Abstract
Genome-wide association studies have shown that the rs340874 single nucleotide polymorphism (SNP) in PROX1 is a genetic susceptibility factor for type 2 diabetes. We conducted genetic and molecular studies to better understand the role of PROX1 in type 2 diabetes. We assessed the impact of the whole common genetic variability of PROX1 (80 SNPs) on type 2 diabetes–related biochemical traits in the HELENA (Healthy Lifestyle in Europe by Nutrition in Adolescence) study (n = 1,155). Three SNPs (rs340838, rs340837, and rs340836) were significantly associated with fasting plasma insulin levels (P ≤ 0.00295). We evaluated the impact of nine PROX1 SNPs (the three insulin-associated SNPs plus six SNPs in strong linkage disequilibrium) on luciferase reporter gene expression. The insulin-lowering alleles of rs340874, rs340873, and rs340835 were associated with lower luciferase activity in MIN6 and HepG2 cells (except for rs340874, which was in HepG2 cells only). Electrophoretic mobility shift assays indicated that specific nuclear protein bindings occur at the three SNPs in HepG2 cells, with allele-binding differences for rs340874. We also showed that the knockdown of Prox1 expression by small interfering RNAs in INS-1E cells resulted in a 1.7-fold reduction in glucose-stimulated insulin secretion. All together, we propose that reduced expression of PROX1 by cis-regulatory variants results in altered β-cell insulin secretion and thereby confers susceptibility to type 2 diabetes.
Type 2 diabetes is a major public health issue that will affect >439 million people worldwide by 2030 (1) and is characterized by impaired β-cell function and insulin resistance, leading to chronic hyperglycemia (2). Hyperglycemia is the main cause of microvascular and macrovascular complications (3). Type 2 diabetes is a complex metabolic disease in which both environmental and genetic factors are involved. To date, ∼40 genetic loci have been found to be associated with type 2 diabetes (4).
The Meta-Analysis of Glucose and Insulin-Related Trait Consortium (MAGIC) combined the results of 21 genome-wide association studies (GWAS) in terms of fasting glucose (FG), fasting insulin (FI), and indices of β-cell function (homeostasis model assessment [HOMA]-B) and insulin resistance (HOMA-IR) in 46,186 nondiabetic subjects (5). The authors reported 2 loci associated with FI and HOMA-IR and 16 associated with FG and HOMA-B, including the rs340874 single nucleotide polymorphism (SNP) in the prospero homeobox 1 (PROX1) gene. They also showed that carriers of the glucose-raising C allele for the rs340874 SNP display a higher risk of type 2 diabetes. Two studies with oral glucose tolerance tests showed that the C allele of rs340874 is associated with reduced insulin sensitivity and secretion (6,7). In children and adolescents, the meta-analysis by Barker et al. (8) showed that the rs340874 is associated with FG level and, to a lesser extent, HOMA-B, with similar effect sizes to those observed in adults.
In humans, PROX1 is located on chromosome 1 and was first identified in mice, thanks to its homology with the Drosophila homeobox protein prospero (9). PROX1 encodes a key transcription factor (TF) involved in the development of tissues, such as endothelial lymphatic vessels, liver, retina, and pancreas (10,11). Expression of PROX1 seems to occur in the specification and proliferation of pancreatic progenitor cells (11). Indeed, the lack of Prox1 activity prevents pancreas development and affects the organ’s cellular structure in mice (11). However, the link between PROX1 and type 2 diabetes in humans has not been established to date.
Detailed characterization of PROX1 genetic variability could help to elucidate the role of PROX1 in type 2 diabetes and to identify potential type 2 diabetes disease pathways. In contrast to previous GWAS focusing on the top hit in PROX1 (rs340874), we assessed the impact of the whole genetic variability of PROX1 (80 SNPs) on type 2 diabetes–related traits in adolescents. Genetic studies in children and adolescents are less susceptible to confounding environmental factors because of the subjects’ young age. We also evaluated the functional impact of SNPs of interest on reporter gene expression in mouse pancreatic β-cells (MIN6) and human hepatocytes (HepG2). Finally, we evaluated the influence of Prox1 in glucose-stimulated insulin secretion (GSIS) in rat pancreatic β-cells (INS-1E).
RESEARCH DESIGN AND METHODS
The HELENA study.
The recruitment and phenotyping of participants in the Healthy Lifestyle in Europe by Nutrition in Adolescence (HELENA) cross-sectional study (www.helenastudy.com) have been described previously (12). A total of 3,865 adolescents (12–18 years of age) were recruited between 2006 and 2007 from nine European countries. Adolescents were randomly selected from schools by proportional cluster sampling, taking age into account. One-third of the classes were randomly selected for blood collection, resulting in 1,155 samples. Data were collected on a detailed case report form and in accordance with standardized procedures. The protocol was approved by the appropriate investigational review board for each center. Written informed consent was obtained from each adolescent and both of his or her parents or legal representatives. Participation in the study was voluntary (13).
Venous blood samples were drawn after a 10-h overnight fast and sent to a central laboratory (IEL, Bonn, Germany) in accordance with standardized protocols (14). Serum triglyceride, total cholesterol, HDL cholesterol, LDL cholesterol, and glucose levels were enzymatically assayed on the Dimension RxL clinical chemistry system (Dade Behring, Schwalbach, Germany). Insulin was measured with an IMMULITE 2000 analyzer (DPC Biermann GmbH, Bad Nauheim, Germany). DNA was extracted from white blood cells with the Gentra Puregene Cell Kit (QIAGEN, Courtaboeuf, France).
Anthropometric measurements were strictly monitored, with participants barefoot and wearing only underwear. Weight (seca 861 electronic scale) and height (seca 225 height rod) were measured and BMI calculated. Waist and hip circumferences were measured with a nonelastic measuring tape (seca 200). Percentage of body fat was estimated from skinfold measurements (15,16).
Gene SNP selection and genotyping.
Using the HapMap database (release 28, August 2010) and applying a minor allele frequency (MAF) >0.025 and r2 ≥ 0.80 in Haploview (17), the region of PROX1 (chromosome 1 212,223,454…212,281,411) described six haplotype blocks (NCBI build 36, block 1 rs366684/rs3767844/rs3754138/rs4282786/rs3754140/rs3767848/rs446175/rs4655480/rs726334, block 2 rs11120242/rs12089523/rs12081352/rs6686424/rs12092859, block 3 rs10494972/rs7543057, block 4 rs4655313/rs4655314, block 5 rs340835/rs340839, block 6 rs340837/rs340873) and six independent SNPs. We selected one tag SNP from each block (rs3754138, rs12092859, rs10494972, rs4655313, rs340835, and rs340837) and the six independent SNPs (rs11802122, rs2289002, rs340877, rs4655482, rs340874, and rs12748973) to cover the whole PROX1. In total, 12 SNPs were genotyped using KASPar technology (KBioscience, Hoddesdon, U.K.) (Supplementary Table 1). The genotyping success rate was between 97.9 and 99.7%. Two SNPs (rs12092859 and rs10494972) did not respect the Hardy-Weinberg equilibrium (Supplementary Table 1) and, therefore, were not used for the imputation analyses.
