Abstract
Background
The conversion of prothrombin to thrombin is one of two non-duplicated enzymatic reactions during coagulation. Thrombin has long been considered an optimal anticoagulant target because it plays a crucial role in fibrin clot formation by catalyzing the cleavage of fibrinogen, upstream coagulation cofactors, and platelet receptors. Although a number of anti-thrombin therapeutics exist, it is challenging to use them clinically due to their propensity to induce bleeding. Previously, we isolated a modified RNA aptamer (R9D-14) that binds prothrombin with high affinity and is a potent anticoagulant in vitro.
Objectives
We sought to explore the structure of R9D-14 and elucidate its anticoagulant mechanism(s). In addition to designing an optimized aptamer (RNAR9D-14T), we also explored whether complementary antidote oligonucleotides can rapidly modulate the optimized aptamer’s anticoagulant activity.
Methods and Results
RNAR9D-14T binds prothrombin and thrombin pro/exosite I with high affinity and inhibits both thrombin generation and thrombin exosite I-mediated activity (i.e., fibrin clot formation, feedback activity, and platelet activation). RNAR9D-14T significantly prolongs the aPTT, PT, and TCT clotting assays, and is a more potent inhibitor than the thrombin exosite I DNA aptamer ARC-183. Moreover, a complementary oligonucleotide antidote can rapidly (<2 min) and durably (>2 hrs) reverse RNAR9D-14T anticoagulation in vitro.
Conclusions
Powerful anticoagulation, in conjunction with antidote reversibility suggests that RNAR9D-14T may be ideal for clinical anticoagulation in settings that require rapid and robust anticoagulation, such as cardiopulmonary bypass, deep vein thrombosis, stroke, or percutaneous coronary intervention.
Keywords: anticoagulant, antidote, aptamer, exosite I, prothrombin, thrombin
Introduction
Coagulation involves a series of enzymatic reactions largely localized on vascular and cellular surfaces that result in fibrin clot formation [1]. Thrombin, the final enzyme formed during the common pathway of coagulation, establishes a positive feedback loop that generates additional thrombin by activating factors V (FV) and VIII (FVIII), as well as by cleaving platelet protease activated receptors (PARs) to activate platelets [2]. Moreover, thrombin cleaves fibrinogen and generates fibrin monomers that form the clot meshwork [3]. Inappropriate or excessive thrombin generation can lead to thrombosis, which is currently the leading cause of morbidity and mortality in the western world [4].
Thrombin’s structure possesses multiple regions that are involved in substrate interactions: the catalytic active site and two extended surfaces, termed exosites, that participate in macromolecular ligand binding [5]. Exosite I has been implicated in binding fibrinogen, FV, FVIII, thrombomodulin (TM), and platelet PARs (for a review see [6]). Recent studies have shown that exosite I exists on prothrombin in a precursor state termed proexosite I, which presumably is involved in FVa/prothrombin interactions within the prothrombinase complex [7–9]. In contrast, exosite II is not structurally mature on prothrombin and is only exposed upon proteolysis and conversion to thrombin; exosite II has been implicated in binding FV, FVIII, platelet GPIbα, and heparin (for a review see [6]).
Aptamers are single-stranded oligonucleotides that form three-dimensional structures that allow them to bind to several types of molecules, including proteins. By binding to their target protein with high affinity and specificity, aptamer anticoagulants can bury a large surface area and directly inhibit protein-protein interactions that are essential for coagulation [10, 11]. Aptamer anticoagulants have been generated against several clotting factors (e.g., factor VIIa, IXa, Xa, and thrombin) that demonstrate high affinity binding for their specific target [11–15]. Importantly, aptamer activity can be modulated with either a matched oligonucleotide antidote or universal antidote and thus represent controllable and potentially safer anticoagulants [13, 16, 17].
A number of aptamers have been previously generated against thrombin, although none have advanced clinically. ARC-183 (HD-1), a DNA aptamer isolated by Bock et al. that binds to pro/thrombin pro/exosite I, demonstrated potential as an anticoagulant in animal cardiopulmonary bypass (CPB) models [15, 18]. The aptamer was originally designed to have a short half-life and rapid clearance in vivo, thus negating the need for antidote control. However, rapid clearance of the therapeutic required continual administration, and as a result ARC-183 clinical trials were terminated due to suboptimal dosing profiles in humans. HD-22 (a DNA aptamer) and Tog-25t (a modified RNA aptamer) both bind to thrombin exosite II and inhibit thrombin-mediated activation of platelets, but only have a nominal effect on fibrinogen cleavage [19, 20]; thus, neither exosite II aptamer has advanced as a clinical anticoagulant.
We sought to develop a high affinity, nuclease resistant and antidote-controllable anticoagulant aptamer that would limit thrombin generation and inhibit thrombin activity. Conversion of prothrombin to thrombin represents an attractive therapeutic target because it is one of two non-redundant reactions within coagulation. By utilizing systematic evolution of ligands by exponential enrichment (SELEX), we previously generated a modified RNA aptamer (R9D-14) that binds to prothrombin and is a powerful anticoagulant [14]. In this study, we examined the structure and function of this compound and characterized the anticoagulant mechanism(s) of an optimized version of this aptamer (RNAR9D-14T). Additionally, we generated an oligonucleotide antidote than can rapidly and stably reverse aptamer-mediated anticoagulation in vitro.
Experimental Procedures
Materials
Human normal and prothrombin deficient citrated plasmas were purchased from George King Biomedical, Inc. (Overland Park, Kansas). The citrated animal plasma was purchased from Lampire Biological Laboratories, Inc. (Pipersville, PA). All of the purified coagulation proteins (i.e., FV, FVIIa, FIXa, FXa, Protein C, Protein S, Protein Z, and prothrombin, as well as α, β, and γ-thrombin) were purchased from Haematologic Technologies, Inc. (Essex Junction, VT). Recombinant hirudin was purchased from Aniara (Manson, OH), unfractionated sodium heparin was purchased from APP Pharmaceuticals, LLC (Schaumburg, IL), and thrombin chromogenic substrate Pefachrome TH 8198 was purchased from Centerchem, Inc. (Norwalk, CT). Bovine serum albumin (BSA) was purchased from Calbiochem (Darmstadt, Germany), PEG-8000 was purchased from Fluka Biochemika (Buchs, Switzerland), and phenol chloroform isoamyl (25:24:1) was purchased from Invitrogen (Carlsbad, CA).
DNA/RNA Aptamers and DNA Antidote Oligonucleotides
The DNA aptamer ARC-183 and modified RNA aptamer Tog-25t were synthesized as previously described [21]. The RNAR9D-14T aptamer sequence is 5'-GG(2'F-C)GG(2'F-U)(2'F-C)GA(2'F-U)(2'F-C)A(2'F-C)A(2'F-C)AG(2'F-U)(2'F-U)(2'F-C)AAA(2'F-C)G(2'F-U)AA(2'F-U)AAG(2'F-C)(2'F-C)AA(2'F-U)G(2'F-U)A(2'F-C)GAGG(2'F-C)AGA(2'F-C)GA(2'F-C)(2'F-U)(2'F-C)G(2'F-C)(2'F-C)-3', and RNAmut is 5'-GG(2'F-C)GG(2'F-U)(2'F-C)GA(2'F-U)(2'F-C)A(2'F-C)A(2'F-C)AG(2'F-U)(2'F-U)(2'F-C)AAA(2'F-C)G(2'F-U)AA(2'F-U)AAG(2'F-C)(2'F-C)GG(2'F-C)G(2'F-U)A(2'F-C)GAGG(2'F-C)AGA(2'F-C)GA(2'F-C)(2'F-U)(2'F-C)G(2'F-C)(2'F-C)-3', where (2’F-C) denotes a 2’Fluorocytosine and (2’F-U) denotes a 2’Fluorouracil. The aptamers were transcribed in vitro and purified as previously described [14]. Biotinylated ARC-183 (5’-Biotin-GGTTGGTGTGGTTGG) and ARC-183mut (5’-Biotin-GGTGGTGGTTGTGGT) were purchased from Integrated DNA Technologies, Inc. (Coralville, IA). The 5’- biotinylated RNAR9D-14T and RNAmut aptamers were generated via in vitro transcription with the addition of a 4-fold molar excess of 5’-Biotin-G-Monophosphate (TriLink Biotechnologies, San Diego, CA) [14]. Aptamer preparations were renatured prior to use by dilution in Hepes-saline buffer (20 mM Hepes, pH 7.4, 150 mM NaCl, and 2 mM CaCl2), unless otherwise indicated, and incubation at 65°C for 5 minutes, followed by cooling to ambient temperature.
The DNA antidotes (AO1: 5’-GAACTGTGTGATCGA-3’; AO2: 5’-CGTTTGAACTGTGTG-3’; AO3: 5’-TATTACGTTTGAACT-3’; AO4: 5’-TGGCTTATTACGTTT-3’; AO5: 5’-CCTCGTACATTGGCTTATTA-3’; AO6: 5’-GTCTGCCTCGTACATTGGCT-3’; and scrAO 5’-TCTAAGCGATGGCTCAAGAC-3’) were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA).
