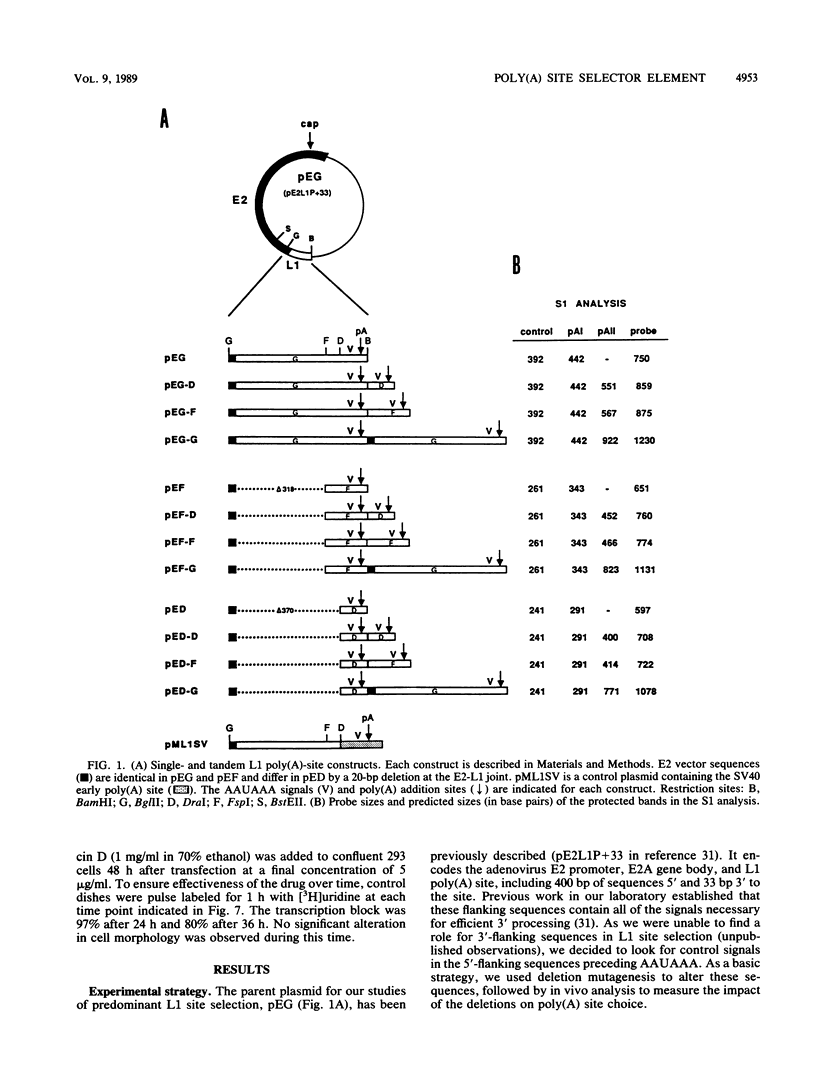
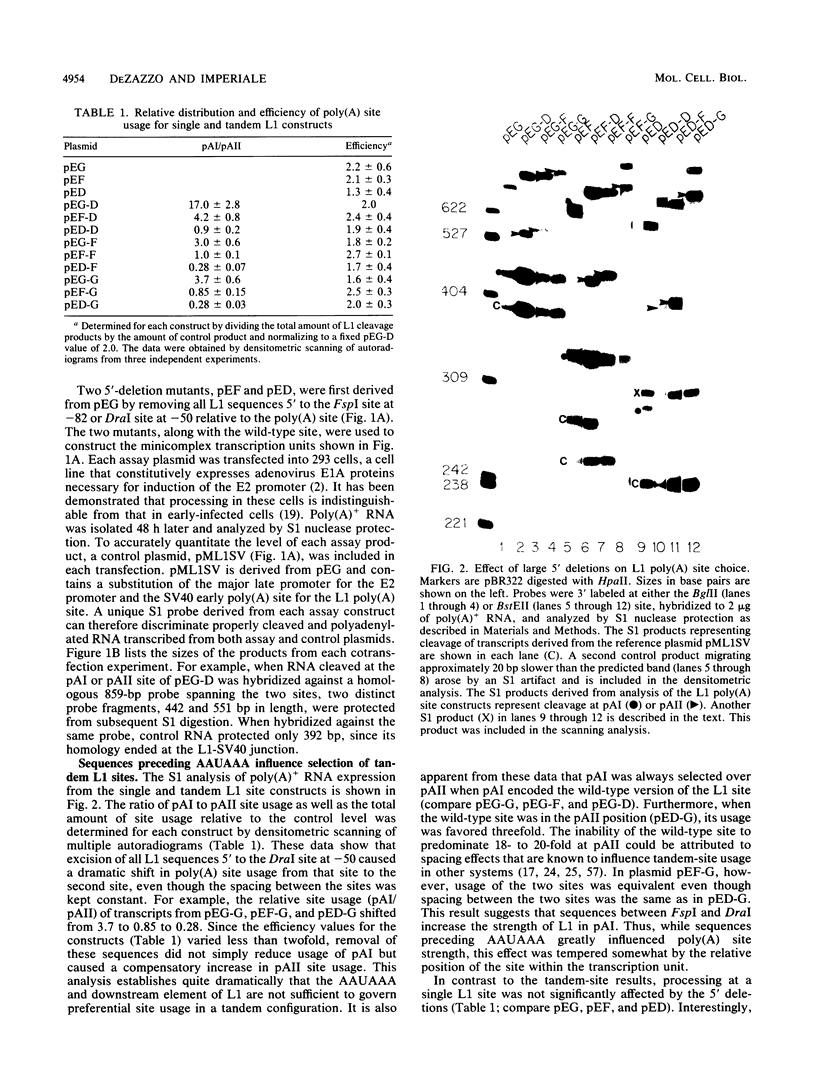
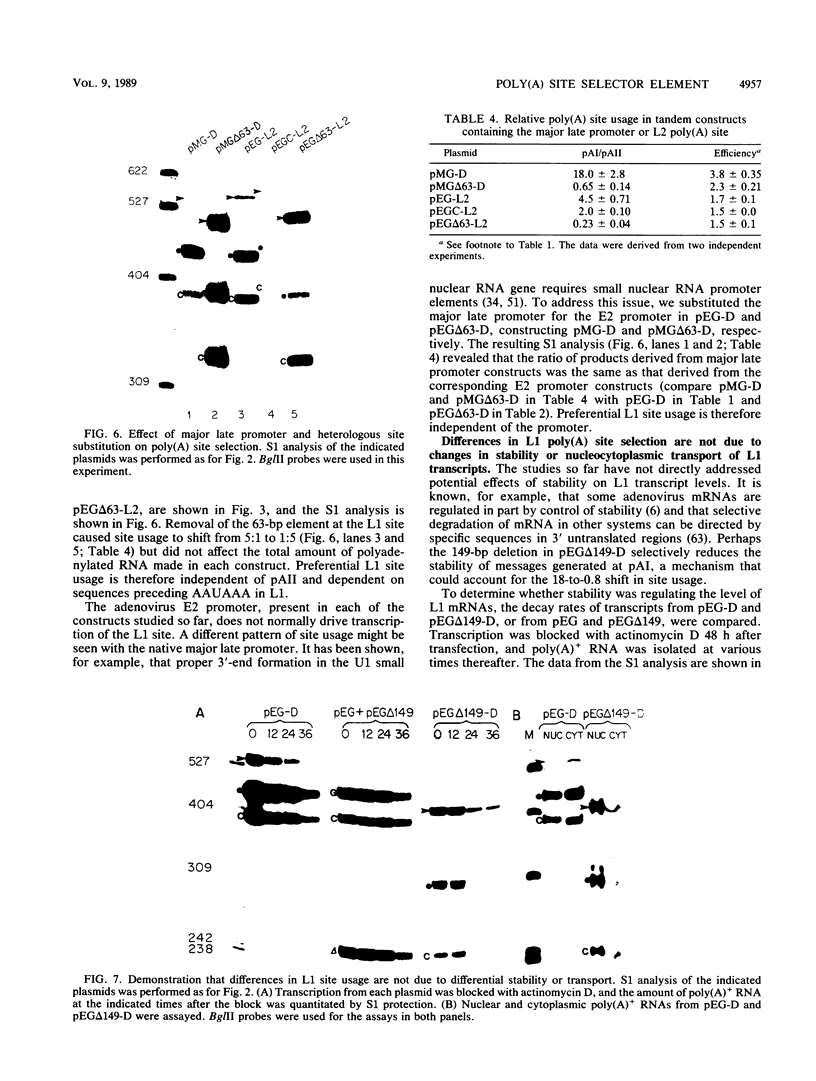
Abstract
The adenovirus major late transcription unit (MLTU) encodes five colinear mRNA families, L1 through L5, each distinguished by a unique poly(A) site. Site selection is regulated during the course of infection, predominating early at the L1 site and late at the L2 through L5 sites. Two general mechanisms can be invoked to explain predominant usage of the L1 site early in infection. MLTU site selection may proceed in a first-come, first-serve manner whereby the L1 site is used most frequently because it is closest to the promoter. Alternatively, specific sequences flanking the L1 site may control predominant L1 site usage in a position-independent manner. To distinguish between these mechanisms, we constructed deletions in the L1 flanking sequences and inserted the mutated sites into either simple transcription units or mini-MLTUs encoding two poly(A) sites. The pattern of site selection for each construct was then quantitated by S1 nuclease analysis after transfection into 293 cells. The results indicated that L1 sequences upstream of AAUAAA define a novel selector element that can cause predominant L1 site usage at either position of a tandem transcription unit. The element did not significantly affect the stability or nucleocytoplasmic transport of L1 transcripts and was not required for efficient 3'-end processing in simple transcription units. Predominant L1 site usage required physical linkage of the processing signals and was independent of the major late promoter.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adami G., Nevins J. R. Splice site selection dominates over poly(A) site choice in RNA production from complex adenovirus transcription units. EMBO J. 1988 Jul;7(7):2107–2116. doi: 10.1002/j.1460-2075.1988.tb03050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiello L., Guilfoyle R., Huebner K., Weinmann R. Adenovirus 5 DNA sequences present and RNA sequences transcribed in transformed human embryo kidney cells (HEK-Ad-5 or 293). Virology. 1979 Apr 30;94(2):460–469. doi: 10.1016/0042-6822(79)90476-8. [DOI] [PubMed] [Google Scholar]
- Akusjärvi G., Persson H. Controls of RNA splicing and termination in the major late adenovirus transcription unit. Nature. 1981 Jul 30;292(5822):420–426. doi: 10.1038/292420a0. [DOI] [PubMed] [Google Scholar]
- Alt F. W., Bothwell A. L., Knapp M., Siden E., Mather E., Koshland M., Baltimore D. Synthesis of secreted and membrane-bound immunoglobulin mu heavy chains is directed by mRNAs that differ at their 3' ends. Cell. 1980 Jun;20(2):293–301. doi: 10.1016/0092-8674(80)90615-7. [DOI] [PubMed] [Google Scholar]
