Abstract
Anticancer therapeutics employing RNA interference mechanism holds promising potentials for sequence-specific silencing of target genes. However targeted delivery of siRNAs to tumor tissues and cells and more importantly, their intracellular release at sites of interest still remains a major challenge that needs to be addressed before this technique could become a clinically viable option. In the current study, we have engineered and screened a series of CD44 targeting hyaluronic acid (HA) based self-assembling nanosystems for targeted siRNA delivery. The HA polymer was functionalized with lipids of varying carbon chain lengths/nitrogen content, as well as polyamines for assessing siRNA encapsulation. From the screens, several HA-derivatives were identified that could stably encapsulate/complex siRNAs and form self-assembled nanosystems, as determined by gel retardation assays and dynamic light scattering. Many HA derivatives could transfect siRNAs into cancer cells overexpressing CD44 receptors. Interestingly, blocking the CD44 receptors on the cells using free excess soluble HA prior to incubation of cy3-labeled-siRNA loaded HA nano-assemblies resulted in >90% inhibition of the receptor mediated uptake, confirming target specificity. In addition, SSB/PLK1 siRNA encapsulated in HA-PEI/PEG nanosystems demonstrated dose dependent and target specific gene knockdown in both sensitive and resistant A549 lung cancer cells overexpressing CD44 receptors. More importantly, these siRNA encapsulated nanosystems demonstrated tumor selective uptake and target specific gene knock down in vivo in solid tumors as well as in metastatic tumors. The HA based nanosystems thus portend to be promising siRNA delivery vectors for systemic targeting of CD44 overexpressing cancers including tumor initiating (stem-) cells and metastatic lesions.
Keywords: Self-assembling nanosystems, siRNA delivery, Multidrug resistance, Tumor targeting, CD44, Hyaluronic acid
1. Introduction
Multi drug resistance (MDR) poses as a great challenge for the treatment of several recurrent cancers including lung cancers in the clinic [1]. MDR develops due to multiple factors that include poor systemic drug delivery efficiency, inefficient drug residence at the tumor site, poor intracellular availability and micro-environmental selection pressures that allow certain cells to survive despite aggressive chemotherapy [2]. RNA interference (RNAi) therapy has emerged as a powerful strategy for combating drug resistance by selective down-regulation of key genes involved in the development of MDR phenotype [3]. However, delivery of small interfering RNAs (siRNAs) to specific tumor tissues and cells followed by their intracellular release from the endosomal/lysosomal compartments into the cytoplasm still remains a major hurdle that needs to be addressed before this experimental technique could become a clinically viable therapeutic option [4,5].
Naked siRNAs are unfavorable for systemic delivery because of their inherent limitations such as negative charge, large molecular weight, size, and instability. They are easily degraded by serum endonucleases and are quickly eliminated by renal excretion [6]. In this regard, engineered nanocarriers that can stably encapsulate/complex, protect and selectively deliver siRNAs to target tumor tissues and cells are highly promising as next generation gene delivery vehicles [7]. The ideal system should preferably be non-toxic, biodegradable, non-immunogenic and able to efficiently deliver siRNAs to target tumors. More importantly, after tumor tissue specific delivery, the nanosystem should facilitate intracellular uptake and promote endosome release/escape of the payload siRNAs into cytoplasm allowing their interaction with the endogenous RISC [8]. Although viral vectors can serve as efficient gene delivery vehicles, the apparent toxicity and safety concerns limit their utility for human applications, necessitating the development of safer delivery technologies. In this regard, non-viral/polymeric nanoparticle based delivery platforms are becoming increasingly viable alternatives for siRNA delivery [9].
For the current study, we selected hyaluronic acid (HA) polymer with a relatively low molecular weight of 20 kDa as our starting building block. HA is a naturally occurring mucopolysaccharide composed of alternating disaccharide units of d-glucuronic acid and N-acetyl d glucosamine (NAG) with β (1–4) interglycosidic linkage [10]. HA is a highly anionic biopolymer present in the extracellular matrix and synovial fluids. It is biodegradable, non-toxic, non-immunogenic and non-inflammatory, which makes it an ideal carrier polymer for systemic drug delivery applications [11]. Also, a relatively simple coupling chemistry allows for modification of the sugar residues on the polymer to arrive at variety of functional macrostructures that facilitates self-assembly and encapsulation of diverse drug and gene payloads for targeted delivery [12]. Of primary importance to tumor selective delivery, the HA backbone in itself is endowed with tumor targeting moieties that specifically recognizes CD44 – an integral membrane glycoprotein overexpressed on several tumors cell surfaces, including tumor initiating stem cells [13–16]. Interestingly, it has already been reported that the CD44 receptors are highly expressed in majority of non-small cell lung cancers (NSCLC) and is a known biomarker for NSCLC tumors [17], however their expression levels are diminished in H69 and H69AR sensitive and resistant small cell lung cancers respectively [17].
Although HA is an attractive carrier polymer, its anionic nature can pose difficulty in encapsulating negatively charged siRNA molecules [18]. However, we propose to overcome this limitation by hydrophobic modification of HA backbone, with fatty amines and cationic polyamines that would not only help reduce the net negative charge density on the backbone polymer but also facilitate siRNA encapsulation via self-assembly [11], and intracellular delivery by endocytosis [19]. High counter-ion density has also been reported to promote efficient endosome release/escape after cellular entry [20,21].
In the current study, we have developed a series of CD44 targeting hyaluronic acid (HA) based versatile self-assembling nanosystems for targeted delivery of siRNA with the aim to treat CD44 overexpressing cancer cells. Furthermore, we evaluated the systemic delivery efficiency and gene silencing activity of the siRNA loaded in select HA nanosystems using drug resistant and sensitive tumors models overexpressing CD44 receptors including metastatic cancer models.
2. Materials and methods
2.1. Materials
Sodium hyaluronate (HA) with an average molecular weight of 20 kDa was obtained from Lifecore Biomedical Co. (Chaska, MN). Polyethyleneimine (bPEI, MW~10,000 Da) was obtained from Polysciences Inc. (Warrington, PA). Sulfo-NHS was purchased from Thermo Scientific Corp (Billerica, MA), mono-functional polyethyleneglycol amine (PEG2k-NH2, MW = 2000 Da) was purchased from Creative PEG Works (Winston Salem, NC). All the fatty amines and other reagents for synthesis were obtained at high purity (>99%) from Sigma–Aldrich Chemical Co (Milwaukee, WI) or Acros Organics (Thermo Fisher, Pittsburgh, PA) and used without further purification.
2.2. Cell lines
The human cell lines used in this study included non-small cell lung cancer (NSCLC) A549 and A549DDP, small cell lung cancer (SCLC) H69 and H69AR, breast cancer MDA-MB468, liver cancer Hep3B, murine melanoma B16F10. Except for A549DDP, all the other cell lines were obtained from ATCC. The cisplatin resistant A549DDP cells were obtained from Massachusetts General Hospital, Boston, MA. Except Hep3B and MDA-MB468 cells, which were cultured using Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% FBS, all the other cell lines were cultured using RPMI-1640 medium supplemented with 10% FBS. All the cells were grown as adherent cultures except H69 which was grown as suspension cells at 37 °C and 5% CO2.
2.3. Synthesis of functional hyaluronic acid based polymers
2.3.1. Modification of hyaluronic acid with mono-functional fatty amines
Mono-functional fatty amines of the general formula CH3(CH2)nNH2 (where n = 3,4,5,7,9,11,13 and 17) such as n-butyl amine, n-pentylamine, n-hexylamine, n-octylamine, n-decylamine, n-dodecylamine, tetradecylamine and stearylamine were chemically conjugated to the backbone of hyaluronic acid by coupling the end group primary amine of the fatty amine with the carboxylic acid or hydroxyl group of hyaluronic acid in the presence of a coupling agent, 1-Ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC) and n-hydroxysuccinimide (NHS). In brief, multiple glass scintillation vials were used to dissolve sodium hyaluronate (MW 20 kDa, 100 mg, 5 mmole) in 5 ml of dry formamide by warming up the reaction mixture up to 50 °C under stirring. After obtaining a clear solution the reaction mixture was allowed to cool to room temperature and then varying feed ratio of fatty amines (~2–25 mg, ~40–100 μmole) was added to each vial containing the hyaluronic acid solution under magnetic stirring. Subsequently, EDC (10–20 mg, 50–100 μmole) and NHS (5–15 mg, 50–150 mmole) mixture in DMF (2 ml) was added dropwise into each of the reaction vial under magnetic stirring. The reaction vials were allowed to stir for 12 h using multi-position magnetic stirrer (IKAMAG, IKA Labs, Germany). The resulting solution was added to a large excess of ethanol (EtOH, 250 ml) to precipitate the derivatized polymer and dissolve the unreacted fatty amines. The polymer precipitate was centrifuged and the washings were discarded. The above step of EtOH precipitation and washing was repeated thrice to purify the polymer. Finally the polymer was re-dissolved in deionized water and subjected to ultrafiltration using Millipore Labscale Tangential Flow Filtration (TFF) system with Biomax 10 kDa MWCO membrane (EMD-Millipore, Billerica, MA). The concentrated and purified polymer was finally lyophilized using a freeze dryer (Freezone-6, Labconco Inc., Kansas, MO) and stored at −20 °C until further use (yield: 85–90 mg, ~81–85%, off-white fibrous product). A 3 mg portion of the lyophilized product was dissolved in 600 μl of D2O and characterized by 400 MHz 1H NMR spectroscopy (Varian Inc., CA) for determining the % lipid modification.