Imputation.
SNP data were mapped to the National Center for Biotechnology Information (NCBI) build 37 coordinates (assembly hg19) to match the genomic position used by the 1000 Genomes European reference panel. IMPUTE2 software was used to phase the observed genotypes and impute missing genotypes in the PROX1 region (chromosome 1 214,156,831–214,214,788, NCBI build 37 coordinates, assembly hg19) (18). We increased the number of hidden Markov model states for phasing the study dataset from 80 to 200 for higher accuracy and increased the number of hidden Markov model states for imputation to take into account all the reference diplotypes in the reference panel. The 1000 Genomes version 2 (March 2012) of the 20110521 release was used. The reference panel comprised 379 reference haplotypes of the European ancestry. Only SNPs with an MAF >0.025 in this reference panel were considered (n = 100). Imputation quality control was assured by looking at the information measure that represents the measure of the observed statistical information associated with the allele frequency estimate. All SNPs with an information measure >0.80 were included for the association analyses. From the 10 genotyped PROX1 SNPs, we imputed 70 other SNPs in the PROX1 region (see linkage disequilibrium [LD] pattern in Supplementary Figure 1).
Statistical analysis.
Departure from Hardy-Weinberg equilibrium within the study groups was tested using the χ2 test. Statistical analyses were performed with SNPTEST software, using the genotype probabilities to take into account the uncertainty (19).
To obtain normal distributions, a log-transformation was applied for plasma triglyceride and insulin levels, HOMA-IR, and HOMA-B. An additive multivariate generalized linear model with a missing data likelihood score test was used for each trait. The covariates were age, sex, BMI, and study center.
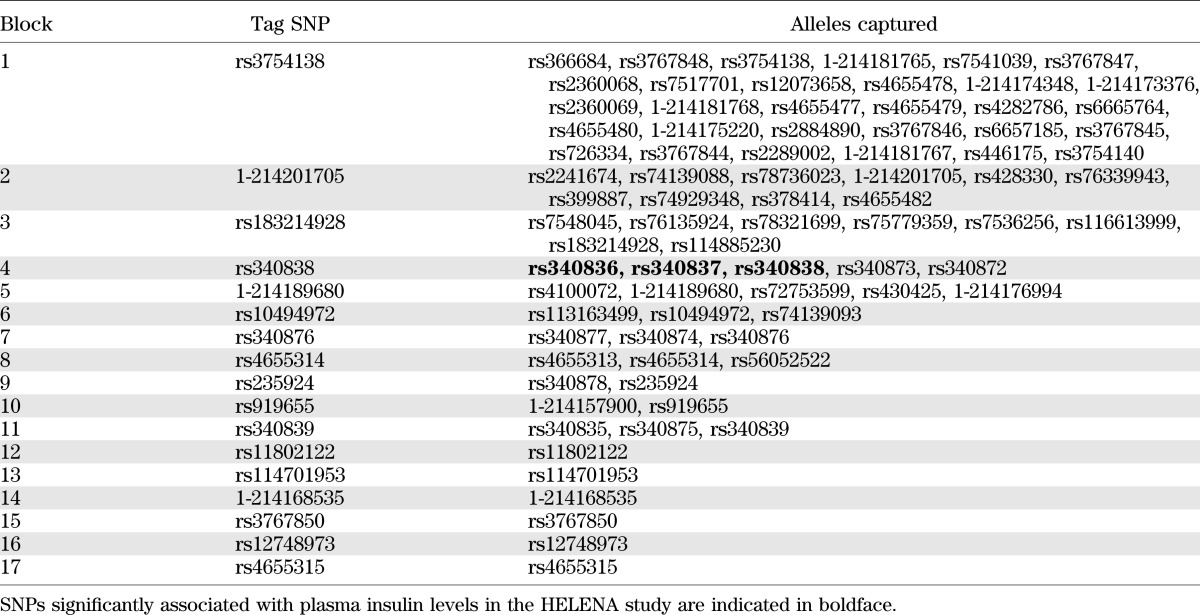
Results with P ≤ 0.05 were considered nominally significant. Correction for multiple testing was applied by dividing 0.05 by the number of independent SNPs (assessed with the SNPtagger tool in Haploview 4.2 [www.broad.mit.edu/mpg/haploview] with an r2 threshold of 0.80). Seventeen tag SNPs were identified (Table 1), and therefore, results with P ≤ 0.00295 were considered statistically significant. Power calculations were performed using Quanto version 1.2.4 (20). For molecular experiments, Mann-Whitney nonparametric tests were performed with SAS statistical software (SAS Institute Inc., Cary, NC).
TABLE 1.
Identification of the tag SNPs among the 70 imputed and 10 genotyped SNPs using SNP tagger (Haploview software) in the 1000 Genomes European reference panel (March 2012 release)
Cell culture.
MIN6 cells (a gift from B. Staels, Lille, France) were cultured in Dulbecco’s modified Eagle’s medium-GlutaMAX medium supplemented with 10% FCS (Life Technologies, Villebon-sur-Yvette, France), 2 mmol/L l-glutamine (Life Technologies), 100 μmol/L β-mercaptoethanol (Sigma, Saint-Quentin-Fallavier, France), and 100 units/mL penicillin/streptomycin (Life Technologies) at 37°C and 5% CO2. INS-1E cells (a gift from B. Staels) were cultured in RPMI medium supplemented with 10% FCS, 1 mmol/L Na pyruvate (Life Technologies), 100 μmol/L β-mercaptoethanol, and 100 units/mL gentamicin (Life Technologies) at 37°C and 5% CO2. HepG2 cells were purchased from ATCC-LGC Standards (Molsheim, France) and were cultured in minimum essential medium (Life Technologies) supplemented with 10% FCS (HyClone, Brebières, France), 2 mmol/L l-glutamine, 100 units/mL penicillin/streptomycin, 2 mmol/L minimum essential medium nonessential amino acids (Life Technologies), and 1 mmol/L Na pyruvate at 37°C and 5% CO2.
Plasmid transfections.
