Abstract
Human immunodeficiency virus type 1 (HIV-1) prevalence and incidence in the fishing communities on Lake Victoria in Uganda are high. This population may play a role in driving the HIV epidemic in Uganda including the spread of transmitted drug resistance (TDR). We report data on TDR in this population among antiretroviral (ARV)-naive, recently infected individuals about 5 years after ARV scaling-up in Uganda. We identified phylogenetic transmission clusters and combined these with volunteer life histories in order to understand the sexual networks within this population. From a prospective cohort of 1,000 HIV-negative individuals recruited from five communities, 51 seroconverters were identified over a period of 2 years. From these, whole blood was collected and population sequencing of the HIV-1 pol gene (protease/reverse transcriptase) was performed from plasma. Drug resistance mutations (DRMs) were scored using the 2009 WHO list for surveillance of TDR. TDR prevalence categories were estimated using the WHO recommended truncated sampling technique for the surveillance of TDR for use in resource-limited settings (RLS). Of the samples 92% (47/51) were successfully genotyped. HIV-1 subtype frequencies were 15/47 (32%) A1, 20/47 (43%) D, 1/47 (2%) C, 1/47 (2%) G, and 10/47 (21%) unique recombinant forms. Nonnucleoside reverse transcriptase inhibitor (NNRTI) drug resistance mutation K103N was identified in two individuals and V106A in one (6%) suggesting that the level of TDR was moderate in this population. No nucleoside/tide reverse transcriptase inhibitor (NRTI) or protease inhibitor (PI) DRMs were detected. In this study, we identified five transmission clusters supported by high bootstrap values and low genetic distances. Of these, one pair included the two individuals with K103N. Two of the genotypic clusters corresponded with reported sexual partnerships as detected through prior in-depth interviews. The level of TDR to NNRTIs in these ARV-naive individuals was moderate by WHO threshold survey categorization. The transmission clusters suggest a high degree of sexual partner mixing between members of these communities.
Introduction
The prevalence and incidence of HIV-1 in the fishing communities around Lake Victoria in East Africa are high.1–3 In a recent study from Uganda, the prevalence was about 29%1 and was associated with a high annual incidence rate of 4.9%.2 The HIV prevalence and incidence rates are high compared to the national prevalence of 7.3%4 and incidence of 0.45%; this was mainly explained by high-risk behavior.2 The location and setting of these study sites have been described elsewhere.1,2 These communities should be high priority groups for HIV intervention programs but the mobile lifestyle of the population is a barrier to accessing them and may pose challenges to acceptable adherence to antiretroviral (ARV) treatment.5 Health service facilities where ARV drugs are dispensed are often inadequate or not available in fishing communities.
Drug resistance mutations (DRMs) are associated with treatment failure. The onward transmission of drug-resistant variants to newly infected individuals is termed transmitted drug resistance (TDR).6,7 The occurrence of TDR is driven by ART service and duration, the rate of newly acquired drug resistance, and the fitness of the drug-resistant strains that evolve.8,9 Initial surveys of TDR in sub-Saharan Africa during early ARV scale-up have shown a prevalence of below 5%.8,10–14 However, recent surveys suggest that TDR increases as these countries scale-up antiretroviral treatment (ART) access.15 There are no data on TDR from fishing communities in Uganda, and given the higher prevalence and incidence it is important to determine the levels of TDR in these highly mobile populations. In this study, we evaluated the prevalence of drug-resistant mutations among newly infected ARV-naive individuals from fishing communities on the shores of Lake Victoria. We refer to the people in these sites as fisher folk, a term commonly used in the literature to refer to all the people, including women and children, who live and work in fishing communities. The fishing communities included in this study are described in more detail elsewhere.1 We also explored changes in sexual partner change (termed sexual mixing) and the pattern of change in HIV-1 transmission in this population through phylogenetic transmission clusters and volunteer life histories. In the course of qualitative data collection, the life histories of participants were gathered through in-depth interviews. These are referred to as “volunteer life histories” in this article.