Binding studies - nitrocellulose filter binding and surface plasmon resonance
Apparent binding affinity constants (Kd) were determined with a double-filter nitrocellulose assay as previously described [14]. Aptamers were 32P 5’-end radiolabeled and incubated with protein diluted in Hepes-saline buffer with 0.01% BSA at 37°C for 5 minutes. The bound and unbound aptamer was separated by passing the mixture over a nitrocellulose filter (Protran BA 85, Whatman Inc., Florham Park, NJ), and the fraction of bound aptamer was quantified with a Storm 825 phosphoimager (GE Healthcare, Piscataway, NY). Nonspecific aptamer binding was subtracted [22], and the data were fit with a non-linear regression analysis for one site binding with the Prism software (GraphPad Software, Inc., La Jolla, CA) to calculate the apparent Kd. For the thrombin competition bindings, the radiolabeled aptamer was pre-bound to human α-thrombin, various concentrations of unlabeled exosite I or II ligands were added, and the assay was incubated for an additional 5 min at 37°C.
Surface plasmon resonance was performed with a BIAcore 3000 instrument, and data analysis was performed with the BIAevaluation 4.1 software (BIAcore Inc, Piscattaway, NJ). Biotinylated aptamers were refolded in Hepes-saline buffer, then immobilized on a streptavidin chip (Biacore) at a flow rate of 5 µL/min at 25°C for 1–2 min to a final immobilization level of 50–150 RU. Prothrombin (0.5–20 µg/mL) was diluted in degassed Hepes-saline buffer and injected over the surface at a flow rate of 30 µL/min at 25°C for 5 minutes. The dissociation responses were monitored for 10 min, and the flow cells were regenerated with a pulse of 0.05% SDS. Non-specific binding responses of prothrombin to the mutant controls (RNAmut or ARC-183mut) were subtracted from the aptamer sensorgrams. The specific binding curves were fitted globally to a 1:1 (Langmuir) binding model to obtain the association and dissociation rate constants (ka and kd respectively), as well as the binding affinity (KD).
Plasma clotting assays (aPTT, PT, TCT)
Activated partial thromboplastin time (aPTT), prothrombin time (PT), and thrombin clot time (TCT) assays were performed on a model ST4 mechanical coagulometer (Diagnostica Stago, Parsnippany, NJ) in Hepes-saline buffer as previously described [21]. For the aPTT, citrated normal human or animal platelet poor plasma (PPP) (50 µL) was incubated with TriniClot aPTT S (Trinity BioTech, Bray, Co Wicklow, Ireland) (50 µL) at 37°C for 5 min. Aptamer (5 µL) was added, the mixture was incubated at 37°C for another 5 min, and CaCl2 (50 µL) was added to initiate the assay. The aPTT DNA antidote reversal assays were run as described, except that antidote (5 µL) was added to the activated, anticoagulated plasma and incubated for various time points (2 min - 2 hrs) at 37°C, which was followed by the addition of CaCl2 and assay initiation [13]. For the PT, human PPP (50 µL) was incubated with aptamer (5 µL) at 37°C for 5 min. TriniClot PT Excel reagent (Trinity BioTech, Bray, Co Wicklow, Ireland) (100 µL) was added to initiate the reaction. For the TCT assay, aptamer (5 µL) was added to purified human α-thrombin (10 nM final, 100 µL) and incubated at 37°C for 200 sec. Normal human PPP or prothrombin deficient PPP (50 µL) was added to the prebound thrombin/aptamer solution (105 µL) to initiate the reaction.
Thrombin small peptide substrate and antithrombin kinetics
The small peptide assays were performed at room temperature in 96-well flat bottom microtiter plates (Corning Incorporated, Corning, NY) in Hepes-saline buffer. Aptamer (300 nM) was incubated with human α-thrombin (10 nM) for 5 min at 37°C, and thrombin chromogenic substrate Pefachrome TH 8198 (0–0.5 mM) was added to a total volume of 100 µL. Substrate cleavage was measured at an absorbance of 405 nm with a Power Wave XS2 kinetic microplate spectrophotometer (BioTek, Winooski, VT). The Km and Kcat were determined by a nonlinear regression analysis with the Prism software according to Michaelis-Menton kinetics (GraphPad Software, Inc., La Jolla, CA).
The antithrombin kinetic (AT) assays were performed as previously described [23] in Hepes-saline buffer with 0.01% BSA and 1 mg/mL PEG 8000. Briefly, aptamer was incubated with thrombin (3 nM final) at ambient temperature for 5 min. Antithrombin (500 nM final) was added (450 µL total), and samples (50 µL) were withdrawn at various time points (0–60 min). Pefachrome TH 8198 (50 µL) was added, and substrate cleavage was measured at 405 nm on a kinetic microplate spectrophotometer at room temperature (BioTek, Winooski, VT). The second order rate constant of inhibition (k2) was calculated as previously described [24].
Thrombin-mediated FV activation
For SDS-PAGE and silver stain analysis, aptamer (5 µM) was incubated with human α-thrombin (0.5 nM) for 5 min at 37°C in Hepes-saline buffer. Human factor V (300 nM) was added (40 µL total), and samples (5 µL) were removed at various time points (0–30 min) and subjected to SDS-PAGE under reducing conditions on a pre-poured 4–15% polyacrylamide gradient gel (Biorad, Hercules, CA). The gels were silver stained with a Bio-Rad Silver Stain kit (Biorad) according to the manufacturer’s instructions.
To assess FVa activity, various concentrations of aptamer (0–500 nM, 5 µL) were diluted in Hepes-saline buffer with 0.01% BSA and incubated with human α-thrombin (3 nM, 5 µL) for 5 min at 37°C. Human factor V (250 ρM, 5 µL) was added, and samples (8 µL) were removed at various time points (0–7 min) and immediately diluted into a prothrombinase mixture of 5 nM human FXa, 40 µM PC/PS/PE lipid vesicles, and 500 µM Pefachrome TH 8198 (42 µL). Large, phosphatidylcholine (PC), phosphatidylserine (PS), and phosphatidylethanolamine (PE) (Avanti Polar Lipids, Alabaster, Alabama) unilamellar vesicles were prepared as previously described [25]. Human prothrombin (500 nM, 50 µL) was added to initiate the reaction, and substrate cleavage was measured at 405 nm on a kinetic microplate spectrophotometer at 37°C (BioTek, Winooski, VT). The data were fit during the linear portion of the reaction to obtain the rate of chromogenic substrate cleavage (ΔA405/time). A control assay was performed in parallel where FV was fully activated with thrombin in the absence of RNA and diluted into prothrombinase containing aptamer to quantify aptamer inhibition of prothrombin cleavage. The experimental rates were normalized to the respective control rates to correct for aptamer inhibition of prothrombin cleavage, and the data are presented as a percentage of FVa cofactor activity of fully activated FV. The lines were arbitrarily drawn with the Prism software (GraphPad Software, Inc., La Jolla, CA).
Thrombin-mediated platelet activation
Platelets were purified from freshly drawn blood from healthy volunteers under a Duke University Institutional Review Board approved protocol. Aptamer inhibition of thrombin-mediated platelet activation was assessed as previously described [21, 26]. Briefly, aptamer (0.39 – 400 nM final, 5 µL) was diluted in Hepes-saline buffer with 0.01% BSA and incubated with α-thrombin (1 nM final, 20 µL) at 37°C for 5 min. Purified platelets (25 µL) were added, and the reaction was incubated at 37°C for 15 min. The platelets were stained with 75 ng of a fluorescent anti-CD62P (P-selectin) or isotype control antibody (eBioscience, San Diego, CA) (50 µL) at room temperature for 30 minutes, then washed and analyzed with a FACScalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ). The data were analyzed with the Cell Quest analysis software, version 3.3 (Becton Dickinson).
Thrombin generation
Thrombin generation in a purified system was measured as previously described [11]. For complete prothrombinase experiments, aptamer (0.3–5 µM, 5 µL) diluted in reaction buffer (Hepes-saline buffer with 0.01% BSA and 1 mg/mL PEG 8000) was incubated with 1.4 µM prothrombin (5 µL) at 37°C for 5 min. FXa (0.2 nM), FVa (30 nM), and PC/PS/PE lipids (40 µM) (10 µL total) were incubated at 37°C for 5 min to allow for complex assembly, and prebound prothrombin/aptamer was added to initiate the reaction. Aliquots (1 µL) were withdrawn at various time points (0–10 min), diluted in quench buffer (20 mM Hepes pH 7.4, 150 mM NaCl, 50 mM EDTA, and 1 mg/mL PEG 8000; 49 µL), and thrombin chromogenic substrate Pefachrome TH 8198 (50 µL) was added at a final concentration of 100 µM. Substrate cleavage was measured at 405 nm on a kinetic microplate spectrophotometer at ambient temperature (BioTek, Winooski, VT). The initial rates of thrombin substrate cleavage were converted to thrombin concentration based upon the specific activity of thrombin Pefachrome TH8198 cleavage, as was determined in a separate assay with known concentrations of purified commercial α-thrombin. The data were fit during the linear portion of the reaction to obtain the thrombin generation rate (µM/min). The experimental rates were normalized to the rate for a mock reaction (no aptamer). Partial prothrombinase experiments contained 100 nM FXa and 40 µM PC/PS/PE lipids (no FVa), or 2 nM FXa and 30 nM FVa (no lipids).
Serum stability
The aptamer was 32P 5’-end radiolabeled as previously described (5,000 cpm/µL, 15 µL) [14] and incubated with human serum (35 µL) at 37°C. Time points (0–24 hr, 3 µL) were diluted in Tris-EDTA buffer (10 mM Tris, pH 7.5 and 0.1 mM EDTA; 40 µL), extracted with phenol chloroform isoamyl (50 µL), heated at 65°C for 5 minutes, run on a 12% denaturing acrylamide gel, and quantified by phosphoimager analysis (Storm 825 phosphoimager, GE Healthcare, Piscataway, NY).