MIN6 and HepG2 cells were grown at 60–70% confluence in 24-well plates and were transfected with 500 ng of empty pGL4.23[luc2/minP] vector (Promega, Charbonnieres, France) or pGL4-minP-PROX1 vector with either lipofectamine 2000 (Life Technologies) using a lipofectamine:DNA ratio of 6:1 for MIN6 cells or FuGENE HD (Roche Applied Science, Meylan, France) using a FuGENE:DNA ratio of 6:1 for HepG2 cells. Luciferase activities were measured 48 h after transfection using the Dual-Luciferase Reporter Assay kit (Promega). The firefly luciferase activity was normalized to the Renilla luciferase activity obtained by cotransfection of 50 ng of the pGL4.74[hRluc/TK] Renilla Luciferase vector (Promega).
Small interfering RNA transfections and insulin immunoassay.
INS-1E cells were grown in 6-well plates at 70% confluence and transfected with either the On-Targetplus nontargeting pool small interfering RNAs (NT siRNAs) (negative control) or a specific On-Targetplus siRNA SMARTpool against rat Prox1 or a specific On-Targetplus siRNA pool against rat cyclophilin B (positive control) (Dharmacon, Thermo Fisher Scientific, Lafayette, CO) using DharmaFECT 2 according to the manufacturer’s instructions. The optimum transfection efficiency (>70% transfection, 75% inhibition of cyclophilin B) was obtained using a cell density of 5 × 106 cells/well, 120 nmol/L siRNAs, and 12 µL DharmaFECT 2 during 48 h (data not shown). After that, INS-1E cells were incubated with 1 mL of Krebs buffer for 1 h and then with 3 or 30 mmol/L glucose diluted in Krebs buffer for 1 h. The insulin concentration was measured in the supernatant (diluted at 1:2,000) using an ultrasensitive rat insulin ELISA kit (Mercodia, Uppsala, Sweden).
Glucose treatment on Prox1 expression.
INS-1E cells were grown in 6-well plates. When the cells reached 60–80% confluence, the culture medium was changed to glucose-free RPMI medium supplemented with 3 mmol/L glucose (Life Technologies) for 24 h. After that time, the medium was changed for glucose-free RPMI medium supplemented with either 3 or 30 mmol/L glucose for 24 or 48 h.
Quantitative real-time PCR.
Total RNA was extracted using the RNeasy Plus Mini Kit (QIAGEN, Les Ulis, France). cDNA was prepared from 0.5 µg total RNA using the SuperScript VILO cDNA Synthesis Kit (Life Technologies). PCRs were performed using the Brilliant SYBR Green QPCR Master Mix and the Mx3000P QPCR System (Agilent Technologies, Massy, France). Quantifications were performed with TFIIB as the internal standard and presented as fold change-over-control values. Primer sequences (forward and reverse, respectively) were rat PROX1 (ACCCGTTACCCCAGCTCCAACATGC, TTGATGGCTTGGCGCGCATACTTC), rat TFIIB (GTTCTGCTCCAACCTTTGCCT, TGTGTAGCTGCCATCTGCACTT), and rat L-PK (liver pyruvate kinase) (TGGACATCATCTTTGCCTCCTT, CTGCTAACACGTCACTGGCTTT).
Western blot analysis.
Nuclear protein extracts were prepared using the NE-PER Kit (Pierce, Rockford, IL). Forty micrograms of nuclear proteins were separated by Novex NuPAGE 4–12% (Life Technologies) and transferred to nitrocellulose membranes. The membranes were immunoblotted overnight with rabbit polyclonal anti-PROX1 (ab38962 diluted 1:250 [Abcam, Cambridge, UK]) or rabbit polyclonal anti-Ras-related nuclear protein (RAN) (ab53775 diluted 1:20,000 [Abcam]). Detection was achieved with horseradish peroxidase-conjugated antirabbit IgG goat immunoglobulin (Bio-Rad, Marnes-la-Coquette, France) at 1:10,000 for 1 h followed by enhanced chemiluminescence using the ECL Western Blotting System detection kit (GE Healthcare, Vélizy-Villacoublay, France). The image was detected with a Chemidoc camera (Bio-Rad).
Electrophoretic mobility shift assays.
Nuclear protein extracts from HepG2 and MIN6 cells were prepared as just described. Probe sequences were 40 bp long, with each SNP allele at the middle. The forward oligonucleotide was labeled with digoxigenin (DIG) at the 3′ end. Electrophoretic mobility shift assays (EMSAs) were performed using the DIG Gel Shift Kit, 2nd generation (Roche Applied Science) with 5 μg of protein extracts and 2 pmol DIG-labeled probe. For competition EMSA, a 50-fold excess of unlabeled oligonucleotide probes was added to the reaction mixtures. Samples underwent electrophoresis on a 6% polyacrylamide gel and were transferred to a nylon membrane. After washes and blocking, the membrane was immunoblotted for 30 min with anti-DIG antibody diluted at 1:10,000 in the blocking solution at room temperature. Detection was achieved with the CDP-Star chemiluminescent substrate (Roche Applied Science) diluted at 1:1,000 in the detection buffer. The image was detected with a Chemidoc camera.
RESULTS
Association studies between SNPs and glucose-related variables.