Materials and Methods
Study population
Our study was nested in a larger study designed to prospectively determine HIV incidence and risk factors for HIV infection, and to describe the molecular epidemiology and the social and behavioral characteristics of fishing populations of three districts in Uganda. This larger study was conducted in preparation for future HIV prevention research including possible HIV vaccine trials1 in five fishing communities selected from three districts of Uganda (Masaka, Wakiso, and Mukono). These sites were chosen based on the population size, the level of fishing activity, mobility, and proximity to Entebbe or Masaka where our research laboratories are based; this was done to ensure that other fishing sites were representative. Details on the process of selection of communities have been explained elsewhere.1,2
A total of 2,074 persons residing in these fishing communities were screened; a cohort of 1,000 seronegative individuals was recruited and samples were taken at six monthly visits over 2 years. These volunteers were chosen based on “high-risk” behavior while also ensuring a gender balance until 1,000 participants had been selected. Of these, 51 individuals subsequently seroconverted. The date of HIV-1 infection was estimated as the mid-point between the last HIV-1 antibody-negative test result and the first antibody-positive test result. This group of 51 recently infected persons with no reported prior exposure to ARVs constituted the study population for the study presented here. Details on HIV-1 prevalence and incidence among this population have been described elsewhere.1,2
Consenting volunteers who were eligible to participate in the prospective study were characterized as follows: aged between 13 and 49, reporting any one of the following: unprotected sex with more than one partner and/or new partners in the past 3 months, being away from home or having a partner who is away from home for at least two nights in the past month, reported or current sexually transmitted infection (STI) in the previous 3 months, and reporting being in an HIV serodiscordant relationship. Multiple sexual partners refers to frequent partner change and having changed a partner in the past 3 months. Demographic and risk behavior data, physical address information, medical examination, HIV testing and counseling, and pregnancy test were collected from study volunteers every 6 months at routine visits. All female study volunteers who seroconverted tested negative for pregnancy.
In addition, a qualitative substudy focused on documenting life histories. We recruited participants from the general population in the fishing communities, as well as a group of volunteers ineligible for the HIV-negative cohort because of seropositivity at the time of enrollment and those who enrolled but later seroconverted during follow-up. Experienced interviewers carried out the interviews under the direction of a senior social scientist. Interviewers were blinded to the HIV status of volunteers but this information was invariably shared spontaneously by the individual during the interview. In total 78 volunteers were interviewed in this way; this included 25% of the incident cases, including the 10 people who were identified as belonging to virological transmission clusters. Interviews were carried out before phylogenetic analysis. The life history data were analyzed manually using thematic content analysis.
The study was approved by the Science and Ethics Committee of the Uganda Virus Research Institute (UVRI) and the Uganda National Council of Science and Technology. Written informed consent was obtained from all volunteers before participation in the study. The consent form indicated that molecular epidemiology results would be compared with social and behavioral data. HIV counseling, STI treatment, and treatment for other ailments were provided by the survey team.
Laboratory procedures
Samples were tested for HIV-1 using a rapid HIV-1 test (Determine HIV-1/2, Inverness Medical, Tokyo, Japan) in the field. HIV-positive results were confirmed using two independent ELISA tests (Vironostika, Biomerieux SA, Marcy-l'Etoile, France and Murex, Murex Biotech Limited, Dartford, UK). Confirmed HIV-1-positive samples were tested for CD4+ T cell counts (Facscalibur Becton-Dickinson, Franklin Lakes, NJ) at the time of diagnosis.
Whole blood was processed to obtain plasma that was stored at −80°C for HIV-1 RNA extraction. RNA was extracted from 140 μl of plasma using the QIAamp Viral RNA extraction Mini-kit (Qiagen, Hilden, Germany). Genotypic analysis was performed through nested polymerase chain reaction (PCR) of protease (codons 1–99) and the amino terminus of reverse transcriptase (codons 1–242) using methods described previously.16 Gag/p24 (460 bp) and env gp41 (460 bp) regions were also amplified using gene-specific primers as described elsewhere.17,18 Amplification products were sequenced at the Medical Research Council (MRC)/UVRI laboratories with an Applied Biosystems 3130 genetic analyzer (Applied Biosystems, Foster City, CA). The MRC/UVRI laboratory is accredited by the WHO for HIV-1 genotyping and HIV drug resistance testing (HIVDR), and is the national and regional reference laboratory conforming to the WHO Virology Quality Assurance (VQA), the TREAT Asia Quality Assurance Scheme (TAQAS), and Quality Control for Molecular Diagnostics (QCMD) external quality assessment for HIVDR genotyping during the course of this study. The external quality control assessments for the CD4 testing were the United Kingdom National External Quality Assessment Service (UKNEQAS) and a proficiency panel from the National Health Laboratory Service (NHLS).
Phylogenetic analysis
Viral sequences were assembled and checked for quality using Sequencer 4.10.1 (Gene Codes Corporation, Ann Arbor, MI); only sequences with clear results with both forward and reverse primers were used. HIV subtype was determined by alignment with standard subtype references (MacGDE) using Clustal X as well as with REGA (www.bioafrica.net/rega-genotype/html/subtypinghiv.html) and RIP (www.hiv.lanl.gov/content/sequence/RIP/RIP.html/). Phylogenetic analysis was conducted using neighbor-joining (MEGA) and maximum parsimony (PAUP) bootstrap computation.19,20 Viral sequences outside subtype clusters were analyzed using SimPlot, v3.421 and SCUEAL22 for intersubtype recombination. Transmission clusters were supported by higher bootstrap values (>99%) with 1,000 resampling and short branch lengths indicating a statistically significant relationship between sequences.
