Abstract
Islet transplantation has proven to be a successful strategy to restore normoglycemia in patients with type 1 diabetes (T1D). However, the dearth of cadaveric islets available for transplantation hampers the widespread application of this treatment option. Although human embryonic stem cells and induced pluripotent stem cells are capable of generating insulin-producing cells in vitro when provided with the appropriate inductive cues, the insulin-expressing cells that develop behave more like immature β-cells with minimal sensitivity to glucose stimulation. Here, we identify a set of signaling factors expressed in mouse embryonic mesenchyme during the time when foregut and pancreatic progenitors are specified and test their activities during in vitro differentiation of human embryonic stem cells. Several of the identified factors work in concert to expand the pancreatic progenitor pool. Interestingly, transforming growth factor (TGF)-β ligands, most potent in inducing pancreatic progenitors, display strong inhibitory effects on subsequent endocrine cell differentiation. Treatment with TGF-β ligands, followed by the addition of a TGF-β receptor antagonist, dramatically increased the number of insulin-producing cells in vitro, demonstrating the need for dynamic temporal regulation of TGF-β signaling during in vitro differentiation. These studies illustrate the need to precisely mimic the in vivo conditions to fully recapitulate pancreatic lineage specification in vitro.
Islet transplantation has opened up the possibility to treat diabetic patients with cell replacement strategies (1–3). Because the demand for pancreatic islet cells far outstrips the supply, concerted efforts have been undertaken to generate a renewable and reliable supply of insulin-producing cells from human embryonic stem cells (hESCs) and induced pluripotent stem cells. Recently, strategies have been devised that aim to recapitulate the stepwise succession of signals that guide the development of pancreatic endocrine and β-cells during embryogenesis. Recapitulation of these embryonic signals in cell culture has led to the production of intermediate pancreatic lineage as well as immature, nonfunctional insulin-producing cells from hESCs and induced pluripotent stem cells in vitro (4–9). More importantly, when transplanted in vivo, pancreatic progenitor cells generated from hESCs gave rise to mature β-cells capable of reverting diabetic phenotypes caused by chemically induced depletion of endogenous β-cells in mice (10). Although these proof-of-principle experiments clearly demonstrated that pancreatic cells generated in vitro were capable of carrying physiological functions in vivo, they also illustrated that critical signals promoting the full differentiation of pancreas endocrine progenitors and mature endocrine cells were missing from the current in vitro conditions.
The pancreas arises from the definitive endoderm, one of the three primary germ layers generated through gastrulation during early embryogenesis (11). Multiple organs, including the notochord and the dorsal aorta, secrete signaling molecules that control the specification of the duodenal part of the posterior foregut from which the pancreatic bud emerges (12–15). At approximately embryonic day 9.5 (E9.5), a layer of mesenchyme condenses around the budding pancreatic epithelium and separates it from the adjacent notochord and dorsal aorta (16,17). Studies initiated by Golosow and Grobstein (18) in the 1960s posed that the pancreatic mesenchyme provides factors necessary for the survival and cytodifferentiation of the developing pancreatic epithelium. Because clean separation of the mesenchyme from the branching pancreas epithelium is difficult at later stages, a full account of all mesenchymal factors expressed throughout pancreas development and how they function individually or in combination is currently missing.
Here, we provide evidence that embryonic pancreas mesenchyme provides temporally distinct signals that promote specific stages of hESC differentiation toward pancreatic progenitors and insulin-producing cells. Microarray analysis of RNAs collected from E11.5 mouse pancreatic mesenchyme and epithelium allowed us to select 11 genes encoding for secreted factors that are preferentially expressed at higher levels in the mesenchyme compared with the pancreatic epithelium. Applying these candidate factors individually and combined with an established in vitro hESC differentiation system revealed that they greatly improved the generation of pancreatic progenitors. In contrast, sustained treatment with these factors beyond the progenitor stage blocked further differentiation into endocrine progenitor and insulin-producing cells. Summarily, our studies illustrate that tight temporal recapitulation of embryonic signaling events is critical for the optimization of hESC-derived pancreatic endocrine cells.
RESEARCH DESIGN AND METHODS
The mice used in this study were maintained according to protocols approved by the University of California, San Francisco Committee on Animal Research. CD1 mice were obtained from Charles River Laboratories. Noon on the day a vaginal plug was detected was considered as E0.5. Whole embryos were fixed using the HEPES glutamic acid buffer–mediated organic solvent protection effect (HOPE; DCS Innovative Diagnostik-systeme, Germany) according to the manufacturer’s protocol. Embryos were embedded in paraffin overnight before being cut into 5-μm sections and mounted onto 2-μm polyethylene naphthalate–membrane slides (Leica).