- Amara S. G., Jonas V., Rosenfeld M. G., Ong E. S., Evans R. M. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature. 1982 Jul 15;298(5871):240–244. doi: 10.1038/298240a0. [DOI] [PubMed] [Google Scholar]
- Babich A., Nevins J. R. The stability of early adenovirus mRNA is controlled by the viral 72 kd DNA-binding protein. Cell. 1981 Nov;26(3 Pt 1):371–379. doi: 10.1016/0092-8674(81)90206-3. [DOI] [PubMed] [Google Scholar]
- Benne R., Van den Burg J., Brakenhoff J. P., Sloof P., Van Boom J. H., Tromp M. C. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986 Sep 12;46(6):819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- Breitbart R. E., Andreadis A., Nadal-Ginard B. Alternative splicing: a ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu Rev Biochem. 1987;56:467–495. doi: 10.1146/annurev.bi.56.070187.002343. [DOI] [PubMed] [Google Scholar]
- Chen S. H., Habib G., Yang C. Y., Gu Z. W., Lee B. R., Weng S. A., Silberman S. R., Cai S. J., Deslypere J. P., Rosseneu M. Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific in-frame stop codon. Science. 1987 Oct 16;238(4825):363–366. doi: 10.1126/science.3659919. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Christofori G., Keller W. 3' cleavage and polyadenylation of mRNA precursors in vitro requires a poly(A) polymerase, a cleavage factor, and a snRNP. Cell. 1988 Sep 9;54(6):875–889. doi: 10.1016/s0092-8674(88)91263-9. [DOI] [PubMed] [Google Scholar]
- Christofori G., Keller W. Poly(A) polymerase purified from HeLa cell nuclear extract is required for both cleavage and polyadenylation of pre-mRNA in vitro. Mol Cell Biol. 1989 Jan;9(1):193–203. doi: 10.1128/mcb.9.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M. F., Gelmann E. P., Reitz M. S., Jr Homology of human T-cell leukaemia virus envelope gene with class I HLA gene. Nature. 1983 Sep 1;305(5929):60–62. doi: 10.1038/305060a0. [DOI] [PubMed] [Google Scholar]
- Conway L., Wickens M. A sequence downstream of A-A-U-A-A-A is required for formation of simian virus 40 late mRNA 3' termini in frog oocytes. Proc Natl Acad Sci U S A. 1985 Jun;82(12):3949–3953. doi: 10.1073/pnas.82.12.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway L., Wickens M. Analysis of mRNA 3' end formation by modification interference: the only modifications which prevent processing lie in AAUAAA and the poly(A) site. EMBO J. 1987 Dec 20;6(13):4177–4184. doi: 10.1002/j.1460-2075.1987.tb02764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossland L. D., Raskas H. J. Identification of adenovirus genes that require template replication for expression. J Virol. 1983 Jun;46(3):737–748. doi: 10.1128/jvi.46.3.737-748.