2.3.2. Modification of hyaluronic acid with bifunctional fatty amines
An aqueous solution of sodium hyaluronate (HA, MW 20 kDa, 100 mg, 5 μmole) prepared in deionized water and mixed with bifunctional fatty amines (~2–25 mg, ~40–100 μmole) having a general formula of NH2(CH2)nNH2 (where n = 4,5,6…) such as 1,4 diaminobutane, 1,6 diaminohexane or 1,8 diaminoocatne. The pH of the reaction mixture was adjusted to 7.5 with 0.1 m NaOH/0.1 m HCl and the pH adjusted solution was added slowly into a mixture of EDC (10–50 mg–50–250 μmole) and NHS (5–25 mg, 50–250 μmole) in deionized water. After mixing the reactants using a magnetic stirrer, the pH of the reaction was maintained at 7.5 by the addition of 0.1 m NaOH for about 2 h and the reaction was allowed to proceed for 12 h. The HA-lipid modified derivatives were purified by extensively dialysis using cellulose membrane (MWCO~12–14 kDa) against deionized water for 48 h with frequent replacement of deionized water. The purified product was then lyophilized and stored at −20 °C until further use (yield: 90 mg, ~86.5%, off-white fibrous product). A 3 mg portion of the lyophilized product was dissolved in 600 μl of D2O and characterized by 400 MHz 1H-NMR spectroscopy (Varian Inc., CA) for determining the % lipid modification.
2.3.3. Chemical modification of hyaluronic acid with polyamines
HA was chemically modified with polyethyleneimine (PEI, 10 kDa) or Poly (l-Lysine) (10–14 kDa) by using a coupling agent, 1-Ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC). In brief, sodium hyaluronate (MW 20 kDa, 100 mg, 5 μmole) was dissolved in 5 ml of dry formamide in a glass scintillation vial by warming up the reaction vial up to 50 °C. After obtaining a clear solution the reaction mixture was allowed to cool to room temperature and then ~3.3 mg of the PEI or PLL (10 kDa–0.33 mmole) was added to the solution. Then EDC (10 mg, 50 μmole) was added into the reaction mixture. The solution turned hazy and then became clear after 1 h. The reaction was stirred overnight or for 12 h using a magnetic stirrer. The resulting solution was added to a large excess of EtOH (250 ml) to precipitate the polymer. The precipitate was centrifuged and collected and the washings were discarded. The above step of EtOH precipitation and washing was repeated thrice to purify the polymer. Finally the precipitate polymer was re-dissolved in deionized water and subjected to ultrafiltration using Labscale Millipore Tangential Flow Filtration (TFF) system with Biomax 10 kDa MWCO membrane (EMD-Millipore, Billerica, MA). The concentrated and purified polymer was then lyophilized using a freeze dryer (Freezone-6, Labconco Inc., Kansas, MO) and stored at −20 °C (yield: 75 mg, ~75%, off-white fibrous product). A 3 mg portion of the lyophilized product was dissolved in 600 μl of D2O and characterized by 400 MHz 1H-NMR spectroscopy (Varian Inc., CA) for determining the % lipid modification.
2.3.4. Synthesis of thiolated and PEGylated hyaluronic acid
The synthesis of thiolated-HA was based on a reported procedure [22]. Briefly, the HA polymer (5 g, MW 20 kDa, 0.25 mmol) was dissolved in 250 ml of PBS solution (pH = 7.0) and stirred well using a magnetic stirrer until the solution became clear. Three molar excess amounts of EDC (1 g, 5 mmol) and N-Hydroxybenzotrizole (HoBt) (750 mg, 5 mmol) were added and stirred for 2 h, and then cystamine dihydrochloride (565 mg, 5 mmol) was added drop wise and mixed overnight. The reaction mixture was dialyzed thoroughly using cellulose dialysis membranes (MW cut off ~ 12–14 kDa) (Spectrapore, Spectrum Labs, CA) for 48 h to remove unreacted reactants. Subsequently, the thiol-cross-linked HA polymer was treated with five molar excess of dithiothreitol (DTT) and stirred for 24 h to cleave the disulfide bridges to yield the water soluble conjugate. The product was dialyzed against deionized water at pH 3.0 for 96 h (MWCO 12–14 kDa) (Spectrapore, Spectrum Labs, CA). The % thiolation was determined using Ellman reagent as described by Habeeb [23]. The final product was lyophilized and stored at −20 °C until use.
For the preparation of the PEGlyate hyaluronic acid, HA was chemically conjugated with polyethyleneglycol amine (PEG-NH2) (PEG, 2 kDa or 5 kDa) using 1-Ethyl-3-[3imethylaminopropyl]carbodiimide hydrochloride (EDC) and n-hydroxy sulfo succinimide (sulfo-NHS). In brief, EDC (19 mg, 100 μmole) and sulfo-NHS (21 mg, 100 μmole) were mixed in 5 ml deionized water and kept for 1 h to form the amine reactive intermediate. Sodium hyaluronate (MW 20 kDa, 200 mg, 10 μmole) was dissolved in 10 ml of deionized water in a glass scintillation vial. After stirring and obtaining a clear solution 50 mg or 125 mg of the PEG (50 μmole, 2 k or 5 k PEG for 5% modification) was added to the solution and stirred. Then EDC/NHS solution prepared earlier was added into the HA/PEG solution dropwise over a period of 1 h. The reaction mixture was stirred for 24 h and then dialyzed using cellulose dialysis membranes (MW cut off ~12–14 kDa) against deionized water/methanol mixture (1v/3v–1v/1v) for 24 h and subsequently with deionized water for 48 h. The purified product was then lyophilized and stored at −20 °C (yield: 80 mg, ~79%, off-white fibrous product). A 3 mg portion of the lyophilized product was dissolved in 600 μl of D2O and characterized by 400 MHz 1H-NMR spectroscopy (Varian Inc., CA) for determining the % modification.
2.4. Assessing self-assembling capability of HA based functional macrostructures and nanosystems
In order to assess the self-assembling ability of the HA based macrostructures (such as HA-butylamine, HA-hexylamine, HA-octylamine, HA-stearylamine, HA-choline, HA-spermine, HA-PLL or HA-PEI), the respective derivative was suspended in deionized water at varying concentrations (1–10 mg/ml) at room temperature. After vortex mixing for 1 min followed by 15 min incubation, their average hydrodynamic size and size distribution were measured by using a dynamic light scattering (DLS) instrument (Malvern Zetasizer Nano-S, Malvern Inc., UK). Further, in order to determine the zeta potentials or the surface charge of the prepared HA derivatives they were transferred to specially designed electrophoretic cells and the zeta potentials were recorded using the same instrument.
2.5. Formation of siRNA loaded HA nanosystems, determination of siRNA encapsulation and release by polyanion competition by gel retardation assay
In order to assess the ability of the series of functionalized-HA derivatives (containing varying lipid content, nitrogen content and polyamine content) to form self-assembling nanosystems with siRNAs, the respective derivatives were incubated with known concentration of siRNA with varying concentrations of the HA-derivatives in solution. The PLK1 and SSB siRNA sequences that were used in this study are:
PLK1: sense: UUGUCUuCAGGuCUuCAGuu antisense: AGAUcacCCUccuuAAAUauu
SSB: sense: AcAAcAGAcuuuAAuGuAAuu antisense:UUAcAuuAAAGucuGuuGuuu
The solution containing the polymer and siRNA was vortexed and mixed for a minute and incubated at room temperature for 15 min to form self-assembly. The nano-assemblies were prepared at several polymer-to-siRNA weight ratios, ranging from 9:1 to 450:1 to study the effect of polymer concentration on siRNA encapsulation. The formation of siRNA loaded self-assembling nanosystems was also studied using the dynamic light scattering (DLS) instrument by measuring the size and charge of the particles formed. Also, transmission electron microscopy (JEOL, JEM-1000, Tokyo, Japan) was performed to assess the formation of siRNA loaded nano-assemblies. A uranyl acetate ribonucleic acid stain was used to demarcate siRNAs from the polymer. The dark staining of siRNA by heavy metals such as uranyl acetate provides a high contrast compared to hyaluronic acid polymer that can help ascertain loading of genes in polymeric nanosystems.