Drug resistance mutation analysis and statistical methods
TDR prevalence categories were estimated using sequencing in keeping with the truncated sampling technique recommended by WHO for surveillance of transmitted HIV TDR, which is based on the testing of at least 47 consecutively collected plasma samples from drug-naive HIV seroconverters. In this study, age was not considered as these were all well-documented seroconverters with the first confirmed HIV-positive sample after a previous seronegative sample within 6 months. The local TDR level is then categorized as low (<5%), moderate (5–15%), or high (>15%) for each of the three main ARV drug classes.23 Sequences were submitted to the Stanford calibrated population resistance (CPR tool) v 5.0beta to identify HIV-1 drug mutations associated with resistance using the WHO SDRM 2009 mutation list.7 Data were imported in MS Access 2003 (Microsoft Corporation, USA) and analyzed using STATA 11 (StataCorp, College Station, TX). Frequencies were obtained for sociodemographic variables and bivariable analyses were done for each of the sociodemographic variables and the outcomes. The discrete variables were compared using Pearson's chi-square test, and the means of the continuous variables were compared using a t-test.
Results
Volunteer characteristics
The study took place between February 2009 and January 2011. The characteristics of the 51 volunteers who had seroconverted and were enrolled in this substudy are shown in Table 1. The study population comprised 24 women (47%) and 27 men (53%). The mean age was 26.5 years (SD 6.32 years). All 51 participants had documented laboratory evidence of seroconversion and had an estimated date of HIV-1 infection between May 2009 and September 2010. The median estimated duration of infection was 2.9 months (IQR: 2.73–3.7 months and range: 1.4–6.4 months).
Table 1.
Patient Characteristics by Study Site
| |
|
Sites |
|
|---|---|---|---|
| Total | Masaka | Wakiso | |
| Volunteers | 51 | 30 (58.8) | 21 (41.2) |
| Sex | |||
| Female | 24 (47.1) | 13 (43.3) | 11 (52.3) |
| Male | 27 (52.9) | 17 (56.7) | 10 (47.6) |
| Median age (IQR) years | 25 (21,31) | 25 (21,31) | 25 (23,33) |
| Resident at the site | |||
| Yes | 44 (86.3) | 27 (90.0) | 17 (81.0) |
| No | 7 (13.7) | 3 (10.0) | 4 (19.0) |
| Number of sexual partners in the past 3 months | |||
| 0–1 | 33 (64.7) | 22 (73.3) | 11 (52.3) |
| 2+ | 18 (35.7) | 8 (26.7) | 10 (47.7) |
| STI | |||
| Yes | 27 (52.9) | 14 (46.7) | 13 (61.9) |
| No | 24 (47.1) | 16 (53.3) | 8 (38.1) |
| CD4 cell count—median cell/μl (IQR) | 656.5 (465,896) | 677.5 (450, 951.5) | 638 (512.5, 830) |
| Marital status | |||
| Not married | 25 (49.0) | 12 (40.0) | 13 (61.9) |
| Married | 26 (51.0) | 18 (60.0) | 8 (38.1) |
| Number of years lived on the site | |||
| 5–49 years | 10 (19.6) | 5 (16.7) | 5 (23.8) |
| 1–<5 years | 25 (49.0) | 15 (50.0) | 10 (47.6) |
| <1 year | 16 (31.4) | 10 (33.3) | 6 (28.6) |
| HIV-1 subtypes in the pol region | |||
| A1 | 15 (31.9) | 8 (29.6) | 7 (35.0) |
| D | 20 (42.6) | 14 (51.9) | 6 (30.0) |
| Othersa | 2 (4.3) | 0 (0.0) | 2 (10.0) |
| Recombinantsb | 10 (21.3) | 5 (18.5) | 5 (25.0) |
Other subtypes include 1G and 1C.
These include A1D, CD.
Data represent n (%), unless otherwise specified.
IQR, interquartile range; STI, sexually transmitted infection.
The median CD4 count at the time of HIV diagnosis was 656.5 cells/μl (IQR 484,874). The majority of the incident volunteers had CD4 counts less than 500 cells/μl and this is not uncommon in our population within 3 to 6 months of diagnosis. Of the participants 86% (44/51) lived within the fishing communities at the time of the study; the remainder were visitors who frequently stayed in the fishing communities for business, e.g., fish traders or women who provided sexual services. Multiple heterosexual partners were reported by 24 (47%) participants, and 27 (53%) reported a recent episode of an STI. Syphilis, genital sores, and genital discharge were the STIs considered as described elsewhere.1
Genotypic profiles
A genotype result was obtained for 92% (47/51) of volunteers from samples collected an average of 90 days after their estimated date of infection (median: 87 days, IQR: 82–111 days). Subtype analysis of the pol region showed that HIV-1 subtype D was most common 42.6% (20/47), followed by subtype A1 31.9% (15/40), subtype G 2% (1/47), subtype C 2% (1/47), and unique recombinants 21.5% (10/47). Using subtype analysis from the gag/p24 region, subtype D was also most commonly identified (48%), followed by subtype A1 (45%), subtype G (5%), and subtype C (2%). Based on the env/gp41 region, HIV-1 subtype A was most commonly identified (59%), followed by subtype D (34%), subtype C (5%), and subtype G (2%). A total of 47% (24/51) had the same subtype between pol, gag/p24, and env/gp41.
