Abstract
Prophylactic and therapeutic vaccines against viral infections have advanced in recent years from attenuated live vaccines to subunit-based vaccines. An ideal prophylactic vaccine should mimic the natural immunity induced by an infection, in that it should generate long-lasting adaptive immunity. To complement subunit vaccines, which primarily target an antibody response, different methodologies are being investigated to develop vaccines capable of driving cellular immunity. T-cell epitope discovery is central to this concept. In this review, the significance of T-cell epitope-based vaccines for prophylactic and therapeutic applications is discussed. Additionally, methodologies for the discovery of T-cell epitopes, as well as recent developments in the clinical testing of these vaccines for various viral infections, are explained.
Keywords: cellular immunity, cytotoxic T cells, epitopes, humoral immunity, immunoproteomics, mass spectrometry, MHC class I, vaccine
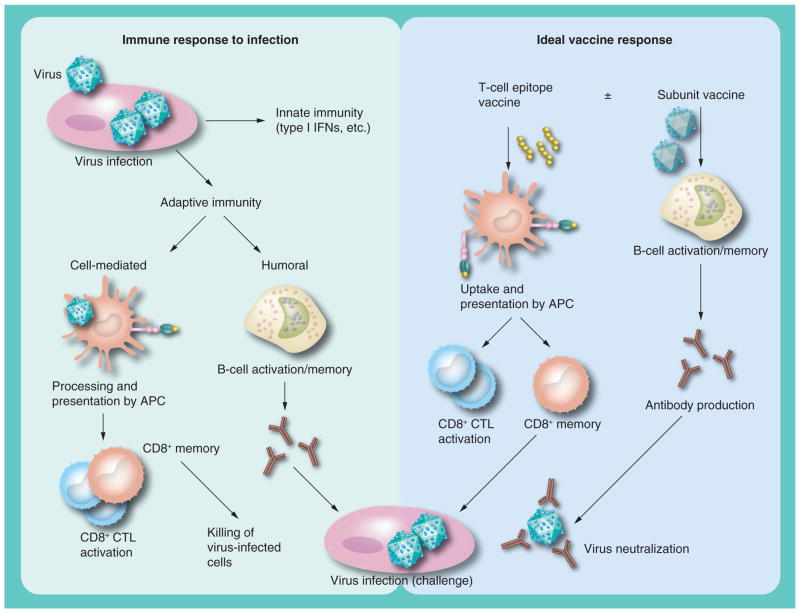
Vaccines play a major role in the control of viral diseases. Prophylactic vaccines are extremely successful and act by preventing diseases caused by several types of viruses. Traditionally, vaccines are developed from live attenuated or inactivated viruses that elicit a strong humoral and cellular immune response leading to long-lasting memory that protects from reinfection. However, recent vaccine development has been focused on driving the neutralizing antibody response by generating subunit vaccines that can be easily synthesized and safely administered. However, these subunit vaccines are poor at developing cellular immunity, as they activate components that do not generate broad T-cell responses. An ideal vaccine should attempt to mimic the natural immunity generated by infection in a manner whereby adaptive humoral and cellular immune memory is generated (Figure 1).
Figure 1. Generation of adaptive immunity in infection and vaccine response.
CTL: Cytotoxic T lymphocyte; IFN: Interferon.
Viruses like influenza and dengue, which differ substantially in their envelope glycoprotein structures between strains, can escape the conventional neutralizing antibody response. To overcome this limitation, one would require a cellular immunity-inducing vaccine that is primarily generated from the internal viral proteins conserved across the genotypes and serotypes of many viruses. T-cell epitope-based vaccines may prove useful in this regard and, indeed, epitopes associated with the MHC class I molecule represent the minimal immunogenic region of a protein antigen and allow for precise direction of the cellular immune response [1]. Considering prophylactic vaccine application further, T-cell epitope-based vaccines may also have a high impact in the therapeutic vaccine field to combat chronic viral infections such as HPV, HCV, HBV and HIV.
This review discusses the importance of the cellular immune response in the control and clearance of viral infections, as well as the advantages and limitations of a T-cell epitope-based vaccine. In addition, methods available for T-cell epitope discovery and an update on the developmental status of several epitope-based vaccines against viral pathogens are discussed.
Importance of T-cell responses in infectious diseases
Current envelope glycoprotein-based subunit vaccines are potent in establishing a strong humoral immune response that neutralizes viral particles [2]. However, many are poor at driving long-lasting cellular immunity due to the lack of internal viral proteins that generate T-cell epitopes, which are essential in eliminating virus-infected cells. For instance, cell-mediated immunity, as elicited by MHC class I-restricted CD8+ cytotoxic T lymphocytes (CTLs), plays a central role in controlling influenza virus infection [3–7]. Indeed, the cell-mediated response generated by primary influenza infection provides substantial protection against serologically distinct viruses due to the recognition of cross-reactive epitopes, often from internal viral proteins conserved between viral subtypes [8–10].
The same theory can also be applied to dengue virus (DENV). While antibodies to DENV infection can be neutralizing and protective against a particular serotype, exposure to another strain increases the risk of dengue hemorrhagic fever (DHF) and dengue shock syndrome [11–14]. It is suggested that this is due to antibody-dependent enhancement (ADE), which is mediated by the Fc receptor of activated macrophages [15]. Gwinn et al. found that patients vaccinated with DENV vaccines generated Th1-based cell-mediated immunity as measured by IFN-γ release in vaccine-stimulated peripheral blood mononuclear cells [16]. Indirect evidence suggests that Th1 cells will contribute to protection by upregulating CD8+ CTLs to destroy virus-infected cells early in infection, therefore limiting disease-associated symptoms [17,18]. It has been suggested that a CTL epitope vaccine that complements the multivalent antibody-producing vaccine is the best option for DENV immunity, as it will elicit a multiple serotype-specific antibody response in addition to cross-reactive protective T-cell responses.
Regarding chronic viral infections, such as HBV, it has been demonstrated that patients who recover fully display strong CD8+ T-cell responses against HBV nucleocapsid, polymerase and envelope proteins. Chronic HBV-infected patients, however, display poor T-cell responses in spite of high viral antigen load [19]. Specifically, chronic hepatitis B patients typically lack effective HBV-specific cellular immunity as defined by functional alteration (tolerance/anergy) or the absence of HBV-specific T cells [20,21]. Unlike acute infection, Th1 cytokine production is usually lower in chronic infection [22]. In addition, CTL responses of equal intensity were observed in chronic patients who resolve infection spontaneously compared with those found in acute patients who have recovered fully or are responding to IFN-γ treatment [23,24].
Immunopathogenesis of antiviral T-cell responses
Antiviral T-cell responses, although beneficial and necessary for eradicating infected cells in the body, have been shown to be detrimental to the host in several instances. Immunopathogenesis due to CD8+ and/or CD4+ T-cell activation occurs in many viral infections such as HBV [25] and dengue [26]. Indeed, cytokines secreted by activated T cells drive the disease phenotype for the acute phase of viral hepatitis [25]. With regard to DENV, T cells have been implicated in the immunopathological disease associated with secondary infection with a different serotype. It is therefore possible that DHF and dengue shock syndrome are not solely mediated by ADE as described above. Upon reinfection with a new serotype, pre-existing heterotypic immunity that is serotype cross-reactive can drive a partial memory response known as ‘original antigenic sin’ [26]. That is, the alteration in the immune response, skewed by the low avidity ‘memory’ of the previous infection, can lead to delayed viral clearance and inadequate killing of virus-infected cells [27]. It has been noted that severe secondary infection is associated with increased inflammatory cytokines and increased viral burden [28]. In what seems to be a downward spiral, DHF could be mediated by increased antigen that drives an upregulation of inflammatory cytokines, such as TNF-α or IL-2, which have been demonstrated to increase vascular permeability [29]. In a similar fashion to DENV, influenza virus clearance is also dependent on the CD4+ and CD8+ T-cell response. However, these very mechanisms can lead to lung immunopathology and severe disease [30]. As mentioned above, inflammatory cytokines in excess can be damaging to the surrounding tissue [31]. In addition, other studies have suggested a role for CD4+ costimulation in influenza-associated lung injury [32]. Therefore, it must understood that T cells, while necessary for efficient viral clearance, can be destructive if improperly activated.
