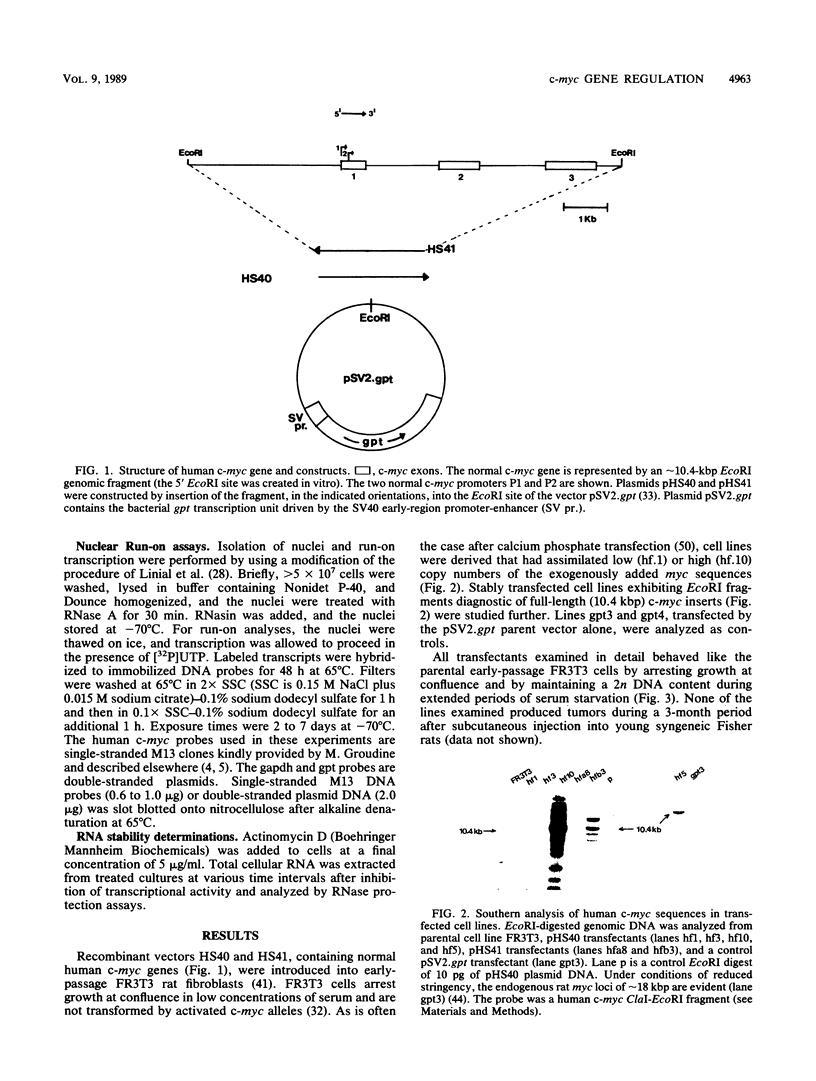
Abstract
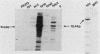
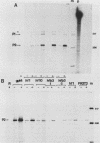
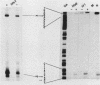
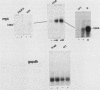
Activation of the c-myc proto-oncogene is implicated in the initiation or progression of many vertebrate cancers. In nontransformed cells, the expression of c-myc is induced by growth factors. Studies have indicated that such induction is effected by multiple mechanisms. To study regulation of c-myc expression, a transfection system has been developed in which introduced c-myc genes exhibit serum-responsive activity. The responsiveness assayed is not mediated by increased transcription initiation. Rather, it is effected at a point(s) between transcription and stabilization of the RNA.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armelin H. A., Armelin M. C., Kelly K., Stewart T., Leder P., Cochran B. H., Stiles C. D. Functional role for c-myc in mitogenic response to platelet-derived growth factor. Nature. 1984 Aug 23;310(5979):655–660. doi: 10.1038/310655a0. [DOI] [PubMed] [Google Scholar]
- Battey J., Moulding C., Taub R., Murphy W., Stewart T., Potter H., Lenoir G., Leder P. The human c-myc oncogene: structural consequences of translocation into the IgH locus in Burkitt lymphoma. Cell. 1983 Oct;34(3):779–787. doi: 10.1016/0092-8674(83)90534-2. [DOI] [PubMed] [Google Scholar]
- Bell L. R., Maine E. M., Schedl P., Cline T. W. Sex-lethal, a Drosophila sex determination switch gene, exhibits sex-specific RNA splicing and sequence similarity to RNA binding proteins. Cell. 1988 Dec 23;55(6):1037–1046. doi: 10.1016/0092-8674(88)90248-6. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986 Jun 12;321(6071):702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. Novel promoter upstream of the human c-myc gene and regulation of c-myc expression in B-cell lymphomas. Mol Cell Biol. 1986 Oct;6(10):3481–3489. doi: 10.1128/mcb.6.10.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. Sequence requirements for premature termination of transcription in the human c-myc gene. Cell. 1988 Apr 22;53(2):245–256. doi: 10.1016/0092-8674(88)90386-8. [DOI] [PubMed] [Google Scholar]
- Blanchard J. M., Piechaczyk M., Dani C., Chambard J. C., Franchi A., Pouyssegur J., Jeanteur P. c-myc gene is transcribed at high rate in G0-arrested fibroblasts and is post-transcriptionally regulated in response to growth factors. Nature. 1985 Oct 3;317(6036):443–445. doi: 10.1038/317443a0. [DOI] [PubMed] [Google Scholar]
- Bond J. F., Farmer S. R. Regulation of tubulin and actin mRNA production in rat brain: expression of a new beta-tubulin mRNA with development. Mol Cell Biol. 1983 Aug;3(8):1333–1342. doi: 10.1128/mcb.3.8.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J., Gray H. E., Pardee A. B., Dean M., Sonenshein G. E. Cell-cycle control of c-myc but not c-ras expression is lost following chemical transformation. Cell. 1984 Feb;36(2):241–247. doi: 10.1016/0092-8674(84)90217-4. [DOI] [PubMed] [Google Scholar]
