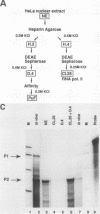
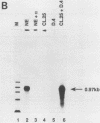
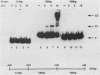
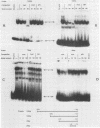
Abstract
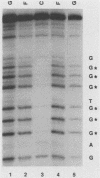
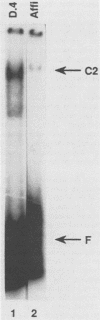
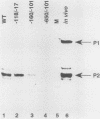
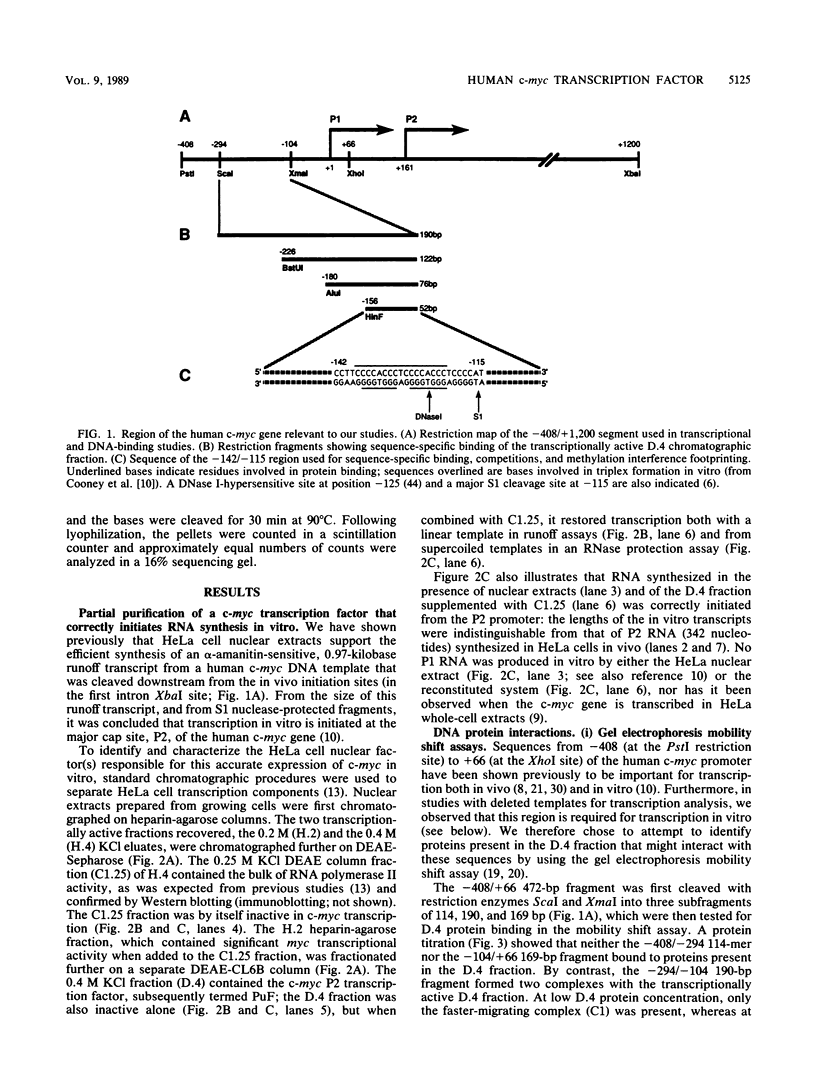
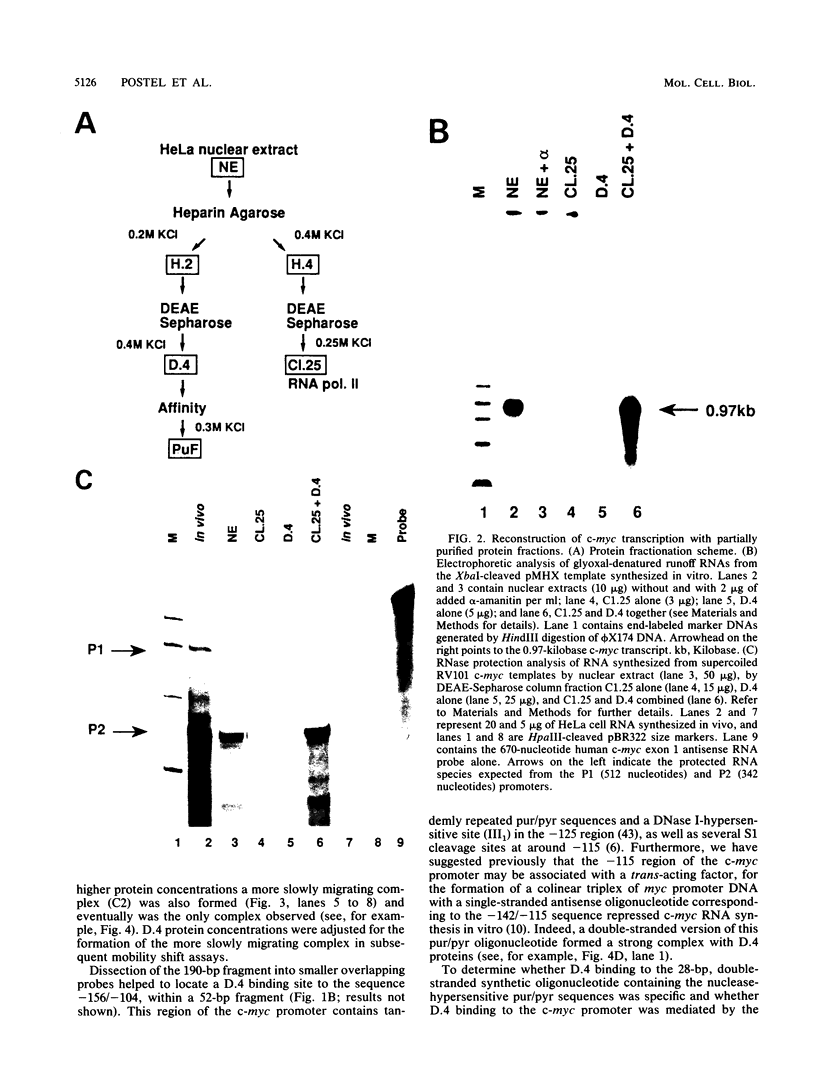
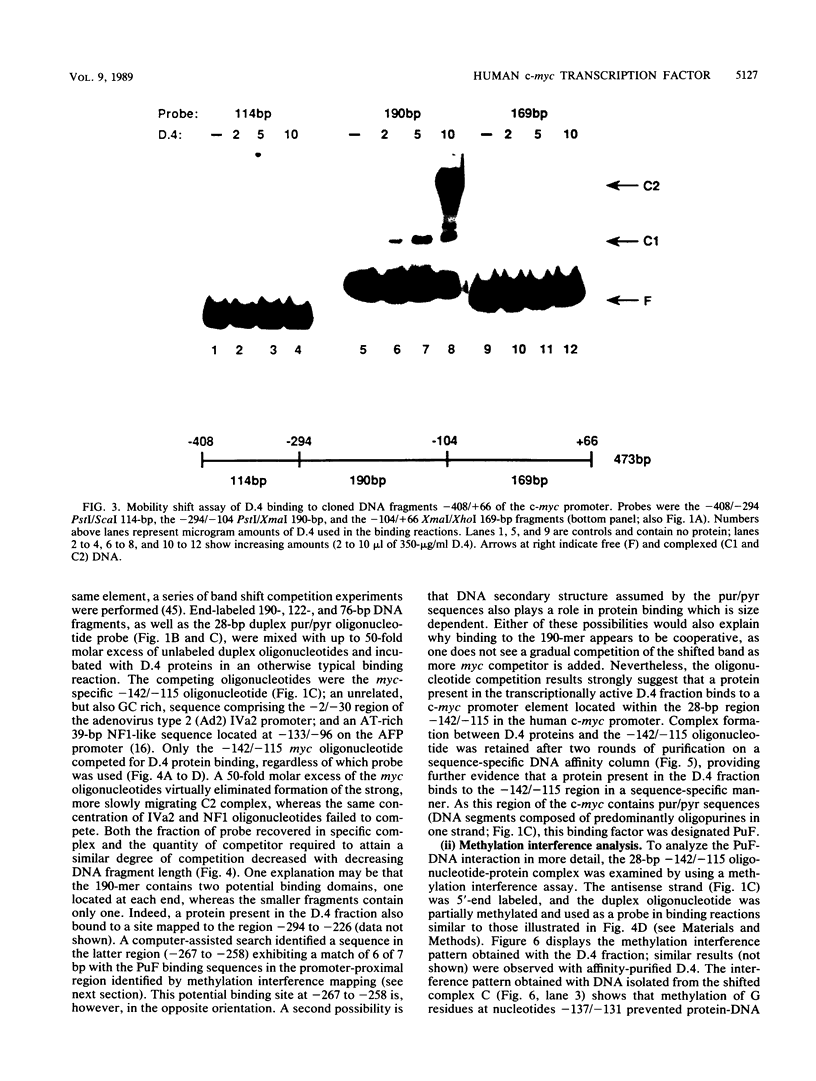
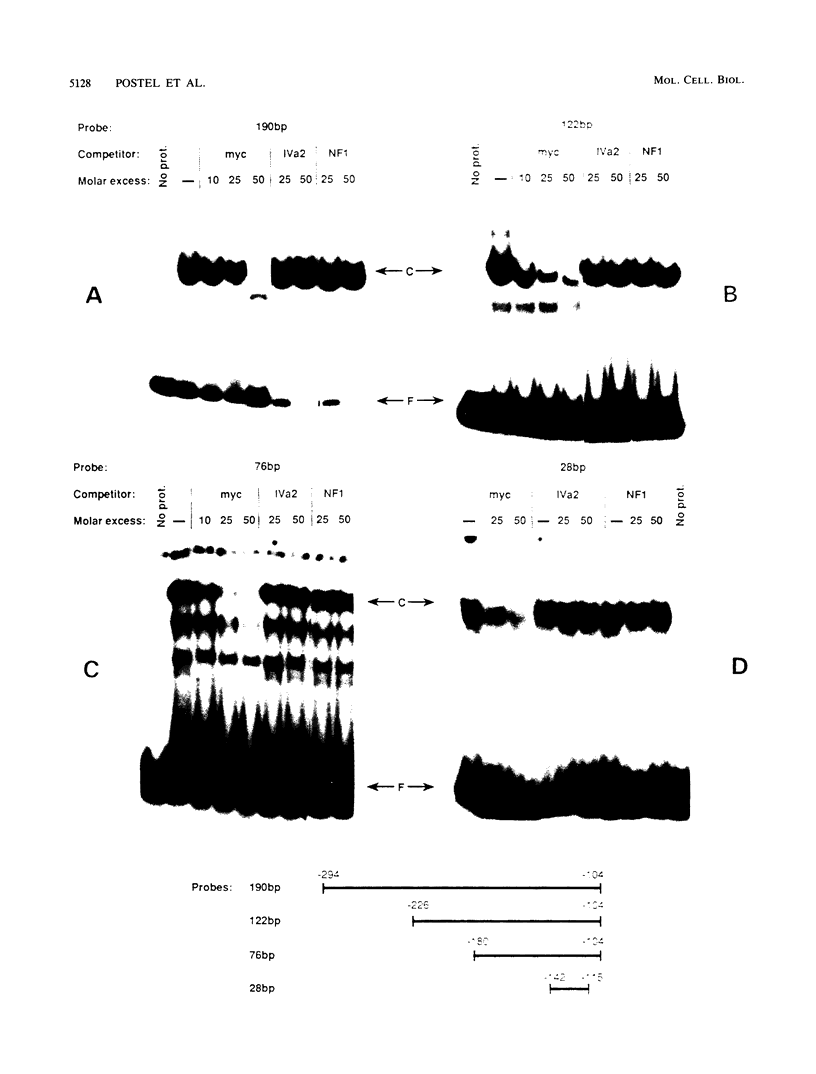
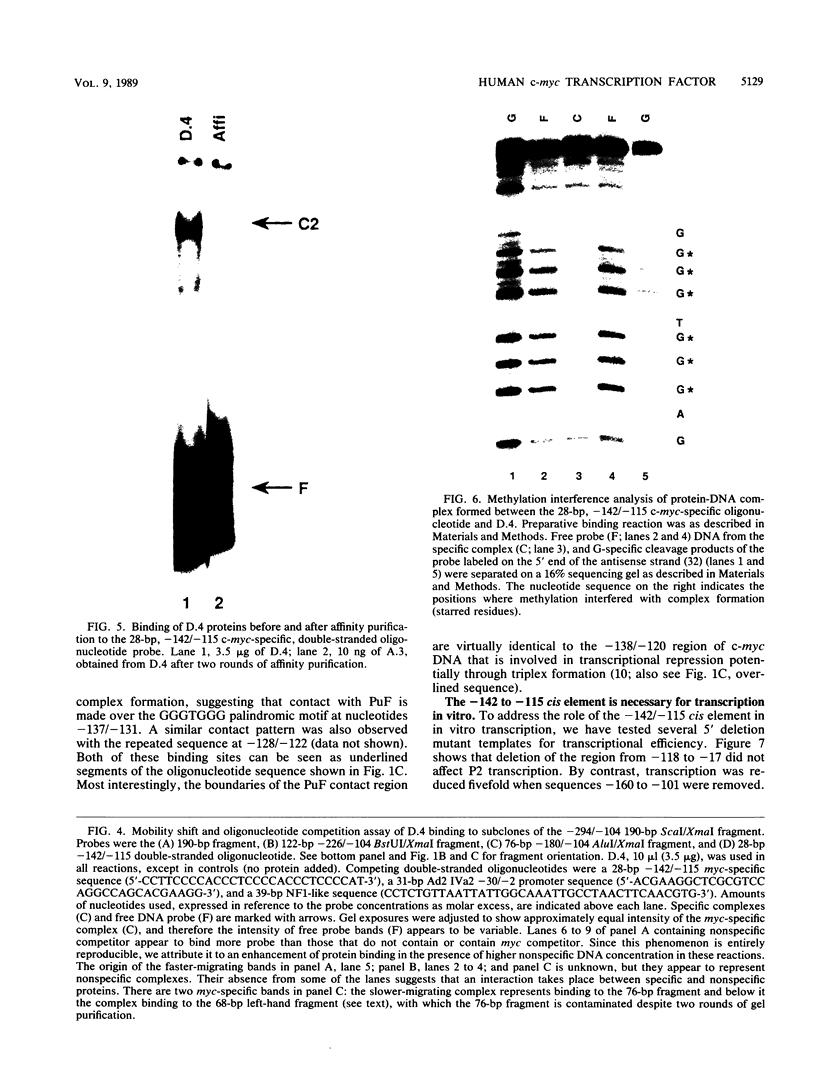
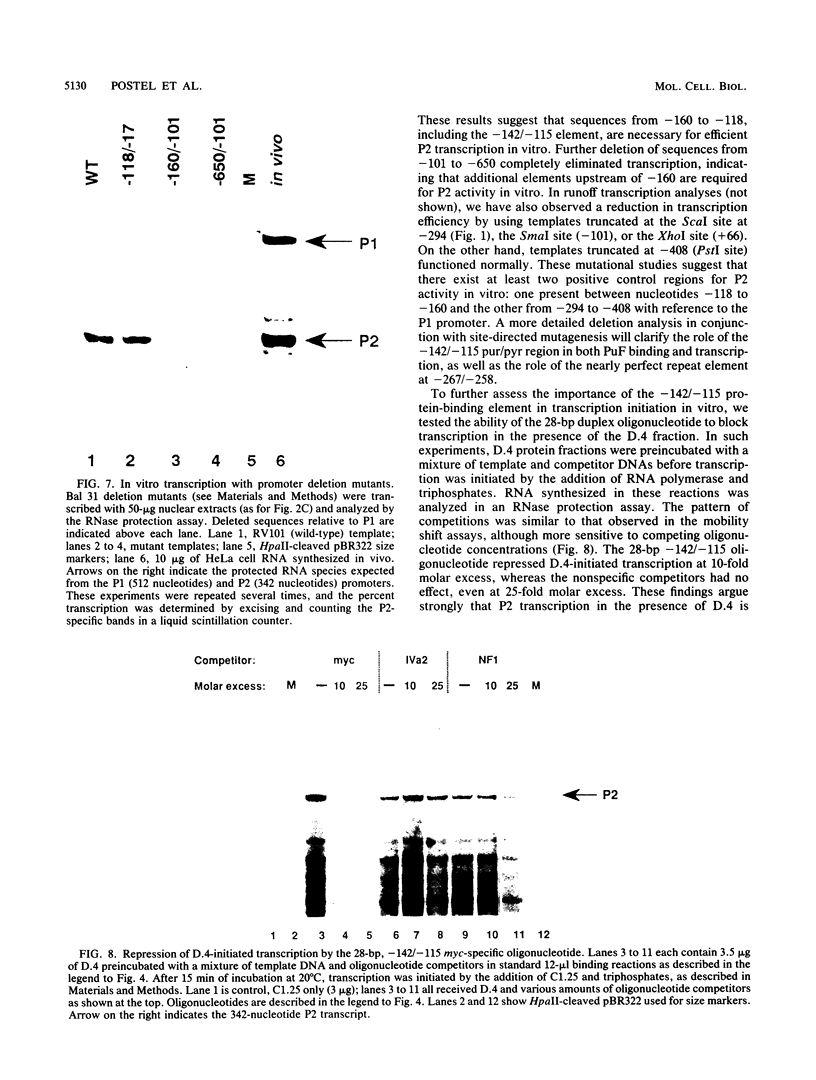
Transcription of the human c-myc oncogene is elaborately regulated, but the relevant molecular mechanisms are not yet understood. To begin to define elements and enzyme systems responsible for c-myc transcription in vitro, we partially purified a transcription factor essential for efficient and accurate in vitro initiation from the principal myc promoter, P2. DNA mobility shift assays located the factor binding domain at -142 to -115 with respect to the P1 promoter. This region contains pur/pyr sequences (predominantly purines in one strand), nuclease-hypersensitive sites (U. Siebenlist, L. Henninghausen, J. Battey, and P. Leder, Cell 37:381-391, 1984; C. Boles and M. Hogan, Biochemistry 26:367-376, 1987), and a triple-helix-forming element (M. Cooney, G. Czernuszewicz, E. Postel, S. Flint, and M. Hogan, Science 241:456-459, 1988). Methylation interference mapping established that the factor, termed PuF, directly contacts the repeated palindromic sequence GGGTGGG of the -142/-115 element. The interaction of PuF with this cis-acting element is necessary for P2 transcription in vitro, for (i) deletion of this 5' region from the myc promoter greatly