Vaccines should mimic natural immunity to infection
The rationale for prophylactic vaccination against any viral infection begins with the knowledge that natural infection protects against exogenous reinfection. Natural infection induces both the innate and adaptive immune responses (Figure 1). The innate immune response is nonspecific and essential to immediately control the spread of the virus; however, the adaptive immune response, which develops within 5–7 days, is characterized by an antibody response to the viral envelope protein and a T-cell response to viral epitopes presented by the MHC molecules on infected cells. If both of these immune response pathways (innate and adaptive) work perfectly, the virus is cleared and the immune system develops a memory response that can be reactivated if the host encounters the virus again. This immune memory, if efficiently developed, could protect the host for a lifetime. An ideal vaccine to any virus must induce this long-lasting immune memory in the form of antibody production to neutralize free virus, as well as the CTL response needed to kill the virus-infected cells.
Vaccines should mimic the antiviral adaptive immune response
Acute viral infection activates adaptive immunity and triggers viral antigen-specific antibody and CTL responses to eliminate circulating virus and virus-infected cells. This process generates a memory population of T and B cells that can be reactivated upon exposure to new infection. Similarly, an ideal vaccine should include all the components of the viral antigens to stimulate both humoral and cell-mediated immune memory capable of protecting the host upon primary and secondary infection.
Current successful vaccines possess the qualities of driving a strong and memory-inducing humoral response. It is important to note, however, that a CD4+ helper response is generally required to develop long-lasting B-cell immunity [25]. Therefore, current antiviral prophylactic vaccines are successful because they can generate a CD4+ response. For viruses that can subvert the immune response, a standard antibody-inducing attenuated vaccine is not sufficient. For example, DENV typically elicits a weak neutralizing antibody response, which upon secondary infection can lead to ADE [33]. For viruses such as influenza, the antibody response is quite protective; however, the virus mutates very quickly by utilizing antigenic drift and antigenic shift, thus allowing secondary infections to evade any existing immunity [34,35]. For chronic viral infections, the option of restoring the functionality of virus-specific T cells is plausible for a therapeutic vaccine. This may, however, be irrelevant for the restoration of B-cell function and the production of neutralizing antibody. Specifically, most chronic viruses are latent within infected cells, leaving cytotoxic killing as the only option for the eradication of virus-infected cells.
Ultimately, the most promising vaccine candidates may come from combining the antigenic subunits for both T- and B-cell immunity. Based on animal models and human studies, combining all possible T- and B-cell ligands to formulate an active synthetic vaccine makes sense. This concept would also be especially promising for a therapeutic vaccine. These types of vaccines would be necessary for driving T-cell immunity to eliminate infected cells left in the body after acute infection. Chronic viral infections, such as HIV, HCV and HBV, are in desperate need of a vaccine that would clear lingering virus-infected cells. The success of the current antibody-inducing HBV prophylactic vaccine has meant that only the need for a therapeutic vaccine for chronic disease remains. A number of anti-HBV therapeutic vaccine approaches are being evaluated in the clinic, including recombinant anti-HBV vaccines, lipopeptide-based vaccines [36] or DNA vaccines, with or without immunomodulatory drugs (reviewed in [37]). Although immunotherapy based on these different vaccine formulations has demonstrated efficient T-cell responses in healthy subjects (prophylaxis), these approaches stimulated only transient clinical responses in patients chronically infected with HBV. Although the diversity of infected patient populations may contribute to the variability of efficacy observed in these vaccine studies, the major unresolved issue involves the composition of the vaccine, including relevant antigens presented by infected cells in vivo and adjuvants that will bias immune responses towards a Th1 profile capable of breaking tolerance in chronic patients. In addition, vaccine delivery systems that can protect the vaccine from degradation, as well as target and facilitate the uptake of both antigens and adjuvant by APCs [38], have to be considered.
Components of an epitope-based T-cell vaccine
T-cell vaccines have much potential for the development of therapeutic vaccines for chronic viral infection or for universal prophylactic vaccines. Ideally, epitope-based vaccines, formulated with an adjuvant to drive a strong immune response with high immunogenicity, will offer a flexible and simple way to synthesize a vaccine. MHC class II epitopes, which elicit a CD4+ helper response, will be necessary in order to get protective CD8+ T-cell memory. To this end, universal MHC class II epitopes that span the entire allelic range of the global population can be used. Indeed, there are many studies that have demonstrated broad CD4+ activity in response to several pan-MHC class II epitopes [39–41].
MHC class I epitopes, however, will have to be antigen-specific to the virus being targeted. Analysis of the memory CTL repertoire at the clonal level in human peripheral blood mononuclear cells revealed a very broad CTL response to epitopes from a wide variety of proteins [42]. This study is significant for the development of synthetic vaccines because it suggests that more clinically relevant epitopes remain to be identified and characterized, and that a variety of novel candidate targets may be found among them. The technical challenge is to identify the few epitopes among a mixture of >10,000 MHC class I-associated epitopes extracted from virus-infected cells that originated from pathogen-associated proteins. Successful identification of the epitopes generated during infection will enable the identification of conserved T-cell epitopes by cross-searching the genomic databases of related viruses, which can be used to develop vaccines for families of viruses.
Genomic and proteomic databases have been generated for many different organisms, and while they contain a vast amount of sequence data, relatively little information is available about the immunological functions of the majority of these genes and proteins. The MHC class I-associated epitopes displayed on the cell surface can be thought of as a ‘finger-print’ because those peptides reflect a ‘readout’ on the physiological status of the cell [43,44]. Identification of the peptides displayed by cells infected with viral pathogens can generate a library of epitopes and, by genomic database searching, the viral proteins of origin that are recognized and processed by the immune system can be identified. These viral epitopes and proteins are associated with adaptive immunity and are therefore putative targets for cross-protective vaccine development.
T-cell epitope identification methods
T-cell epitope-based vaccines are promising for many vaccine applications, including cross-serotype prophylactic vaccines as well as therapeutic vaccines. Therefore, several methods exist for the sole purpose of identifying T-cell epitopes. Indeed, many academic investigators and for-profit companies have implemented the methods discussed below for the production of epitope-based vaccines, despite the risk associated with the development of a cellular vaccine. Currently, there is no example of a T-cell peptide-based vaccine that is US FDA-approved. However, several are in late-stage clinical trials, and display very promising results [45]. The remainder of this review focuses on the methods used for epitope discovery as well as the epitope-based vaccines currently in clinical trials.
Overlapping peptide library
A commonly used method for the identification of T-cell epitopes involves the use of an overlapping peptide library derived from a protein of interest, and has been used successfully for a variety of pathogen-associated epitopes [46,47]. Typically, such pools contain peptides that are between nine and 20 amino acids in length, and overlap to a degree that ensures that every epitope can be presented. An advantage of this method is that it allows for the discovery of both MHC class I and II epitopes in the context of several alleles. Rather than testing each peptide individually, peptide libraries are generally set up in a matrix so as to reduce the amount of testing necessary [48,49]. Furthermore, software exists that can optimize the peptide pools used so that libraries of up to 1200 peptides can be used for epitope determination [50]. Additionally, a methodology called AnTigen Lead Acquisition System (ATLAS™) uses overlapping peptide libraries in conjunction with CTL screening from patients with various allelic backgrounds [51–53]. Patients are then divided and analyzed based on whether or not their immunity is protective. While the overlapping peptide library method may be able to identify epitopes in an active T-cell response, it is still be unable to uncover the exact epitopes presented on the cell surface of infected cells. This could be potentially problematic, as protective epitopes may not necessarily be shaped by what active CTLs are present, especially in chronic disease.