- Caudy M., Vässin H., Brand M., Tuma R., Jan L. Y., Jan Y. N. daughterless, a Drosophila gene essential for both neurogenesis and sex determination, has sequence similarities to myc and the achaete-scute complex. Cell. 1988 Dec 23;55(6):1061–1067. doi: 10.1016/0092-8674(88)90250-4. [DOI] [PubMed] [Google Scholar]
- Cole M. D. The myc oncogene: its role in transformation and differentiation. Annu Rev Genet. 1986;20:361–384. doi: 10.1146/annurev.ge.20.120186.002045. [DOI] [PubMed] [Google Scholar]
- Coppola J. A., Cole M. D. Constitutive c-myc oncogene expression blocks mouse erythroleukaemia cell differentiation but not commitment. Nature. 1986 Apr 24;320(6064):760–763. doi: 10.1038/320760a0. [DOI] [PubMed] [Google Scholar]
- Cory S. Activation of cellular oncogenes in hemopoietic cells by chromosome translocation. Adv Cancer Res. 1986;47:189–234. doi: 10.1016/s0065-230x(08)60200-6. [DOI] [PubMed] [Google Scholar]
- Dani C., Blanchard J. M., Piechaczyk M., El Sabouty S., Marty L., Jeanteur P. Extreme instability of myc mRNA in normal and transformed human cells. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7046–7050. doi: 10.1073/pnas.81.22.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M., Levine R. A., Ran W., Kindy M. S., Sonenshein G. E., Campisi J. Regulation of c-myc transcription and mRNA abundance by serum growth factors and cell contact. J Biol Chem. 1986 Jul 15;261(20):9161–9166. [PubMed] [Google Scholar]
- Early P., Rogers J., Davis M., Calame K., Bond M., Wall R., Hood L. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell. 1980 Jun;20(2):313–319. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- Fort P., Marty L., Piechaczyk M., el Sabrouty S., Dani C., Jeanteur P., Blanchard J. M. Various rat adult tissues express only one major mRNA species from the glyceraldehyde-3-phosphate-dehydrogenase multigenic family. Nucleic Acids Res. 1985 Mar 11;13(5):1431–1442. doi: 10.1093/nar/13.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Greenberg M. E., Ziff E. B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984 Oct 4;311(5985):433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- Hay N., Takimoto M., Bishop J. M. A FOS protein is present in a complex that binds a negative regulator of MYC. Genes Dev. 1989 Mar;3(3):293–303. doi: 10.1101/gad.3.3.293. [DOI] [PubMed] [Google Scholar]
- Hayday A. C., Gillies S. D., Saito H., Wood C., Wiman K., Hayward W. S., Tonegawa S. Activation of a translocated human c-myc gene by an enhancer in the immunoglobulin heavy-chain locus. 1984 Jan 26-Feb 1Nature. 307(5949):334–340. doi: 10.1038/307334a0. [DOI] [PubMed] [Google Scholar]
- Hodgkin J. Drosophila sex determination: a cascade of regulated splicing. Cell. 1989 Mar 24;56(6):905–906. doi: 10.1016/0092-8674(89)90619-3. [DOI] [PubMed] [Google Scholar]
- Imagawa M., Chiu R., Karin M. Transcription factor AP-2 mediates induction by two different signal-transduction pathways: protein kinase C and cAMP. Cell. 1987 Oct 23;51(2):251–260. doi: 10.1016/0092-8674(87)90152-8. [DOI] [PubMed] [Google Scholar]
