Abstract
High interindividual pharmacokinetic variability was observed in phase 1 studies of vorinostat (suberoylanilide hydroxamic acid), an oral histone deacetylase inhibitor. Thus, we hypothesized that the variability can be explained by genetic variants of the uridine 5′-diphosphate-glucuronosyltransferases (UGTs) involved in vorinostat metabolism. Baculosomes expressing human UGTs and 52 human liver microsomes were screened for vorinostat glucuronidation activity to identify the potential enzymes and functional variants. UGT2B17 had the largest activity. Human liver microsomes with at least one copy of UGT2B17 showed significantly greater enzymatic activity than those with UGT2B17 null genotype (P <0.004). UGT2B17 plays an important role in vorinostat hepatic glucuronidation and the gene deletion polymorphism may influence vorinostat biotransformation and clearance. The clinical impact of this UGT2B17 genetic variant on vorinostat metabolism and drug effect is unknown.
Keywords: pharmacogenetics, suberoylanilide hydroxamic acid, uridine diphosphate-glucuronosyltransferases
Vorinostat (suberoylanilide hydroxamic acid, SAHA) is an oral histone deacetylase inhibitor indicated for the treatment of cutaneous T-cell lymphoma. Studies have shown promising activities of vorinostat in various types of cancer, and there are ongoing investigations in a wide range of cancers [1]. High interindividual variability in pharmacokinetics was observed during vorinostat development, with a coefficient of variation of clearance reaching up to 61% [2]. This extent of interindividual variability presents challenges to clinicians as a uniform dose of such agent can place subgroups of patients at the risk of excessive toxicities or ineffective treatment; therefore, factors contributing to the variability should be identified. However, the sources of high interindividual variability in vorinostat pharmacokinetics are left unidentified so far.
Vorinostat is extensively metabolized by the liver, primarily glucuronidated to an inactive metabolite vorinostat glucuronide (SAHA-G) [2]. Recently, Balliet et al. [3] showed that several uridine 5′-diphosphate-glucuronosyltransferases (UGTs) contribute to vorinostat glucuronidation, and suggested that the UGT2B17 copy number variant is of potential importance. This paper is the second report characterizing UGT enzymes that are significant in vorinostat glucuronidation and identifying genetic variants in UGTs that would result in altered metabolism of vorinostat.
Human liver microsomes (HLMs) and DNA from 52 donors were processed in Dr Mary Relling’s laboratory at St Jude Children’s Research Hospital (Memphis, Tennessee, USA). The liver tissue was obtained from the Liver Tissue Procurement and Distribution System (funded by #NO1-DK-9-2310) and by the Cooperative Human Tissue Network.
Screening of UGT isoforms and HLMs for vorinostat glucuronidation activity was carried out with microsomes expressing specific human UGTs and pooled HLMs (positive control) purchased from BD Biosciences (San Jose, California, USA). Incubation mixtures contained 5 mmol/l of uridine diphosphate glucuronic acid, 5 mmol/l of MgCl2, 5 mmol/l of saccharolactone, 10 μg of alamethicin (5 μg/mg of protein), 50mmol/l of Tris–HCl (pH 7.1), 0.5 mg/ml of all UGT1A and UGT2B isoforms, and pooled HLMs in a final volume of 200 μl. A vorinostat concentration of 2.4 μmol/l was selected to reflect serum concentrations seen in clinical trials with the recommended dose of vorinostat [2]. Reactions were stopped after 60 min with 200 μl of cold acetonitrile with internal standard (triamcinolone 1 μg/ml), and all samples were centrifuged at 20 817 relative centrifugal force for 10 min at 4°C. Supernatants were dried under nitrogen gas at 37°C, before they were reconstituted in 200 μl of mobile phase. Assay mixtures, after incubation, were analyzed using a liquid chromatography tandem mass spectrometry method adopted from a published study [4]. Vorinostat, SAHA-G (multiple reaction monitoring transition pair 265.2/232.3 and 441.2/232.3, respectively), and the internal standard, triamcinolone (multiple reaction monitoring transition pair 395.3/357.2) were monitored. Retention times for vorinostat, SAHA-G, and triamcinolone were 17, 18, and 15 min, respectively.