Motif prediction by algorithm
Additionally, T-cell epitope identification has taken the motif prediction or epitope-mapping algorithm approach [201]. These algorithms screen protein sequences for nine to ten amino acid-long peptide segments predicted to bind to one or more HLA alleles. HLA alleles can also be selected to maximize cross-HLA coverage [54–56]. It has been demonstrated that epitope-based vaccines containing epitopes restricted by selected HLA ‘supertypes’ can provide the broadest possible coverage of the human population [57]. Thus, computationally identified cross-reactive epitopes are a good starting point, even though further extensive immunological studies must be performed to qualify these epitopes as vaccine candidates.
Although a vast number of predicted and immunologically characterized epitopes are reported in the literature (immune epitope database analysis resource [IEDB-AR] [202]), successful vaccines based on these predicted epitopes have not yet been developed. Clinical evidence shows that predictive analysis may not reliably generate antigens that are naturally processed and presented by an infected cell. Although the motif-based predicted epitopes may activate CTLs in in vitro assays, not all activated CTLs recognize the naturally processed antigenic peptides on infected cells due to differences between the motif-predicted epitopes and the cellular endogenously presented epitopes (antigenic mismatch). Indeed, subtle structural differences can have profound effects on CTL recognition in vivo. Furthermore, the data generated by in silico approaches may overestimate the number of conserved epitopes and may not necessarily identify all immunoreactive naturally presented epitopes.
As demonstrated in influenza, the motif-prediction approach may be limited in identifying subdominant epitopes that are reported to activate T cells in secondary influenza virus-specific responses [58]. Furthermore, motif-predicted epitopes are validated by screening circulating CTLs from virus-infected individuals. Thomas et al. have recently demonstrated that there is a CD8+ T-cell immunodominance hierarchy, which may be dependent on the concentration of the presented epitope, the size of the available CD8+ T-cell repertoire, activation after the initial priming and the competition and cooperation between different epitope-specific responses [58]. Additionally, Zhong et al. compared the motif-prediction method with direct mass spectrometry analysis of endogenously presented epitopes isolated from virus-infected cells using a large number of naturally processed and in vivo-presented viral CTL epitopes in a mouse model system [59]. The study revealed that a high number of predicted epitopes were not processed and presented by infected cells and that the complexity of the CD8+ T-cell-based screening of functional epitopes by this approach may miss hidden subdominant epitopes. Therefore, an advantage of working ‘in reverse’ and using an empirical approach to identify naturally presented HLA-associated T-cell epitopes – the end products of MHC class I processing – is that the peptides that stimulate CTLs are verified as being presented by infected cells and are available for recognition in vivo by the CTLs activated by the vaccine.
Immunoproteomic approach
In the last decade, direct identification of MHC class I-presented epitopes from infected cells has emerged as an alternative to the motif-prediction method and is termed ‘immunoproteomics’ [60–63]. One of the challenges of this methodology is the identification of the few infection-related peptides among a majority of nondisease peptides derived from cellular proteins that are presented on the infected cell surface [64]. Several strategies have been developed to analyze MHC class I-associated peptides on APCs. These approaches are generally based on the isolation of the MHC-associated peptides, followed by mass spectrometry analysis [62,65–69]. A stable isotope tagging method has also been developed to distinguish viral epitopes from the epitopes derived from cellular proteins [67,70]. Although immunogenic MHC class I-associated viral epitopes are a less-abundant species present in a highly complex mixture of directly extracted peptides, stable isotope tagging of epitopes is a powerful strategy for the rapid, unambiguous identification of naturally processed and presented epitopes, which are induced by viral infection and are of strategic importance for vaccine development. Recently, several cross-serotype conserved DENV-specific MHC class I-associated T-cell epitopes from DENV-infected cells [71] and cross-strain conserved influenza epitopes have been reported by our laboratory using an immunoproteomics approach, underlining the significance and feasibility of T-cell epitopes for vaccine development [72].
Adjuvant & delivery systems
A successful T-cell vaccine must be delivered in a system that is capable of inducing a robust peptide-specific T-cell response against virus-infected cells. These delivery systems must be in the appropriate size range to mimic viral particles to allow uptake, as well as include built-in danger signals (adjuvant) to target APCs. Promising delivery systems for T-cell epitopes include gold nanoparticles [73,74], liposomes [75–77] and virus-like particles [78,79], all of which have been shown to induce efficient vaccine responses [80]. There are advantages and disadvantages to each system. For instance, peptides and molecular adjuvants, such as Toll-like receptor agonists, can be easily incorporated in nanoparticles or liposomes by chemical means. On the other hand, virus-like particles would require the cloning of a target antigen and the use of mammalian cell culture systems to synthesize them, but their virus-like nature would offer excellent adjuvancy [80]. It is also feasible to incorporate the above delivery systems into current vaccines, which would drive T-cell immunity in the presence of an active and proven humoral response.
Peptide vaccines: successes in infectious diseases
While the T-cell vaccine field is in its infancy, there are a few ongoing early-stage clinical studies that are relevant. In general, peptide vaccines have many advantages, as they are considered to be safe, easy to produce and stable [81]. Regardless of how epitopes are identified, several peptide vaccines are in development due to their early successes in animal studies [1]. It is feasible that within the next 5–10 years, several therapeutic peptide vaccines against chronic viral infections will be on the market. Examples of peptide vaccines in development include those designed towards HSV-2, CMV, HCV, HPV and HIV.
A therapeutic peptide vaccine against HCV (IC41, Intercell) is currently in clinical trials and includes four highly conserved HLA-A2 CTL epitopes in addition to three MHC class II helper epitopes [82]. Upon analysis of the CTL response in chronically infected patients, it would seem it is multivalent; however, the response is weak compared with an acute infection [83]. Present treatment for chronic HCV infection consists of a combination of pegylated interferon and ribavirin, which is marginally successful depending on tolerability and costs [84]. Fortunately, IC41 is progressing through Phase II clinical trials and has a promising outlook with regard to immunogenicity and sustaining reduced viral loads in chronic HCV-infected patients [84,85]. This is the first significant antiviral effect of a peptide vaccine in HCV-infected patients.
Another therapeutic peptide-based vaccine that is being studied is against HPV, the leading cause of cervical cancer. It is focused on T-cell determinants derived from the two oncoproteins E6 and E7, expression of which is required for the transformation of cervical cells [86,87]. In a recent murine study, Wu and colleagues demonstrated that MHC class I peptides, in addition to pan-MHC class II helper peptides and TLR-3 agonists, were very effective [87]. Indeed, intratumoral injection with this formulation demonstrated better survival and more efficient CTL responses. Several clinical trials are underway that involve CTL epitopes with helper peptides [88]. Interestingly, a peptide vaccine that went into Phase II clinical trials was not made up of known epitopes, but of overlapping long peptides that covered the E6 and E7 proteins [89]. Inoculation with this formulation, comprising 13 peptides and Montanide™-51 (Seppic) adjuvant, demonstrated strong CD4+ and CD8+ T-cell responses in addition to memory development. Furthermore, the vaccine displayed low toxicity and high immunogenicity [90], and demonstrated promising efficacy.