Laser capture microdissection.
Slides were stained with hematoxylin (Sigma-Aldrich), followed by staining with eosin (Protocol). Slides were then microdissected using the Leica Laser Micro Dissection Microscope LMD 2000 (Leica).
Microarray analysis.
RNA was processed by the Gladstone Genome Core. RNAs collected from laser capture microdissection (LCM) was extracted using the PicoPure RNA isolation kit (Arturus) and further amplified using the NuGEN WT-Ovation Pico RNA Amplification kit (NuGEN), following the manufacturer’s instruction. Gene array was performed using Mouse Gene 1.0 ST arrays, following the manufacturer’s instruction (Affymetrix).
Cell culture.
Undifferentiated Cythera 49 and MEL 1 INS GFP/w cells (19) were maintained on mouse embryo fibroblast feeder layers (Millipore). Standard differentiation was carried out as described previously (4,10).
Test of mesenchymal factors.
Growth factors (R&D Systems) were used at the following concentrations: angiotensin-1, 20 ng/mL; CXC chemokine ligand-13, 5 ng/mL; fibroblast growth factor-9 (FGF-9), 4 ng/mL; hepatocyte growth factor, 50 ng/mL; IGF, 50 ng/mL; platelet-derived growth factor, 10 ng/mL; transforming growth factor (TGF)-β2, 2 ng/mL; TGF-β3, 5 ng/mL; and vascular endothelial growth factor (VEGF), 100 ng/mL. For the TGF-β2/3 combination, 1 ng/mL TGF-β2 was mixed with 2 ng/mL TGF-β3.
Immunofluorescence.
Cells were fixed using an adapted protocol published previously (4). Antibodies used are listed in Supplementary Table 2. Images were taken with Olympus IX70 fluorescence microscope (Olympus), ApoTome.2 widefield microscope (Carl Zeiss, Inc), or InCell Analyzer 2000 for quantification (GE Healthcare).
Ten fields from each well were picked at random for quantification analysis. The pancreatic duodenal homeobox-1 (PDX1)-positive area was calculated in relation to the area occupied by DAPI-positive nuclei. For the proliferation index, phospho-Histone-H3–positive cells were normalized to the total number of PDX1- or DAPI-positive cells. The percentage of insulin-positive cells was calculated by dividing the number of insulin-expressing cells by the total cell number.
Real-time quantitative PCR.
RNA extraction was performed using RNeasy micro kit, as instructed by the manufacturer (Qiagen), then reverse-transcribed with the iScript cDNA synthesis kit (Bio-Rad). Human islet cDNA was obtained from The Hebrew University of Jerusalem. Real-time PCR was performed on the 7900 HT Fast Real-Time PCR System (Applied Biosystems) using a protocol described previously (4). Primer sequences are listed in Supplementary Table 3.
C-peptide content and release assays.
C-peptide levels in culture supernatants or cell lysates were measured using ultrasensitive C-peptide ELISA (Mercodia AB). For total C-peptide content analysis, day 15 cell cultures were scraped and sonicated in 300 μL water. DNA content was measured using PicoGreen (Invitrogen). C-peptide release was measured by a protocol described previously (4).
Statistics.
P values were determined using the unpaired, two-tailed Student t test. Error bars in the bar diagrams represent the standard deviation of the samples.
Transplantation.
Cells collected from day 9 culture were transplanted under the kidney capsule of NOD severe combined immune-deficient γ (NSG) mice, as described previously (20). Kidney capsules were surgically extracted, fixed, and stained as previously described (10).
RESULTS
Identification of candidate factors from mouse embryonic pancreas mesenchyme.