Enzyme kinetic parameters were determined by kinetic analyses of pooled HLMs and individual UGTs as described above using a range of vorinostat concentrations (0.05–1000 μmol/l). The vorinostat glucuronidation rates were characterized in 52 normal HLMs using the same assay conditions and liquid chromatography tandem mass spectrometry method described above.
HLMs were screened for their activities against probe substrates for UGT2B17 (testosterone), UGT2B7 (morphine), and UGT1A9 (mycophenolic acid). Testosterone glucuronidation was determined in 47 HLMs as follows: incubation (90 min at 37°C) mixtures containing 5mmol/l of uridine diphosphate glucuronic acid, 1mmol/l of MgCl2, 100mmol/l of Tris–HCl (pH 7.1), 0.1 mg of HLMs, and 100 μmol/l of testosterone in a final volume of 200 μl. Samples were analyzed by reversed-phase high-performance liquid chromatography (250 nm) with a mobile phase consisting of acetonitrile and NH4H2PO4, pH 4.5. HLMs were screened for morphine (n=44) and mycophenolic acid (n=41) glucuronidation as described earlier [5,6].
UGT mRNA levels were measured by two-step real-time PCR using IQSYBR Green Supermix (Bio-Rad Laboratories, Hercules, California, USA) on the Mx3000P system (Stratagene, Cedar Creek, Texas, USA). TATA binding protein was used as the control gene. Two-color TaqMan Assays (Gene Expression Assay, ID: hs00854486_sH for UGT2B17 and Copy Number Reference Assay RNase P; Applied Biosystems, Foster City, California, USA) with 6-carboxy-fluorescein-labeled probe for the gene of interest and a VIC-labeled probe for the endogenous control were used to determine copy number variations in 52 genomic DNA samples.
The screening of vorinostat glucuronidation activity in commercially available microsomes expressing specific UGT isoforms identified three enzymes with the highest vorinostat glucuronidation rates at 2.4 μmol/l (mean± standard error in pmol/min/mg, UGT2B17: 352±1.1, UGT2B7: 405±5, UGT1A9: 504±0.5). UGT1A9, UGT2B7, and UGT2B17 showed at least six-fold higher activities compared with those of UGT1A1, UGT1A3, UGT1A7, UGT1A8, and UGT2B15. UGT1A4, UGT1A6, UGT1A10, and UGT2B4 showed no detectable activity.
The SAHA-G formation rates as a function of substrate concentration were measured in pooled HLMs and supersomes expressing human UGT2B17, UGT2B7, and UGT1A9. The vorinostat glucuronidation of pooled HLMs showed biphasic kinetics consistent with multiple enzyme involvement observed in supersome screening. The catalytic efficiencies (Vmax/Km) of UGT1A9, UGT2B7, and UGT2B17 were 0.07, 0.58, and 0.09 ml/min/mg respectively.
Phenotype screening of 52 HLMs from unrelated individuals showed that the mean rate±standard deviation of vorinostat glucuronidation was 912±540 pmol/min/mg. The vorinostat glucuronidation rate was correlated with the formation rates of testosterone glucuronide (r=0.68, P=0.0001, n=47) and morphine-6-glucuronide (r=0.38, P=0.01, n=44). There was no significant correlation between SAHA-G and mycophenolic acid glucuronide formation rates in our sample set (r=0.29, P=0.59, n=41). A multivariate analysis using activities of UGT2B17, UGT2B7, and UGT1A9 as covariates showed that a significant portion (53%) of interindividual variability in vorinostat glucuronidation can be attributed to enzymatic activities of both UGT2B17 and UGT2B7 (r2=0.53).