reduced transcription efficiency and (ii) a synthetic duplex oligonucleotide corresponding to the -142/-115 sequence completely repressed c-myc transcription in the presence of the partially purified factor. These observations lend support to the hypothesis that pur/pyr sequences perform important biological roles in the regulation of c-myc gene expression, most likely by serving as transcription factor binding sites.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker C. C., Ziff E. B. Promoters and heterogeneous 5' termini of the messenger RNAs of adenovirus serotype 2. J Mol Biol. 1981 Jun 25;149(2):189–221. doi: 10.1016/0022-2836(81)90298-9. [DOI] [PubMed] [Google Scholar]
- Battey J., Moulding C., Taub R., Murphy W., Stewart T., Potter H., Lenoir G., Leder P. The human c-myc oncogene: structural consequences of translocation into the IgH locus in Burkitt lymphoma. Cell. 1983 Oct;34(3):779–787. doi: 10.1016/0092-8674(83)90534-2. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986 Jun 12;321(6071):702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. Novel promoter upstream of the human c-myc gene and regulation of c-myc expression in B-cell lymphomas. Mol Cell Biol. 1986 Oct;6(10):3481–3489. doi: 10.1128/mcb.6.10.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles T. C., Hogan M. E. DNA structure equilibria in the human c-myc gene. Biochemistry. 1987 Jan 27;26(2):367–376. doi: 10.1021/bi00376a006. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chung J., Sinn E., Reed R. R., Leder P. Trans-acting elements modulate expression of the human c-myc gene in Burkitt lymphoma cells. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7918–7922. doi: 10.1073/pnas.83.20.7918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J., Sussman D. J., Zeller R., Leder P. The c-myc gene encodes superimposed RNA polymerase II and III promoters. Cell. 1987 Dec 24;51(6):1001–1008. doi: 10.1016/0092-8674(87)90586-1. [DOI] [PubMed] [Google Scholar]
- Cooney M., Czernuszewicz G., Postel E. H., Flint S. J., Hogan M. E. Site-specific oligonucleotide binding represses transcription of the human c-myc gene in vitro. Science. 1988 Jul 22;241(4864):456–459. doi: 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]
- Dierks P., van Ooyen A., Cochran M. D., Dobkin C., Reiser J., Weissmann C. Three regions upstream from the cap site are required for efficient and accurate transcription of the rabbit beta-globin gene in mouse 3T6 cells. Cell. 1983 Mar;32(3):695–706. doi: 10.1016/0092-8674(83)90055-7. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell. 1983 Mar;32(3):669–680. doi: 10.1016/0092-8674(83)90053-3. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A., Posakony J. W., Maniatis T., Lawn R. M., O'Connell C., Spritz R. A., DeRiel J. K., Forget B. G., Weissman S. M., Slightom J. L. The structure and evolution of the human beta-globin gene family. Cell. 1980 Oct;21(3):653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- Elgin S. C. DNAase I-hypersensitive sites of chromatin. Cell. 1981 Dec;27(3 Pt 2):413–415. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- Feuerman M. H., Godbout R., Ingram R. S., Tilghman S. M. Tissue-specific transcription of the mouse alpha-fetoprotein gene promoter is dependent on HNF-1. Mol Cell Biol. 1989 Oct;9(10):4204–4212. doi: 10.1128/mcb.9.10.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finer M. H., Aho S., Gerstenfeld L. C., Boedtker H., Doty P. Unusual DNA sequences located within the promoter region and the first intron of the chicken pro-alpha 1(I) collagen gene. J Biol Chem. 1987 Sep 25;262(27):13323–13332. [PubMed] [Google Scholar]