Peptide vaccine formulations targeting HIV proteins are also under development. Peptides derived from the major core protein Gag p24 were combined with GM-CSF as an adjuvant into a vaccine termed Vacc-4x (Bionor Pharma, Norway) [91]. Phase II clinical trials demonstrated significant immunogenicity to the peptide formulation in addition to decreased viral burden in the high responders [92,93]. Fortunately, viral escape mutants, as measured by amino acid alterations in postvaccinated patients, remained low [94]. Furthermore, Vacc-4x has also been shown to elicit an efficient memory response, as vaccine-recipients displayed a strong delayed-type hypersensitivity reaction 4 years after immunization [95]. However, another CTL multiepitope vaccine underwent a Phase I clinical trial and displayed weak immunogenicity, with only six of 80 volunteers generating specific responses as measured by IFN-γ ELISPOT assay [96]. Unfortunately, peptide plus adjuvant alone can be poorly immunogenic, although peptides delivered through a more natural mechanism may have a better outcome [97]. For instance, a vaccine is under development that incorporates peptides derived from the envelope glycoprotein gp41 into a virosome [1]. Taken together, a peptide vaccine for HIV is plausible, but immunogenicity can only be driven by a few major determinants, including the peptides themselves, adjuvant and delivery systems.
Recently, a T-cell vaccine based on peptide-mapping technology (ATLAS) designed against HSV-2 (GEN-003, Genocea Biosciences, MA, USA) has entered into a Phase I clinical study. The GEN-003 formulation includes viral antigens ICP4 and gD2, as well as a novel saponin-derived adjuvant that has demonstrated a strong immunostimulatory profile. Thus far, preclinical proof-of-concept studies are promising, as they illustrate protection against viral diseases, in addition to reduced duration and severity of clinical symptoms [98].
Limitations of T-cell epitope-based vaccines
A limitation of a vaccine primarily dependent on T-cell epitopes, without any complementary subunit components, is the immune evasion by the viruses [99]. Many viruses have immune evasion strategies ranging from alterations in the amino acid sequences of the epitopes themselves, to those of the flanking residues that may hinder recognition or epitope processing and presentation. Mutations present in the viral genome represent another mechanism by which the virus can abolish the epitope-binding properties to the MHC molecule or cause a failure to generate the epitope altogether. In addition to mutation, many viruses interfere with the antiviral immune response by expressing specific molecules that inhibit or hinder antigen processing and presentation of antigenic epitopes. This specific immune impairment is a controversial one, because antiviral responses are oligoclonal and several experimental studies indicate that not all the epitope-specific responses are inhibited by virus-induced immune evasion mechanisms [100]. The key for a successful T-cell vaccine is to generate an oligoclonal antiviral response that can overcome virus strategies for immune evasion.
Conclusion
It is clear that the vaccines of the future will require more than simply inactivating a pathogen strain. Fortunately, technology has progressed enough to allow us to identify immunodominant and memory-inducing peptides that are presented by the MHC class I molecules of virus-infected cells. Armed with these peptides, vaccine formulations will now have to incorporate antigens that activate both humoral and cellular immunity with various adjuvants to drive a strong immune response with high immunogenicity. Additionally, the use of peptides offers a flexible and simple way to synthesize a vaccine. It is, therefore, highly likely that peptide vaccines will play a large part in overall vaccination strategies and should offer hope for universal prophylactics as well as therapeutic vaccines for protection against infection and therapy for chronic infections, respectively.
Future perspective
T-cell vaccines could play a major role in viral infections such as influenza and DENV, where the antibody-targeted vaccines have limited clinical efficacy due to significant variations in the envelope protein between various strains. Significant efforts are being directed to find conserved regions of envelope proteins to generate broad humoral immunity to influenza virus, since the current conventional influenza vaccines are generally not cross-protective [101]. While there has been some success with this approach, especially with regards to conserved antibody epitopes in hemagglutinin [102–108], a two-pronged humoral and cellular approach is most likely going to be the best option. Additionally, some efforts are aimed at targeting conserved proteins within the virus, such as influenza virus nucleoprotein. Since antibodies cannot reach these proteins to prevent infection, peptides derived from intracellular processed protein presented in the context of MHC class I molecules must be utilized. The concept behind this approach is to stimulate T cells to quickly kill virus-infected cells before the cells can produce new virions, thus limiting disease severity. A vaccine based on nucleoprotein from human H1N1 virus was shown to protect animals from a lethal dose of the H5N1 bird influenza [109], and generated increased T-cell responses in healthy adults in a clinical study [110].
Vaccines that have built-in cross-subtype efficacy could prevent significant spread of an emerging or re-emerging strain. A cross-subtype vaccine containing immunogenic consensus sequence epitopes could achieve this goal. Mounting evidence suggests that an efficient universal influenza vaccine must induce activation of both cell-mediated immunity and humoral immunity. Along those lines, vaccines based on defined epitopes presented by infected cells would be far superior to viral protein subunit or motif-predicted epitope-based vaccines because protein processing by the immune system may alter native viral epitopes [72].
Another cross-serotype vaccine that is needed is one against DENV. Early vaccine studies demonstrated T-cell responses to the virus, but they were largely serotype-specific [16]. This may suggest that the level of presentation of MHC class I and II antigens differs among serotypes [111]. Beneficial effects of the vaccine-induced Th1 response further underscore the significance of the T-cell response in vaccine development [16,17]. Another important step may be the addition of a peptide vaccine to other formulations currently in clinical trials that can elicit a strong humoral response, but poor T-cell responses [112]. Recent reports on conserved T-cell epitopes identified from DENV-infected cells by direct immunoproteomic analysis of DENV-infected cells [72] further highlight the feasibility of the T-cell vaccine concept.
Executive summary.
Importance of T-cell responses in infectious diseases
It is well established that cell-mediated immune response plays a critical role in combating viral infections.
T-cell responses eliminate virus-infected cells, while humoral responses neutralize the virus in vivo.
Immunopathogenesis of antiviral T-cell responses
Improper activation of T cells during infection has been shown to induce pathological effects in the host.
Care is to be taken to consider the balance between protection and pathogenesis when developing T-cell vaccines.
Vaccines should mimic natural immunity to infection
Prophylactic vaccines should mimic natural immunity to infection in generating both humoral and cell-mediated adaptive immune memory.
A universal vaccine for viral infections such as dengue and influenza will be achievable when antibody-inducing vaccines are combined with T-cell vaccines.
The cell-mediated immune response plays a major role in therapeutic vaccine applications by eliminating chronically infected cells from the body.
Cross-strain conserved T-cell epitopes are essential for therapeutic vaccines against chronic infections such as HBV, HCV and HIV.
Components of an epitope-based T-cell vaccine
Cell-mediated immune responses generated against conserved antigenic epitopes are crucial in viral infections such as dengue and influenza, where variations in the viral envelope protein are common.
The identities of the T-cell epitopes presented by the MHC class I molecules on the infected cells are essential for the development of a T-cell vaccine.
T-cell epitope-based vaccines require efficient adjuvants and epitope-delivery systems that target APCs.
T-cell epitope identification methods
Methods for the identification of T-cell epitopes include overlapping peptide analysis, motif-prediction algorithms and direct immunoproteomic analysis.
Direct identification of T-cell epitopes from infected cells using immunoproteomics has the most advantages compared with various other methods used in the field.
Peptide vaccines: successes in infectious diseases
Major advances have been made in the generation of T-cell responses to peptide immunization for prophylactic and therapeutic vaccine applications.
Several therapeutic vaccines for infectious diseases, including HBV, HCV, HIV and HPV infections, are being clinically tested.
Preclinical and early-stage clinical studies are being conducted for T-cell epitope-based vaccines for infections including dengue, influenza and HSV.
Limitations of T-cell epitope-based vaccine
Viruses have developed strategies to evade T-cell immune response development.