Of the 47 samples sequenced, two had K103N and one V106A nonnucleoside reverse transcriptase inhibitor (NNRTI) mutation (6.4%; 95% CI: 0.6–13%) indicating a moderate level of NNRTI TDR (5–15%). The lack of other resistance mutations suggests that the level of TDR for protease inhibitor (PI) and nucleoside reverse transcriptase inhibitor (NRTI) was low (<5%) (Table 2). The three volunteers with TDR were identified as subtype D/A1/D in the pol region, subtype A1/A1/D in the gag region, and subtype A1/A1/D in the env region, respectively. To account for the variability in HIV incidence, we treated the denominator of the prevalence of drug resistance (denominator=number of incident cases) as a random variable and estimated prevalence as a ratio. This resulted in a smaller standard error. The estimated prevalence was 6.4% with standard error 0.0022563 and 95% CI (0 .0594022–0.0682574). Thus the result, which does not treat the denominator as a random variable, is more conservative and is to be preferred.
Table 2.
Demographic Characteristics of Three Patients Who Had Drug Resistance Mutations
| Number | Age | Gender | Profession | Reported STI | Number of sexual partners | Date of enrollment mm.dd.yy | Estimated date of infection | CD4 counts cells/μl | Subtype (gag/pol/env) | NNRTI SDRM |
|---|---|---|---|---|---|---|---|---|---|---|
| 1a | 20 | Female | Fish processing | Yes | 2 | 1/6/10 | 7/30/09 | 461 | A1/D/A1 | K103N |
| 2a | 22 | Male | Fisherman | Yes | 2 | 2/17/10 | 10/29/09 | 1122 | D/D/D | K103N |
| 3 | 24 | Male | Fishmonger | Yes | 1 | 5/5/10 | 12/7/09 | 616 | A1/A1/A1 | V106A |
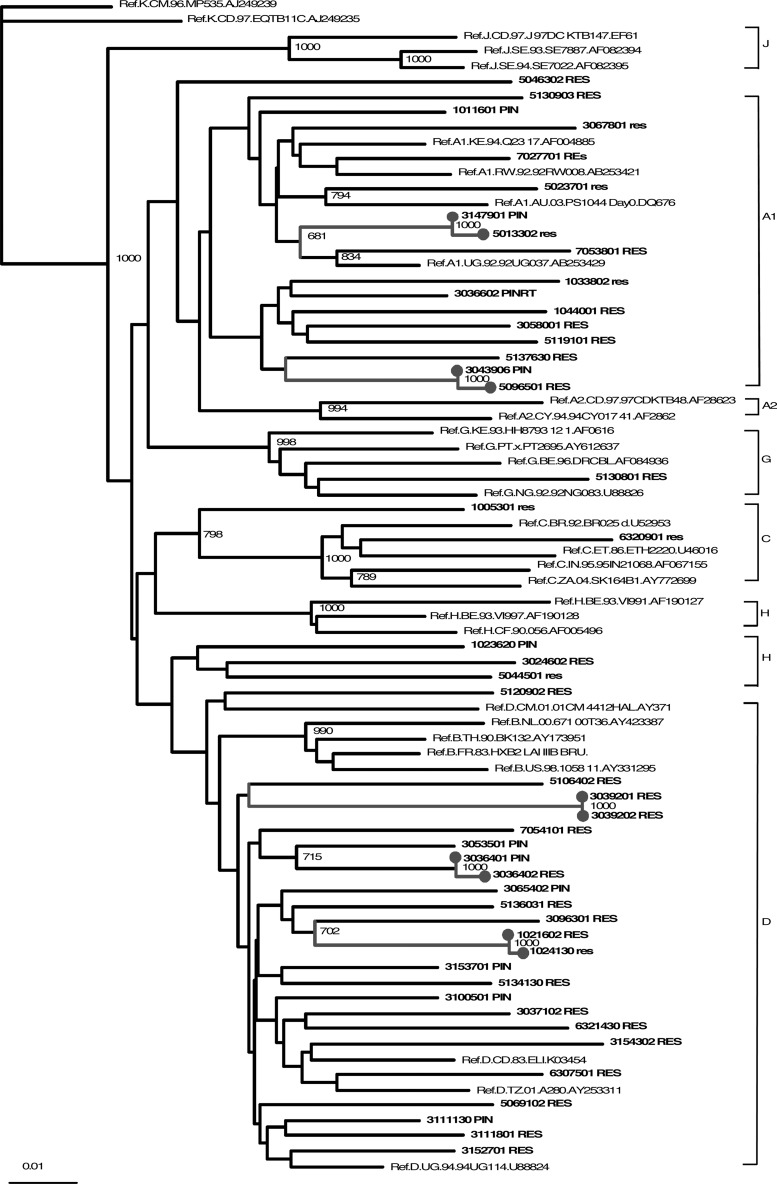
Patients 1 and 2 clustered together in the phylogenetic tree (Fig. 1).