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denome R. M., Cole C. N. Patterns of polyadenylation site selection in gene constructs containing multiple polyadenylation signals. Mol Cell Biol. 1988 Nov;8(11):4829–4839. doi: 10.1128/mcb.8.11.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early P., Rogers J., Davis M., Calame K., Bond M., Wall R., Hood L. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell. 1980 Jun;20(2):313–319. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- Falck-Pedersen E., Logan J. Regulation of poly(A) site selection in adenovirus. J Virol. 1989 Feb;63(2):532–541. doi: 10.1128/jvi.63.2.532-541.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feagin J. E., Jasmer D. P., Stuart K. Developmentally regulated addition of nucleotides within apocytochrome b transcripts in Trypanosoma brucei. Cell. 1987 May 8;49(3):337–345. doi: 10.1016/0092-8674(87)90286-8. [DOI] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Fraser N. W., Sehgal P. B., Darnell J. E., Jr Multiple discrete sites for premature RNA chain termination late in adenovirus-2 infection: enhancement by 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2571–2575. doi: 10.1073/pnas.76.6.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D. I., Imperiale M. J., Adhya S. L. RNA 3' end formation in the control of gene expression. Annu Rev Genet. 1987;21:453–488. doi: 10.1146/annurev.ge.21.120187.002321. [DOI] [PubMed] [Google Scholar]
- Galli G., Guise J. W., McDevitt M. A., Tucker P. W., Nevins J. R. Relative position and strengths of poly(A) sites as well as transcription termination are critical to membrane versus secreted mu-chain expression during B-cell development. Genes Dev. 1987 Jul;1(5):471–481. doi: 10.1101/gad.1.5.471. [DOI] [PubMed] [Google Scholar]
- Galli G., Guise J., Tucker P. W., Nevins J. R. Poly(A) site choice rather than splice site choice governs the regulated production of IgM heavy-chain RNAs. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2439–2443. doi: 10.1073/pnas.85.8.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor R. B., Berk A. J. Cis-acting induction of adenovirus transcription. Cell. 1983 Jul;33(3):683–693. doi: 10.1016/0092-8674(83)90011-9. [DOI] [PubMed] [Google Scholar]
- Gil A., Proudfoot N. J. A sequence downstream of AAUAAA is required for rabbit beta-globin mRNA 3'-end formation. 1984 Nov 29-Dec 5Nature. 312(5993):473–474. doi: 10.1038/312473a0. [DOI] [PubMed] [Google Scholar]
- Gilmartin G. M., McDevitt M. A., Nevins J. R. Multiple factors are required for specific RNA cleavage at a poly(A) addition site. Genes Dev. 1988 May;2(5):578–587. doi: 10.1101/gad.2.5.578. [DOI] [PubMed] [Google Scholar]