Antidote gel mobility shift
32P 5’-end radiolabeled aptamer (5,000 cpm/µL; 6 µL) was mixed with unlabeled aptamer (125 nM), diluted in Hepes-saline buffer with 0.01% BSA, and incubated with various concentrations of DNA antidote (62.5–500 nM) at 37°C for 5 minutes. Aptamer/antidote control duplex formation was ensured by heating the aptamer/antidote mixture to 95°C and slow cooling to ambient temperature. Sample aliquots (6 µL) were subjected to native-PAGE on an 8% polyacrylamide gel at 4°C and quantified by phosphoimager analysis.
Statistical analysis
The data were statistically analyzed with a two-tailed, paired t-test with the Prism software (GraphPad Software, Inc., La Jolla, CA). P-values less than 0.05 were considered statistically significant.
Thrombin peptide alignment and sequence analysis
Thrombin beta-chain peptide sequences were downloaded from the NCBI protein HomoloGene database (http://www.ncbi.nlm.nih.gov) and aligned with the MacVector 7.2 software (MacVector, Inc., Cary NC). The percent sequence similarity (percent homology) to the human sequence was calculated with the Clustal W2 software (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
Results
Delineating the essential RNA sequence within aptamer R9D-14
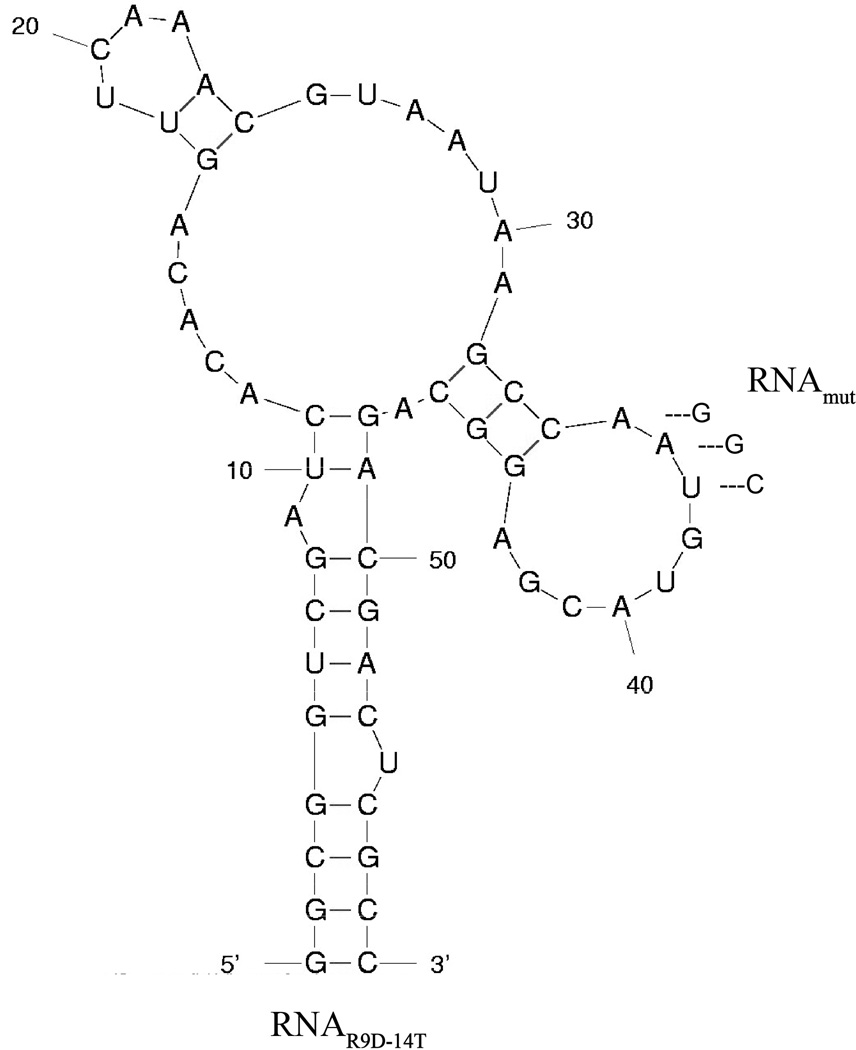
The full-length prothrombin aptamer R9D-14 is an 80 nucleotide long, 2’Fluoro-pyrimidine modified RNA aptamer that we originally generated by systematic evolution of ligands with exponential enrichment (SELEX) against the gamma carboxylic glutamic acid (GLA) proteome of human plasma [14]. To determine the minimal RNA sequence required for high affinity aptamer binding and functionality, R9D-14 was truncated by systematic deletion of the 5’ and 3’ termini. The shortest RNA derivative that retains high affinity binding and anticoagulant activity is 58 nucleotides and was renamed RNAR9D-14T. A point mutant control aptamer (RNAmut) was created by mutating three nucleotides (Figure 1).
Figure 1. The M-fold predicted secondary structure of RNAR9D-14T and RNAmut.
Aptamer-protein binding affinity and specificity
The majority of anticoagulant aptamers bind both the zymogen and enzyme [11–13, 27]. Nitrocellulose filter binding indicates that RNAR9D-14T binds with high affinity to both human prothrombin (apparent Kd=10 nM) and α-thrombin (apparent Kd=1 nM), while RNAmut shows weak binding toward either protein (Figure 2a). Additional nitrocellulose filter binding studies with structurally related human coagulation proteins (e.g., FVIIa, FIXa, FXa and Protein C, S, and Z) indicate that RNAR9D-14T shows very weak binding toward these coagulation factors (apparent Kd>1,000 nM) and thus specifically recognizes a surface that is unique to prothrombin/thrombin (Figure 2b). Surface plasmon resonance (SPR) was also performed to obtain more information regarding the kinetics of RNAR9D-14T prothrombin binding, and the data were compared to the pro/thrombin binding DNA aptamer ARC-183. Compared to ARC-183, RNAR9D-14T has a greater than 40-fold higher affinity for prothrombin (KD RNAR9D-14T=1.4 nM and ARC-183=60.7 nM), which is a result of both a faster association rate (Ka=8.97×105 vs. 5.03×104 1/Ms, respectively) and a slower dissociation rate (Kd=1.26×10−3 vs. 3.05×10−3 1/s, respectively) compared to ARC-183 (Supplemental Figure 1).
Figure 2. RNAR9D-14T binds both human prothrombin and thrombin with high affinity and specificity.
A) Nitrocellulose filter binding of the aptamer and mutant with human prothrombin and thrombin. Circles (●) represent RNAR9D-14T with α-thrombin, squares (■) are RNAR9D-14T with prothrombin, diamonds (◆) are RNAmut with α-thrombin, and triangles (▲) are RNAmut with prothrombin. The data represent the average ± the SEM of triplicates. B) Nitrocellulose filter binding of RNAR9D-14T to human coagulation factors. Squares (■) represent FVIIa, triangles (▲) are FIXa, inverted triangles (▼) are FXa, diamonds (◆) are Protein C, circles (●) are Protein S, and open squares (□) are Protein Z. The data represent the average ± the SEM of duplicates.
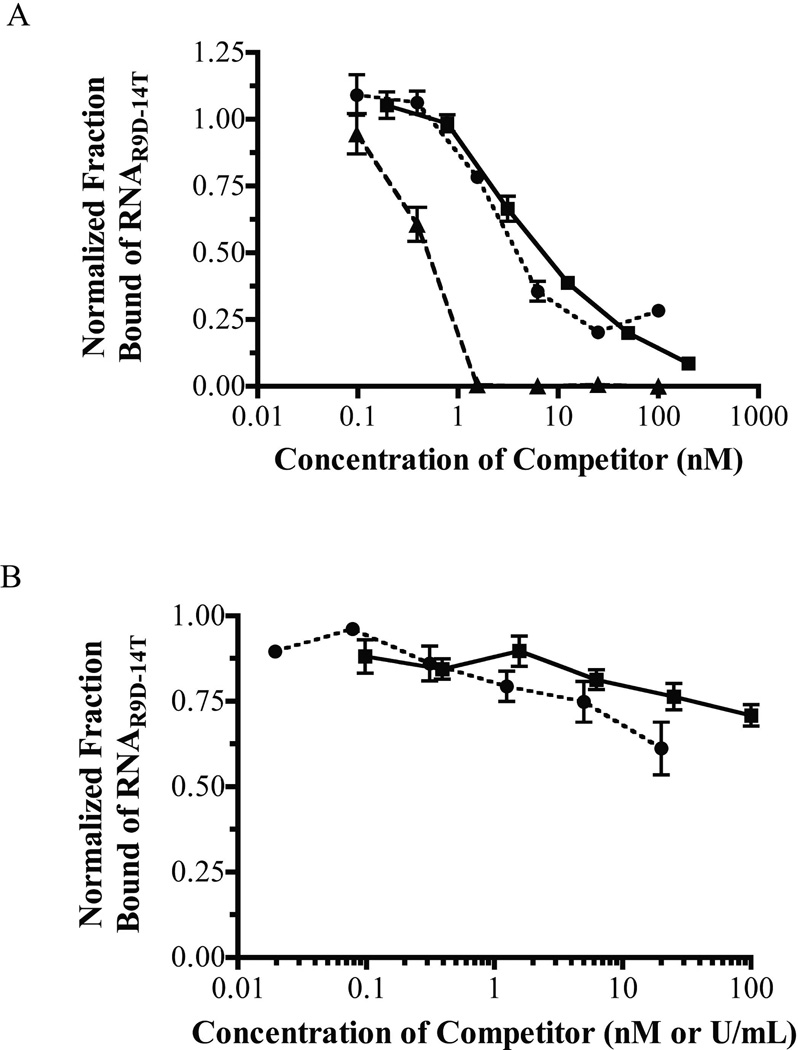
Binding competition experiments with several thrombin exosite ligands were performed to localize the RNAR9D-14T binding site. Hirudin, thrombomodulin (TM), and ARC-183 all compete with RNAR9D-14T for human α-thrombin binding, while heparin and the RNA aptamer Tog-25t do not (Figure 3a,b). Additionally, RNAR9D-14T has a decreased affinity for β-thrombin (apparent Kd=20 nM) and very weak affinity for γ-thrombin (apparent Kd>1,000 nM), which have altered exosite I surfaces and decreased activity toward fibrinogen (data not shown) [28, 29]. Collectively these data indicate that RNAR9D-14T binds specifically to (pro)thrombin (pro)exosite I.