- Flint S. J., Beltz G. A., Linzer D. I. Synthesis and processing of simian virus 40-specific RNA in adenovirus-infected, simian virus 40-transformed human cells. J Mol Biol. 1983 Jun 25;167(2):335–359. doi: 10.1016/s0022-2836(83)80339-8. [DOI] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N., Bishop J. M., Levens D. Regulatory elements that modulate expression of human c-myc. Genes Dev. 1987 Sep;1(7):659–671. doi: 10.1101/gad.1.7.659. [DOI] [PubMed] [Google Scholar]
- Imagawa M., Chiu R., Karin M. Transcription factor AP-2 mediates induction by two different signal-transduction pathways: protein kinase C and cAMP. Cell. 1987 Oct 23;51(2):251–260. doi: 10.1016/0092-8674(87)90152-8. [DOI] [PubMed] [Google Scholar]
- Johnson A. C., Jinno Y., Merlino G. T. Modulation of epidermal growth factor receptor proto-oncogene transcription by a promoter site sensitive to S1 nuclease. Mol Cell Biol. 1988 Oct;8(10):4174–4184. doi: 10.1128/mcb.8.10.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. R., Cole M. D. Rapid cytoplasmic turnover of c-myc mRNA: requirement of the 3' untranslated sequences. Mol Cell Biol. 1987 Dec;7(12):4513–4521. doi: 10.1128/mcb.7.12.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakkis E., Calame K. A plasmacytoma-specific factor binds the c-myc promoter region. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7031–7035. doi: 10.1073/pnas.84.20.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick M. W., Torri A., Kang D. S., Engler J. A., Wells R. D. Unusual DNA structures in the adenovirus genome. J Biol Chem. 1986 Aug 25;261(24):11350–11354. [PubMed] [Google Scholar]
- Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982 Jun;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- Leong K., Flint S. J. Specific transcription of an adenoviral gene that possesses no TATA sequence homology in extracts of HeLa cells. J Biol Chem. 1984 Sep 25;259(18):11527–11533. [PubMed] [Google Scholar]
- Lewis C. D., Clark S. P., Felsenfeld G., Gould H. An erythrocyte-specific protein that binds to the poly(dG) region of the chicken beta-globin gene promoter. Genes Dev. 1988 Jul;2(7):863–873. doi: 10.1101/gad.2.7.863. [DOI] [PubMed] [Google Scholar]
- Lipp M., Schilling R., Wiest S., Laux G., Bornkamm G. W. Target sequences for cis-acting regulation within the dual promoter of the human c-myc gene. Mol Cell Biol. 1987 Apr;7(4):1393–1400. doi: 10.1128/mcb.7.4.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKeon C., Schmidt A., de Crombrugghe B. A sequence conserved in both the chicken and mouse alpha 2(I) collagen promoter contains sites sensitive to S1 nuclease. J Biol Chem. 1984 May 25;259(10):6636–6640. [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H., Sobell H. M. A molecular model for gene repression. Proc Natl Acad Sci U S A. 1966 May;55(5):1201–1205. doi: 10.1073/pnas.55.5.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. J., Wang C., Tjian R. Positive and negative regulation of transcription in vitro: enhancer-binding protein AP-2 is inhibited by SV40 T antigen. Cell. 1987 Sep 11;50(6):847–861. doi: 10.1016/0092-8674(87)90512-5. [DOI] [PubMed] [Google Scholar]