NNRTI, nonnucleoside reverse transcriptase inhibitor; SDRM, surveillance drug resistance mutation.
We detected five (10/47, 21%) transmission clusters. Two clusters of two volunteers each were documented among partners from the same household and one of these had the K103N mutation as shown in Fig. 1. Another transmission cluster was detected linking two volunteers from recruitment sites separated by 120 miles. All these volunteers reported that they had had multiple sex partners, that they had rarely used condoms, and that they had lived in more than three villages or towns in the past 12 months.
FIG. 1.
Phylogenetic tree of HIV strains detected in 47 recent seroconverters, based on pol sequences. Transmission clusters are marked in gray.
Discussion
In an effort to contribute to the surveillance of HIV TDR in East Africa by applying the WHO TDR threshold survey method,8 we have shown that TDR was about 6% in this cohort of 47 recent HIV seroconverters, recruited from Ugandan fishing communities with a known high prevalence and incidence of HIV infection. According to WHO surveillance criteria this level is moderate, i.e., the prevalence of TDR has reached the second of three categories of severity (<5% low, 5–15% moderate, and >15% high). To our knowledge, this was the first study of TDR among fishing communities on a large inland lake in East Africa.
Previous studies of TDR in Uganda showed a prevalence of less than 5% among 46 antenatal care attending women from Entebbe in 2006,14 3% among 40 female sex workers from Kampala in 2009,24 and 1.5% among 66 volunteers living in discordant couple (DC) partnerships in rural SW Uganda in 2010.15 However, it was 9% among 70 participants of Voluntary Counseling and Testing (VCT) services in Kampala in 200925 and 19% among 26 DC and VCT participants from Entebbe in 2010.15 While these studies differed with respect to populations and sampling strategies and while some did not meet all the criteria recommended by WHO for TDR surveillance,8 they are consistent with trends reported from elsewhere in East Africa and other parts of sub-Saharan Africa where a prevalence between 7% and 13% has recently been reported.15,26–29
The predominant HIV subtypes were strains D, A1, and unique recombinant forms; but a few other strains were also observed (subtypes C and G). Over half of the viruses sequenced had different subtypes assigned across the three gene regions analyzed, which may suggest that a high degree of viral recombination occurs within this population. These findings were in line with those from other populations in SW Uganda.30 The mutations seen in our study were associated with resistance to NNRTIs (K103N and V106A). These mutations confer resistance to efavirenz and nevirapine, which form part of standard first line ART regimens in Uganda. Some NNRTI mutants are relatively fit and may therefore be more likely to be transmitted and to persist over time.31 In our study, no mutations were found that are associated with resistance to other classes of ARV drugs. However, mutations to NRTIs and PIs have been reported from elsewhere in Uganda since 2009.15,25
The implications of the results from this work and similar studies in the wider area include concerns about growing levels of TDR in the region. These observations are also worrying because ART has been rolled out in Uganda only from 2004 onward and levels of HIVDR seem to increase with time since the introduction of ART,32 although such trends have not been confirmed everywhere.33
The results from our and other studies call for intensified efforts to monitor the development and propagation of drug resistance. So far the monitoring of TDR is often opportunistic and depends on individual research initiatives that may or may not be available in an area. Surveillance strategies are urgently required that systematically screen populations at risk in the various countries that currently roll out ART. For this study, all volunteers with CD4 counts below 250 were referred to the nearest health care centers providing ART that were 5–12 km from the fishing communities. However, only 10% of those eligible for ART were able to make their appointment attributed to distance and time of scheduled appointment. Limited outreach programs also provide ART to these communities.
All possible efforts must be made to reduce the development of ART-resistant strains. Well-implemented intensive health education, counseling, and monitoring of patients on ART are required to achieve maximal treatment adherence. Strategies are required that already at the time of treatment initiation help to identify those most likely to be poor adherers.34 ART recipients who are highly mobile such as those found among migrant laborers or fishermen working on the East African inland lakes are likely to require additional efforts to support them in achieving good adherence. The same is the case for HIV-infected individuals who carry a high risk of HIV onward transmission due to their life styles, such as HIV-infected male and female sex workers. In our own research we experienced that these groups sometimes meet almost insurmountable barriers when they seek HIV care2,5 and this is mainly attributed to the distances between the service providers and the fishing communities as well as limited outreach programs.