Figure 3. RNAR9D-14T competes with exosite I, but not exosite II ligands for human α-thrombin binding.
A) Binding competition between RNAR9D-14T and various thrombin exosite I ligands. Squares (■) represent the ARC-183 DNA aptamer (0–200 nM), triangles (▲) represent recombinant hirudin (0–100 nM), and circles (●) represent rabbit thrombomodulin (0–100 nM). The data represent the mean ± SEM of triplicates. B) Binding competition between RNAR9D-14T and various exosite II ligands. Squares (■) represent the Tog-25t RNA aptamer (0–100 nM), while circles (●) represent heparin (0–20 U/mL). The data represent the mean ± SEM of triplicates.
Nuclease resistance and serum stability
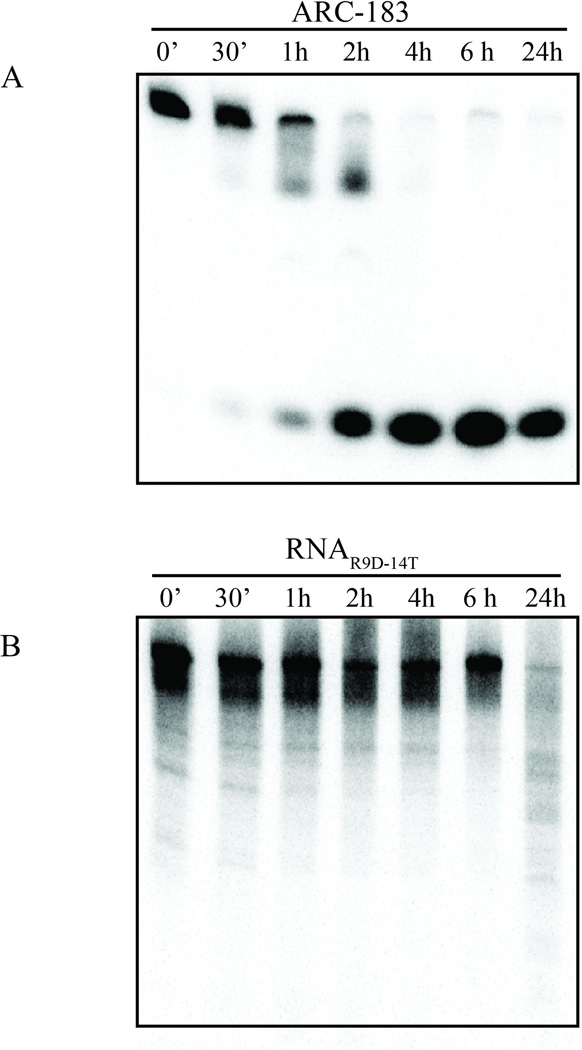
The data so far indicate that RNAR9D-14T and ARC-183 both bind to (pro)thrombin (pro)exosite I, although RNAR9D-14T has a higher binding affinity. Additionally, the two aptamers differ in their chemical composition - ARC-183 is comprised of unmodified DNA, while RNAR9D-14T is comprised of 2’Fluoro-pyrimidine modified RNA [14, 15]. To compare the stability of these oligonucleotides, the aptamers were radiolabeled and incubated with human serum over a period of 24 hours. Unlike the DNA aptamer ARC-183, which has a half-life of less than 2 hr and was completely degraded within 4 hr, RNAR9D-14T has an extended half-life of greater than 6 hr (Figure 4). Thus, RNAR9D-14T is much more resistant to human serum nucleases than ARC-183.
Figure 4. The modified RNA aptamer RNAR9D-14T is more stable in human serum compared to the DNA aptamer ARC-183.
Denaturing gel analysis of A) ARC-183 and B) RNAR9D-14T stability in human serum. The aptamers were 32P 5’-end radiolabeled (5,000 cpm/µL), incubated with human serum, and samples were withdrawn at the indicated times and run on a 12% denaturing acrylamide gel; min (’) and hours (h).
Clinical anticoagulation assays
The ability of RNAR9D-14T to anticoagulate human plasma was tested with several clinical clotting assays. RNAR9D-14T dose-dependently prolonged the activated partial thromboplastin (aPTT) and prothrombin time (PT) and was more robust than ARC-183, while RNAmut had no effect (Figure 5a). At 4 µM, RNAR9D-14T increased the aPTT clotting time from 31 sec to 664 sec (21-fold), and increased the PT clotting time to from 13 sec to greater than 15 min (exceeded the standard assay limit). In contrast, 4 µM of ARC-183 only increased the aPTT time to 86 sec (2.7-fold) and the PT time to 62 sec (4.8-fold) (Figure 5a). Thus, the clinical clotting data indicate that RNAR9D-14T anticoagulation is specific and much more potent than ARC-183.
Figure 5. RNAR9D-14T prolongs the aPTT, PT, and TCT clotting time and is a more potent anticoagulant than ARC-183.
A) Effect of RNAR9D-14T, RNAmut, and ARC-183 on the aPTT and PT in normal human plasma. Squares (■) represent ARC-183 in the aPTT, triangles (▲) are RNAR9D-14T in the aPTT, inverted triangles (▼) are ARC-183 in the PT, circles (●) are RNAR9D-14T in the PT, pluses (+) are RNAmut in the aPTT, and X’s (×) are RNAmut in the PT. The data were normalized to the baseline clot time and represent the mean ± SEM of triplicates. B) Effect of RNAR9D-14T and RNAmut on the thrombin clot time (TCT). Squares (■) represent RNAR9D-14T in normal plasma, triangles (▲) are RNAmut in normal plasma, circles (●) are RNAR9D-14T in prothrombin deficient plasma (<3% activity), and diamonds (◆) are RNAmut in prothrombin deficient plasma. The data were normalized to the baseline clot time and represent the mean ± the SEM of triplicates.
Fibrinogen binding to thrombin exosite I is essential for fibrin generation, which forms the meshwork of a blood clot. Because RNAR9D-14T binds exosite I, we tested its impact on fibrinogen cleavage in both normal and prothrombin deficient (<3% activity) plasma with a thrombin clot time assay (TCT). While RNAmut had no effect, 1 µM RNAR9D-14T increased the clot time from approximately 53 sec to greater than 15 minutes (exceeded the standard assay limit) (Figure 5b). Thus, RNAR9D-14T binding to thrombin exosite I impairs fibrin clot formation.
Thrombin active site function
Several exosite I ligands have been reported to have allosteric effects on thrombin active site activity [30–34]. We therefore studied the impact of RNAR9D-14T binding to exosite I on the thrombin active site by monitoring small peptide chromogenic substrate cleavage. At super-saturating concentrations, RNAR9D-14T binding, compared to RNAmut binding, only slightly decreased the Km (9 vs. 24 µM, respectively; p=0.06) and slightly decreased the kcat (72.7 vs. 107.9 sec−1, respectively; p=0.04) (Table 1). Further studies regarding the aptamer’s impact on the thrombin active site were performed with a physiological, active site-binding macromolecule – the serine protease inhibitor antithrombin (AT). Previous aptamer studies have shown that Tog-25t exosite II binding decreased the second order rate constant (k2) of AT inhibition by approximately 3-fold [23], while ARC-183 exosite I binding had little effect [35]. Similar to ARC-183, a super-saturating concentration of RNAR9D-14T had a non-significant effect on AT inhibition and increased the k2 constant 1.3-fold compared to RNAmut (3.43×105 vs. 2.56×105 M−1 min−1, respectively; p=0.09) (Table 1). Collectively, the kinetic data suggest that RNAR9D-14T has only a subtle impact on thrombin active site structure and activity. Thus, we hypothesized that RNAR9D-14T anticoagulation is primarily due to inhibition of exosite I mediated protein-protein interactions.
Table 1.
Thrombin small peptide and Antithrombin kinetics with RNAR9D-14T and RNAmut.
| Small peptide kinetics | AT kinetics | ||
|---|---|---|---|
| Km (mM) | kcat (sec_1) | k2 (M1 min1) | |
| No aptamer | 0.020 ± 0.002 | 100.5 ±6.7 | 2.18 ± 0.29×105 |
| RNAR9D-14T | 0.009 ±0.001 | 72.7 ±6.1 | 3.43 ± 0.35×105 |
| RNAraut | 0.024 ± 0.004 | 107.9 ±8.6 | 2.56 ± 0.34×105 |
AT=Antithrombin; the data represent the mean ± the SEM of three independent experiments performed in triplicate.