The ability of these complexes to release siRNA was then determined by treating them with a highly charged polyanionic polymer, polyacrylic acid (PAA) and running on gel. The anionic polymer (PAA) would compete with the anionic polymer and release the siRNA which then appears as a free band in the gel. For this purpose, a 5 μl polymer-siRNA complex was diluted up to 10 μl with deionized water. Then 10 μl of 2% PAA was added to this mixture and vortexed. The 20 μl sample was then run on Agarose gel along with the PAA untreated samples (with same amount of total siRNA content) to compare their ability for de-complexation of siRNA by monitoring the released/free siRNA bands.
2.6. Assessment of siRNA uptake by confocal microscopy and flow cytometry
To assess the ability of the polymeric nanosystems to release the siRNA into cells, a NIR dye cy3-labeled siRNA was formulated using functionally variant HA derivatives at optimal polymer:siRNA weight ratios according to the above mentioned protocol. These particles were then incubated with 2e4 MDA-MB468 cells containing 100 nM siRNA at 37 °C. After incubation for 12 h, the cells were washed with PBS and trypsinized to detach the cells. The uptake of cy3-labeled siRNA was determined by using a flow cytometer (BD FACSCalibur, BD Biosciences, San Jose, CA). The data was analyzed with BD Cell Quest software. In parallel, identical polymer-siRNA nanosystems were incubated with cancer cells overexpressing CD44 receptors grown on the glass cover slips for 12 h to assess the intracellular trafficking of siRNA. At the end of the incubation period, the cells were washed thrice with PBS, fixed in paraformaldehyde in PBS for 10 min. The localization of siRNA within the cells was visualized by a confocal microscope (Carl Zeiss Microscopy GmbH, Jena, Germany). Flow cytometry was also used to determine the CD44 receptor levels in the cells. For this purpose 200 μl of 1e6 A549, A549DDP, H69, H69AR, Hep3B, MDA-MB468 and B16F10 cells were incubated separately with antibody against CD44 with PE tag for 1 h and washed thrice with PBS. Cells were then reconstituted in PBS and ran through flow cytometer to assess the CD44 receptor levels. The anti-CD44 monoclonal antibodies used in this study was obtained from R&D systems.
2.7. Competition assays to assess receptor mediated cellular entry
In order to determine the receptor mediated cellular entry of the engineered HA-nanosystems, cy3-labeled siRNA encapsulated HA-nanosystems were incubated in MDA-MB468 cells for 12 h. In parallel a competitive inhibition (or blocking control) study was also undertaken. For the blocking studies the cell culture medium was replaced with serum free media containing excess free soluble HA polymer at a concentration of 10 mg/ml for an hour to block the receptors prior to incubation with the cy3-labeled-siRNA/HA-nanosystem for 12 h under identical conditions. Both sets of cells were then washed with 1 × PBS and visualized under the confocal fluorescence microscope (Zeiss, Germany). In parallel, two other sets of cells were treated similarly and harvested as described previously for flow cytometric analysis to quantitate the cy3 positive cells. For negative control studies, a cell line that does not express CD44 (or expresses very low levels of CD44) was treated with cy3-labeled-siRNA encapsulated HA-nanosystems.
2.8. Transfection of PLK1 targeted siRNA to assess silencing of target gene expression
PLK1 or CTL siRNA was encapsulated in a series of HA-derivatives at mass ratios ranging from 450:1 to 9:1 (polymer: siRNA) as previously described. These siRNA loaded nanosystems were then reverse transfected into MDA-MB468 cells and incubated for 48 h at siRNA concentrations of 300, 200 and 100 nM. After incubation, cells were harvested and the RNA was extracted. The extracted RNA was used to run quantitative PCR to assess the mRNA levels. mRNA knockdown was determined by normalizing the PLK1 expression levels to the endogenous GAPDH levels.
2.9. Assessment of siRNA release using endosome-disrupting agents
To determine if chloroquine, an endosome-disrupting agent could release the siRNA/nanosystem trapped in the endosome, the MDA-MB468 cells were incubated with HA-PEI/PLK1 complex prepared using a mass ratio of 27:1 in the presence or absence of chloroquine in the medium and incubated for 48 h. After the incubation, the RNA was extracted from the cells as previously described and the PCR was run to determine siRNA release by measuring the PLK1 mRNA knockdown.
2.10. Establishment of subcutaneous, metastatic and syngeneic tumor models
Animal procedures were performed according to a protocol approved by Northeastern University, Institutional Animal Care and Use Committee (NU-IACUC). The tumor models for this study were developed in nude mice obtained from Harlan Laboratories, South Easton, MA. 5–6 week old nude mice were injected subcutaneously (s.c.) with tumor cells A549 (5 × 106 cells + matrigel), A549DDP (1 × 107 cells), H69 (1 × 106 cells + matrigel), H69AR (1 × 106 cells + matrigel), Hep3B (7 × 106 cells + matrigel), MDA-MB468 (5 × 106 cells + matrigel) and B16F10 (1 × 106 cells) under the right shoulder. Tumor sizes were measured at least once or twice a week to monitor the tumor growth. In addition, the B16F10 cells were intravenously (i.v.) injected (5 × 105 cells/mouse) into nude mice to generate an experimental metastatic lung or syngeneic model with the idea of checking out the delivery and activity when the tumors are located on lungs as metastatic lesions. Since these cells do not express luciferase, several mice were opened up on day 7 and 10 to make sure that they developed tumors in the lung. In a similar trend, the A549-luc cells (5e6) were also i.v. injected to generate lung metastatic tumor models in mice. Tumor growth progression was monitored using an in vivo imaging system (IVIS, Carestream Molecular Imaging Inc., Rochester, NY) and measuring the luciferase activity in mice at different time points.
2.11. Evaluating target gene knockdown in tumors with differential CD44 expression and vascularity using ‘tool’ siRNA
In order to correlate the CD44 expression levels and the vascularity of the tumors, we examined several tumor types in mice as described above. Typically, when A549 subcutaneous tumors were approximately ~150–200 mm3 in size, the animals were randomized into groups such that each group had similar tumor size. These groups of mice with A549 tumors then received either PLK1 or SSB formulated HA-PEI/siRNA, HA-PEI/HA-PEG/siRNA or HA-PEI/HA-PEG/HA-SH/siRNA at a dose of 0.5 mg/kg every day for three consecutive days. Subsequently, 24 h after the last dose, the tumors were excised, RNA was extracted and PCR was run to determine the PLK1 or SSB knockdown. Likewise, similar doses were evaluated in other tumor models as well (A549DDP (s.c.), H69 (s.c.), H69AR (s.c.), B16F10 (s.c. and met), Hep3B (s.c.), MDA-MB468 (s.c.) and A549 (s.c. and met) as described.
3. Results and discussion
Development of new and versatile non-toxic and biodegradable polymers that are safe in systemic circulation, efficient in encapsulation of therapeutic genes, and can selectively deliver the payload to the sites of interest (such as tumors tissues and cells), is urgently needed for successful translation of RNAi based mechanisms from bench to the bedside [24]. In the current study, we have synthesized a series of functionally variant library of self-assembling CD44 targeting HA based macrostructures by varying the carbon chain length, nitrogen content and polyamine side chain grafting onto the HA backbone. The modification of anionic HA polymer resulted in effective lowering/shielding of its negative charge density that could enable siRNA encapsulation. The hydrophobic modification of HA polymer could be efficiently achieved by chemical conjugation/grafting of fatty amines with varying alkyl chains lengths onto the hydrophilic HA backbone by using EDC/NHS coupling chemistry (Fig. 1). It was observed that by varying the molar feed ratio of the fatty amines and the polyamine content, the degree of HA modification could be controlled in the range of 5–20% as determined by 1H-NMR spectroscopy (Fig. S1). As described, several different approaches were undertaken to arrive at HA based functional macrostructures that varied in nitrogen content, carbon chain lengths (or lipid content), polyamine content, and other variables to understand the structure–activity relationship. Some of the different classes of functional HA blocks that were synthesized included HA derivatized with monofunctional fatty amines (namely, n-butylamine, C4; n-hexylamine, C6; n-octylamine, C8; dodecylamine, C12; and stearylamine, C18); bifunctional fatty amines (namely, 1–6 diaminohexane, C6N2; 1–8 diaminohexane, C8N2); and multiple nitrogen containing derivatives and polyamines such as pipirazine, spermine, choline, poly (l-lysine) and polyethyleneimine (PEI).
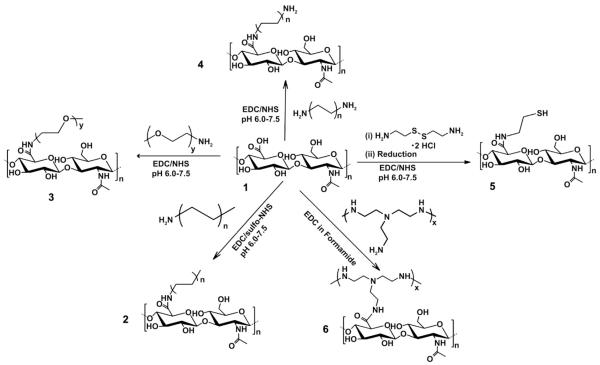
Fig. 1.