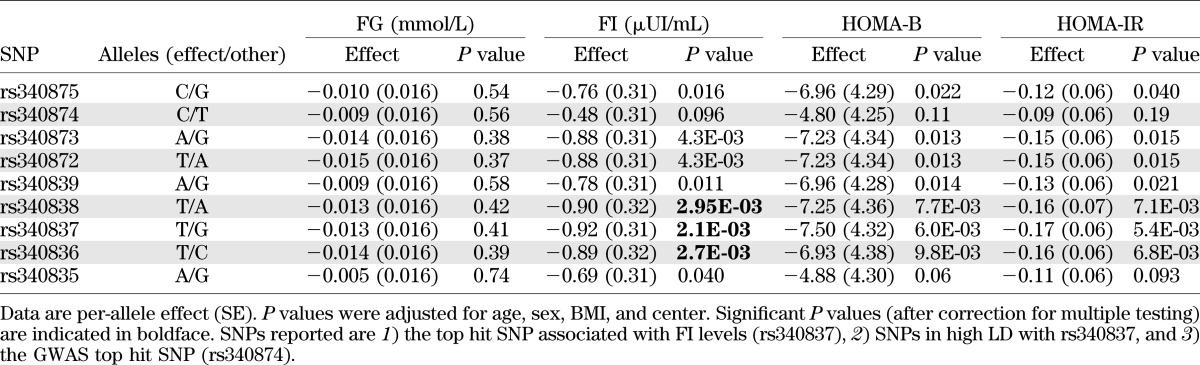
Table 2 shows the clinical characteristics of the HELENA study participants. The mean age was 14.7 ± 1.4 years, and 48% were boys. We tested the associations between the 17 tag SNPs (3 genotyped and 14 imputed) and anthropometric parameters, biochemical parameters, and the HOMA-B and HOMA-IR indices. No significant associations were detected with the anthropometric phenotypes or with the plasma lipid variables (data not shown). In contrast, significant associations were observed between rs340838 and fasting plasma insulin levels (P = 2.95 × 10−3). Carriers of the major T allele of rs340838 had lower insulin levels than those homozygous for the minor allele (β = −0.90 ± 0.32 µIU/mL). To better define this association signal, all 80 SNPs were tested with plasma insulin levels (Fig. 1). Two other SNPs (rs340837 and rs340836) were significantly associated with plasma insulin levels, with the best association detected for rs340837 (P = 2.1 × 10−3) (Fig. 1 and Table 3). The rs340873 and rs340872 SNPs also tended to be associated with insulin levels (P = 0.0043). Consistently, the insulin-lowering alleles of the rs340838, rs340837, and rs340836 SNPs tended to be associated with lower HOMA-B (0.0060 ≤ P ≤ 0.0098) and lower HOMA-IR (0.0054 ≤ P ≤ 0.0071) (Table 3). Of note, the GWAS top hit SNP (rs340874) was not significantly associated with plasma glucose level, insulin level, HOMA-B, or HOMA-IR (P = 0.56, 0.096, 0.11, and 0.19, respectively) (Table 3), but the data are in line with those of Barker et al. (8) because the carriers of the rs340874 C glucose-raising allele had lower HOMA-B than the carriers of the other allele (P = 0.11) (Table 3), especially in girls (P = 0.033) (data not shown).
TABLE 2.
Characteristics of the participants in the HELENA study
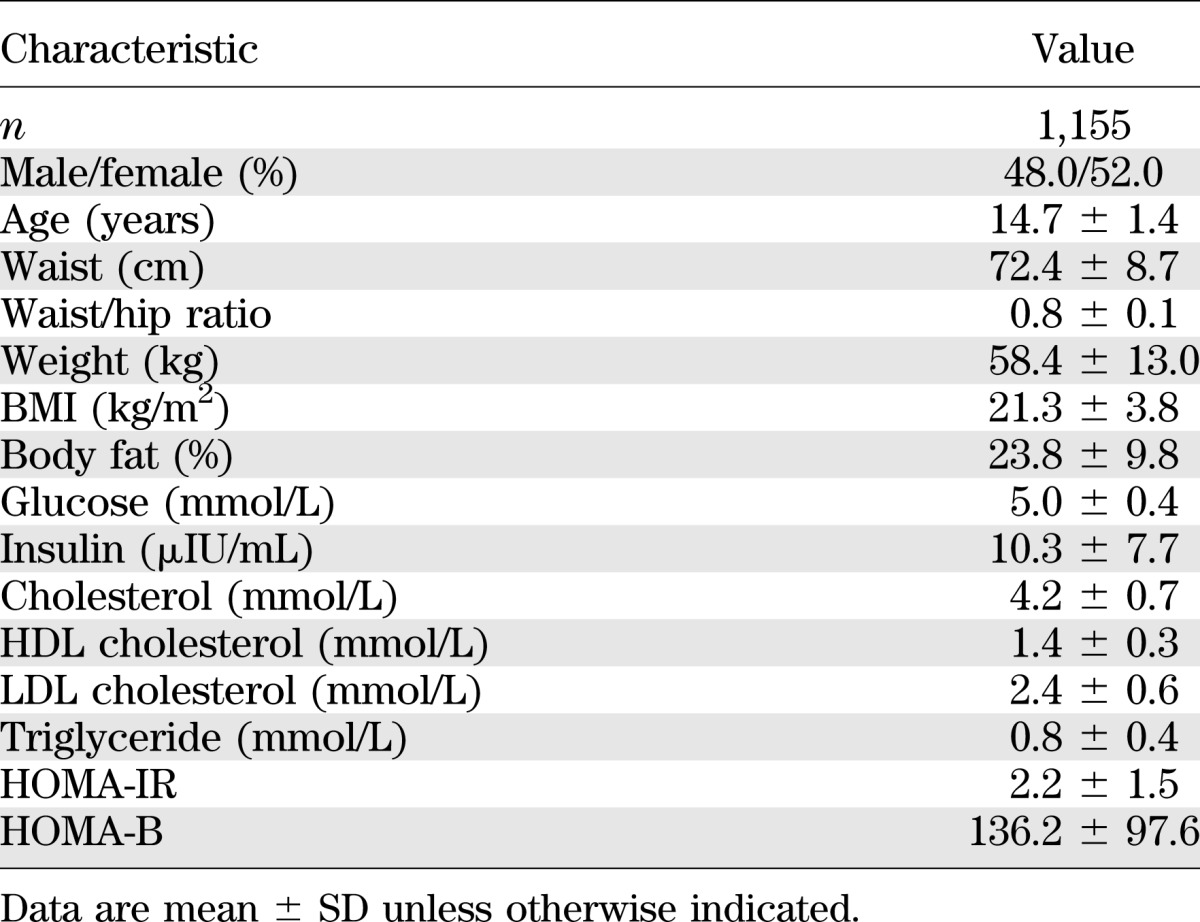
FIG. 1.

Degree of association between the 80 PROX1 SNPs and FI levels in the HELENA study. The P values (as –log10 values) of the associations between the 10 genotyped (▲) or 70 imputed (●) SNPs and fasting plasma insulin levels are plotted as a function of genomic position (NCBI build 37) with LocusZoom. Estimated recombination rates (taken from the 1000 Genomes European reference panel) are plotted to reflect the local LD structure around the associated SNPs and their correlated proxies (according to a blue-to-red scale from r2 = 0 to 1 based on pairwise r2 values from the 1000 Genomes European reference panel and taking the rs340837 SNP [best hit] as the reference).
TABLE 3.
Association between PROX1 SNPs and glucose-related traits in the HELENA study
Luciferase assays.
To identify the functional SNPs responsible of the association with FI levels, we characterized the functionality of the three insulin-associated SNPs as well as of all the SNPs in strong LD (r2 > 0.80) with them (i.e., rs340875, rs340873, rs340872, rs340839, and rs340835) (SNPs shown in red in Fig. 1 and in blocks 4 and 11 in Table 1). We also tested the functional impact of the GWAS top hit SNP (rs340874). Because these nine SNPs are located in the 5′ region or in intron 1 of PROX1, they may modulate PROX1 transcription. To investigate this hypothesis, we subcloned into the pGL4-minP luciferase reporter vector oligonucleotides (60 bp) encompassing the alleles of the nine SNPs and assessed in vitro the impact of these SNPs on luciferase activity using transient transfection assays in two different cell lines. MIN6 cells were used as a model of pancreatic β-cells, and because PROX1 plays a role in liver morphogenesis (21), an organ involved in glucose homeostasis, we also used HepG2 cells.