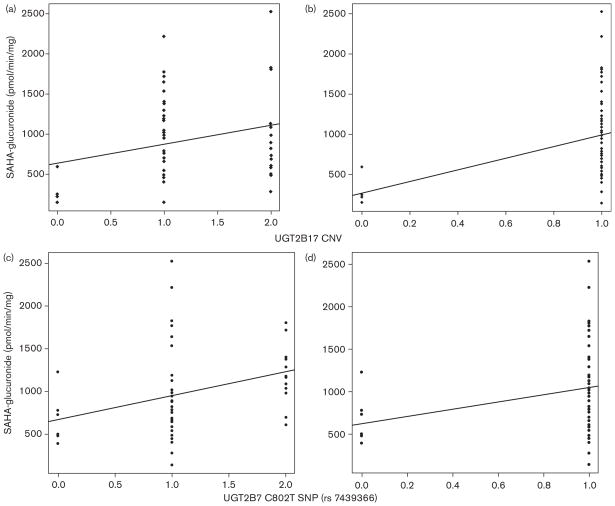
There was a statistically significant correlation between UGT2B17 copy number and SAHA-G formation rates (dominant model; r=0.4, P=0.004). HLMs with UGT2B17 null genotype (0 copy number of UGT2B17, n=5) had significantly decreased activities compared with HLMs with one or two UGT2B17 alleles (n=30 and n=16, respectively). There was no significant difference between groups with one or two UGT2B17 alleles (additive model; r=0.26, P=0.06). The UGT2B7 C802T single nucleotide polymorphism (rs 7439366) was associated, to a lesser extent than UGT2B17 copy number variation, with SAHA-G formation rates (additive: r=0.35, P=0.02; dominant r=0.3, P=0.04). Figure 1 in the subset of our cohort with available mRNA data, only UGT2B17 mRNA levels statistically significantly correlated with vorinostat glucuronidation rates (r=0.37, P= 0.02, n=44). UGT1A9 genotype (*1b,rs3832043) did not show a significant association with SAHA-G formation rates. Multivariate analysis was conducted to examine the effect of the combination of the variables (UGT2B17 and 2B7 genotypes). Addition of UGT2B7 genotype (rs 7439366) did not improve the correlation coefficient of UGT2B17 and SAHA-G formation (r=0.34).
Fig. 1.
Linear regression analysis of genotype versus suberoylanilide hydroxamic acid-glucuronide (SAHA-G). UGT2B17 copy number variation (CNV) and SAHA-G formation rates: (a) additive model, r=0.26, P=0.062. (b) dominant model (null genotype vs. at least one copy) r=0.4, P=0.0036. UGT2B7 C802T single nucleotide polymorphism (SNP) (rs 7439366) and SAHA-G formation rates, (c) additive model, C/C=0; C/T =1; T/T=2 r=0.35, P=0.0156. (d) dominant model, C/C=0; C/T or T/T=1, r=0.30, P=0.0353.
This paper is the second report illustrating a significant contribution of UGT2B17 and its polymorphism to the interindividual variability of vorinostat glucuronidation [3]. The findings of this study offer new insights to vorinostat metabolism while corroborating recent findings by Balliet et al. [3]. In our study, UGT2B17 enzyme exhibited considerable affinity and catalytic efficiency, which support the existing data [2,3]. However, the significance of UGT2B7 and insignificant activities of UGT1A8 and UGT1A10 in vorinostat metabolism were inconsistent with the data published by Balliet et al. [3]. Different assay environments and different substrate concentrations used may explain the difference between this study and the earlier published study. In addition, different sources of microsomes (i.e. insect versus human embryonic kidney cell lines) may have contributed to the inconsistencies between the two studies. Although these systems have distinct strengths and limitations, there is no evidence that one is superior to the other. Rather, they both remain valuable and widely used systems supplementing HLMs in studying glucuronidation [3,7]. As most UGT substrates are not entirely specific for any given UGT isoenzyme, a significant role of UGT2B7, in addition to, UGT2B17, is a plausible hypothesis.
Associations between genetic variants of drug metabolizing enzymes and pharmacokinetics of several anticancer agents have been well described [8]. Statistically significant correlations between genotypes of UGTs (UGT2B17 and UGT2B7) and glucuronidation of vorinostat suggest that a portion of interindividual variability in vorinostat pharmacokinetics may be attributed to polymorphisms in UGTs. This study confirms that the UGT2B17 null genotype may result in considerably reduced vorinostat glucuronidation in HLMs [3]. In contrast, this is the first report showing a correlation between a UGT2B7 polymorphism (rs 7439366) and SAHA-G formation rates in HLMs. Given the relatively weak association, we would have liked to perform kinetic studies of HLMs from each genotype group. However, such studies were not conducted because of the limited amount of available microsomes, and it is one of the limitations of this study. The role of UGT2B7 and its polymorphisms in vorinostat metabolism should be interrogated further with follow-up studies.