- Morgan A. R., Wells R. D. Specificity of the three-stranded complex formation between double-stranded DNA and single-stranded RNA containing repeating nucleotide sequences. J Mol Biol. 1968 Oct 14;37(1):63–80. doi: 10.1016/0022-2836(68)90073-9. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Tilly K., Maniatis T. Fine structure genetic analysis of a beta-globin promoter. Science. 1986 May 2;232(4750):613–618. doi: 10.1126/science.3457470. [DOI] [PubMed] [Google Scholar]
- Nishikura K., Goldflam S., Vuocolo G. A. Accurate and efficient transcription of human c-myc genes injected into Xenopus laevis oocytes. Mol Cell Biol. 1985 Jun;5(6):1434–1441. doi: 10.1128/mcb.5.6.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelicci P. G., Knowles D. M., 2nd, Magrath I., Dalla-Favera R. Chromosomal breakpoints and structural alterations of the c-myc locus differ in endemic and sporadic forms of Burkitt lymphoma. Proc Natl Acad Sci U S A. 1986 May;83(9):2984–2988. doi: 10.1073/pnas.83.9.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinberg D., Horikoshi M., Roeder R. G. Factors involved in specific transcription in mammalian RNA polymerase II. Functional analysis of initiation factors IIA and IID and identification of a new factor operating at sequences downstream of the initiation site. J Biol Chem. 1987 Mar 5;262(7):3322–3330. [PubMed] [Google Scholar]
- Reinberg D., Roeder R. G. Factors involved in specific transcription by mammalian RNA polymerase II. Purification and functional analysis of initiation factors IIB and IIE. J Biol Chem. 1987 Mar 5;262(7):3310–3321. [PubMed] [Google Scholar]