Several mechanisms, including impeding antigen processing and epitope presentation and expression of immune response inhibitory molecules, have been reported.
Induction of multiepitope-specific T-cell responses are essential to overcome virus-induced immune evasion.
Acknowledgments
The authors wish to thank J Hafner for her critical editing of the article.
Footnotes
Financial & competing interests disclosure
This work is supported by NIH SBIR grants 1R43AI062177 and R43AI091232 from the National Institute of Allergy and Infectious Diseases. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Purcell AW, Mccluskey J, Rossjohn J. More than one reason to rethink the use of peptides in vaccine design. Nat Rev Drug Discov. 2007;6(5):404–414. doi: 10.1038/nrd2224. [DOI] [PubMed] [Google Scholar]
- 2.Zakay-Rones Z. Human influenza vaccines and assessment of immunogenicity. Expert Rev Vaccines. 2010;9(12):1423–1439. doi: 10.1586/erv.10.144. [DOI] [PubMed] [Google Scholar]
- 3.Doherty PC, Allan W, Eichelberger M, Carding SR. Roles of alpha beta and gamma delta T cell subsets in viral immunity. Ann Rev Immunol. 1992;10:123–151. doi: 10.1146/annurev.iy.10.040192.001011. [DOI] [PubMed] [Google Scholar]
- 4.Eichelberger M, Allan W, Zijlstra M, Jaenisch R, Doherty PC. Clearance of influenza virus respiratory infection in mice lacking class I major histocompatibility complex-restricted CD8+ T cells. J Exp Med. 1991;174(4):875–880. doi: 10.1084/jem.174.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Epstein SL, Lo CY, Misplon JA, Bennink JR. Mechanism of protective immunity against influenza virus infection in mice without antibodies. J Immunol. 1998;160(1):322–327. [PubMed] [Google Scholar]
- 6.Graham MB, Braciale TJ. Resistance to and recovery from lethal influenza virus infection in B lymphocyte-deficient mice. J Exp Med. 1997;186(12):2063–2068. doi: 10.1084/jem.186.12.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Subbarao K, Murphy BR, Fauci AS. Development of effective vaccines against pandemic influenza. Immunity. 2006;24(1):5–9. doi: 10.1016/j.immuni.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 8.Rimmelzwaan GF, Osterhaus AD. Cytotoxic T lymphocyte memory: role in cross-protective immunity against influenza? Vaccine. 1995;13(8):703–705. doi: 10.1016/0264-410x(94)00030-q. [DOI] [PubMed] [Google Scholar]
- 9.Yewdell JW, Bennink JR, Smith GL, Moss B. Influenza A virus nucleoprotein is a major target antigen for cross-reactive anti-influenza A virus cytotoxic T lymphocytes. Proc Natl Acad Sci USA. 1985;82(6):1785–1789. doi: 10.1073/pnas.82.6.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan PT, Khan AM, August JT. Highly conserved influenza A sequences as T cell epitopes-based vaccine targets to address the viral variability. Hum Vaccin. 2011;7(4):402–409. doi: 10.4161/hv.7.4.13845. [DOI] [PubMed] [Google Scholar]
- 11.Endy TP, Nisalak A, Chunsuttitwat S, et al. Relationship of preexisting dengue virus (DV) neutralizing antibody levels to viremia and severity of disease in a prospective cohort study of DV infection in Thailand. J Infect Dis. 2004;189(6):990–1000. doi: 10.1086/382280. [DOI] [PubMed] [Google Scholar]
- 12.Halstead SB. Antibody, macrophages, dengue virus infection, shock, and hemorrhage: a pathogenetic cascade. Rev Infect Dis. 1989;11(Suppl 4):S830–S839. doi: 10.1093/clinids/11.supplement_4.s830. [DOI] [PubMed] [Google Scholar]
- 13.Malasit P. Complement and dengue haemorrhagic fever/shock syndrome. Southeast Asian J Trop Med Public Health. 1987;18(3):316–320. [PubMed] [Google Scholar]
- 14.Falconar AK. The dengue virus nonstructural-1 protein (NS1) generates antibodies to common epitopes on human blood clotting, integrin/adhesin proteins and binds to human endothelial cells: potential implications in haemorrhagic fever pathogenesis. Arch Virol. 1997;142(5):897–916. doi: 10.1007/s007050050127. [DOI] [PubMed] [Google Scholar]
- 15.Wilder-Smith A, Ooi EE, Vasudevan SG, Gubler DJ. Update on dengue: epidemiology, virus evolution, antiviral drugs, and vaccine development. Curr Infect Dis Rep. 2010;12(3):157–164. doi: 10.1007/s11908-010-0102-7. [DOI] [PubMed] [Google Scholar]
- 16.Gwinn W, Sun W, Innis BL, Caudill J, King AD. Serotype-specific Th1 responses in recipients of two doses of candidate live-attenuated dengue virus vaccines. Am J Trop Med Hyg. 2003;69(6 Suppl):39–47. doi: 10.4269/ajtmh.2003.69.39. [DOI] [PubMed] [Google Scholar]
- 17.Innis BL, Eckels KH, Kraiselburd E, et al. Virulence of a live dengue virus vaccine candidate: a possible new marker of dengue virus attenuation. J Infect Dis. 1988;158(4):876–880. doi: 10.1093/infdis/158.4.876. [DOI] [PubMed] [Google Scholar]
- 18.Webster DP, Farrar J, Rowland-Jones S. Progress towards a dengue vaccine. Lancet Infect Dis. 2009;9(11):678–687. doi: 10.1016/S1473-3099(09)70254-3. [DOI] [PubMed] [Google Scholar]
- 19.Webster GJ, Reignat S, Brown D, et al. Longitudinal analysis of CD8+ T cells specific for structural and nonstructural hepatitis B virus proteins in patients with chronic hepatitis B: implications for immunotherapy. J Virol. 2004;78(11):5707–5719. doi: 10.1128/JVI.78.11.5707-5719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertoletti A, Gehring AJ. The immune response during hepatitis B virus infection. J Gen Virol. 2006;87(Pt 6):1439–1449. doi: 10.1099/vir.0.81920-0. [DOI] [PubMed] [Google Scholar]
- 21.Chisari FV, Isogawa M, Wieland SF. Pathogenesis of hepatitis B virus infection. Pathol Biol (Paris) 2010;58(4):258–266. doi: 10.1016/j.patbio.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung MC, Hartmann B, Gerlach JT, et al. Virus-specific lymphokine production differs quantitatively but not qualitatively in acute and chronic hepatitis B infection. Virology. 1999;261(2):165–172. doi: 10.1006/viro.1999.9833. [DOI] [PubMed] [Google Scholar]
- 23.Rehermann B, Lau D, Hoofnagle JH, Chisari FV. Cytotoxic T lymphocyte responsiveness after resolution of chronic hepatitis B virus infection. J Clin Invest. 1996;97(7):1655–1665. doi: 10.1172/JCI118592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Han Y, Jin K, et al. Dynamic changes of cytotoxic T lymphocytes (CTLs), natural killer (NK) cells, and natural killer T (NKT) cells in patients with acute hepatitis B infection. Virol J. 2011;8:199. doi: 10.1186/1743-422X-8-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zajac AJ, Harrington LE. Immune response to viruses: cell-mediated immunity. In: Mahy BWJ, van Regenmortel MHV, editors. Encyclopedia of Virology. Vol. 3. Elsevier; AL, USA: 2008. pp. 70–77. [Google Scholar]
- 26.Rothman AL. Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nat Rev Immunol. 2011;11(8):532–543. doi: 10.1038/nri3014. [DOI] [PubMed] [Google Scholar]
- 27.Mongkolsapaya J, Dejnirattisai W, Xu XN, et al. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat Med. 2003;9(7):921–927. doi: 10.1038/nm887. [DOI] [PubMed] [Google Scholar]