- Graham F. L., Smiley J., Russell W. C., Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977 Jul;36(1):59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- Green T. L., Hart R. P. Mutations in poly(A) site downstream elements affect in vitro cleavage activity. Mol Cell Biol. 1988 Apr;8(4):1839–1841. doi: 10.1128/mcb.8.4.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales K. H., Birk J. M., Imperiale M. J. Analysis of adenovirus type 2 L1 RNA 3'-end formation in vivo and in vitro. J Virol. 1988 Apr;62(4):1464–1468. doi: 10.1128/jvi.62.4.1464-1468.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampson R. K., La Follette L., Rottman F. M. Alternative processing of bovine growth hormone mRNA is influenced by downstream exon sequences. Mol Cell Biol. 1989 Apr;9(4):1604–1610. doi: 10.1128/mcb.9.4.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart R. P., McDevitt M. A., Nevins J. R. Poly(A) site cleavage in a HeLa nuclear extract is dependent on downstream sequences. Cell. 1985 Dec;43(3 Pt 2):677–683. doi: 10.1016/0092-8674(85)90240-5. [DOI] [PubMed] [Google Scholar]
- Hernandez N., Weiner A. M. Formation of the 3' end of U1 snRNA requires compatible snRNA promoter elements. Cell. 1986 Oct 24;47(2):249–258. doi: 10.1016/0092-8674(86)90447-2. [DOI] [PubMed] [Google Scholar]
- Higgs D. R., Goodbourn S. E., Lamb J., Clegg J. B., Weatherall D. J., Proudfoot N. J. Alpha-thalassaemia caused by a polyadenylation signal mutation. Nature. 1983 Nov 24;306(5941):398–400. doi: 10.1038/306398a0. [DOI] [PubMed] [Google Scholar]
- Humphrey T., Christofori G., Lucijanic V., Keller W. Cleavage and polyadenylation of messenger RNA precursors in vitro occurs within large and specific 3' processing complexes. EMBO J. 1987 Dec 20;6(13):4159–4168. doi: 10.1002/j.1460-2075.1987.tb02762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperiale M. J., Feldman L. T., Nevins J. R. Activation of gene expression by adenovirus and herpesvirus regulatory genes acting in trans and by a cis-acting adenovirus enhancer element. Cell. 1983 Nov;35(1):127–136. doi: 10.1016/0092-8674(83)90215-5. [DOI] [PubMed] [Google Scholar]
- Imperiale M. J., Hart R. P., Nevins J. R. An enhancer-like element in the adenovirus E2 promoter contains sequences essential for uninduced and E1A-induced transcription. Proc Natl Acad Sci U S A. 1985 Jan;82(2):381–385. doi: 10.1073/pnas.82.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto S., Eggerding F., Falck-Pederson E., Darnell J. E., Jr Transcription unit mapping in adenovirus: regions of termination. J Virol. 1986 Jul;59(1):112–119. doi: 10.1128/jvi.59.1.112-119.