Lastly, the results presented indicate that people living in fishing communities are at a particularly high risk of acquiring and transmitting HIV drug-resistant strains. Factors contributing to this include high HIV prevalence and incidence, intensive sexual mixing patterns, and a lack of access to good quality primary health care services nearby. These populations deserve much more intensive interventions targeted to prevent both HIV infection and the development of ART resistance than currently available. Various strategies could be used to improve both access to and quality of care. These may include mobile outreach services to fishing communities including those on islands. As fishing crews often operate at night and rest during the day, health services may need to be provided at unconventional hours; peer educators could help to achieve effective mobilization and health education.
There are some limitations to our study that may possibly result in inaccurate data on TDR. Minority resistance variants are likely to remain undetected by population sequencing as used in our research. Also, NNRTI minor variants may compromise treatment effectiveness. On the other hand, transmitted drug resistance may be overestimated due to natural sequence polymorphisms. Such compromises are necessary as surveillance programs are struggling with resource restrictions, resulting in small sample sizes and results with wide confidence intervals.
In conclusion, we found that the level of TDR in this vulnerable population was moderate (6%), which is a concern in this population that has limited access to healthcare services. The transmission clusters seen suggest a high degree of sexual partner mixing. Available HIV prevention and care services are still insufficient. There are all the ingredients in place that are likely to keep HIV transmission at a high rate and to lead to much higher future levels of HIV TDR in this population and beyond. Targeted interventions to reduce high-risk behavior and TDR are urgently needed including improved provision of ART as well as drug adherence. ART program issues leading to increased rates of TDR and sexual networks in these communities should be investigated.
Sequence Data
The sequences have been submitted to GenBank, accession numbers JX498971–JX499018.
The CHIVTUM Study Team
The Masaka MRC/UVRI clinic, data, community, and field team: Joseph Sembatya, Baker Ssebunya, Ventus Kwigenga, Razia Nassuna, Nasser Abdu Kivumbi, Ignatius Kamulegeya, Vincent Basajja, Frederick Makaire, Hellen Nabaleera, Michael Sebayinda, and Emanuel Aling.
The UVRI-IAVI field team: Mathias Wambuuzi, Mathias Ssekitoleko, Allan Bucyana, Simpson Nuwamanya, David Nyende, and Lilian Wampande.
The social science team: Collins Agaba, Bessie Kalina, Georgina Nabaggala, Zubayiri Sebyala, and Richard Muhumuza.
Contributor Information
Collaborators: the CHIVTUM Study Team
Acknowledgments
The authors thank the study participants and the staff of the MRC/UVRI Uganda Research Unit on AIDS, the UVRI-IAVI HIV Vaccine program, and Dr. Raph Hamers, Dr. Chris Parry, and Dr. Matt Price for carefully reviewing the manuscript.
This study was funded by the European and Developing Countries Clinical Trials Partnership (EDCTP) grant CT.2006.33111 011, IAVI and the MRC-UVRI Uganda Research Unit on AIDS. The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of any of the institutions mentioned above.
Author Disclosure Statement
The authors have no conflicts of interest.
References
- 1.Asiki G. Mpendo J. Abaasa A. Agaba C. Nanvubya A. Nielsen L, et al. HIV and syphilis prevalence and associated risk factors among fishing communities of Lake Victoria, Uganda. Sex Transm Infect. 2011;87(6):511–515. doi: 10.1136/sti.2010.046805. [DOI] [PubMed] [Google Scholar]
- 2.Seeley J. Kamali A. Mpendo J. Asiki G. Abaasa A. De Bont J. Nielsen L. Kaleebu P on behalf of the CHIVTUM study team. High HIV incidence and socio-behavioural risk patterns in fishing communities on the shores of Lake Victoria. Sex Trans Dis. 2012;39(6):433–439. doi: 10.1097/OLQ.0b013e318251555d. [DOI] [PubMed] [Google Scholar]
- 3.Kwena ZA. Bukusi EA. Ng'ayo MO. Buffardi AL. Nguti R. Richardson B, et al. Prevalence and risk factors for sexually transmitted infections in a high-risk occupational group: The case of fishermen along Lake Victoria in Kisumu, Kenya. Int J STD AIDS. 2010;21(10):708–713. doi: 10.1258/ijsa.2010.010160. [DOI] [PubMed] [Google Scholar]