Synthesis of HA based functional macrostructures. A series of functional HA based derivatives were synthesized using a simple and versatile EDC/NHS conjugation chemistry as shown. 1, Hyaluronic acid (HA) of 20 kDa molecular weight; 2, HA conjugated to monofunctional fatty amines with the general formula CH3(CH2)nNH2 (where n = 3,4,5…); 3, HA conjugated to PEG of 2000 Da molecular weight; 4, HA conjugated to bifunctional fatty amines with the general formula NH2(CH2)nNH2 (where n = 4,5…); 5, Thiolated-HA (HA-SH) formed by conjugating HA with cysteamine dihydrochloride followed by reduction; and 6, HA conjugated to polyamines such as polyethyleneimine (PEI) of 10 kDa molecular weight.
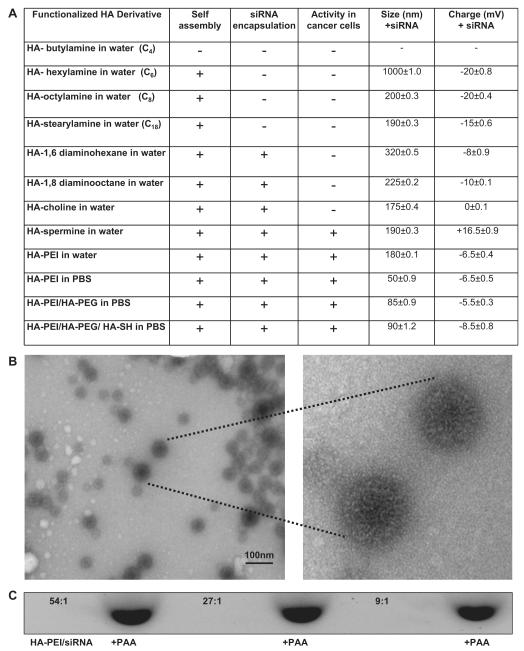
In order to assess the ability of the modified HA macrostructures to form self-assembly, the free polymers (without the payload siRNA) were suspended in water at concentrations ranging from 1 to 10 mg/ml. It was observed that the critical micellar concentration (as determine by DLS) varied for different classes of HA-lipids and polyamine containing HA derivatives. Notably, the HA conjugated to fatty amines with one or two amine residues (such as HA-hexylamine, HA-octylamine, HA-dodecylamine, HA-stearlyamine, HA-diaminohexane and HA-diaminooctane), with a lipid modification of ~8–10% required at least a concentration of ~5 mg/ml to form stable self-assembling nanosystems. However a lower polymer concentration of 3 mg/ml was sufficient enough and in fact found ideal for self-assembly of polyamine modified HA systems (namely HA-spermine, HA-PLL and HA-PEI). As noticed, the derivatives with very short alkyl chain lengths such as butyl amine modified HA with lipid content of ~10–12% or lower did not promote self-assembly, whereas HA-derivatives modified with fatty amines comprising alkyl chains with six or more carbon atoms (with ~10–15% lipid modification) could self-assemble to form nanosized particles in deionized water or 1×PBS (Fig. 2A). This data suggests that certain amount of lipid modification/hydrophobicity is essential for the formation of self-assembly with the tested molecular weight of HA.
Fig. 2.
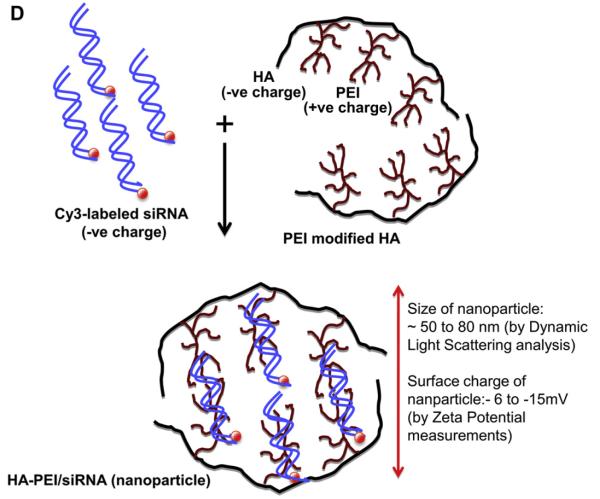
Characteristics of HA derivative/siRNA particles. Illustrative examples from each class of lipid chain that was used for derivatization (A). The HA-PEI/siRNA particles made at 54:1 ratio in PBS were visualized by TEM (B). Electrophoretic retardation analysis of siRNA binding by HA-PEI derivatives at different mass ratios (54:1, 27:1 and 9:1). The release of intact siRNA by polyacrylic acid was shown in each case (C). Schematic representation of HA-PEI/siRNA nanosystem suggesting a positive core containing the siRNA with a negatively charged HA shell structure (D).
As a next step, we assessed siRNA payload encapsulation using the identified HA-derivatives. For this purpose, the siRNA in solution was physically mixed with the functionalized-HA derivatives in DI water or 1 × PBS at varying polymer-to-siRNA mass ratios (ranging from 450:1, 270:1, 180:1, 90:1, 54:1, 45:1, 27:1 to 9:1 respectively) and at the optimum polymer concentrations found earlier. Many classes of the HA derivatives containing hydrophobic/lipid domains with a net negative charge of (~−10 mV or lower) facilitated siRNA encapsulation as well as formation of self-assembled nanosystems as determined by dynamic light scattering (DLS) studies (Fig. 2A). However, it was realized that the HA-derivatives with varying degrees of modification and nitrogen content required individual optimization. Interestingly, an optimal encapsulation efficiency of ~90–100% with an optimal particle size in the range of 150–300 nm was obtained only at certain polymer-to-siRNA mass ratios. Also, the parameters changed from one class of lipid to another, probably because the net charge contribution and the nitrogen content in the polymer played a significant role in the siRNA encapsulation/complexation and self-assembly. Zeta potential values indeed varied from one set of formulation to the other (from −20 mV to +16 mV) confirming the proposed role of charge on complexation and formation of self-assembled nano-systems. For instance, the experimentally observed results for siRNA encapsulation in mono-functional fatty amine modified HA were not encouraging although not surprising. This observation can in part be explained by the absence of positively charged domains to promote siRNA complexation. However, the addition of extra nitrogen atoms to the fatty acid chains (such as diamine containing HA) favorably enhanced the encapsulation of siRNAs. In this class of HA-polymers, under mild acidic conditions (pH = 5.0–5.5), the free primary amine residues were protonated to generate a net positive charge on the polymer to facilitate siRNA encapsulation. For this class of HA-polymers namely HA-1,6 diaminohexane, HA-1,8-diaminooctane and HA-choline (with ~8–10% lipid modification), the best observed polymer-to-siRNA mass ratio was found to be 450:1 (at a –5 mg/ml polymer and 0.5 mg/ml siRNA respectively). However, for the polyamine modified HA variants such as HA-spermine, HA-PLL and HA-PEI systems, the best polymer-to-siRNA mass ratio was found to be much lower at 54:1 (at 3 mg/ml of polymer and 0.5 mg/ml of siRNA respectively). Interestingly, in this class of polymers, except for HA-spermine, which needed a lower pH (~5.0–5.5) environment for encapsulation, we could achieve 100% siRNA encapsulation efficiencies even at neutral pH, which could be attributed to the self-condensing ability of the charged PEI/PLL units as reported else-where [25]. Another interesting observation was that the HA-PEI/siRNA prepared in 1× PBS (pH = 7.4) were found to be smaller in size (of the order of 50–60 nm) when compared to the ones made using water as the solvent (size ~150–200 nm) (Fig. 2A). This may probably be due to tighter binding between the siRNA and the HA-PEI polymer due to the buffering of counter-ions and a shift in charge balance in 1× PBS resulting in formation of smaller size particles. Also, these HA-PEI/siRNA nanoparticles surprisingly exhibited negative surface charge despite the presence of positively charged PEI (Fig. 2A). These results were thought to reflect a positive “core” containing the PEI/PLL that holds the intact siRNA, while displaying a hydrophilic “shell” of negatively charged HA as revealed by DLS and TEM (Fig. 2B and D). In fact, the transmission microscopy clearly shows the dark core due to high contrast arising from the uranyl acetate stained siRNA [26] loaded in the HA/PEI polymer. The proposed mechanism is similar to the ones reported by Han et al. earlier [11]. Although their systems had high PEI content compared to HA and differed from the ones we synthesized with much higher HA content, the mechanism of self-assembly remains identical. We then selected the HA polymer systems that formed self-assembling nanostructures with particle size ranging from 150 to 300 nm for further elucidation of cellular uptake and gene silencing activity.