Among the nine SNPs, three presented significant allelic expression differences (Fig. 2). The rs340873 A allele was associated with a 1.5-fold (P = 0.004) and twofold (P = 0.02) lower luciferase activity in MIN6 and HepG2 cells, respectively. The rs340835 A allele was associated with a 3.4-fold (P = 0.05) and 4.2-fold (P = 0.02) lower luciferase activity in MIN6 and HepG2 cells, respectively. In addition, the rs340874 C allele was associated with a 1.65-fold (P = 0.05) lower luciferase activity in HepG2 cells only. These results suggest that PROX1 expression may be lower in the pancreas and liver of individuals carrying the insulin-lowering alleles.
FIG. 2.
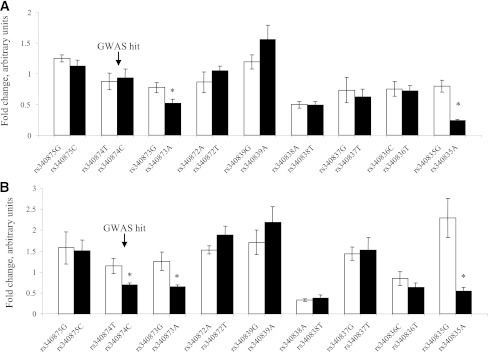
Impact of nine PROX1 SNPs on luciferase activities in MIN6 and HepG2 cells. MIN6 (A) and HepG2 (B) cells were transiently transfected for 48 h with either empty pGL4-minP vector or pGL4-minP-PROX1 vectors. Firefly luciferase activities were normalized to the Renilla luciferase activities. Activity of the construct is expressed as fold activity compared with empty pGL4-minP vector. Data are mean ± SEM of three independent experiments. *P < 0.05 (Mann-Whitney nonparametric test).
EMSAs.
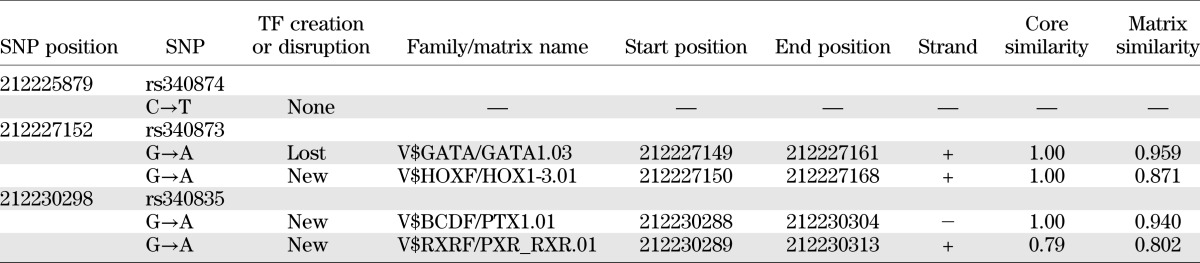
To investigate whether the differences in luciferase activities observed were a result of differential TF binding to PROX1 risk alleles, we performed EMSAs with nuclear protein extracts of HepG2 and MIN6 cells. Probes that contained both alleles of the three SNPs were used. No DNA-protein complex could be detected with MIN6 or INS-1E extracts (data not shown). In contrast, DNA-protein complexes were observed for the rs340874, rs340873, and rs340835 probes with HepG2 extracts (Fig. 3). Each DNA-protein complex was specific because it was inhibited by specific unlabeled oligonucleotide probes. Allele binding differences could be detected for the rs340874 SNP only, with a 60% higher affinity for the rs340874 C allele than for the T allele. We conducted an in silico search with Genomatix software to examine whether the SNP modified any TF binding sites, but no putative TF was described at rs340874. Two TF binding sites for rs340873 and rs340835 could be detected, but neither seemed particularly relevant in type 2 diabetes (Table 4).
FIG. 3.
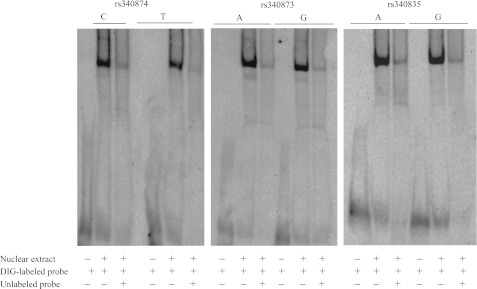
EMSAs of PROX1 SNPs. The DIG-labeled probes carrying alleles of rs340874, rs340873, and rs340835 were incubated with reaction mixtures containing no nuclear extracts or 5 μg of nuclear extracts from HepG2 cells. Specific competition was assessed using 50-fold molar excess of unlabeled probes.
TABLE 4.
TF binding sites identified by Genomatix software at the rs340874, rs340873, and rs340835 SNPs
Prox1 and GSIS.