- 4.Report: GARP. Uganda AIDS Indicator Survey 2011. www.unaidsorg/en/dataanalysis/monitoringcountryprogress/progressreports/2012countries/ce_UG_Narrative_Report%5B1%5Dpdf. [Jul 20;2012 ]. www.unaidsorg/en/dataanalysis/monitoringcountryprogress/progressreports/2012countries/ce_UG_Narrative_Report%5B1%5Dpdf
- 5.Seeley JA. Allison EH. HIV/AIDS in fishing communities: Challenges to delivering antiretroviral therapy to vulnerable groups. AIDS Care. 2005;17(6):688–697. doi: 10.1080/09540120412331336698. [DOI] [PubMed] [Google Scholar]
- 6.Shafer RW. Rhee SY. Pillay D. Miller V. Sandstrom P. Schapiro JM, et al. HIV-1 protease and reverse transcriptase mutations for drug resistance surveillance. AIDS. 2007;21(2):215–223. doi: 10.1097/QAD.0b013e328011e691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennett DE. Camacho RJ. Otelea D. Kuritzkes DR. Fleury H. Kiuchi M, et al. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One. 2009;4(3):e4724. doi: 10.1371/journal.pone.0004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennett DE. Myatt M. Bertagnolio S. Sutherland D. Gilks CF. Recommendations for surveillance of transmitted HIV drug resistance in countries scaling up antiretroviral treatment. Antivir Ther. 2008;13(Suppl 2):25–36. [PubMed] [Google Scholar]
- 9.Baggaley RF. Garnett GP. Ferguson NM. Modelling the impact of antiretroviral use in resource-poor settings. PLoS Med. 2006;3(4):e124. doi: 10.1371/journal.pmed.0030124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Somi GR. Kibuka T. Diallo K. Tuhuma T. Bennett DE. Yang C, et al. Surveillance of transmitted HIV drug resistance among women attending antenatal clinics in Dar es Salaam, Tanzania. Antivir Ther. 2008;13(Suppl 2):77–82. [PubMed] [Google Scholar]
- 11.Kamoto K. Aberle-Grasse J. Surveillance of transmitted HIV drug resistance with the World Health Organization threshold survey method in Lilongwe, Malawi. Antivir Ther. 2008;13(Suppl 2):83–87. [PubMed] [Google Scholar]
- 12.Abegaz WE. Grossman Z. Wolday D. Ram D. Kaplan J. Sibide K, et al. Threshold survey evaluating transmitted HIV drug resistance among public antenatal clinic clients in Addis Ababa, Ethiopia. Antivir Ther. 2008;13(Suppl 2):89–94. [PubMed] [Google Scholar]
- 13.Maphalala G. Okello V. Mndzebele S. Gwebu P. Mulima N. Dlamini S, et al. Surveillance of transmitted HIV drug resistance in the Manzini-Mbabane corridor, Swaziland, in 2006. Antivir Ther. 2008;13(Suppl 2):95–100. [PubMed] [Google Scholar]
- 14.Ndembi N. Lyagoba F. Nanteza B. Kushemererwa G. Serwanga J. Katongole-Mbidde E, et al. Transmitted antiretroviral drug resistance surveillance among newly HIV type 1-diagnosed women attending an antenatal clinic in Entebbe, Uganda. AIDS Res Hum Retroviruses. 2008;24(6):889–895. doi: 10.1089/aid.2007.0317. [DOI] [PubMed] [Google Scholar]
- 15.Price MA. Wallis CL. Lakhi S. Karita E. Kamali A. Anzala O, et al. Transmitted HIV type 1 drug resistance among individuals with recent HIV infection in East and Southern Africa. AIDS Res Hum Retroviruses. 2011;27(1):5–12. doi: 10.1089/aid.2010.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ndembi N. Goodall RL. Dunn DT. McCormick A. Burke A. Lyagoba F, et al. Viral rebound and emergence of drug resistance in the absence of viral load testing: A randomized comparison between zidovudine-lamivudine plus nevirapine and zidovudine-lamivudine plus abacavir. J Infect Dis. 2010;201(1):106–113. doi: 10.1086/648590. [DOI] [PubMed] [Google Scholar]
- 17.Chunfu Yang DP. Owen S. Fridlund C, et al. Detection of phylogenetically diverse human immunodeficiency virus type 1 groups M and O from plasma by using highly sensitive and specific generic primers. J Clin Microbiol. 1999;37(8):2581–2586. doi: 10.1128/jcm.37.8.2581-2586.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ssemwanga D. Lyagoba F. Ndembi N. Mayanja BN. Larke N. Wang S, et al. Multiple HIV-1 infections with evidence of recombination in heterosexual partnerships in a low risk rural clinical cohort in Uganda. Virology. 2011;411(1):113–131. doi: 10.1016/j.virol.2010.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 20.Saitou N. Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 21.Lole KS. Bollinger RC. Paranjape RS. Gadkari D. Kulkarni SS. Novak NG, et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73(1):152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosakovsky Pond SL. Posada D. Stawiski E. Chappey C. Poon AF. Hughes G, et al. An evolutionary model-based algorithm for accurate phylogenetic breakpoint mapping and subtype prediction in HIV-1. PLoS Comput Biol. 2009;5(11):e1000581. doi: 10.1371/journal.pcbi.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myatt M. Bennett DE. A novel sequential sampling technique for the surveillance of transmitted HIV drug resistance by cross-sectional survey for use in low resource settings. Antivir Ther. 2008;13(Suppl 2):37–48. [PubMed] [Google Scholar]
- 24.Ssemwanga D. Lyagoba F. Magambo B, et al. Transmitted antiretroviral drug resistance among drug-naïve commercial sex workers with recent infection in Kampala, Uganda. Clin Infect Dis. 2012;54(Supp l4):S339–342. doi: 10.1093/cid/cir937. [DOI] [PubMed] [Google Scholar]
- 25.Ndembi N. Hamers RL. Sigaloff KC. Lyagoba F. Magambo B. Nanteza B, et al. Transmitted antiretroviral drug resistance among newly HIV-1 diagnosed young individuals in Kampala. AIDS. 2011;25(7):905–910. doi: 10.1097/QAD.0b013e328346260f. [DOI] [PubMed] [Google Scholar]
- 26.Sigaloff KC. Mandaliya K. Hamers RL. Otieno F. Jao IM. Lyagoba F, et al. High prevalence of transmitted antiretroviral drug resistance among newly HIV type 1 diagnosed adults in Mombasa, Kenya. AIDS Res Hum Retroviruses. 2012;28(9):833–837. doi: 10.1089/AID.2011.0348. [DOI] [PubMed] [Google Scholar]
- 27.Aghokeng AF. Kouanfack C. Laurent C. Ebong E. Atem-Tambe A. Butel C, et al. Scale-up of antiretroviral treatment in sub-Saharan Africa is accompanied by increasing HIV-1 drug resistance mutations in drug-naive patients. AID. 2011;25(17):2183–2188. doi: 10.1097/QAD.0b013e32834bbbe9. [DOI] [PubMed] [Google Scholar]
- 28.Hamers RL. Wallis CL. Kityo C. Siwale M. Mandaliya K. Conradie F, et al. HIV-1 drug resistance in antiretroviral-naive individuals in sub-Saharan Africa after rollout of antiretroviral therapy: A multicentre observational study. Lancet Infect Dis. 2011;11(10):750–759. doi: 10.1016/S1473-3099(11)70149-9. [DOI] [PubMed] [Google Scholar]
- 29.Nwobegahay JM. Bessong PO. Masebe TM. Mavhandu LG. Iweriebor BC. Selabe G. Prevalence of antiretroviral drug resistance mutations and HIV-I subtypes among newly-diagnosed drug-naive persons visiting a voluntary testing and counselling centre in northeastern South Africa. J Health Popul Nutr. 2011;29(4):303–309. doi: 10.3329/jhpn.v29i4.8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ssemwanga D. Ndembi N. Lyagoba F. Bukenya J. Seeley J. Vandepitte J, et al. HIV type 1 subtype distribution, multiple infections, sexual networks, and partnership histories in female sex workers in Kampala, Uganda. AIDS Res Hum Retroviruses. 2012;28(6):558–565. doi: 10.1089/aid.2011.0024. [DOI] [PubMed] [Google Scholar]
- 31.Koval CE. Dykes C. Wang J. Demeter LM. Relative replication fitness of efavirenz-resistant mutants of HIV-1: Correlation with frequency during clinical therapy and evidence of compensation for the reduced fitness of K103N+L100I by the nucleoside resistance mutation L74V. Virology. 2006;353(1):184–192. doi: 10.1016/j.virol.2006.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta RK SB HA. Haley T. Hamers RL. Pillay D, et al. Transmitted Drug Resistance in Low and Middle Income Settings—a Meta Regression Analysis. XX International Drug Resistance Workshop Mexico; Los Cabos. Jun 7–11;2011 . [Google Scholar]
- 33.Manasa J. Katzenstein D. Cassol S. Newell ML de Oliveira for the Southern Africa Treatment and Resistance Network (SATuRN) Primary drug resistance in South Africa: Data from 10 years of surveys. AIDS Res Hum Retroviruses. 2012;28(6):558–565. doi: 10.1089/aid.2011.0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muyingo SK. Walker AS. Reid A. Munderi P. Gibb DM. Ssali F, et al. Patterns of individual and population-level adherence to antiretroviral therapy and risk factors for poor adherence in the first year of the DART trial in Uganda and Zimbabwe. J Acquir Immune Defic Syndr. 2008;48(4):468–475. doi: 10.1097/QAI.0b013e31817dc3fd. [DOI] [PubMed] [Google Scholar]