In order to confirm if siRNA is indeed encapsulated in the HA-derivatives, an agarose gel electrophoresis was utilized as described. At the optimum mass ratios (polymer: siRNA), the encapsulation efficiency for most of the screened HA derivatives were found to be almost 100%. However, HA-1, 6 diaminohexane, HA-1, 8-diaminooctane and HA-choline needed a ratio of 450:1 for optimal siRNA encapsulation whereas the HA-PEI encapsulated siRNA with 100% efficiency starting from 54:1 to 9:1 polymer-to-siRNA ratios. The ability of these complexes to release siRNA was then determined by treating the samples with competing charged polymer such as polyanionic polyacrylic acid (PAA) and running them on gel (Fig. 2C). The % binding was calculated based on the relative intensity of free siRNA band that appeared after PAA treatment with respect to the PAA untreated sample. When there was complete complexation or encapsulation of siRNA inside the self-assembled nanosystem, the band completely disappeared and it re-appeared with the same intensity as the free siRNA band after treating with PAA, suggesting that the nanosystem released the entire siRNA when it was challenged with excess PAA.
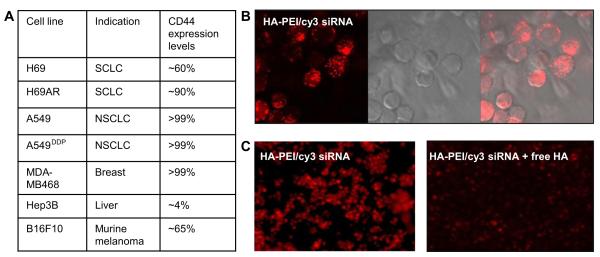
It is well known that the receptor expression levels are critical for cellular entry for systems that are internalized based on receptor recognition [27]. Thus prior to testing the siRNA encapsulated nanosystems in cells for gene silencing activity, the relative amounts of CD44 receptor levels on the surface of different types of cells including MDA-MB468 were determined by flow cytometric analysis, as described in the materials and methods. The results are summarized in Fig. 3A. The derivatives that formed nanosized particles and demonstrated good siRNA encapsulation were taken forward for in vitro cell uptake studies. The Cy3-labeled siRNA loaded (50 nM) HA nanosystems were incubated in CD44 expressing MDA-MB468 cells for 12 h. Subsequent confocal microscopic evaluations revealed bright fluorescence signal in cytoplasm of cells due to the cy3-siRNA localization within the cells that were treated with cy3-siRNA loaded HA-PEI nanosystems (Fig. 3B). Flow cytometry of the duplicate cells showed that there was 100% uptake (data not shown). Cy3-siRNA encapsulated HA-octylamine, HA-stearylamine, HA-choline and HA-spermine also demonstrated compelling results with high cellular uptake when tested in the same cell line (data not shown). However, no detectable signal was observed with any of those derivatives in Hep3B cells that do not or minimally express CD44 receptors (data not shown). These data clearly suggests that several of the HA-derivatives were able to internalize into cells that overexpress CD44 receptors. In order to establish that the HA nanosystems was selectively internalized by receptor mediated endocytosis into CD44 overexpressing cancer cells, a competition assay (blocking control study) was then performed by pre-treating the cells with free soluble HA containing serum free medium prior to incubating the cells with Cy3-labeled siRNA/HA-PEI nanosystems to possibly block all the receptors expressed on the cell surface. Indeed, a highly diminished cellular localization/uptake (>90% inhibition) of the cy3-labeled siRNA was observed in the cells that were pre-treated with excess HA, suggesting that these particles are predominantly trafficked into the cells via receptor mediated endocytosis pathway (Fig. 3C). Flow cytometry also suggests that the uptake was reduced by >85% when the cells were pre-treated with excess HA corroborating the confocal microscopy results.
Fig. 3.
CD44 receptor levels in different type of cells and the corresponding cell uptake. NSCLC (non-small cell lung cancer), SCLC (small cell lung cancer), breast, liver and murine melanoma cells were treated with monoclonal antibody against CD44 receptors to determine the receptor levels on the surface of the cells by flow cytometry (A). To demonstrate that the particles enter the cells via the receptors, MDA-MB468 cells were treated with HA-PEI/siRNA nanosystems at 50 nM for 12 h and observed under confocal microscope to capture images. The internalized cy3-labeled-siRNA appears as red (B). For competitive inhibition study, the cells were incubated with HA-PEI/Cy3-loaded-siRNA in the presence and absence of excess free soluble HA (C) and observed under confocal microscope as shown.
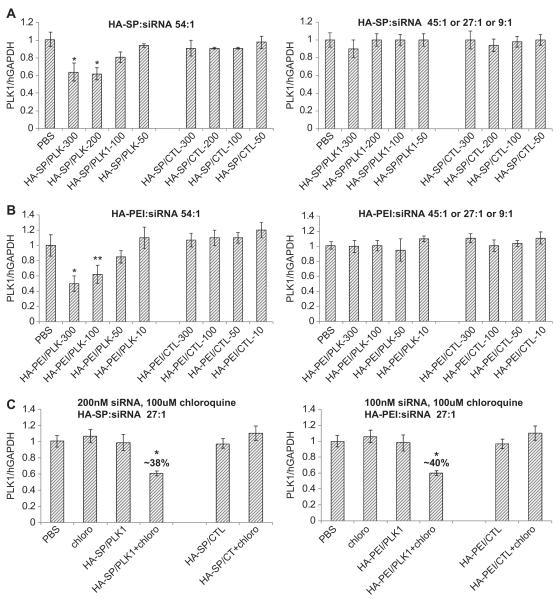
After confirming the cellular uptake and pathway of internalization, the ability of these nanosystems to deliver a functional siRNA was evaluated using PLK1 targeted siRNA to silence the PLK1 gene expression in cells overexpressing CD44 receptors. For this purpose, breast cancer cells, MDA-MB468 (with >99% CD44 expression levels) were transfected with varying HA derivative/siRNA at concentrations ranging from 50 to 300 nM. Although, all the fatty amine modified HAs derivatives demonstrated good cellular uptake, most of them were unsuccessful in down regulating PLK1 gene expression in vitro. The spermine derivatized HA demonstrated about 40% target gene knock down at 100, 200 and 300 nM while the control siRNA/HA-spermine in the same study did not demonstrate any silencing (Fig. 4A). It is interesting to note that the HA-spermine demonstrated activity only at the mass ratio of 54:1 (polymer: siRNA) and failed to demonstrate gene silencing at other ratios of 45:1, 27:1 or 9:1 (polymer: siRNA) or lower (Fig. 4A). It is also worth noting that the nanoassembly formed using 54:1 polymer-to-siRNA ratio had zeta potentials of around +16.5 mV whereas the others with ratios of 27:1 or 9:1 revealed a lower surface charge of +5–6 mV or close to neutral. In addition to HA-spermine, the PEI modified HA also demonstrated comparable gene silencing activity in the CD44 expressing MDA-MB 468 cells (Fig. 4B). In this case too the 54:1 of polymer-to-siRNA demonstrated dose dependent response and good gene silencing activity. The most interesting observation was, unlike HA-spermine/siRNA nanosystem, the HA-PEI/siRNA revealed a completely negative surface charge, and despite of this, it exhibited the best gene silencing activity in cells, indicative of a positive core that holds the intact siRNA, with a negatively charged HA shell/surface. Indeed DLS, zeta potentials measurements and transmission electron microscopy all indicated the formation of a siRNA-PEI condensed “core” (which stains dark in the presence of uranyl acetate stain) with a hydrophilic HA polymer shell (Fig. 2D).
Fig. 4.
HA-spermine and HA-PEI/siRNA mediated PLK1 gene silencing in MDA-MB 468 cells. Cells were treated with PLK1 or CTL siRNA formulated HA-spermine (A) or HA-PEI (B) at polymer-to-siRNA mass ratios of (1) 54:1 or (2) 45:1 or 27:1 or 9:1 respectively for 48 h. The PLK1 gene expression was measured by qPCR. Data represented as a mean ± SD (n = 3). * P = 0.01 compared to PBS and CTL treatment groups for A and B. ** P = 0.02 compared to PBS and CTL treatment groups for B. In order to assess if the nanosystems formulated at polymer-to-siRNA ratios other than 54:1 were trapped in the endosome without being released, the HA-spermine and HA-PEI/PLK1 siRNA nanosystems were formulated at 27:1 ratio. The PLK1 gene silencing study was carried out in the presence and absence of chloroquine in MDA-MB 468 cells for 48 h (C). The PLK1 gene expression was measured by qPCR. Data represented as a mean ± SD (n = 3). * P = 0.01 compared to PBS and all the other CTL groups.