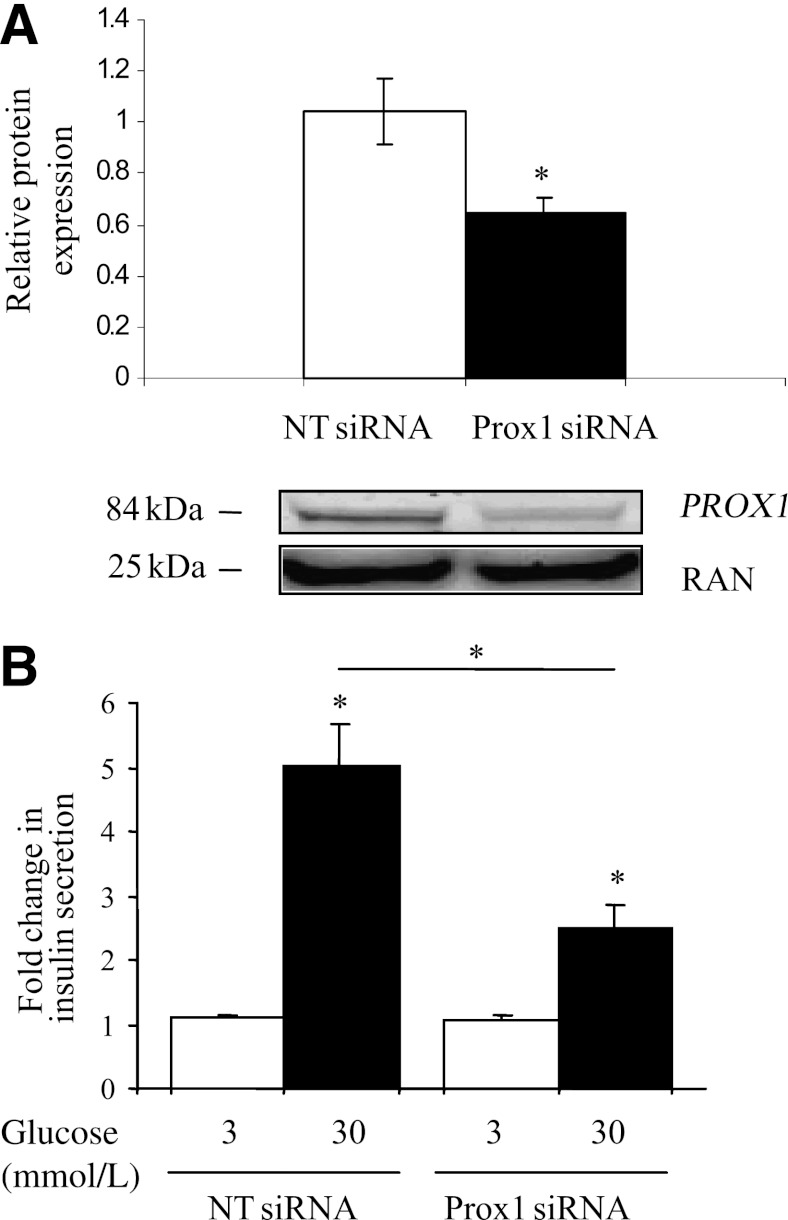
To investigate the possible consequence of a lower expression of Prox1 in β-cells and to assess whether Prox1 could play a role in GSIS, we knocked down Prox1 expression by siRNAs in INS-1E cells. After transfection with Prox1 siRNAs or NT siRNAs for 48 h, INS-1E cells were treated for 1 h with 3 or 30 mmol/L glucose. Knock down of Prox1 expression was verified at the mRNA and protein level. Cells transfected with Prox1 siRNAs showed 40% less Prox1 mRNA (data not shown) and fewer proteins (Fig. 4A) than cells transfected with NT siRNAs. Only 40% of the inhibition of Prox1 could be obtained despite the use of 1) the optimal conditions based on the cyclophilin B positive control siRNA data and the Prox1 siRNAs and 2) a pool of four different Prox1 siRNAs. Of note, Prox1 siRNAs had no effect on cyclophilin B mRNA expression levels. After the cells were stimulated with either 3 or 30 mmol/L glucose, insulin level was measured in the supernatant. Knock down of Prox1 mRNA expression levels resulted in a 1.7-fold decreased GSIS in Prox1 siRNA cells compared with NT siRNA cells (P = 0.029) (Fig. 4B), indicating that Prox1 stimulates GSIS in β-cells in normal conditions.
FIG. 4.
The siRNA knock down of Prox1 reduces the glucose-stimulated insulin secretion in INS-1E cells. A: INS-1E cells were transfected with control NT siRNAs or Prox1 siRNAs. The effects of siRNAs on Prox1 expression were measured by Western blot analysis 48 h later. RAN expression was used as a reference. The results represent at least three experiments. *P < 0.05 (Mann-Whitney nonparametric test). B: INS-1E cells were transfected with control NT siRNAs or Prox1 siRNAs for 48 h and then stimulated with 3 or 30 mmol/L glucose in Krebs buffer for 1 h. Secreted insulin was measured by ELISA. Data show relative insulin secretion obtained for Prox1 siRNA- relative to control NT siRNA-treated cells at 3 compared with 30 mmol/L glucose. The results represent at least three experiments. *P < 0.05 (Mann-Whitney nonparametric test).
Regulation of Prox1 expression by glucose.
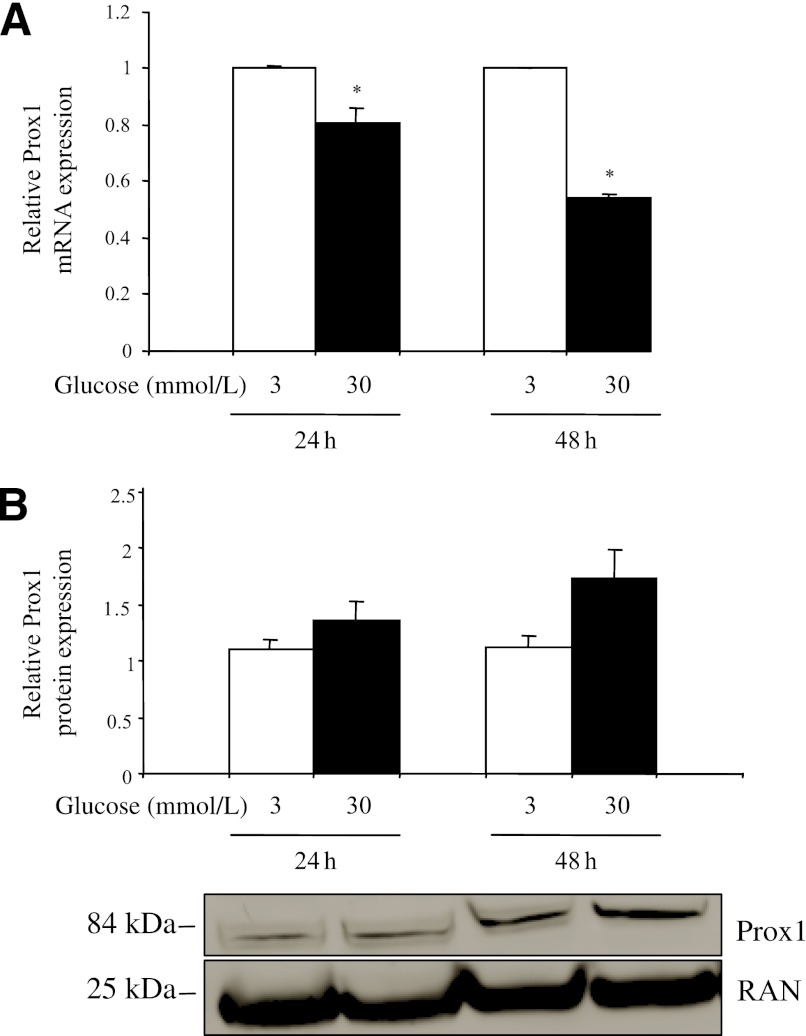
Because type 2 diabetes is characterized by hyperglycemia, we investigated whether the expression of Prox1 could be regulated by glucose concentration in INS-1E cells. These cells were cultured for 24 or 48 h in medium containing 3 or 30 mmol/L glucose. Total RNA was extracted, and Prox1 mRNA level was quantified relative to that of TFIIB. High concentrations of glucose (30 mmol/L) repressed Prox1 expression to ∼20 and 45% (both P = 0.029) of the expression at 3 mmol/L glucose at 24 and 48 h, respectively (Fig. 5A). As a positive control of the treatment (22), L-PK mRNA expression was quantified and, as expected, was induced approximately sevenfold (P < 0.05) on exposure to 30 mmol/L glucose (data not shown). To check that a high glucose concentration would also modify Prox1 protein levels, nuclear protein extracts were prepared from the same samples (Prox1 protein being too weakly detected in total protein extracts). In contrast to the mRNA level, at the protein level, a high concentration of glucose upregulated Prox1 expression by ∼20 (P = 0.20) and 50% (P = 0.11) at 24 and 48 h, respectively, relative to the values at 3 mmol/L (Fig. 5B).