- 28.Libraty DH, Young PR, Pickering D, et al. High circulating levels of the dengue virus nonstructural protein NS1 early in dengue illness correlate with the development of dengue hemorrhagic fever. J Infect Dis. 2002;186(8):1165–1168. doi: 10.1086/343813. [DOI] [PubMed] [Google Scholar]
- 29.Yang G, Hamacher J, Gorshkov B, et al. The dual role of TNF in pulmonary edema. J Cardiovasc Dis Res. 2010;1(1):29–36. doi: 10.4103/0975-3583.59983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Damjanovic D, Small CL, Jeyananthan M, McCormick S, Xing Z. Immunopathology in influenza virus infection: uncoupling the friend from foe. Clin Immunol. 2012;144(1):57–69. doi: 10.1016/j.clim.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura R, Maeda N, Shibata K, Yamada H, Kase T, Yoshikai Y. Interleukin-15 is critical in the pathogenesis of influenza a virus-induced acute lung injury. J Virol. 2010;84(11):5574–5582. doi: 10.1128/JVI.02030-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teijaro JR, Njau MN, Verhoeven D, et al. Costimulation modulation uncouples protection from immunopathology in memory T cell responses to influenza virus. J Immunol. 2009;182(11):6834–6843. doi: 10.4049/jimmunol.0803860. [DOI] [PubMed] [Google Scholar]
- 33.Wahala WM, Silva AM. The human antibody response to dengue virus infection. Viruses. 2011;3(12):2374–2395. doi: 10.3390/v3122374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hensley SE, Das SR, Bailey AL, et al. Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science. 2009;326(5953):734–736. doi: 10.1126/science.1178258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J, Deng YM. Influenza virus antigenic variation, host antibody production and new approach to control epidemics. Virol J. 2009;6:30. doi: 10.1186/1743-422X-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vitiello A, Ishioka G, Grey HM, et al. Development of a lipopeptide-based therapeutic vaccine to treat chronic HBV infection I Induction of a primary cytotoxic T lymphocyte response in humans. J Clin Invest. 1995;95(1):341–349. doi: 10.1172/JCI117662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pol S, Michel ML. Therapeutic vaccination in chronic hepatitis B virus carriers. Expert Rev Vaccines. 2006;5(5):707–716. doi: 10.1586/14760584.5.5.707. [DOI] [PubMed] [Google Scholar]
- 38.Donnelly JJ, Wahren B, Liu MA. DNA vaccines: progress and challenges. J Immunol. 2005;175(2):633–639. doi: 10.4049/jimmunol.175.2.633. [DOI] [PubMed] [Google Scholar]
- 39.Alexander J, Sidney J, Southwood S, et al. Development of high potency universal DR-restricted helper epitopes by modification of high affinity DR-blocking peptides. Immunity. 1994;1(9):751–761. doi: 10.1016/s1074-7613(94)80017-0. [DOI] [PubMed] [Google Scholar]
- 40.Panina-Bordignon P, Tan A, Termijtelen A, Demotz S, Corradin G, Lanzavecchia A. Universally immunogenic T cell epitopes: promiscuous binding to human MHC class II and promiscuous recognition by T cells. Eur J Immunol. 1989;19(12):2237–2242. doi: 10.1002/eji.1830191209. [DOI] [PubMed] [Google Scholar]
- 41.Wilson CC, Palmer B, Southwood S, et al. Identification and antigenicity of broadly cross-reactive and conserved human immunodeficiency virus type 1-derived helper T-lymphocyte epitopes. J Virol. 2001;75(9):4195–4207. doi: 10.1128/JVI.75.9.4195-4207.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jameson J, Cruz J, Ennis FA. Human cytotoxic T-lymphocyte repertoire to influenza A viruses. J Virol. 1998;72(11):8682–8689. doi: 10.1128/jvi.72.11.8682-8689.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shastri N, Schwab S, Serwold T. Producing nature’s gene-chips: the generation of peptides for display by MHC class I molecules. Ann Rev Immunol. 2002;20:463–493. doi: 10.1146/annurev.immunol.20.100301.064819. [DOI] [PubMed] [Google Scholar]
- 44.Janeway C, Travers P, Walport M, Schlomchik M. Immunobiology the Immune System in Health and Disease. 5. Garland Publishing; NY, USA: 2001. T-cell-mediated immunity; pp. 295–340. [Google Scholar]
- 45.Dudek NL, Perlmutter P, Aguilar MI, Croft NP, Purcell AW. Epitope discovery and their use in peptide based vaccines. Curr Pharm Des. 2010;16(28):3149–3157. doi: 10.2174/138161210793292447. [DOI] [PubMed] [Google Scholar]
- 46.Yu XG, Addo MM, Rosenberg ES, et al. Consistent patterns in the development and immunodominance of human immunodeficiency virus type 1 (HIV-1)-specific CD8+ T-cell responses following acute HIV-1 infection. J Virol. 2002;76(17):8690–8701. doi: 10.1128/JVI.76.17.8690-8701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osorio Y, Mott KR, Jabbar AM, et al. Epitope mapping of HSV-1 glycoprotein K (gK) reveals a T cell epitope located within the signal domain of gK. Virus Res. 2007;128(1–2):71–80. doi: 10.1016/j.virusres.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kern F, Surel IP, Faulhaber N, et al. Target structures of the CD8+-T-cell response to human cytomegalovirus the 72-kilodalton major immediate–early protein revisited. J Virol. 1999;73(10):8179–8184. doi: 10.1128/jvi.73.10.8179-8184.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Betts MR, Casazza JP, Patterson BA, et al. Putative immunodominant human immunodeficiency virus-specific CD8+ T-cell responses cannot be predicted by major histocompatibility complex class I haplotype. J Virol. 2000;74(19):9144–9151. doi: 10.1128/jvi.74.19.9144-9151.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roederer M, Koup RA. Optimized determination of T cell epitope responses. J Immunol Meth. 2003;274(1–2):221–228. doi: 10.1016/s0022-1759(02)00423-4. [DOI] [PubMed] [Google Scholar]
- 51.Bouwer HG, Alberti-Segui C, Montfort MJ, Berkowitz ND, Higgins DE. Directed antigen delivery as a vaccine strategy for an intracellular bacterial pathogen. Proc Natl Acad Sci USA. 2006;103(13):5102–5107. doi: 10.1073/pnas.0509381103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Picard MD, Cohane KP, Gierahn TM, Higgins DE, Flechtner JB. High-throughput proteomic screening identifies Chlamydia trachomatis antigens that are capable of eliciting T cell and antibody responses that provide protection against vaginal challenge. Vaccine. 2012;30(29):4387–4393. doi: 10.1016/j.vaccine.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 53.Roan NR, Gierahn TM, Higgins DE, Starnbach MN. Monitoring the T cell response to genital tract infection. Proc Natl Acad Sci USA. 2006;103(32):12069–12074. doi: 10.1073/pnas.0603866103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Groot AS, Berzofsky JA. From genome to vaccine – new immunoinformatics tools for vaccine design. Methods. 2004;34(4):425–428. doi: 10.1016/j.ymeth.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 55.De Groot AS, Rivera DS, Mcmurry JA, Buus S, Martin W. Identification of immunogenic HLA-B7 “Achilles’ heel” epitopes within highly conserved regions of HIV. Vaccine. 2008;26(24):3059–3071. doi: 10.1016/j.vaccine.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Erup Larsen M, Kloverpris H, Stryhn A, et al. HLArestrictor – a tool for patient-specific predictions of HLA restriction elements and optimal epitopes within peptides. Immunogenetics. 2011;63(1):43–55. doi: 10.1007/s00251-010-0493-5. [DOI] [PubMed] [Google Scholar]