Thrombin activation of FV
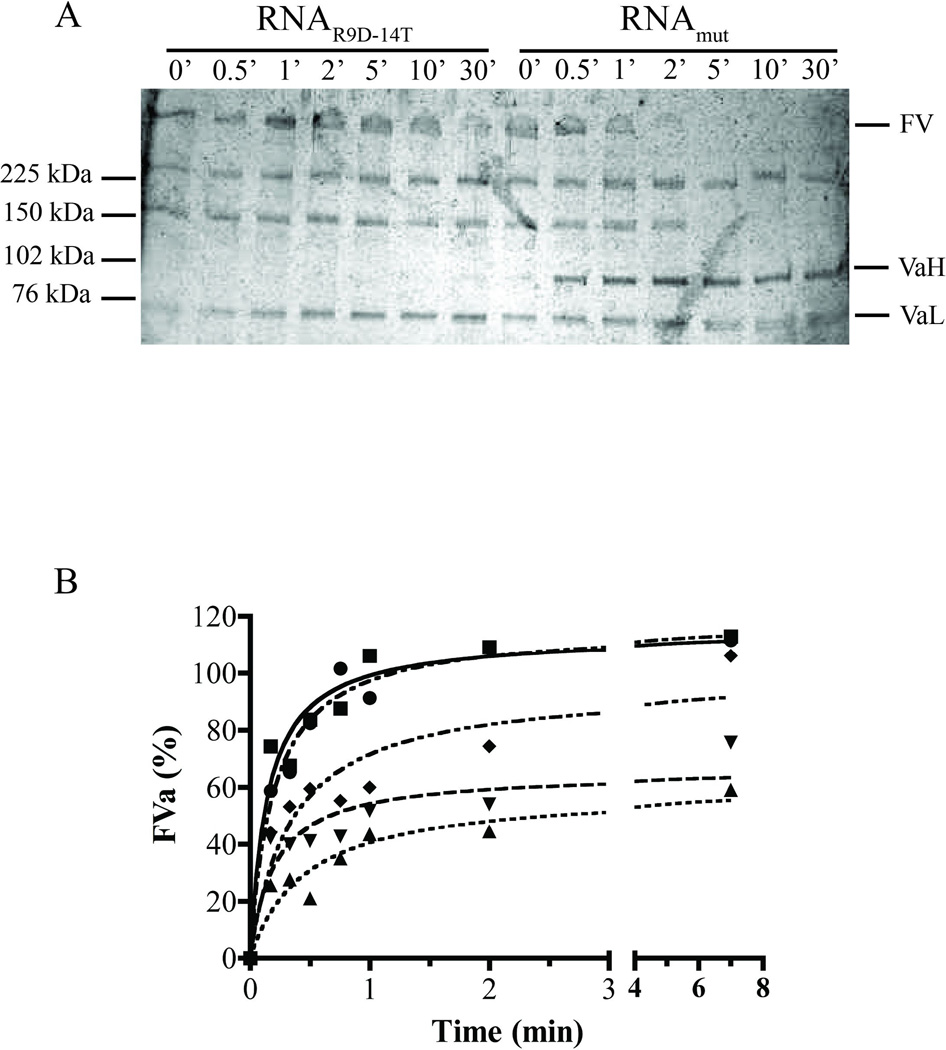
Previous studies have shown that exosite I ligands inhibit FV and FVIII cleavage and decrease thrombin feedback activity [27, 36]. To test the impact of RNAR9D-14T on cofactor activation, thrombin-mediated cleavage of purified FV was analyzed via silver stain analysis and FVa cofactor activity in a purified prothrombinase system. Compared to RNAmut, RNAR9D-14T delayed FV cleavage for greater than 10 minutes and inhibited Arg709 cleavage for greater than 30 minutes, thereby preventing the appearance of the FVa heavy chain (VaH) (Figure 6a). RNAR9D-14T also dose-dependently impaired FVa cofactor activity compared to a similar concentration of RNAmut (Figure 6b). Interestingly, a previous study by Thorelli et al. reported that a FV mutant that cannot be cleaved at Arg709 has similarly decreased FVa activity [37]. Thus, RNAR9D-14T binding to thrombin exosite I inhibits cleavage of Arg709 and decreases FVa cofactor activity.
Figure 6. RNAR9D-14T inhibits factor V activation by thrombin.
A) Silver stain analysis of FV activation by thrombin in the presence of saturating RNAR9D-14T or RNAmut. FV denotes uncleaved factor V, VaH is the factor Va heavy chain, and VaL is the factor Va light chain. The numbers above the lanes indicate the assay incubation time in minutes (’), and the numbers on the left represent the protein standard sizes. B) Chromogenic assay for FVa cofactor activity. FV (250 ρM) was activated by thrombin (3 nM) in the presence of RNAR9D-14T or RNAmut and time points were diluted into a prothrombinase chromogenic assay where the rate of thrombin generation was dependent upon the concentration of FVa formed. Squares (■) represent no aptamer, triangles (▲) are 500 nM RNAR9D-14T, inverted triangles (▼) are 100 nM RNAR9D-14T, diamonds (◆) are 20 nM RNAR9D-14T, and circles (●) are 500 nM RNAmut. The data were normalized to a parallel control experiment where FV was fully activated in the absence of RNA and are expressed as a percentage of the FVa activity of fully activated FV. The data are single experiments that are representative of three independent experiments.
Thrombin-mediated platelet activation
The initial small amounts of thrombin formed activate platelets and prime the system for a rapid burst of thrombin generation [1]. Thrombin exosite I binds and mediates active site cleavage of platelet protease activated receptors (PARs), which results in platelet degranulation and upregulation of the P-selectin (CD62P) surface receptor [38, 39]. RNAR9D-14T prevented thrombin-mediated platelet activation in a dose-dependent manner, and at saturating concentrations prevented greater than 90% of platelet activation; in contrast, RNAmut had no effect (Figure 7). Therefore, RNAR9D-14T exosite I binding also potently inhibits thrombin exosite I-mediated platelet activation.
Figure 7. RNAR9D-14T inhibits thrombin-mediated platelet activation.
Thrombin was incubated with purified human platelets in the presence of aptamer, and platelet degranulation was measured via flow cytometry with an anti-CD62P (P-selectin) antibody. Unstim represents resting platelets (no thrombin added), the black bars represent RNAR9D-14T preincubated with 1 nM of human thrombin, and the gray bars represent RNAmut preincubated with 1 nM human thrombin. The data represent the mean ± SEM of duplicates and are representative of data with three different platelet donors.
Thrombin generation
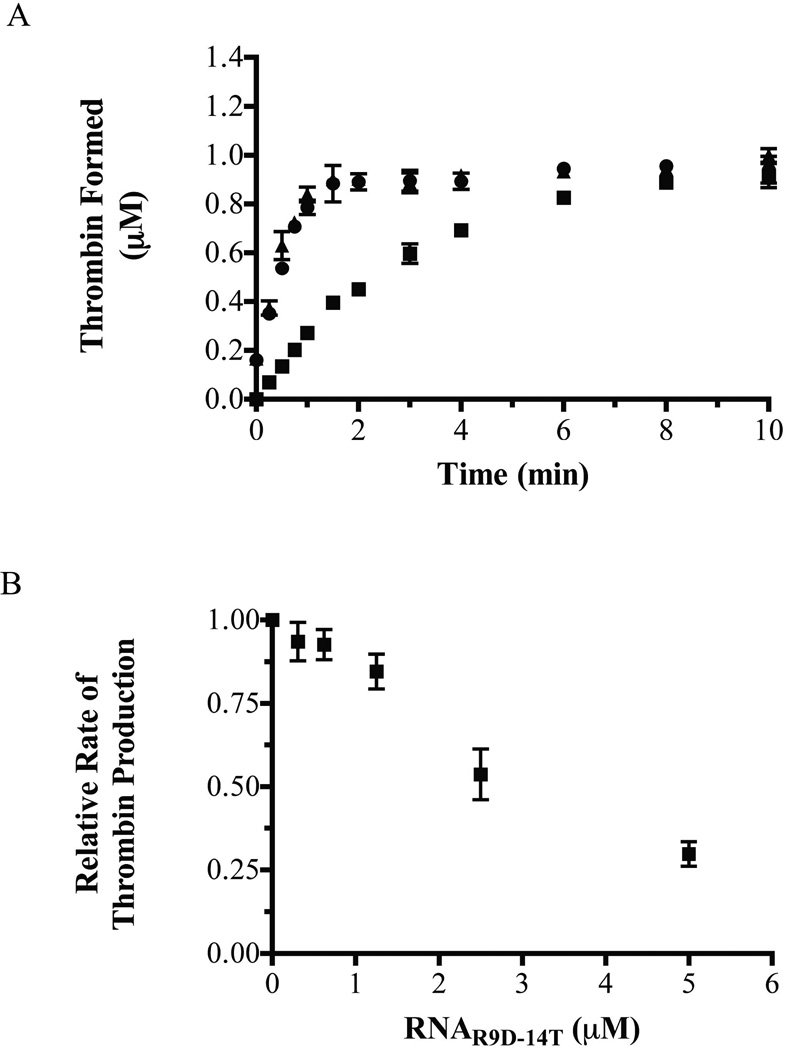
Because RNAR9D-14T binds to both human prothrombin and thrombin, the aptamer may function as an anticoagulant by inhibiting both thrombin generation and thrombin activity. The impact of the aptamer on prothrombin cleavage was assessed by monitoring thrombin generation in a purified system with the prothrombinase complex (purified FXa and FVa) pre-assembled on lipid vesicles. At physiological concentrations of prothrombin, RNAR9D-14T caused a dose dependent decrease in the rate of thrombin generation and decreased the rate of thrombin generation by greater than 70% at high concentrations (5 µM); similar concentrations of RNAmut had no effect (Figure 8a,b). Thus, RNAR9D-14T inhibits both thrombin generation and thrombin exosite I function.