FIG. 5.
Glucose represses Prox1 expression at the mRNA level but enhances Prox1 expression at the protein levels. INS-1E cells were precultured for 24 h in RPMI medium before addition of 3 or 30 mmol/L glucose and were incubated for 24 or 48 h. After cells were harvested, total RNAs and proteins were extracted, and expression of Prox1 was quantified by real-time PCR (A) or Western blot (B) analysis. The data were normalized to TFIIB (A) or RAN (B) expression. The results represent at least three experiments. *P < 0.05 (Mann-Whitney nonparametric test).
DISCUSSION
To our knowledge, this study is the first to 1) test the impact of the whole common genetic variability of PROX1 (80 SNPs) on type 2 diabetes–related traits in adolescents, 2) analyze the sequence activity of the associated SNPs in HepG2 and MIN6 cells, and 3) evaluate the role of Prox1 in GSIS in β-cells. In the HELENA study, three PROX1 SNPs (rs340838, rs340837, and rs340836) were significantly associated, and two SNPs (rs340872 and rs340873) tended to be associated with plasma insulin levels. Consistently, the rs340838, rs340837, and rs340836 SNPs tended to be associated with HOMA-B and HOMA-IR. Haplotype analyses did not add any further information (data not shown).
The results are in line with those previously reported in adults and adolescents. The MAGIC study (5) reported significant associations between the C allele of rs340874 and higher FG level (effect size +0.013 mmol/L, P = 6.6 × 10−6) and type 2 diabetes (OR 1.07, P = 7.2 × 10−10) and nominal associations with lower HOMA-B (effect size −0.008, P = 0.020). Unfortunately, this study did not assess the impact of the PROX1 rs340838, rs340837, or rs340836 SNPs (or proxies) (MAGIC consortium, July 2011 personal communication). The meta-analysis on children and adolescents by Barker et al (8) showed that the C allele of rs340874 was associated with higher FG levels (effect size +0.013 mmol/L, P = 0.042) and, to a lesser extent, with lower HOMA-B (effect size −0.016, P = 0.067). Conversely, in the present study, we were unable to detect a significant association between rs340874 and FG levels; the sample size was not large enough to detect a difference of 0.013 mmol/L (power 12%). Nevertheless, the present results agree with these meta-analysis data because carriers of the rs340874 glucose-raising allele had lower HOMA-B than carriers of the other allele. It is noteworthy that rs340874 is in modest LD with the rs340838, rs340837, and rs340836 SNPs (r2 ∼0.60). The results indicate that this group of SNPs has a larger effect than rs340874 on insulin-related traits in adolescents. Replication studies in larger samples are needed to confirm these findings.
We next tried to identify the functional variant responsible for the association signal. Although the best association with insulin levels was observed for rs340837, other SNPs in strong LD with rs340837 could explain the statistical association. Using luciferase reporter assays in HepG2 and MIN6 cells, we show that the insulin-lowering alleles of rs340873, rs340835, and rs340874 are associated with lower luciferase activity in both cells (except for rs340874 in HepG2 cells only), especially rs340835 (around fourfold lower), suggesting that PROX1 expression could be reduced in hepatocytes and β-cells of individuals carrying these alleles. The observed association signal, therefore, could be explained by one of these three functional SNPs, even if they are not themselves statistically associated with glucose-related phenotypes (because of a slightly lower MAF and lower effect size). Although the whole regulatory and genomic context was not assessed with 60-bp inserts, this method helped to identify functional effects of isolated SNPs. The specific effect of rs340874 in HepG2 cells suggests the activity of liver-specific regulatory elements at this site. EMSA indicated that some TFs are able to bind to the three SNPs in HepG2 cells, with some allele affinity differences for the rs340874 SNP. Genomatix could not detect alterations at rs340874 in putative TF binding sites. In the pancreas and, more specifically, in β-cells, nothing is known about the regulation of PROX1 expression. Other experiments, therefore, are needed to determine the exact mechanisms underlying the reduced expression of PROX1.
We show that Prox1 plays a role in GSIS in pancreatic β-cells because the inhibition of its expression was associated with a nearly twofold reduction in GSIS. As a TF, Prox1 could regulate the expression of genes involved in the insulin secretion pathway. Because Prox1 is known to be expressed in endocrine islets in late gestation embryos and in adults (11), subtle modifications in its expression caused by one or several SNPs may mediate alterations in GSIS and lead to type 2 diabetes. This result is in accordance with the GWAS data showing that the majority of loci associated with type 2 diabetes points to primary defects in the β-cell (4).
Finally, we show that high glucose levels downregulate PROX1 mRNA expression but upregulate PROX1 protein expression in INS-1E cells. Although consistency between mRNA and protein expression often is assumed, examples of divergent trends are frequent. Jayapal et al. (23) showed that >30% of the analyzed genes in Streptomyces coelicolor exhibited divergent patterns and that nearly one-third of them showed opposing trends. In mice, divergent trends were observed, for example, for IL-18 in a model of cutaneous wound repair (24). It is not known which posttranscriptional mechanism (improved Prox1 mRNA stability, increased translation efficiency, etc.) is responsible for this effect. This situation may also reflect a negative transcriptional feedback for the avoidance of an excess of Prox1 protein. Nevertheless, glucose appears to modulate Prox1 expression in β-cells.
In conclusion, the rs340838, rs340837, and rs340836 SNPs in PROX1 appear to play an important role in the regulation of insulin secretion at the population level. These SNPs probably tag functional cis-regulatory SNPs, such as rs340874, rs340873, and rs340835, that are associated with a lower reporter gene expression in vitro. We also show that Prox1 plays a role in GSIS in pancreatic β-cells. Although the exact mechanisms are not completely understood, the present results bring us a step closer to a better understanding of the involvement of PROX1 in type 2 diabetes.