- 57.Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thomas PG, Brown SA, Keating R, et al. Hidden epitopes emerge in secondary influenza virus-specific CD8+ T cell responses. J Immunol. 2007;178(5):3091–3098. doi: 10.4049/jimmunol.178.5.3091. [DOI] [PubMed] [Google Scholar]
- 59.Zhong W, Reche PA, Lai CC, Reinhold B, Reinherz EL. Genome-wide characterization of a viral cytotoxic T lymphocyte epitope repertoire. J Biol Chem. 2003;278(46):45135–45144. doi: 10.1074/jbc.M307417200. [DOI] [PubMed] [Google Scholar]
- 60.York IA, Rock KL. Antigen processing and presentation by the class I major histocompatibility complex. Ann Rev Immunol. 1996;14:369–396. doi: 10.1146/annurev.immunol.14.1.369. [DOI] [PubMed] [Google Scholar]
- 61.Wiesmuller KH, Fleckenstein B, Jung G. Peptide vaccines and peptide libraries. Biol Chem. 2001;382(4):571–579. doi: 10.1515/BC.2001.070. [DOI] [PubMed] [Google Scholar]
- 62.Berzofsky JA, Ahlers JD, Belyakov IM. Strategies for designing and optimizing new generation vaccines. Nat Rev Immunol. 2001;1(3):209–219. doi: 10.1038/35105075. [DOI] [PubMed] [Google Scholar]
- 63.Purcell AW, Gorman JJ. Immunoproteomics: mass spectrometry-based methods to study the targets of the immune response. Mol Cell Proteomics. 2004;3(3):193–208. doi: 10.1074/mcp.R300013-MCP200. [DOI] [PubMed] [Google Scholar]
- 64.De Jong A. Contribution of mass spectrometry to contemporary immunology. Mass Spectrom Rev. 1998;17(5):311–335. doi: 10.1002/(SICI)1098-2787(1998)17:5<311::AID-MAS1>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 65.Hunt DF, Henderson RA, Shabanowitz J, et al. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science. 1992;255(5049):1261–1263. doi: 10.1126/science.1546328. [DOI] [PubMed] [Google Scholar]
- 66.Di Marzo Veronese F, Arnott D, Barnaba V, et al. Autoreactive cytotoxic T lymphocytes in human immunodeficiency virus type 1-infected subjects. J Exp Med. 1996;183(6):2509–2516. doi: 10.1084/jem.183.6.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Els CA, Herberts CA, Van Der Heeft E, et al. A single naturally processed measles virus peptide fully dominates the HLA-A*0201-associated peptide display and is mutated at its anchor position in persistent viral strains. Eur J Immunol. 2000;30(4):1172–1181. doi: 10.1002/(SICI)1521-4141(200004)30:4<1172::AID-IMMU1172>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 68.Hickman HD, Luis AD, Bardet W, et al. Cutting edge: class I presentation of host peptides following HIV infection. J Immunol. 2003;171(1):22–26. doi: 10.4049/jimmunol.171.1.22. [DOI] [PubMed] [Google Scholar]
- 69.Lemmel C, Weik S, Eberle U, et al. Differential quantitative analysis of MHC ligands by mass spectrometry using stable isotope labeling. Nat Biotechnol. 2004;22(4):450–454. doi: 10.1038/nbt947. [DOI] [PubMed] [Google Scholar]
- 70.Meiring HD, Soethout EC, Poelen MC, et al. Stable isotope tagging of epitopes A highly selective strategy for the identification of major histocompatibility complex class I-associated peptides induced upon viral infection. Mol Cell Proteomics. 2006;5(5):902–913. doi: 10.1074/mcp.T500014-MCP200. [DOI] [PubMed] [Google Scholar]
- 71.Testa JS, Shetty V, Sinnathamby G, et al. Conserved MHC class I-presented dengue virus epitopes identified by immunoproteomics analysis are targets for cross-serotype reactive T-cell response. J Infect Dis. 2012;205(4):647–655. doi: 10.1093/infdis/jir814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Testa JS, Hafner J, Nickens Z, Kamal S, Sinnathamby G, Philip R. MHC class I-presented T cell epitopes identified by immunoproteomics analysis are targets for a cross reactive influenza-specific T cell response. PLoS One. 2012;205(4):647–655. doi: 10.1371/journal.pone.0048484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fifis T, Gamvrellis A, Crimeen-Irwin B, et al. Size-dependent immunogenicity: therapeutic and protective properties of nano-vaccines against tumors. J Immunol. 2004;173(5):3148–3154. doi: 10.4049/jimmunol.173.5.3148. [DOI] [PubMed] [Google Scholar]
- 74.Reddy ST, Van Der Vlies AJ, Simeoni E, et al. Exploiting lymphatic transport and complement activation in nanoparticle vaccines. Nat Biotechnol. 2007;25(10):1159–1164. doi: 10.1038/nbt1332. [DOI] [PubMed] [Google Scholar]
- 75.Ohno S, Kohyama S, Taneichi M, et al. Synthetic peptides coupled to the surface of liposomes effectively induce SARS coronavirus-specific cytotoxic T lymphocytes and viral clearance in HLA-A*0201 transgenic mice. Vaccine. 2009;27(29):3912–3920. doi: 10.1016/j.vaccine.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Espuelas S, Roth A, Thumann C, Frisch B, Schuber F. Effect of synthetic lipopeptides formulated in liposomes on the maturation of human dendritic cells. Mol Immunol. 2005;42(6):721–729. doi: 10.1016/j.molimm.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 77.Steers NJ, Peachman KK, Mcclain S, Alving CR, Rao M. Liposome-encapsulated HIV-1 Gag p24 containing lipid A induces effector CD4+ T-cells, memory CD8+ T-cells, and pro-inflammatory cytokines. Vaccine. 2009;27(49):6939–6949. doi: 10.1016/j.vaccine.2009.08.105. [DOI] [PubMed] [Google Scholar]
- 78.Grgacic EV, Anderson DA. Virus-like particles: passport to immune recognition. Methods. 2006;40(1):60–65. doi: 10.1016/j.ymeth.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Herzog C, Hartmann K, Kunzi V, et al. Eleven years of Inflexal V – a virosomal adjuvanted influenza vaccine. Vaccine. 2009;27(33):4381–4387. doi: 10.1016/j.vaccine.2009.05.029. [DOI] [PubMed] [Google Scholar]
- 80.Black M, Trent A, Tirrell M, Olive C. Advances in the design and delivery of peptide subunit vaccines with a focus on toll-like receptor agonists. Expert Rev Vaccines. 2010;9(2):157–173. doi: 10.1586/erv.09.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rudolf MP, Man S, Melief CJ, Sette A, Kast WM. Human T-cell responses to HLA-A-restricted high binding affinity peptides of human papillomavirus type 18 proteins E6 and E7. Clin Cancer Res. 2001;7(3 Suppl):788S–795S. [PubMed] [Google Scholar]
- 82.Firbas C, Jilma B, Tauber E, et al. Immunogenicity and safety of a novel therapeutic hepatitis C virus (HCV) peptide vaccine a randomized, placebo controlled trial for dose optimization in 128 healthy subjects. Vaccine. 2006;24(20):4343–4353. doi: 10.1016/j.vaccine.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 83.Cerny A, Chisari FV. Pathogenesis of chronic hepatitis C: immunological features of hepatic injury and viral persistence. Hepatology. 1999;30(3):595–601. doi: 10.1002/hep.510300312. [DOI] [PubMed] [Google Scholar]
- 84.Klade CS, Schuller E, Boehm T, Von Gabain A, Manns MP. Sustained viral load reduction in treatment-naive HCV genotype 1 infected patients after therapeutic peptide vaccination. Vaccine. 2012;30(19):2943–2950. doi: 10.1016/j.vaccine.2012.02.070. [DOI] [PubMed] [Google Scholar]