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Moullec J. M., Akusjärvi G., Stålhandske P., Pettersson U., Chambraud B., Gilardi P., Nasri M., Perricaudet M. Polyadenylic acid addition sites in the adenovirus type 2 major late transcription unit. J Virol. 1983 Oct;48(1):127–134. doi: 10.1128/jvi.48.1.127-134.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leff S. E., Evans R. M., Rosenfeld M. G. Splice commitment dictates neuron-specific alternative RNA processing in calcitonin/CGRP gene expression. Cell. 1987 Feb 13;48(3):517–524. doi: 10.1016/0092-8674(87)90202-9. [DOI] [PubMed] [Google Scholar]
- Leff S. E., Rosenfeld M. G., Evans R. M. Complex transcriptional units: diversity in gene expression by alternative RNA processing. Annu Rev Biochem. 1986;55:1091–1117. doi: 10.1146/annurev.bi.55.070186.005303. [DOI] [PubMed] [Google Scholar]
- Malim M. H., Hauber J., Le S. Y., Maizel J. V., Cullen B. R. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989 Mar 16;338(6212):254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- Manley J. L., Sharp P. A., Gefter M. L. Rna synthesis in isolated nuclei processing of adenovirus serotype 2 late messenger rna precursors. J Mol Biol. 1982 Aug 25;159(4):581–599. doi: 10.1016/0022-2836(82)90102-4. [DOI] [PubMed] [Google Scholar]
- McDevitt M. A., Gilmartin G. M., Reeves W. H., Nevins J. R. Multiple factors are required for poly(A) addition to a mRNA 3' end. Genes Dev. 1988 May;2(5):588–597. doi: 10.1101/gad.2.5.588. [DOI] [PubMed] [Google Scholar]
- McDevitt M. A., Imperiale M. J., Ali H., Nevins J. R. Requirement of a downstream sequence for generation of a poly(A) addition site. Cell. 1984 Jul;37(3):993–999. doi: 10.1016/0092-8674(84)90433-1. [DOI] [PubMed] [Google Scholar]
- McGrogan M., Raskas H. J. Two regions of the adenovirus 2 genome specify families of late polysomal RNAs containing common sequences. Proc Natl Acad Sci U S A. 1978 Feb;75(2):625–629. doi: 10.1073/pnas.75.2.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLauchlan J., Gaffney D., Whitton J. L., Clements J. B. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3' termini. Nucleic Acids Res. 1985 Feb 25;13(4):1347–1368. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C., Fisher E. F., Caruthers M. H., Berk A. J. Control of adenovirus E1B mRNA synthesis by a shift in the activities of RNA splice sites. Mol Cell Biol. 1984 May;4(5):966–972. doi: 10.1128/mcb.4.5.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins J. R., Darnell J. E. Groups of adenovirus type 2 mRNA's derived from a large primary transcript: probable nuclear origin and possible common 3' ends. J Virol. 1978 Mar;25(3):811–823. doi: 10.1128/jvi.25.3.811-823.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins J. R., Darnell J. E., Jr Steps in the processing of Ad2 mRNA: poly(A)+ nuclear sequences are conserved and poly(A) addition precedes splicing. Cell. 1978 Dec;15(4):1477–1493. doi: 10.1016/0092-8674(78)90071-5. [DOI] [PubMed] [Google Scholar]