As such, it has been demonstrated clearly that the cell entry was receptor mediated and it is independent of the charge on the surface. All the HA derivatives demonstrated cellular uptake but showed no gene down-regulation except the HA-spermine and HA-PEI derivatives at a specific ratio (Fig. 4A and B). The nanosystems that were unable to exhibit gene silencing activity in cells were most likely due to lack of entry into cells or more critically due to lack of release from endosomes or a combination of both. This issue of endosome escape was then addressed by proper selection of functional derivatives to conjugate onto the HA backbone, namely by introducing small % of polyamines (such as spermine, PLL and PEI), in which the polyamines contribute to more positive charges that can not only afford efficient encapsulation and self-assembly of the siRNA payload but also facilitated endosome release, resulting in pronounced gene silencing activity. Interestingly, the activity in cells was noticed for both HA-spermine and HA-PEI only at a particular polymer-to-siRNA ratio suggesting that a critical balance of charge is essential in order to design nanosystems that can promote endosome escape and at the same time remain as a safe carrier for in vivo delivery.
In order to confirm that the nanosystems loaded with siRNA that demonstrated cellular uptake but no gene silencing were indeed trapped in the endosome, the transfection was done in the presence of a weak base, chloroquine. As literature suggests [28,29], this small molecule helps to disrupt the endosome in addition to inhibiting endosome–lysosome fusion. Indeed, treatment of cells with HA-PEI/siRNA or HA-spermine/siRNA at 27:1 ratio with chloroquine demonstrated activity in cells (Fig. 4C) whereas the same complex without chloroquine failed to show gene silencing activity in cells (Fig. 4C). Also, the nanosystems constructed with other class of functional HA macrostructures that failed to show activity in cells in the absence of chloroquine showed activity in the presence of chloroquine (data not shown) again suggesting that these self-assembled nanosystems efficiently enter the cells but get trapped in the endosome without the ability to release the siRNA payload. As a result of our study, we were able to identify one class of HA derivatives, namely the polyamine modified functional HA macrostructures such as HA-spermine, HA-PEI and HA-PLL that showed favorable cellular entry in cells as well as endosome escape to demonstrate pronounced gene silencing activity. These HA based polymers were thus carried forward for more thorough in vitro and in vivo evaluation.
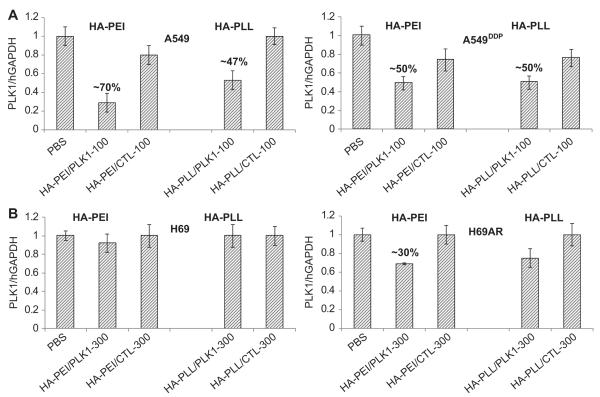
Since our ultimate goal of engineering such nanosystems was to test their utility in reversing drug resistance in lung cancers, we evaluated their activity in both resistant and sensitive NSCLC and SCLC lung cancer cells. When the PLK1/CTL siRNA encapsulated HA-PEI particles were tested (100 nM for 24 h) in A549/A549DDP cells (sensitive/resistance NSCLC pair) that express greater than 99% CD44 expression levels, the gene knockdown was close to 70% and 50% respectively (Fig. 5A). The silencing was down to 30% at 300 nM siRNA concentration in H69AR cells that express ~92% CD44 receptor levels. However, we observed no down-regulation activity in H69 cells that express only 60% receptors even at a high doses of (~300 nM) siRNA (Fig. 5B) suggesting that the CD44 receptors need to be at a certain threshold level for these particles to actively enter and mediate gene silencing. Similar trend was noticed with PLL (poly-l-lysine) modified HA nanosystems as well.
Fig. 5.
Target (PLK1) knockdown in sensitive and resistant lung cancer cells. A549/A549DDP (A, NSCLC) and H69/H69AR (B, SCLC) cells were transfected with PLK1 or SSB encapsulated HA-PEI or HA-PLL nanosystems at 100 or 300 nM concentrations. Cells were harvested and RNA was extracted after 48 h. qPCR was run to determine the target knockdown.
Finally, to translate the observed in vitro activity in vivo, we tested their potentials in animal tumor models. Our choice of tumors mainly included the lung cancer models with and without resistance to chemotherapeutic drugs. A pair of subcutaneous tumors from NSCLC indication (A549 and cisplatin resistant A549 also called A549DDP) and a pair from SCLC indication (H69 and doxorubicin resistance H69 also called H69AR) were thus used. These subcutaneous xenografts are human tumors and therefore they would grow only in immune deficient mice, an animal model with no humoral immune system [30]. More importantly, these tumors mimic the heterogeneity and complexity of the actual human tumors. However, in addition to these xenograft models, it is also useful to look at other types of relevant tumor models such as metastatic tumor models, which mimic the more realistic advanced tumors in clinical settings. As most of the patients with late stage cancers succumb to metastasis and in majority of cases, and these are the tumors being treated in the clinic, it is important to evaluate these tumor models as well in the pre-clinical settings [31]. Primary tumors are generally surgically removed at the time of treatment. Apart from these two types of models, understanding the delivery to syngeneic mouse models is also important, as these tumors are developed in normal mice with fully developed immune system to mimic what is present in human patients. As noted, these different tumor types provide different benefit, so collective information from these models will be more predictive of a future clinical outcome.
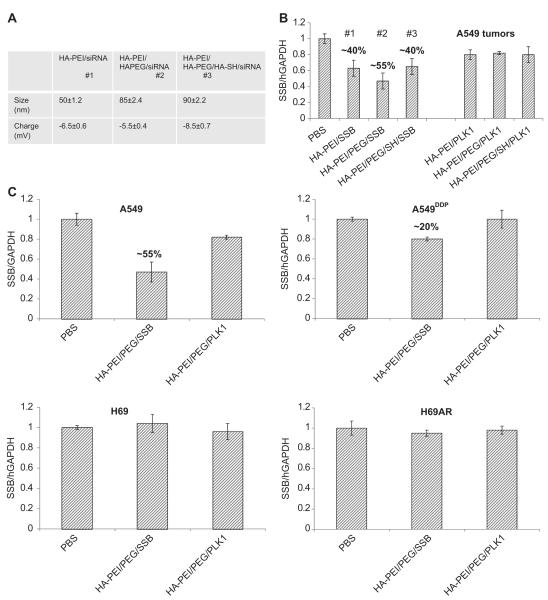
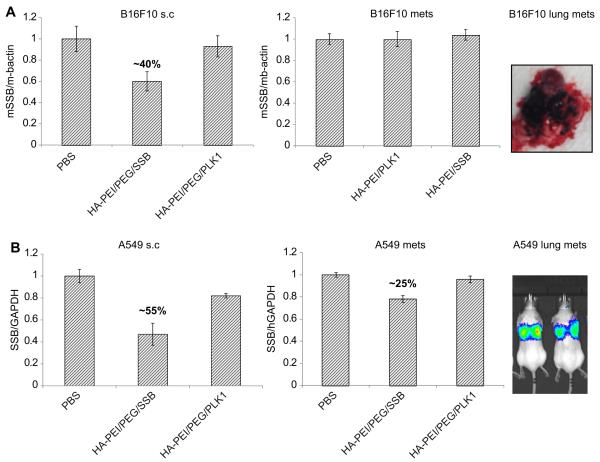
For the in vivo studies, we thought it to be relevant to introduce two additional functional HA blocks, namely a PEGlylated-HA functional block and a thiolated-HA block that can self-assemble with the siRNA condensing HA-PEI functional block, forming multifunctional nanoparticles as described [32]. Our hypothesis was that the HA-PEI block would bind with siRNA, the sulfhydryl cross-linked HA-thiol blocks apart from stabilizing the nanosystem in circulation would facilitate endosome escape and the amphiphilic HA-PEG block will enhance in vivo circulation time and facilitate passive tumor targeting based on the EPR effect as observed with several other nanosystems [32–34] (Fig. 6A). The multifunctional HA-blocks in solution when mixed together would form self-assembled nanosystems [32,35]. The nanosystems with and without all the HA-blocks were first tested in subcutaneous A549 tumors to identify the best combination for developing the ideal siRNA delivery system. The outcome of our evaluation indicated that the HA-nanosystem using HA-PEI/HA-PEG/siRNA was found to be the ideal system that could deliver the siRNA most efficiently and showed the highest target gene knockdown (55%) compared to the other groups that had either HA-PEI alone with siRNA or with all the three HA components with siRNA proposed earlier (Fig. 6B). Based on these results, the HA-PEI/HA-PEG/siRNA system was selected for further testing in other tumor models. Similarly, the mice with A549DDP, H69 and H69AR tumors were given the same doses and mRNA knockdown was examined 24 h after the last dose (Fig. 6C). There was only marginal activity detected in A549DDP tumors at the time point tested. However, no activity was seen in the other two s.c. SCLC tumors at the doses administered to mice. Given that the CD44 levels were not that high in H69/H69AR pair compared to A549 pair, it is not very surprising to see this outcome. However, as the doses used in these studies were very low, it is possible that the siRNA delivered to these tumors might also be very low. In order to understand if other factors such as vascularity also play a role in tumor delivery apart from the receptors, these particles with PLKI/SSB siRNA were also tested in other tumor types that either expresses CD44 receptors at higher or lower levels with varied levels of vascularity, to understand the correlation between gene silencing activity, CD44 expression levels and vascularity. To address this, B16F10, a murine melanoma type tumor that expresses reasonable levels of CD44 receptors (~65%) were implanted subcutaneously into nude mice. These tumors were treated the same way as described before. Reasonable gene knockdown activity of ~40% was seen in these tumors confirming the role of CD44 levels. It is also worthwhile to note that these tumors seem to be highly vascularized. In order to see if the HA nanosystems could still deliver siRNA to the same tumor type when it is present in lung as metastatic lesions, they were tested in the metastatic B16F10 model (Fig. 7A). The study results suggested that there was no demonstrable activity in these metastatic tumors. In a similar trend to B16F10 tumors, the metastatic A549-luc lung lesions were also developed by intravenous injection of the cancer cells. As this cell line express luciferase, the mice were imaged to monitor the tumor growth. Both s.c. and metastatic tumors were treated with PLK1 or SSB encapsulated HA-PEI/PEG particles at the same doses (3 doses of 0.5 mg/kg each). We observed a target knockdown of ~55% in s.c tumors whereas the metastatic lesions in lung exhibited only 25% target knockdown (Fig. 7B). The interesting observation from these studies was that the difference in activity that was noted in s.c. tumors versus the activity in metastatic tumors of the same tumor type. It was surprising to see no knockdown in the B16F10 lung lesions and lower knockdown in A549 lung lesions compared to their corresponding s.c tumors at the same doses used. One explanation would be that these metastatic tumors that are formed in the lung may not have the same architecture or tumor micro-environment compared to the ones that are developed at the subcutaneous space. Previous studies have also identified the heterogeneity within the same tumor mass as well as the distinct molecular differences between primary tumors and metastatic tumors based on meta-analysis and profiling experiments [36,37], indicating that tumor micro-environment and location can also play an important role in responding to treatment.
