, Hitter E, Crowe B, Gerencer M, et al. A double-inactivated whole virus candidate SARS coronavirus vaccine stimulates neutralising and protective antibody responses. Vaccine. 2006;24:652–661. doi: 10.1016/j.vaccine.2005.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YV, Massare MJ, Barnard DL, Kort T, Nathan M, Wang L, et al. Chimeric severe acute respiratory syndrome coronavirus (SARS-CoV) S glycoprotein and influenza matrix 1 efficiently form virus-like particles (VLPs) that protect mice against challenge with SARS-CoV. Vaccine. 2011;29:6606–6613. doi: 10.1016/j.vaccine.2011.06.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Huang Y, Huang L, Li B, Zheng Z, Chen Z, et al. Effect of mucosal and systemic immunization with virus-like particles of severe acute respiratory syndrome coronavirus in mice. Immunology. 2010;130:254–261. doi: 10.1111/j.1365-2567.2010.03231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- See RH, Zakhartchouk AN, Petric M, Lawrence DJ, Mok CP, Hogan RJ, et al. Comparative evaluation of two severe acute respiratory syndrome (SARS) vaccine candidates in mice challenged with SARS coronavirus. J Gen Virol. 2006;87:641–650. doi: 10.1099/vir.0.81579-0. [DOI] [PubMed] [Google Scholar]
- Bisht H, Roberts A, Vogel L, Subbarao K, Moss B. Neutralizing antibody and protective immunity to SARS coronavirus infection of mice induced by a soluble recombinant polypeptide containing an N-terminal segment of the spike glycoprotein. Virology. 2005;334:160–165. doi: 10.1016/j.virol.2005.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Lau YF, Lamirande EW, Paddock CD, Bartlett JH, Zaki SR, et al. Cellular immune responses to severe acute respiratory syndrome coronavirus (SARS-CoV) infection in senescent BALB/c mice: CD4+ T cells are important in control of SARS-CoV infection. J Virol. 2010;84:1289–1301. doi: 10.1128/JVI.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Perlman S. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J Virol. 2010;84:9318–9325. doi: 10.1128/JVI.01049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Van Rooijen N, Perlman S. Evasion by stealth: inefficient immune activation underlies poor T cell response and severe disease in SARS-CoV-infected mice. PLoS Pathog. 2009;5:e1000636. doi: 10.1371/journal.ppat.1000636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Quan Y, Xin ZT, Wrammert J, Ma MJ, Lv H, et al. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J Immunol. 2011;186:7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- Khanolkar A, Hartwig SM, Haag BA, Meyerholz DK, Epping LL, Haring JS, et al. Protective and pathologic roles of the immune response to mouse hepatitis virus type 1: implications for severe acute respiratory syndrome. J Virol. 2009;83:9258–9272. doi: 10.1128/JVI.00355-09. [DOI] [PMC free article] [PubMed] [Google Scholar]