- Nevins J. R., Wilson M. C. Regulation of adenovirus-2 gene expression at the level of transcriptional termination and RNA processing. Nature. 1981 Mar 12;290(5802):113–118. doi: 10.1038/290113a0. [DOI] [PubMed] [Google Scholar]
- Pelham H. R. A regulatory upstream promoter element in the Drosophila hsp 70 heat-shock gene. Cell. 1982 Sep;30(2):517–528. doi: 10.1016/0092-8674(82)90249-5. [DOI] [PubMed] [Google Scholar]
- Peterson M. L., Perry R. P. Regulated production of mu m and mu s mRNA requires linkage of the poly(A) addition sites and is dependent on the length of the mu s-mu m intron. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8883–8887. doi: 10.1073/pnas.83.23.8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson M. L., Perry R. P. The regulated production of mu m and mu s mRNA is dependent on the relative efficiencies of mu s poly(A) site usage and the c mu 4-to-M1 splice. Mol Cell Biol. 1989 Feb;9(2):726–738. doi: 10.1128/mcb.9.2.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell L. M., Wallis S. C., Pease R. J., Edwards Y. H., Knott T. J., Scott J. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987 Sep 11;50(6):831–840. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986 Aug 29;46(5):681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- Rogers J., Early P., Carter C., Calame K., Bond M., Hood L., Wall R. Two mRNAs with different 3' ends encode membrane-bound and secreted forms of immunoglobulin mu chain. Cell. 1980 Jun;20(2):303–312. doi: 10.1016/0092-8674(80)90616-9. [DOI] [PubMed] [Google Scholar]
- Sadofsky M., Connelly S., Manley J. L., Alwine J. C. Identification of a sequence element on the 3' side of AAUAAA which is necessary for simian virus 40 late mRNA 3'-end processing. Mol Cell Biol. 1985 Oct;5(10):2713–2719. doi: 10.1128/mcb.5.10.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw G., Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986 Aug 29;46(5):659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Shaw J. M., Feagin J. E., Stuart K., Simpson L. Editing of kinetoplastid mitochondrial mRNAs by uridine addition and deletion generates conserved amino acid sequences and AUG initiation codons. Cell. 1988 May 6;53(3):401–411. doi: 10.1016/0092-8674(88)90160-2. [DOI] [PubMed] [Google Scholar]
- Skolnik-David H., Moore C. L., Sharp P. A. Electrophoretic separation of polyadenylation-specific complexes. Genes Dev. 1987 Sep;1(7):672–682. doi: 10.1101/gad.1.7.672. [DOI] [PubMed] [Google Scholar]