Fig. 6.
In vivo gene knockdown activity mediated by siRNA loaded HA-nanosystems. Size and charge characterization (A) and gene silencing activity (B) of HA-nanosystems engineered with various functional blocks in A549 tumors following 3, i.v doses of 0.5 mg/kg followed by further evaluation of HA-PEI/PEG/siRNA nanosystem in both sensitive and resistant tumors. Sensitive/resistant tumor (A549/A549DDP and H69/H69AR) bearing mice were injected with PLK1 or SSB siRNA encapsulated HA-PEI/HA-PEG nanosystem at 0.5 mg/kg for 3 days. 24 h after the last injection, tumors were harvested and RNA was extracted. qPCR was run to determine the target gene knockdown (C).
Fig. 7.
Target gene knockdown in metastatic lung models vs. subcutaneous models. B16F10 cells (A) and A549 cells (B) were subcutaneously implanted in nude mice to get s.c. tumors. These cells were also injected intravenously to generate experimental metastatic lung tumors. Mice with both types of tumors were injected with SSB/PLK1 encapsulated HA-PEI/PEG nanosystem at 0.5 mg/kg for 3 days. 24 h after the last dose, tumors were harvested and the target gene knockdown was measured.
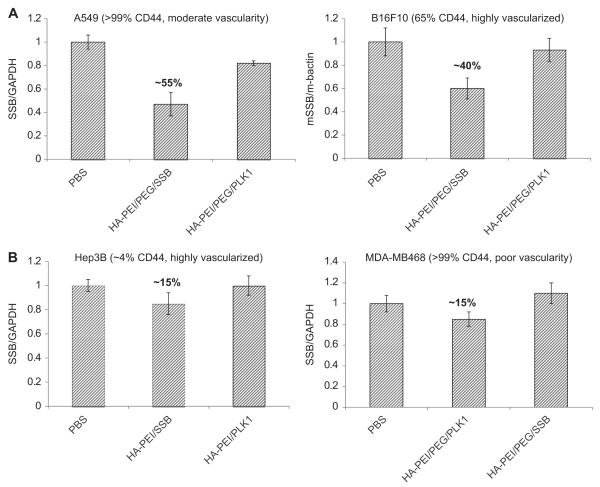
In order to further understand the correlation between the CD44 expression levels and vascularity of tumors for siRNA delivery (and gene silencing potential), we used a Hep3B tumor model in mice, a highly vascularized human hepatic tumor which has relatively very low expression of CD44 receptors (~4%). When these tumors were treated with the similar doses of HA-PEI/PEG/siRNA nanosystems, the activity was very minimal (~15%), but not completely absent. These results suggest that both vascularity and CD44 expression play an important role in tumor delivery and activity. Judge et al. reported efficient delivery of siRNA to these highly vascularized Hep3B tumors using a liposomal non-targeted delivery system named as SNLAP [38,39]. It was also reported by Yan et al. that siRNA could be delivered to few other vascularized hepatic tumors other than Hep3B using the same liposomal system mainly by the EPR effect [40]. However, these non-targeted systems failed to deliver siRNA to tumors that are not vascularized enough (un-published data), indicating that targeting may be necessary to penetrate hypovascular or solid tumors. Yet in another study, we used a hypovascular MDA-MB468 tumor model in mice that expresses very high levels of CD44 (>99%) (Fig. 8). In this case as well, we observed only 15% gene knockdown activity in the tumors at the same doses of HA-PEI/siRNA used for other tumor studies. These results substantiate that the CD44 expression level may not serve as the only factor for achieving gene silencing in tumors. In other words, receptors and favorable vascular architecture are crucial to facilitate tumor selective delivery followed by intracellular uptake and endosome escape/release to show necessary gene silencing activity. In selection of the ideal candidate tumors obviously the ones with higher levels of CD44 (that promotes receptor mediated internalization) and higher levels of vascularity that facilitate tumor accumulation based on the EPR effect will show pronounced effect [34,41].
Fig. 8.
Target gene knockdown in tumors with differential CD44 expression levels and vascularity. Different types of subcutaneous tumors (A549, B16F10, Hep3B and MDA-MB468) with varied levels of CD44 expression and varied levels of vascularity were used to test the delivery efficiency of the HA based nanosystems is presented.
4. Conclusions
In summary, in this study we have synthesized and evaluated a series of HA based functional macrostructures that can form self-assembled nanosystems encapsulating siRNA payload as discussed. Several HA based nanosystems were effective in entering tumor cells overexpressing CD44 receptors and selected candidate HA derivatives showed gene silencing activity in vitro and in vivo. A linear correlation between CD44 expression levels and activity in cells was observed, however, it was not exactly translated in vivo in solid tumor models in mice. Although the heterogeneity presented by tumors of same and different origins, their sites of localization, region of metastasis and the impact of tumor vasculature present on tumors poses serious challenge in the development of ideal delivery systems, the comprehensive screening of modular HA based self-assembling nanosystems developed in this study portend to be promising candidates and the way forward for effective treatment of sensitive and resistant tumors overexpressing CD44 receptors, including tumor initiating stem cells and metastatic lesions warranting further evaluations.
Supplementary Material
Acknowledgments
The authors wish to thank Dr. Amit Singh for assistance with TEM image acquisition. The authors also wish to gratefully acknowledge the funding support from the National Cancer Institute’s (NCI’s) Alliance for Nanotechnology in Cancer, Center for Cancer Nanotechnology Excellence (CCNE) grant U54-CA151881 and the Cancer Nanotechnology Platform Partnership (CNPP) grant U01-CA151452.
Footnotes
Appendix A. Supplementary material Supplementary material related to this article can be found online at http://dx.doi.org/10.1016/j.biomaterials.2013.01.077.