- Siebenlist U., Bressler P., Kelly K. Two distinct mechanisms of transcriptional control operate on c-myc during differentiation of HL60 cells. Mol Cell Biol. 1988 Feb;8(2):867–874. doi: 10.1128/mcb.8.2.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Hennighausen L., Battey J., Leder P. Chromatin structure and protein binding in the putative regulatory region of the c-myc gene in Burkitt lymphoma. Cell. 1984 Jun;37(2):381–391. doi: 10.1016/0092-8674(84)90368-4. [DOI] [PubMed] [Google Scholar]
- Strauss F., Varshavsky A. A protein binds to a satellite DNA repeat at three specific sites that would be brought into mutual proximity by DNA folding in the nucleosome. Cell. 1984 Jul;37(3):889–901. doi: 10.1016/0092-8674(84)90424-0. [DOI] [PubMed] [Google Scholar]
- Suck D., Lahm A., Oefner C. Structure refined to 2A of a nicked DNA octanucleotide complex with DNase I. Nature. 1988 Mar 31;332(6163):464–468. doi: 10.1038/332464a0. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Collier D. A., Hanvey J. C., Shimizu M., Wohlrab F. The chemistry and biology of unusual DNA structures adopted by oligopurine.oligopyrimidine sequences. FASEB J. 1988 Nov;2(14):2939–2949. [PubMed] [Google Scholar]
- Wells R. D. Unusual DNA structures. J Biol Chem. 1988 Jan 25;263(3):1095–1098. [PubMed] [Google Scholar]
- Wu C., Wilson S., Walker B., Dawid I., Paisley T., Zimarino V., Ueda H. Purification and properties of Drosophila heat shock activator protein. Science. 1987 Nov 27;238(4831):1247–1253. doi: 10.1126/science.3685975. [DOI] [PubMed] [Google Scholar]
- Yu Y. T., Manley J. L. Structure and function of the S1 nuclease-sensitive site in the adenovirus late promoter. Cell. 1986 Jun 6;45(5):743–751. doi: 10.1016/0092-8674(86)90788-9. [DOI] [PubMed] [Google Scholar]