Supplementary Material
ACKNOWLEDGMENTS
The HELENA study received funding from the European Union’s Sixth Framework Programme for Research and Technological Development (contract FOOD-CT-2005-007034), the Spanish Ministry of Education (AGL2007-29784-E/ALI, AP-2005-3827), and the Universidad Politécnica de Madrid (CH/018/2008).
No potential conflicts of interest relevant to this article were reported.
The funder played no role in the conduct of the study, collection of data, management of the study, analysis of data, interpretation of data, or preparation of the manuscript.
S.L. performed the in vitro experiments and statistical analyses and wrote the manuscript. G.P. and X.H. performed the in vitro experiments. B.G.-B. performed the imputation and statistical analyses. M.G.-G., S.D.H., D.M., P.S., L.B., and L.A.M. collected the HELENA data and revised the manuscript. P.A. and J.D. revised the manuscript. A.M. designed the study, interpreted the data, and wrote the manuscript. A.M. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
The authors thank Julie Dumont, INSERM U744, Institut Pasteur de Lille, Université Lille 2, for help with the manuscript.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db12-0864/-/DC1.
REFERENCES
- 1.Chamnan P, Simmons RK, Forouhi NG, et al. Incidence of type 2 diabetes using proposed HbA1c diagnostic criteria in the European Prospective Investigation of Cancer-Norfolk cohort: implications for preventive strategies. Diabetes Care 2011;34:950–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferrannini E, Camastra S. Relationship between impaired glucose tolerance, non-insulin-dependent diabetes mellitus and obesity. Eur J Clin Invest 1998;28(Suppl. 2):3–6; discussion 6–7 [DOI] [PubMed] [Google Scholar]
- 3.Fowler MJ. Microvascular and macrovascular complications of diabetes. Clin Diabetes 2011;29:116–122 [Google Scholar]
- 4.Billings LK, Florez JC. The genetics of type 2 diabetes: what have we learned from GWAS? Ann N Y Acad Sci 2010;1212:59–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dupuis J, Langenberg C, Prokopenko I, et al. DIAGRAM Consortium. GIANT Consortium. Global BPgen Consortium. Anders Hamsten on behalf of Procardis Consortium. MAGIC Investigators New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 2010;42:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boesgaard TW, Grarup N, Jørgensen T, Borch-Johnsen K, Hansen T, Pedersen O, Meta-Analysis of Glucose and Insulin-Related Trait Consortium (MAGIC) Variants at DGKB/TMEM195, ADRA2A, GLIS3 and C2CD4B loci are associated with reduced glucose-stimulated beta cell function in middle-aged Danish people. Diabetologia 2010;53:1647–1655 [DOI] [PubMed] [Google Scholar]
- 7.Wagner R, Dudziak K, Herzberg-Schäfer SA, et al. Glucose-raising genetic variants in MADD and ADCY5 impair conversion of proinsulin to insulin. PLoS ONE 2011;6:e23639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barker A, Sharp SJ, Timpson NJ, et al. Association of genetic loci with glucose levels in childhood and adolescence: a meta-analysis of over 6,000 children. Diabetes 2011;60:1805–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliver G, Sosa-Pineda B, Geisendorf S, Spana EP, Doe CQ, Gruss P. Prox 1, a prospero-related homeobox gene expressed during mouse development. Mech Dev 1993;44:3–16 [DOI] [PubMed] [Google Scholar]
- 10.Dyer MA, Livesey FJ, Cepko CL, Oliver G. Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat Genet 2003;34:53–58 [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Kilic G, Aydin M, Burke Z, Oliver G, Sosa-Pineda B. Prox1 activity controls pancreas morphogenesis and participates in the production of “secondary transition” pancreatic endocrine cells. Dev Biol 2005;286:182–194 [DOI] [PubMed] [Google Scholar]
- 12.Moreno LA, De Henauw S, González-Gross M, et al. HELENA Study Group Design and implementation of the Healthy Lifestyle in Europe by Nutrition in Adolescence Cross-Sectional Study. Int J Obes (Lond) 2008;32(Suppl. 5):S4–S11 [DOI] [PubMed] [Google Scholar]
- 13.Béghin L, Castera M, Manios Y, et al. HELENA Study Group Quality assurance of ethical issues and regulatory aspects relating to good clinical practices in the HELENA Cross-Sectional Study. Int J Obes (Lond) 2008;32(Suppl. 5):S12–S18 [DOI] [PubMed] [Google Scholar]
- 14.González-Gross M, Breidenassel C, Gómez-Martínez S, et al. Sampling and processing of fresh blood samples within a European multicenter nutritional study: evaluation of biomarker stability during transport and storage. Int J Obes (Lond) 2008;32(Suppl. 5):S66–S75 [DOI] [PubMed] [Google Scholar]
- 15.Nagy E, Vicente-Rodriguez G, Manios Y, et al. HELENA Study Group Harmonization process and reliability assessment of anthropometric measurements in a multicenter study in adolescents. Int J Obes (Lond) 2008;32(Suppl. 5):S58–S65 [DOI] [PubMed] [Google Scholar]
- 16.Slaughter MH, Lohman TG, Boileau RA, et al. Skinfold equations for estimation of body fatness in children and youth. Hum Biol 1988;60:709–723 [PubMed] [Google Scholar]
- 17.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–265 [DOI] [PubMed] [Google Scholar]
- 18.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009;5:e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 2007;39:906–913 [DOI] [PubMed] [Google Scholar]
- 20.Gauderman WJ, Morrison JM. QUANTO 1.1: a computer program for power and sample size calculations for genetic-epidemiology studies [Internet], 2006. Available from http://hydra.usc.edu/gxe Accessed May 2009
- 21.Burke Z, Oliver G. Prox1 is an early specific marker for the developing liver and pancreas in the mammalian foregut endoderm. Mech Dev 2002;118:147–155 [DOI] [PubMed] [Google Scholar]
- 22.Eckert DT, Zhang P, Collier JJ, O’Doherty RM, Scott DK. Detailed molecular analysis of the induction of the L-PK gene by glucose. Biochem Biophys Res Commun 2008;372:131–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jayapal KP, Philp RJ, Kok YJ, et al. Uncovering genes with divergent mRNA-protein dynamics in Streptomyces coelicolor. PLoS ONE 2008;3:e2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kämpfer H, Kalina U, Mühl H, Pfeilschifter J, Frank S. Counterregulation of interleukin-18 mRNA and protein expression during cutaneous wound repair in mice. J Invest Dermatol 1999;113:369–374 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.