- Kelly K., Cochran B. H., Stiles C. D., Leder P. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell. 1983 Dec;35(3 Pt 2):603–610. doi: 10.1016/0092-8674(83)90092-2. [DOI] [PubMed] [Google Scholar]
- Kelly K., Siebenlist U. The regulation and expression of c-myc in normal and malignant cells. Annu Rev Immunol. 1986;4:317–338. doi: 10.1146/annurev.iy.04.040186.001533. [DOI] [PubMed] [Google Scholar]
- Kindy M. S., Sonenshein G. E. Regulation of oncogene expression in cultured aortic smooth muscle cells. Post-transcriptional control of c-myc mRNA. J Biol Chem. 1986 Sep 25;261(27):12865–12868. [PubMed] [Google Scholar]
- Levine R. A., McCormack J. E., Buckler A., Sonenshein G. E. Transcriptional and posttranscriptional control of c-myc gene expression in WEHI 231 cells. Mol Cell Biol. 1986 Nov;6(11):4112–4116. doi: 10.1128/mcb.6.11.4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linial M., Gunderson N., Groudine M. Enhanced transcription of c-myc in bursal lymphoma cells requires continuous protein synthesis. Science. 1985 Dec 6;230(4730):1126–1132. doi: 10.1126/science.2999973. [DOI] [PubMed] [Google Scholar]
- Martinez R., Mathey-Prevot B., Bernards A., Baltimore D. Neuronal pp60c-src contains a six-amino acid insertion relative to its non-neuronal counterpart. Science. 1987 Jul 24;237(4813):411–415. doi: 10.1126/science.2440106. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mougneau E., Cerni C., Tillier F., Cuzin F. Tumorigenic transformation of rat FR3T3 fibroblasts carrying an activated myc oncogene requires subsequent mutational events. Oncogene Res. 1988;2(2):177–188. [PubMed] [Google Scholar]
- Mulligan R. C., Berg P. Expression of a bacterial gene in mammalian cells. Science. 1980 Sep 19;209(4463):1422–1427. doi: 10.1126/science.6251549. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Nepveu A., Marcu K. B. Intragenic pausing and anti-sense transcription within the murine c-myc locus. EMBO J. 1986 Nov;5(11):2859–2865. doi: 10.1002/j.1460-2075.1986.tb04580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepveu A., Marcu K. B., Skoultchi A. I., Lachman H. M. Contributions of transcriptional and post-transcriptional mechanisms to the regulation of c-myc expression in mouse erythroleukemia cells. Genes Dev. 1987 Nov;1(9):938–945. doi: 10.1101/gad.1.9.938. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Cole M. D. Posttranscriptional regulation of cellular gene expression by the c-myc oncogene. Mol Cell Biol. 1989 Jan;9(1):124–134. doi: 10.1128/mcb.9.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Saito H., Hayday A. C., Wiman K., Hayward W. S., Tonegawa S. Activation of the c-myc gene by translocation: a model for translational control. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7476–7480. doi: 10.1073/pnas.80.24.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seif R., Cuzin F. Temperature-sensitive growth regulation in one type of transformed rat cells induced by the tsa mutant of polyoma virus. J Virol. 