- 85.Firbas C, Boehm T, Buerger V, et al. Immunogenicity and safety of different injection routes and schedules of IC41, a hepatitis C virus (HCV) peptide vaccine. Vaccine. 2010;28(12):2397–2407. doi: 10.1016/j.vaccine.2009.12.072. [DOI] [PubMed] [Google Scholar]
- 86.Crook T, Morgenstern JP, Crawford L, Banks L. Continued expression of HPV-16 E7 protein is required for maintenance of the transformed phenotype of cells co-transformed by HPV-16 plus EJ-ras. EMBO J. 1989;8(2):513–519. doi: 10.1002/j.1460-2075.1989.tb03405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu CY, Monie A, Pang X, Hung CF, Wu TC. Improving therapeutic HPV peptide-based vaccine potency by enhancing CD4+ T help and dendritic cell activation. J Biomed Sci. 2010;17:88. doi: 10.1186/1423-0127-17-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ma B, Xu Y, Hung C-F, Wu T. HPV and therapeutic vaccines: where are we in 2010? Curr Cancer Ther Rev. 2010;6:81–103. [Google Scholar]
- 89.Welters MJ, Kenter GG, Piersma SJ, et al. Induction of tumor-specific CD4+ and CD8+ T-cell immunity in cervical cancer patients by a human papillomavirus type 16 E6 and E7 long peptides vaccine. Clin Cancer Res. 2008;14(1):178–187. doi: 10.1158/1078-0432.CCR-07-1880. [DOI] [PubMed] [Google Scholar]
- 90.Kenter GG, Welters MJ, Valentijn AR, et al. Phase I immunotherapeutic trial with long peptides spanning the E6 and E7 sequences of high-risk human papillomavirus 16 in end-stage cervical cancer patients shows low toxicity and robust immunogenicity. Clin Cancer Res. 2008;14(1):169–177. doi: 10.1158/1078-0432.CCR-07-1881. [DOI] [PubMed] [Google Scholar]
- 91.Asjo B, Stavang H, Sorensen B, Baksaas I, Nyhus J, Langeland N. Phase I trial of a therapeutic HIV type 1 vaccine, Vacc-4x, in HIV type 1-infected individuals with or without antiretroviral therapy. AIDS Res Hum Retroviruses. 2002;18(18):1357–1365. doi: 10.1089/088922202320935438. [DOI] [PubMed] [Google Scholar]
- 92.Kran AM, Sommerfelt MA, Sorensen B, et al. Reduced viral burden amongst high responder patients following HIV-1 p24 peptide-based therapeutic immunization. Vaccine. 2005;23(31):4011–4015. doi: 10.1016/j.vaccine.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 93.Kran AM, Sorensen B, Nyhus J, et al. HLA- and dose-dependent immunogenicity of a peptide-based HIV-1 immunotherapy candidate (Vacc-4x) AIDS. 2004;18(14):1875–1883. doi: 10.1097/00002030-200409240-00003. [DOI] [PubMed] [Google Scholar]
- 94.Kran AM, Jonassen TO, Sommerfelt MA, Lovgarden G, Sorensen B, Kvale D. Low frequency of amino acid alterations following therapeutic immunization with HIV-1 Gag p24-like peptides. AIDS. 2010;24(17):2609–2618. doi: 10.1097/QAD.0b013e32833e502b. [DOI] [PubMed] [Google Scholar]
- 95.Kran AM, Sommerfelt MA, Baksaas I, Sorensen B, Kvale D. Delayed-type hypersensitivity responses to HIV Gag p24 relate to clinical outcome after peptide-based therapeutic immunization for chronic HIV infection. APMIS. 2012;120(3):204–209. doi: 10.1111/j.1600-0463.2011.02843.x. [DOI] [PubMed] [Google Scholar]
- 96.Spearman P, Kalams S, Elizaga M, et al. Safety and immunogenicity of a CTL multiepitope peptide vaccine for HIV with or without GM-CSF in a Phase I trial. Vaccine. 2009;27(2):243–249. doi: 10.1016/j.vaccine.2008.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Westerfeld N, Zurbriggen R. Peptides delivered by immunostimulating reconstituted influenza virosomes. J Pept Sci. 2005;11(11):707–712. doi: 10.1002/psc.700. [DOI] [PubMed] [Google Scholar]
- 98.Foon KA, Chakraborty M, John WJ, Sherratt A, Kohler H, Bhattacharya-Chatterjee M. Immune response to the carcinoembryonic antigen in patients treated with an anti-idiotype antibody vaccine. J Clin Invest. 1995;96(1):334–342. doi: 10.1172/JCI118039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Alcami A, Koszinowski UH. Viral mechanisms of immune evasion. Immunol Today. 2000;21(9):447–455. doi: 10.1016/S0167-5699(00)01699-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Finlay BB, McFadden G. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell. 2006;124(4):767–782. doi: 10.1016/j.cell.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 101.Johansson BE, Bucher DJ, Kilbourne ED. Purified influenza virus hemagglutinin and neuraminidase are equivalent in stimulation of antibody response but induce contrasting types of immunity to infection. J Virol. 1989;63(3):1239–1246. doi: 10.1128/jvi.63.3.1239-1246.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Thomson CA, Wang Y, Jackson LM, et al. Pandemic H1N1 influenza infection and vaccination in humans induces cross-protective antibodies that target the hemagglutinin stem. Front Immunol. 2012;3:87. doi: 10.3389/fimmu.2012.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li GM, Chiu C, Wrammert J, et al. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc Natl Acad Sci USA. 2012;109(23):9047–9052. doi: 10.1073/pnas.1118979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Corti D, Voss J, Gamblin SJ, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333(6044):850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 105.Ekiert DC, Friesen RH, Bhabha G, et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011;333(6044):843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Corti D, Suguitan AL, Jr, Pinna D, et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J Clin Invest. 2010;120(5):1663–1673. doi: 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ekiert DC, Bhabha G, Elsliger MA, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324(5924):246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sui J, Hwang WC, Perez S, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16(3):265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Epstein SL, Tumpey TM, Misplon JA, et al. DNA vaccine expressing conserved influenza virus proteins protective against H5N1 challenge infection in mice. Emerg Infect Dis. 2002;8(8):796–801. doi: 10.3201/eid0808.010476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kreijtz JH, De Mutsert G, Van Baalen CA, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. Cross-recognition of avian H5N1 influenza virus by human cytotoxic T-lymphocyte populations directed to human influenza A virus. J Virol. 2008;82(11):5161–5166. doi: 10.1128/JVI.02694-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rothman AL, Kanesa-Thasan N, West K, Janus J, Saluzzo JF, Ennis FA. Induction of T lymphocyte responses to dengue virus by a candidate tetravalent live attenuated dengue virus vaccine. Vaccine. 2001;19(32):4694–4699. doi: 10.1016/s0264-410x(01)00236-5. [DOI] [PubMed] [Google Scholar]
- 112.Philip R, Testa JS. Synthetic universal dengue vaccine: a reality. Dengue Digest. 2011;7(2):3–4. [Google Scholar]
Websites
- 201.Greenbaum J. Knowledgebase and Forums/Epitope analysis in emerging H1N1 swine flu viruses/Analysis version 1.0. IEDB. IEDB Analysis Resource; 2009. p. 202. http://iedb.zendesk.com/forums/45499/entries/35037. http://tools.immuneepitope.org. [Google Scholar]