Figure 8. RNAR9D-14T inhibits thrombin generation in a purified prothrombinase system.
A) Thrombin formation curves, as measured via a discontinuous purified prothrombinase assay with a physiological concentration of prothrombin (1.4 µM). Circles (●) represent no addition of aptamer, squares (■) are 5 µM RNAR9D-14T, and triangles (▲) are 5 µM RNAmut. The data represent the mean ± SEM of two independent preparations of prothrombinase. B) Dose dependent response of the rate of prothrombin activation with increasing concentrations of RNAR9D-14T. The data were normalized to the rate in the absence of aptamer and represent the mean ± SEM for three independent experiments.
The current literature suggests that FVa binds to prothrombin within the prothrombinase complex via proexosite I [7–9], and several proexosite I-binding ligands prevent prothrombin cleavage in a FVa-dependent manner [27, 40]. To determine if RNAR9D-14T has a similar mechanism of anticoagulation, thrombin generation assays were repeated with various combinations of the prothrombinase components. A partial prothrombinase complex of FXa with FVa (no lipids) was sensitive to RNAR9D-14T inhibition, while FXa with lipids (no FVa) was not sensitive (data not shown). Therefore, similar to other proexosite I ligands, RNAR9D-14T blocks thrombin generation in a FVa-dependent manner.
Antidote reversibility
Anticoagulant RNA aptamers can be formulated to circulate in vivo for 24–30 hours [41]. Therefore, in the event of clinical complications, such as hemorrhage, physicians need to rapidly and safely reverse anticoagulation. We have previously described the design of complementary oligonucleotide antidotes to modulate anticoagulant aptamers [13, 16]. Because RNAR9D-14T is an extremely potent and stable anticoagulant, we sought to design antidotes that can rapidly control its anticoagulant activity and improve its safety profile.
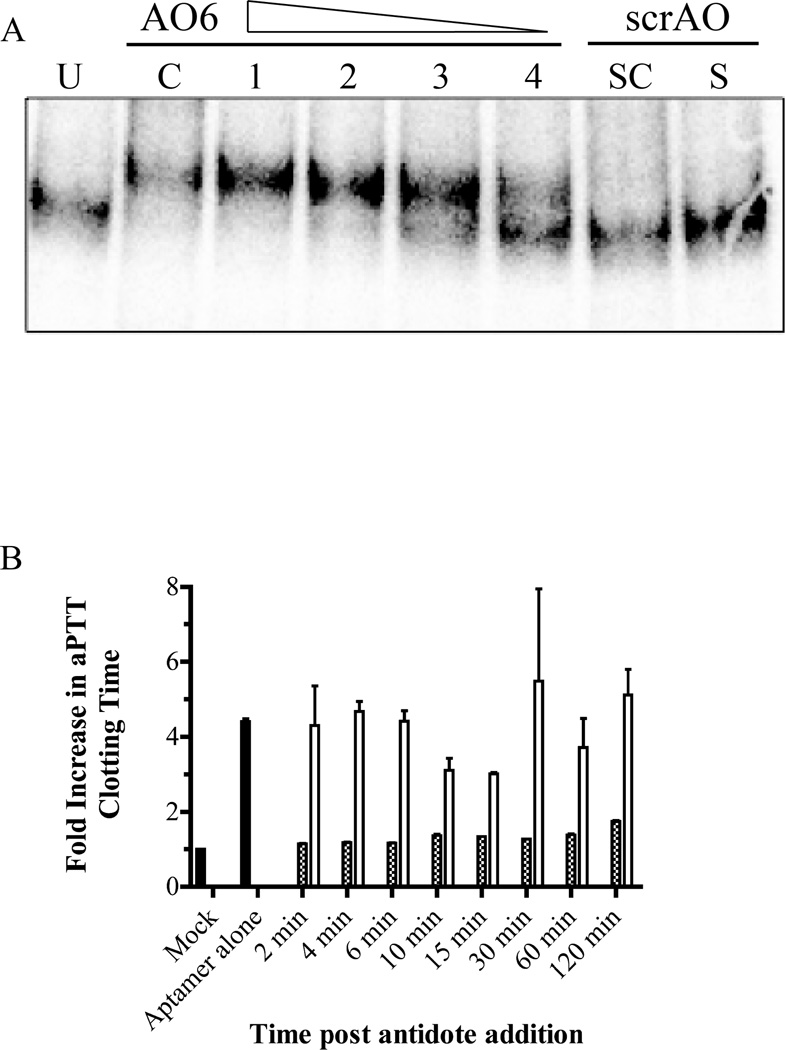
Six complementary DNA antidote sequences were designed to bind to various regions across RNAR9D-14T via Watson-Crick base pairing and tested for their ability to disrupt aptamer binding [16]. Antidotes 5 and 6 completely reversed RNAR9D-14T binding to human prothrombin in a nitrocellulose filter binding assay, while antidotes 1–4 only partially reversed binding (data not shown). Native gel electrophoresis showed that a four-fold molar excess of AO6 formed a duplex with more than 80% of free RNAR9D-14T aptamer, while a four-fold molar excess of a scrambled antidote control (scrAO) did not form a duplex (Figure 9a). Additionally, a four-fold molar excess of AO6 was able to reverse 96% of RNAR9D-14T anticoagulation in an aPTT within two minutes, and 78% of RNAR9D-14T anticoagulation after two hours; in contrast, a scrambled antidote did not reverse RNAR9D-14T anticoagulation (Figure 9b). Thus, for clinical applications, RNAR9D-14T can be formulated to have an extended half-life, yet can be rapidly controlled with an antidote.
Figure 9. DNA antidote oligonucleotide 6 forms a duplex with RNAR9D-14T and reverses anticoagulation in vitro.
A) Native gel analysis of antidote oligonucleotide 6 (AO6) binding to radiolabeled RNAR9D-14T. Radiolabeled aptamer and unlabeled aptamer (125 nM) were mixed and incubated with various concentrations of DNA antidote (62.5–500 nM) at 37°C for 5 minutes. Untreated radiolabeled RNAR9D-14T (U), and RNAR9D-14T denatured and complexed with AO6 before gel loading (C). Lanes 1–4 show a decrease in the AO6:RNAR9D-14T molar ratio from 4:1 to 0.5:1 in two-fold increments. A scrambled antidote (scrAO) was incubated with RNAR9D-14T at a 4:1 ratio at 37°C (S) or heated to 95°C and slow cooled (SC) prior to loading. B) aPTT time course for AO6 reversal of RNAR9D-14T anticoagulation. Normal human plasma was anticoagulated with RNAR9D-14T, antidote was added and incubated for various time points at 37°C, and the aPTT clotting time was measured. Mock is a standard aPTT assay with no aptamer (baseline); aptamer alone is a standard aPTT assay with 1 µM of RNAR9D-14T and no antidote. The gray bars represent addition of 4 µM AO6, while white bars represent addition of 4 µM scrAO. The data were normalized to the baseline clotting time and represent the mean ± SEM of duplicates.
Animal cross-reactivity and anticoagulation
Because aptamers can bind to their therapeutic targets with extremely high specificity, aptamers have been isolated that do not cross-react with orthologous proteins from other animals [20]. RNAR9D-14T was tested for its ability to anticoagulate plasma from several species in an aPTT assay. At 2 µM, RNAR9D-14T robustly anticoagulated human (19-fold increase in clotting time), rhesus (7.6-fold increase), and pig plasma (4.2-fold increase), but showed decreased activity in dog, rabbit, sheep, mouse, rat, and cow plasma (Supplemental Figure 2a). The apparent binding affinity of RNAR9D-14T toward purified thrombin or prothrombin from several of these species was also assessed to confirm the species cross-reactivity results. RNAR9D-14T binds to porcine thrombin (apparent Kd=5 nM) with a similar binding affinity as human thrombin (apparent Kd=1 nM), but shows little binding affinity toward cow thrombin, rat thrombin, or mouse prothrombin (apparent Kd s>1,000 nM; Supplemental Figure 2b). Thus, the aptamer-binding epitope on exosite I appears to be conserved in humans, primates, and pigs, but not cows or rodents. Moreover, RNAR9D-14T anticoagulation appears to be specific for pro/thrombin binding because bovine plasma was not anticoagulated by a high dose of RNAR9D-14T, which has a very weak affinity toward bovine thrombin.
Discussion
Because thrombin is the final enzyme formed during the coagulation cascade and plays a number of key roles in fibrin clot formation, thrombin has been an important target for anticoagulant therapy. Although several direct thrombin inhibitors are clinically available (e.g., lepirudin, argatroban, and bivalrudin), they can be challenging to use because they are associated with an increased risk of bleeding and cannot be easily reversed with an antidote [42]. Additionally, several aptamers have been developed against thrombin (e.g., ARC-183, HD-22, and Tog-25t), although none have advanced clinically due to chemical instability, rapid clearance in vivo, and/or limited anticoagulant activity [15, 19, 20]. Here we describe the structure and function of a 2’F-modified nuclease resistant RNA aptamer (RNAR9D-14T) that binds to prothrombin/thrombin and is a potent anticoagulant. RNAR9D-14T binds prothrombin at proexosite I and prevents thrombin generation in a FVa-dependent manner. Additionally, although RNAR9D-14T binding to thrombin has only a minor impact on active site function, it severely impairs coagulation by inhibiting exosite I mediated function (i.e., fibrin clot formation, FV activation, and platelet activation).