References
- [1].Saad M, Garbuzenko OB, Minko T. Co-delivery of siRNA and an anticancer drug for treatment of multidrug-resistant cancer. Nanomedicine (Lond) 2008;3:761–76. doi: 10.2217/17435889.3.6.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Tredan O, Galmarini CM, Patel K, Tannock IF. Drug resistance and the solid tumor microenvironment. J Natl Cancer Inst. 2007;99:1441–54. doi: 10.1093/jnci/djm135. [DOI] [PubMed] [Google Scholar]
- [3].Pai SI, Lin YY, Macaes B, Meneshian A, Hung CF, Wu TC. Prospects of RNA interference therapy for cancer. Gene Ther. 2006;13:464–77. doi: 10.1038/sj.gt.3302694. [DOI] [PubMed] [Google Scholar]
- [4].Aagaard L, Rossi JJ. RNAi therapeutics: principles, prospects and challenges. Adv Drug Deliv Rev. 2007;59:75–86. doi: 10.1016/j.addr.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Aigner A. Applications of RNA interference: current state and prospects for siRNA-based strategies in vivo. Appl Microbiol Biotechnol. 2007;76:9–21. doi: 10.1007/s00253-007-0984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bumcrot D, Manoharan M, Koteliansky V, Sah DW. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol. 2006;2:711–9. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Weinstein S, Peer D. RNAi nanomedicines: challenges and opportunities within the immune system. Nanotechnology. 2010;21:232001. doi: 10.1088/0957-4484/21/23/232001. [DOI] [PubMed] [Google Scholar]
- [8].Oh YK, Park TG. siRNA delivery systems for cancer treatment. Adv Drug Deliv Rev. 2009;61:850–62. doi: 10.1016/j.addr.2009.04.018. [DOI] [PubMed] [Google Scholar]
- [9].Ditto AJ, Shah PN, Yun YH. Non-viral gene delivery using nanoparticles. Expert Opin Drug Deliv. 2009;6:1149–60. doi: 10.1517/17425240903241796. [DOI] [PubMed] [Google Scholar]
- [10].Ossipov DA. Nanostructured hyaluronic acid-based materials for active delivery to cancer. Expert Opin Drug Deliv. 2010;7:681–703. doi: 10.1517/17425241003730399. [DOI] [PubMed] [Google Scholar]
- [11].Han SE, Kang H, Shim GY, Kim SJ, Choi HG, Kim J, et al. Cationic derivatives of biocompatible hyaluronic acids for delivery of siRNA and antisense oligonucleotides. J Drug Target. 2009;17:123–32. doi: 10.1080/10611860802472461. [DOI] [PubMed] [Google Scholar]
- [12].Yadav AK, Mishra P, Agrawal GP. An insight on hyaluronic acid in drug targeting and drug delivery. J Drug Target. 2008;16:91–107. doi: 10.1080/10611860802095494 . [DOI] [PubMed] [Google Scholar]
- [13].Platt VM, Szoka FC., Jr Anticancer therapeutics: targeting macromolecules and nanocarriers to hyaluronan or CD44, a hyaluronan receptor. Mol Pharm. 2008;5:474–86. doi: 10.1021/mp800024g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Rivkin I, Cohen K, Koffler J, Melikhov D, Peer D, Margalit R. Paclitaxel-clusters coated with hyaluronan as selective tumor-targeted nanovectors. Biomaterials. 2010;31:7106–14. doi: 10.1016/j.biomaterials.2010.05.067. [DOI] [PubMed] [Google Scholar]
- [15].Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci USA. 2007;104:973–8. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–51. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- [17].Coradini D, Pellizzaro C, Abolafio G, Bosco M, Scarlata I, Cantoni S, et al. Hyaluronic-acid butyric esters as promising antineoplastic agents in human lung carcinoma: a preclinical study. Invest New Drugs. 2004;22:207–17. doi: 10.1023/B:DRUG.0000026247.72656.8a. [DOI] [PubMed] [Google Scholar]
- [18].Ruponen M, Honkakoski P, Ronkko S, Pelkonen J, Tammi M, Urtti A. Extracellular and intracellular barriers in non-viral gene delivery. J Control Release. 2003;93:213–7. doi: 10.1016/j.jconrel.2003.08.004. [DOI] [PubMed] [Google Scholar]
- [19].Cullis PM, Green RE, Merson-Davies L, Travis N. Probing the mechanism of transport and compartmentalisation of polyamines in mammalian cells. Chem Biol. 1999;6:717–29. doi: 10.1016/s1074-5521(00)80019-8. [DOI] [PubMed] [Google Scholar]
- [20].Akinc A, Thomas M, Klibanov AM, Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J Gene Med. 2005;7:657–63. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- [21].Xiong XB, Uludag H, Lavasanifar A. Biodegradable amphiphilic poly(ethylene oxide)-block-polyesters with grafted polyamines as supramolecular nanocarriers for efficient siRNA delivery. Biomaterials. 2009;30:242–53. doi: 10.1016/j.biomaterials.2008.09.025. [DOI] [PubMed] [Google Scholar]
- [22].Lee H, Choi SH, Park TG. Direct visualization of hyaluronic acid polymer chain by self-assembled one-dimensional array of gold nanoparticles. Macromolecules. 2006;39:23–5. [Google Scholar]
- [23].Habeeb AF. A sensitive method for localization of disulfide containing peptides in column effluents. Anal Biochem. 1973;56:60–5. doi: 10.1016/0003-2697(73)90169-3. [DOI] [PubMed] [Google Scholar]
- [24].Shim MS, Kwon YJ. Efficient and targeted delivery of siRNA in vivo. FEBS J. 2010;277:4814–27. doi: 10.1111/j.1742-4658.2010.07904.x. [DOI] [PubMed] [Google Scholar]
- [25].Jiang G, Park K, Kim J, Kim KS, Oh EJ, Kang H, et al. Hyaluronic acid-polyethyleneimine conjugate for target specific intracellular delivery of siRNA. Biopolymers. 2008;89:635–42. doi: 10.1002/bip.20978. [DOI] [PubMed] [Google Scholar]
- [26].Huxley HE, Zubay G. Preferential staining of nucleic acid-containing structures for electron microscopy. J Biophys Biochem Cytol. 1961;11:273–96. doi: 10.1083/jcb.11.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kim TH, Nah JW, Cho MH, Park TG, Cho CS. Receptor-mediated gene delivery into antigen presenting cells using mannosylated chitosan/DNA nanoparticles. J Nanosci Nanotechnol. 2006;6:2796–803. doi: 10.1166/jnn.2006.434. [DOI] [PubMed] [Google Scholar]
- [28].Murphy EA, Waring AJ, Murphy JC, Willson RC, Longmuir KJ. Development of an effective gene delivery system: a study of complexes composed of a peptide-based amphiphilic DNA compaction agent and phospholipid. Nucleic Acids Res. 2001;29:3694–704. doi: 10.1093/nar/29.17.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Canine BF, Wang Y, Hatefi A. Evaluation of the effect of vector architecture on DNA condensation and gene transfer efficiency. J Control Release. 2008;129:117–23. doi: 10.1016/j.jconrel.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Richmond A, Su Y. Mouse xenograft models vs GEM models for human cancer therapeutics. Dis Model Mech. 2008;1:78–82. doi: 10.1242/dmm.000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Steeg PS, Theodorescu D. Metastasis: a therapeutic target for cancer. Nat Clin Pract Oncol. 2008;5:206–19. doi: 10.1038/ncponc1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Abeylath SC, Ganta S, Iyer AK, Amiji M. Combinatorial-designed multifunctional polymeric nanosystems for tumor-targeted therapeutic delivery. Acc Chem Res. 2011;44:1009–17. doi: 10.1021/ar2000106. [DOI] [PubMed] [Google Scholar]
- [33].Susa M, Iyer AK, Ryu K, Choy E, Hornicek FJ, Mankin H, et al. Inhibition of ABCB1 (MDR1) expression by an siRNA nanoparticulate delivery system to overcome drug resistance in osteosarcoma. PLoS One. 2010;5:e10764. doi: 10.1371/journal.pone.0010764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Iyer AK, Khaled G, Fang J, Maeda H. Exploiting the enhanced permeability and retention effect for tumor targeting. Drug Discov Today. 2006;11:812–8. doi: 10.1016/j.drudis.2006.07.005. [DOI] [PubMed] [Google Scholar]
- [35].Abeylath SC, Amiji MM. ‘Click’ synthesis of dextran macrostructures for combinatorial-designed self-assembled nanoparticles encapsulating diverse anticancer therapeutics. Bioorg Med Chem. 2011;19:6167–73. doi: 10.1016/j.bmc.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Dexter DL, Kowalski HM, Blazar BA, Fligiel Z, Vogel R, Heppner GH. Heterogeneity of tumor cells from a single mouse mammary tumor. Cancer Res. 1978;38:3174–81. [PubMed] [Google Scholar]
- [37].Poste G, Tzeng J, Doll J, Greig R, Rieman D, Zeidman I. Evolution of tumor cell heterogeneity during progressive growth of individual lung metastases. Proc Natl Acad Sci USA. 1982;79:6574–8. doi: 10.1073/pnas.79.21.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Judge AD, Robbins M, Tavakoli I, Levi J, Hu L, Fronda A, et al. Confirming the RNAi-mediated mechanism of action of siRNA-based cancer therapeutics in mice. J Clin Invest. 2009;119:661–73. doi: 10.1172/JCI37515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Li L, Wang R, Wilcox D, Zhao X, Song J, Lin X, et al. Tumor vasculature is a key determinant for the efficiency of nanoparticle-mediated siRNA delivery. Gene Ther. 2012;19:775–80. doi: 10.1038/gt.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lee YH, Andersen JB, Song HT, Judge AD, Seo D, Ishikawa T, et al. Definition of ubiquitination modulator COP1 as a novel therapeutic target in human hepatocellular carcinoma. Cancer Res. 2010;70:8264–9. doi: 10.1158/0008-5472.CAN-10-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Matsumura Y, Maeda H. A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 1986;46:6387–92. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.