Despite the limitations, our results combined with existing data provide a basis for investigating the clinical implications of UGT2B17 null genotype. Once validated, genotype screening before administration may help to identify patients who are predisposed to unusually high drug exposure and excessive toxicities. Studies showed that UGT2B17 null genotype is present in 11% and 12% of Caucasians and African–Americans, respectively, whereas it can reach up to 92% in Asian populations [9,10]. Such high variability between ethnicities in the frequency of UGT2B17 null genotype may add to the significance of this genotype as we are striving to individualize cancer treatments.
Acknowledgments
This study was supported by the Pharmacogenetics of Anticancer Agents Research Group (http://pharmacogenetics.org) (NIH/NIGMS grant U01GM61393). Data will be made available to PharmGKB (supported by NIH/NIGMS U01GM61374, http://pharmgkb.org/). S.P.K. was supported by Clinical Therapeutics T32 GM007019 from NIH.
References
- 1.Richon VM, Garcia-Vargas J, Hardwick JS. Development of vorinostat: current applications and future perspectives for cancer therapy. Cancer Lett. 2009;280:201–210. doi: 10.1016/j.canlet.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 2.FDA. Zolinza, Clinical pharmacology and pharmaceutics review. 2006:2006. http://www.accessdata.fda.gov/scripts/cder/drugsatfda/index.cfm?fuseaction=Search.Label_ApprovalHistory#apphist.
- 3.Balliet RM, Chen G, Gallagher CJ, Dellinger RW, Sun D, Lazarus P. Characterization of UGTs active against SAHA and association between SAHA glucuronidation activity phenotype with UGT genotype. Cancer Res. 2009;69:2981–2989. doi: 10.1158/0008-5472.CAN-08-4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel K, Guichard SM, Jodrell DI. Simultaneous determination of decitabine and vorinostat (suberoylanalide hydroxamic acid, SAHA) by liquid chromatography tandem mass spectrometry for clinical studies. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;863:19–25. doi: 10.1016/j.jchromb.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 5.Ramirez J, Liu W, Mirkov S, Desai AA, Chen P, Das S, et al. Lack of association between common polymorphisms in UGT1A9 and gene expression and activity. Drug Metab Dispos. 2007;35:2149–2153. doi: 10.1124/dmd.107.015446. [DOI] [PubMed] [Google Scholar]
- 6.Innocenti F, Iyer L, Ramirez J, Green MD, Ratain MJ. Epirubicin glucuronidation is catalyzed by human UDP-glucuronosyltransferase 2B7. Drug Metab Dispos. 2001;29:686–692. [PubMed] [Google Scholar]
- 7.Hutzler JM, Hauer MJ, Tracy TS. Dapsone activation of CYP2C9-mediated metabolism: evidence for activation of multiple substrates and a two-site model. Drug Metab Dispos. 2001;29:1029–1034. [PubMed] [Google Scholar]
- 8.Huang RS, Ratain MJ. Pharmacogenetics and pharmacogenomics of anticancer agents. CA Cancer J Clin. 2009;59:42–55. doi: 10.3322/caac.20002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park J, Chen L, Ratnashinge L, Sellers TA, Tanner JP, Lee JH, et al. Deletion polymorphism of UDP-glucuronosyltransferase 2B17 and risk of prostate cancer in African American and Caucasian men. Cancer Epidemiol Biomarkers Prev. 2006;15:1473–1478. doi: 10.1158/1055-9965.EPI-06-0141. [DOI] [PubMed] [Google Scholar]
- 10.Xue Y, Sun D, Daly A, Yang F, Zhou X, Zhao M, et al. Adaptive evolution of UGT2B17 copy-number variation. Am J Hum Genet. 2008;83:337–346. doi: 10.1016/j.ajhg.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]