Binding studies indicate that RNAR9D-14T and ARC-183 bind to the same region on thrombin (exosite I). An analysis of RNAR9D-14T animal cross-reactivity highlights key residues within exosite I that may be important for aptamer binding. An alignment of the human, rhesus, porcine, bovine, rat, and mouse thrombin beta chain amino acid sequence shows that two exosite I amino acids (Ile24 and Ile79) are conserved in only the species that RNAR9D-14T robustly anticoagulates (i.e., human, rhesus, and pig) (Supplemental Figures 2a and 3). Interestingly, ARC-183 binding forms direct contacts with these surface residues in thrombin exosite I [43], and the species cross reactivity data suggest that these residues may be important for RNAR9D-14T binding as well.
Although RNAR9D-14T and ARC-183 have a similar mechanism of anticoagulation (i.e., pro/exosite I inhibition), RNAR9D-14T has a different chemical composition that results in a higher binding affinity and increased plasma stability (Figure 4, and Supplemental Figure 1). Moreover, RNAR9D-14T (58 nucleotides) is much larger than ARC-183 (15 nucleotides) and may therefore cover a larger area on the protein surface. Consequentially, RNAR9D-14T appears to be a more powerful anticoagulant in plasma clotting assays (Figure 5a). Although RNAR9D-14T binds prothrombin/thrombin with high affinity, a high concentration is required to saturate prothrombin, which has an estimated concentration of 1.4 µM in human blood. Additionally, only approximately 30–40% of the aptamer folds into the high affinity conformation that can bind to prothrombin and thrombin (Figure 2a and data not shown); the combination of inefficient aptamer folding and a high target concentration mean that the saturating dose of RNAR9D-14T is greater than 5 µM. Future experiments should be performed to see if aptamer folding can be improved, thereby reducing the dose required for robust anticoagulation, or to determine if combining RNAR9D-14T with another anticoagulant aptamer (i.e., FIXa and Xa aptamers) can reduce the dose required.
Because RNAR9D-14T is such a robust anticoagulant, we designed several complementary DNA oligonucleotide antidotes to bind to the aptamer and alter its three-dimensional conformation, thus resulting in an inactive stable complex. RNAR9D-14T was rapidly and durably reversed with a complementary antidote in vitro, which makes this compound an attractive candidate for further development as a robust, yet rapidly controllable anticoagulant. Future in vivo studies of anticoagulation with RNAR9D-14T alone, or in combination with other RNA aptamers and their antidotes are needed to assess the applicability of aptamer/antidote anticoagulation in clinical settings where safe yet effective anticoagulation is essential, such as cardiopulmonary bypass, deep vein thrombosis, stroke, or percutaneous coronary artery intervention.
Supplementary Material
The interaction between A) RNAR9D-14T and B) ARC-183 with various concentrations of human prothrombin (0.5–15 µg/mL). The chi-square values were 1.1 and 0.575, respectively. The numbers inside the brackets [ ] represent the concentration of prothrombin in µg/mL. C) Kinetic parameters for ARC-183 and RNAR9D-14T binding to human prothrombin.
A) aPTT assay showing the species cross reactivity of RNAR9D-14T. Gray bars represent the addition of 2 µM RNAR9D-14T and black bars are addition of 2 µM RNAmut. The data were normalized to the baseline aPTT time for each individual species and represent the mean ± SEM of duplicates. B) Binding affinity of RNAR9D-14T toward purified protein from various species. Squares (■) represent human thrombin, triangles (▲) are cow thrombin, inverted triangles (▼) represent porcine thrombin, diamonds (◆) are rat thrombin, circles (●) are human prothrombin, and open squares (□) are mouse prothrombin. The data represent single experiments that are representative of three independent experiments.
The thrombin β-chain peptide sequences were downloaded from the NCBI HomoloGene protein database, aligned to the human sequence, and the percent similarity to the human sequence was calculated. Exosite I residues Ile24 and Ile79 (residues 9 and 75 in this alignment, enclosed within a box) are only conserved in human, rhesus, and pig, which are the species that RNAR9D-14T robustly anticoagulates.
Acknowledgements
We are grateful to Dr. Shahid Nimjee for his critical review of this manuscript and David Boczkowski for his help with flow cytometry. We would also like to the DHVI shared resource facilities under the direction of Dr. Munir Alam for help with the SPR, Kara Anasti for SPR technical assistance, and Dr. S. Moses Dennison for SPR data analysis. This work was supported by an American Heart Association Predoctoral Fellowship to KMB (10PRE3260011) and an NIH grant to BAS (R01HL65222).
Footnotes
Conflict of Interest Disclosure: Dr. Sullenger is a scientific founder of Regado Biosciences Inc., which is a biotechnology company that was spun out of Duke University to commercialize therapeutic aptamers.
References
- 1.Hoffman M, Monroe DM., 3rd A cell-based model of hemostasis. Thromb Haemost. 2001;85:958–965. [PubMed] [Google Scholar]
- 2.Coughlin SR. Thrombin signalling and protease-activated receptors. Nature. 2000;407:258–264. doi: 10.1038/35025229. [DOI] [PubMed] [Google Scholar]
- 3.Ferry JD, Morrison PR. Preparation and properties of serum and plasma proteins the conversion of human fibrinogen to fibrin under various conditions. J Am Chem Soc. 1947;69:388–400. doi: 10.1021/ja01194a066. [DOI] [PubMed] [Google Scholar]
- 4.Lloyd-Jones D, Adams R, Carnethon M, De Simone G, Ferguson TB, Flegal K, Ford E, Furie K, Go A, Greenlund K, Haase N, Hailpern S, Ho M, Howard V, Kissela B, Kittner S, Lackland D, Lisabeth L, Marelli A, McDermott M, Meigs J, Mozaffarian D, Nichol G, O'Donnell C, Roger V, Rosamond W, Sacco R, Sorlie P, Stafford R, Steinberger J, Thom T, Wasserthiel-Smoller S, Wong N, Wylie-Rosett J, Hong Y. Heart Disease and Stroke Statistics--2009 Update. A Report From the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2008 doi: 10.1161/CIRCULATIONAHA.108.191261. [DOI] [PubMed] [Google Scholar]
- 5.Stubbs MT, Bode W. A player of many parts: the spotlight falls on thrombin's structure. Thromb Res. 1993;69:1–58. doi: 10.1016/0049-3848(93)90002-6. [DOI] [PubMed] [Google Scholar]
- 6.Bock PE, Panizzi P, Verhamme IM. Exosites in the substrate specificity of blood coagulation reactions. J Thromb Haemost. 2007;5(Suppl 1):81–94. doi: 10.1111/j.1538-7836.2007.02496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson PJ, Nesset A, Dharmawardana KR, Bock PE. Role of proexosite I in factor Va-dependent substrate interactions of prothrombin activation. J Biol Chem. 2000;275:16435–16442. doi: 10.1074/jbc.M001255200. [DOI] [PubMed] [Google Scholar]
- 8.Anderson PJ, Nesset A, Dharmawardana KR, Bock PE. Characterization of proexosite I on prothrombin. J Biol Chem. 2000;275:16428–16434. doi: 10.1074/jbc.M001254200. [DOI] [PubMed] [Google Scholar]
- 9.Chen L, Yang L, Rezaie AR. Proexosite-1 on prothrombin is a factor Va-dependent recognition site for the prothrombinase complex. J Biol Chem. 2003;278:27564–27569. doi: 10.1074/jbc.M302707200. [DOI] [PubMed] [Google Scholar]
- 10.Long SB, Long MB, White RR, Sullenger BA. Crystal structure of an RNA aptamer bound to thrombin. RNA. 2008;14:1–9. doi: 10.1261/rna.1239308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buddai SK, Layzer JM, Lu G, Rusconi CP, Sullenger BA, Monroe DM, Krishnaswamy S. An anticoagulant RNA aptamer that inhibits proteinase-cofactor interactions within prothrombinase. J Biol Chem. 2010;285:5212–5223. doi: 10.1074/jbc.M109.049833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rusconi CP, Yeh A, Lyerly HK, Lawson JH, Sullenger BA. Blocking the initiation of coagulation by RNA aptamers to factor VIIa. Thromb Haemost. 2000;84:841–848. [PubMed] [Google Scholar]
- 13.Rusconi CP, Scardino E, Layzer J, Pitoc GA, Ortel TL, Monroe D, Sullenger BA. RNA aptamers as reversible antagonists of coagulation factor IXa. Nature. 2002;419:90–94. doi: 10.1038/nature00963. [DOI] [PubMed] [Google Scholar]
- 14.Layzer JM, Sullenger BA. Simultaneous generation of aptamers to multiple gamma-carboxyglutamic acid proteins from a focused aptamer library using DeSELEX and convergent selection. Oligonucleotides. 2007;17:1–11. doi: 10.1089/oli.2006.0059. [DOI] [PubMed] [Google Scholar]