- Somasekhar M. B., Mertz J. E. Exon mutations that affect the choice of splice sites used in processing the SV40 late transcripts. Nucleic Acids Res. 1985 Aug 12;13(15):5591–5609. doi: 10.1093/nar/13.15.5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano J. E., Adams D. E. Assembly of a polyadenylation-specific 25S ribonucleoprotein complex in vitro. Mol Cell Biol. 1988 May;8(5):2052–2062. doi: 10.1128/mcb.8.5.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streuli M., Saito H. Regulation of tissue-specific alternative splicing: exon-specific cis-elements govern the splicing of leukocyte common antigen pre-mRNA. EMBO J. 1989 Mar;8(3):787–796. doi: 10.1002/j.1460-2075.1989.tb03439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G. P., Mathews M. B. DNA replication and the early to late transition in adenovirus infection. Cell. 1980 Nov;22(2 Pt 2):523–533. doi: 10.1016/0092-8674(80)90362-1. [DOI] [PubMed] [Google Scholar]
- Thomas S. M., Lamb R. A., Paterson R. G. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell. 1988 Sep 9;54(6):891–902. doi: 10.1016/S0092-8674(88)91285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M., Stephenson P. Role of the conserved AAUAAA sequence: four AAUAAA point mutants prevent messenger RNA 3' end formation. Science. 1984 Nov 30;226(4678):1045–1051. doi: 10.1126/science.6208611. [DOI] [PubMed] [Google Scholar]
- Zarkower D., Stephenson P., Sheets M., Wickens M. The AAUAAA sequence is required both for cleavage and for polyadenylation of simian virus 40 pre-mRNA in vitro. Mol Cell Biol. 1986 Jul;6(7):2317–2323. doi: 10.1128/mcb.6.7.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkower D., Wickens M. A functionally redundant downstream sequence in SV40 late pre-mRNA is required for mRNA 3'-end formation and for assembly of a precleavage complex in vitro. J Biol Chem. 1988 Apr 25;263(12):5780–5788. [PubMed] [Google Scholar]
- Zarkower D., Wickens M. Specific pre-cleavage and post-cleavage complexes involved in the formation of SV40 late mRNA 3' termini in vitro. EMBO J. 1987 Dec 20;6(13):4185–4192. doi: 10.1002/j.1460-2075.1987.tb02765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Cole C. N. Identification of a complex associated with processing and polyadenylation in vitro of herpes simplex virus type 1 thymidine kinase precursor RNA. Mol Cell Biol. 1987 Sep;7(9):3277–3286. doi: 10.1128/mcb.7.9.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vegvar H. E., Lund E., Dahlberg J. E. 3' end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell. 1986 Oct 24;47(2):259–266. doi: 10.1016/0092-8674(86)90448-4. [DOI] [PubMed] [Google Scholar]