1977 Dec;24(3):721–728. doi: 10.1128/jvi.24.3.721-728.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Bressler P., Kelly K. Two distinct mechanisms of transcriptional control operate on c-myc during differentiation of HL60 cells. Mol Cell Biol. 1988 Feb;8(2):867–874. doi: 10.1128/mcb.8.2.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Steffen D. Proviruses are adjacent to c-myc in some murine leukemia virus-induced lymphomas. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2097–2101. doi: 10.1073/pnas.81.7.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott S. J., Davis R. L., Thayer M. J., Cheng P. F., Weintraub H., Lassar A. B. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science. 1988 Oct 21;242(4877):405–411. doi: 10.1126/science.3175662. [DOI] [PubMed] [Google Scholar]
- Treisman R. Identification and purification of a polypeptide that binds to the c-fos serum response element. EMBO J. 1987 Sep;6(9):2711–2717. doi: 10.1002/j.1460-2075.1987.tb02564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R. Identification of a protein-binding site that mediates transcriptional response of the c-fos gene to serum factors. Cell. 1986 Aug 15;46(4):567–574. doi: 10.1016/0092-8674(86)90882-2. [DOI] [PubMed] [Google Scholar]
- Treisman R. Transient accumulation of c-fos RNA following serum stimulation requires a conserved 5' element and c-fos 3' sequences. Cell. 1985 Oct;42(3):889–902. doi: 10.1016/0092-8674(85)90285-5. [DOI] [PubMed] [Google Scholar]
- Villares R., Cabrera C. V. The achaete-scute gene complex of D. melanogaster: conserved domains in a subset of genes required for neurogenesis and their homology to myc. Cell. 1987 Jul 31;50(3):415–424. doi: 10.1016/0092-8674(87)90495-8. [DOI] [PubMed] [Google Scholar]
- Wigler M., Pellicer A., Silverstein S., Axel R., Urlaub G., Chasin L. DNA-mediated transfer of the adenine phosphoribosyltransferase locus into mammalian cells. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1373–1376. doi: 10.1073/pnas.76.3.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T., Treisman R. Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3' AU-rich sequences. Nature. 1988 Nov 24;336(6197):396–399. doi: 10.1038/336396a0. [DOI] [PubMed] [Google Scholar]
- Wiman K. G., Clarkson B., Hayday A. C., Saito H., Tonegawa S., Hayward W. S. Activation of a translocated c-myc gene: role of structural alterations in the upstream region. Proc Natl Acad Sci U S A. 1984 Nov;81(21):6798–6802. doi: 10.1073/pnas.81.21.6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerberg J., Parsegian V. A. Polymer inaccessible volume changes during opening and closing of a voltage-dependent ionic channel. Nature. 1986 Sep 4;323(6083):36–39. doi: 10.1038/323036a0. [DOI] [PubMed] [Google Scholar]
- Zinn K., DiMaio D., Maniatis T. Identification of two distinct regulatory regions adjacent to the human beta-interferon gene. Cell. 1983 Oct;34(3):865–879. doi: 10.1016/0092-8674(83)90544-5. [DOI] [PubMed] [Google Scholar]