- 15.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 16.Oney S, Nimjee SM, Layzer J, Que-Gewirth N, Ginsburg D, Becker RC, Arepally G, Sullenger BA. Antidote-controlled platelet inhibition targeting von Willebrand factor with aptamers. Oligonucleotides. 2007;17:265–274. doi: 10.1089/oli.2007.0089. [DOI] [PubMed] [Google Scholar]
- 17.Oney S, Lam RT, Bompiani KM, Blake CM, Quick G, Heidel JD, Liu JY, Mack BC, Davis ME, Leong KW, Sullenger BA. Development of universal antidotes to control aptamer activity. Nat Med. 2009;15:1224–1228. doi: 10.1038/nm.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeAnda A, Jr, Coutre SE, Moon MR, Vial CM, Griffin LC, Law VS, Komeda M, Leung LL, Miller DC. Pilot study of the efficacy of a thrombin inhibitor for use during cardiopulmonary bypass. The Annals of thoracic surgery. 1994;58:344–350. doi: 10.1016/0003-4975(94)92206-3. [DOI] [PubMed] [Google Scholar]
- 19.Tasset DM, Kubik MF, Steiner W. Oligonucleotide inhibitors of human thrombin that bind distinct epitopes. J Mol Biol. 1997;272:688–698. doi: 10.1006/jmbi.1997.1275. [DOI] [PubMed] [Google Scholar]
- 20.White R, Rusconi C, Scardino E, Wolberg A, Lawson J, Hoffman M, Sullenger B. Generation of species cross-reactive aptamers using "toggle" SELEX. Mol Ther. 2001;4:567–573. doi: 10.1006/mthe.2001.0495. [DOI] [PubMed] [Google Scholar]
- 21.Nimjee SM, Oney S, Volovyk Z, Bompiani KM, Long SB, Hoffman M, Sullenger BA. Synergistic effect of aptamers that inhibit exosites 1 and 2 on thrombin. RNA. 2009;15:2105–2111. doi: 10.1261/rna.1240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong I, Lohman TM. A double-filter method for nitrocellulose-filter binding: application to protein-nucleic acid interactions. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeter ML, Vy LV, Fortenbery YM, Whinna HC, White RR, Rusconi CP, Sullenger BA, Church FC. RNA aptamer to thrombin binds anion-binding exosite-2 and alters protease inhibition by heparin-binding serpins. FEBS Letters. 2004;568:10–14. doi: 10.1016/j.febslet.2004.04.087. [DOI] [PubMed] [Google Scholar]
- 24.Church FC, Griffith MJ. Evidence for essential lysines in heparin cofactor II. Biochem Biophys Res Commun. 1984;124:745–751. doi: 10.1016/0006-291x(84)91021-0. [DOI] [PubMed] [Google Scholar]
- 25.Hope MJ, Bally MB, Webb G, Cullis PR. Production of large unilamellar vesicles by a rapid extrusion procedure. Characterization of size distribution, trapped volume and ability to maintain a membrane potential. Biochim Biophys Acta. 1985;812:55–65. doi: 10.1016/0005-2736(85)90521-8. [DOI] [PubMed] [Google Scholar]
- 26.Hoffman M, Monroe DM, Roberts HR. A rapid method to isolate platelets from human blood by density gradient centrifugation. American journal of clinical pathology. 1992;98:531–533. doi: 10.1093/ajcp/98.5.531. [DOI] [PubMed] [Google Scholar]
- 27.Kretz CA, Stafford AR, Fredenburgh JC, Weitz JI. HD1, a thrombin-directed aptamer, binds exosite 1 on prothrombin with high affinity and inhibits its activation by prothrombinase. J Biol Chem. 2006;281:37477–37485. doi: 10.1074/jbc.M607359200. [DOI] [PubMed] [Google Scholar]
- 28.Lundblad RL, Kingdon HS, Mann KG. Thrombin. Methods in enzymology. 1976;45:156–176. doi: 10.1016/s0076-6879(76)45017-6. [DOI] [PubMed] [Google Scholar]
- 29.Mann KG, Heldebrant CM, Fass DN. Multiple active forms of thrombin. I. Partial resolution, differential activities, and sequential formation. J Biol Chem. 1971;246:5994–6001. [PubMed] [Google Scholar]
- 30.Petrera NS, Stafford AR, Leslie BA, Kretz CA, Fredenburgh JC, Weitz JI. Long range communication between exosites 1 and 2 modulates thrombin function. J Biol Chem. 2009;284:25620–25629. doi: 10.1074/jbc.M109.000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naski MC, Shafer JA. Alpha-thrombin-catalyzed hydrolysis of fibrin I. Alternative binding modes and the accessibility of the active site in fibrin I-bound alpha-thrombin. J Biol Chem. 1990;265:1401–1407. [PubMed] [Google Scholar]
- 32.Liu LW, Ye J, Johnson AE, Esmon CT. Proteolytic formation of either of the two prothrombin activation intermediates results in formation of a hirugen-binding site. J Biol Chem. 1991;266:23633–23636. [PubMed] [Google Scholar]
- 33.Hortin GL, Trimpe BL. Allosteric changes in thrombin's activity produced by peptides corresponding to segments of natural inhibitors and substrates. J Biol Chem. 1991;266:6866–6871. [PubMed] [Google Scholar]
- 34.Liu LW, Vu TK, Esmon CT, Coughlin SR. The region of the thrombin receptor resembling hirudin binds to thrombin and alters enzyme specificity. J Biol Chem. 1991;266:16977–16980. [PubMed] [Google Scholar]
- 35.Holland CA, Henry AT, Whinna HC, Church FC. Effect of oligonucleotide thrombin aptamer on thrombin inhibition by heparin cofactor II and antithrombin. FEBS Letters. 2000;484:87–91. doi: 10.1016/s0014-5793(00)02131-1. [DOI] [PubMed] [Google Scholar]
- 36.Segers K, Dahlback B, Bock PE, Tans G, Rosing J, Nicolaes GA. The role of thrombin exosites I and II in the activation of human coagulation factor V. J Biol Chem. 2007;282:33915–33924. doi: 10.1074/jbc.M701123200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thorelli E, Kaufman RJ, Dahlback B. Cleavage requirements for activation of factor V by factor Xa. Eur J Biochem. 1997;247:12–20. doi: 10.1111/j.1432-1033.1997.00012.x. [DOI] [PubMed] [Google Scholar]
- 38.Ayala YM, Cantwell AM, Rose T, Bush LA, Arosio D, Di Cera E. Molecular mapping of thrombin-receptor interactions. Proteins. 2001;45:107–116. doi: 10.1002/prot.1130. [DOI] [PubMed] [Google Scholar]
- 39.Stenberg PE, McEver RP, Shuman MA, Jacques YV, Bainton DF. A platelet alpha-granule membrane protein (GMP-140) is expressed on the plasma membrane after activation. J Cell Biol. 1985;101:880–886. doi: 10.1083/jcb.101.3.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monteiro RQ, Zingali RB. Bothrojaracin, a proexosite I ligand, inhibits factor Va-accelerated prothrombin activation. Thromb Haemost. 2002;87:288–293. [PubMed] [Google Scholar]
- 41.Chan MY, Cohen MG, Dyke CK, Myles SK, Aberle LG, Lin M, Walder J, Steinhubl SR, Gilchrist IC, Kleiman NS, Vorchheimer DA, Chronos N, Melloni C, Alexander JH, Harrington RA, Tonkens RM, Becker RC, Rusconi CP. Phase 1b randomized study of antidote-controlled modulation of factor IXa activity in patients with stable coronary artery disease. Circulation. 2008;117:2865–2874. doi: 10.1161/CIRCULATIONAHA.107.745687. [DOI] [PubMed] [Google Scholar]
- 42.Yavari M, Becker RC. Anticoagulant therapy during cardiopulmonary bypass. J Thromb Thrombolysis. 2008;26:218–228. doi: 10.1007/s11239-008-0280-4. [DOI] [PubMed] [Google Scholar]
- 43.Padmanabhan K, Padmanabhan KP, Ferrara JD, Sadler JE, Tulinsky A. The structure of alpha-thrombin inhibited by a 15-mer single-stranded DNA aptamer. J Biol Chem. 1993;268:17651–17654. doi: 10.2210/pdb1hut/pdb. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The interaction between A) RNAR9D-14T and B) ARC-183 with various concentrations of human prothrombin (0.5–15 µg/mL). The chi-square values were 1.1 and 0.575, respectively. The numbers inside the brackets [ ] represent the concentration of prothrombin in µg/mL. C) Kinetic parameters for ARC-183 and RNAR9D-14T binding to human prothrombin.
A) aPTT assay showing the species cross reactivity of RNAR9D-14T. Gray bars represent the addition of 2 µM RNAR9D-14T and black bars are addition of 2 µM RNAmut. The data were normalized to the baseline aPTT time for each individual species and represent the mean ± SEM of duplicates. B) Binding affinity of RNAR9D-14T toward purified protein from various species. Squares (■) represent human thrombin, triangles (▲) are cow thrombin, inverted triangles (▼) represent porcine thrombin, diamonds (◆) are rat thrombin, circles (●) are human prothrombin, and open squares (□) are mouse prothrombin. The data represent single experiments that are representative of three independent experiments.
The thrombin β-chain peptide sequences were downloaded from the NCBI HomoloGene protein database, aligned to the human sequence, and the percent similarity to the human sequence was calculated. Exosite I residues Ile24 and Ile79 (residues 9 and 75 in this alignment, enclosed within a box) are only conserved in human, rhesus, and pig, which are the species that RNAR9D-14T robustly anticoagulates.