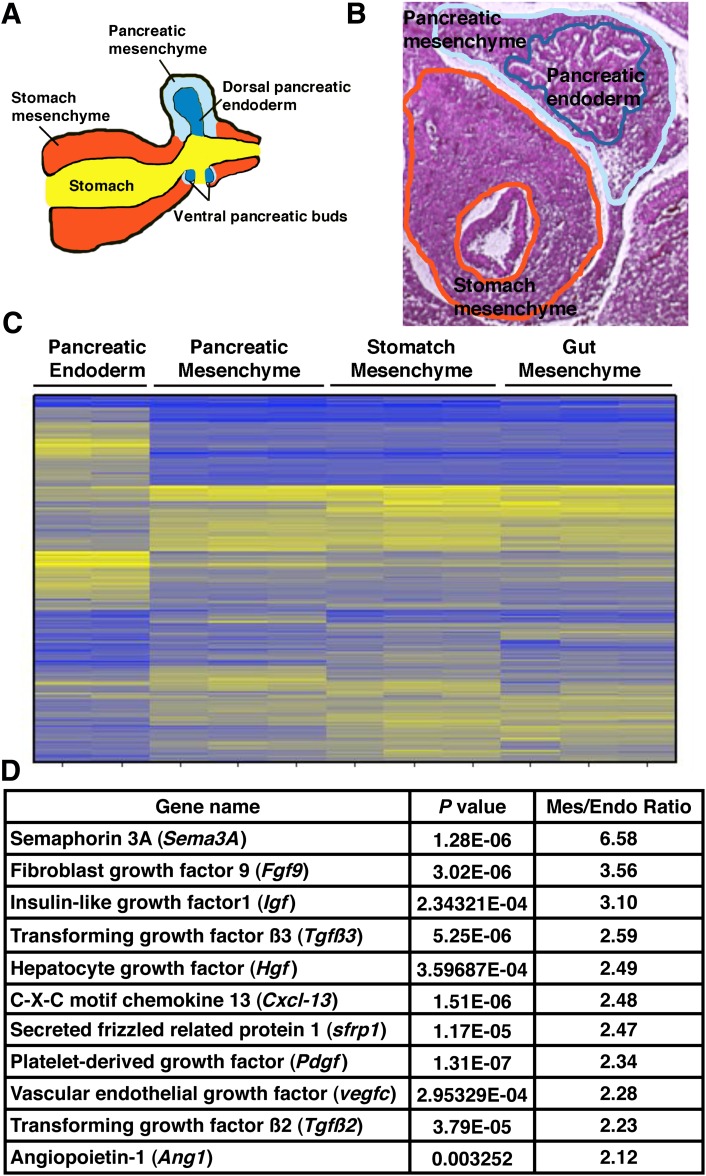
To identify soluble factors secreted by the pancreatic mesenchyme, we separated pancreatic epithelium and mesenchyme from wild-type mouse embryos at E11.5 via LCM (Fig. 1A and B and Supplementary Fig. 1). Differential gene expression arrays were performed to compare cDNA samples collected from pancreatic endoderm and pancreatic mesenchyme. To determine potential tissue specific factors unique to pancreas mesenchyme, we included cDNAs from stomach and intestinal mesenchyme as well. Microarray analysis revealed that pancreatic mesenchyme has a distinct gene expression pattern compared with pancreatic endoderm (Fig. 1C). In contrast, the transcriptional signature for all of the mesenchymal tissues analyzed was remarkably similar (Fig. 1C). From this dataset, we identified 11 secreted factors that belong to several signaling pathways, including the TGF-β, FGF, and Wnt pathways, and are preferentially expressed in the mesenchymal compared with the pancreatic endoderm for further analysis (Fig. 1D).
FIG. 1.
Identification of candidate mesenchymal factors. A: Diagram of the developing pancreas. B: Isolation of different mouse tissues using LCM. Representative images are shown for isolating pancreatic endoderm, pancreatic mesenchyme, and stomach mesenchyme. C: Microarray data show gene expression patterns across different tissues. Yellow indicates genes with high expression levels; blue indicates genes with low expression level. D: List of secreted factors upregulated in pancreatic mesenchyme compared with pancreatic epithelium.
Combinations of mesenchymal factors elicit different effects on distinct hESC populations.
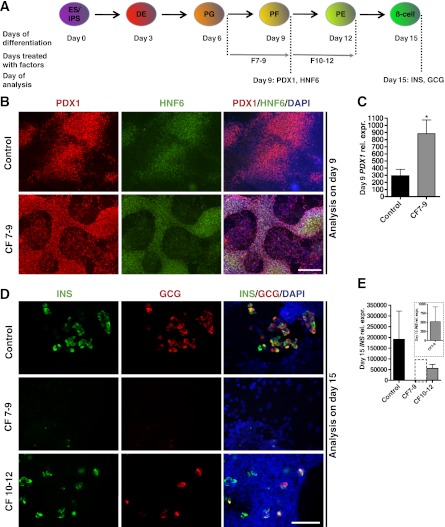
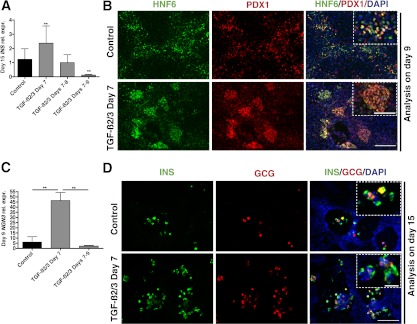
Considering that all factors are expressed concomitantly during embryogenesis, we first investigated their combined effects during hESC to β-cell differentiation using the protocol originally described by Kroon and colleagues (4,9,10). Under the conditions used for the original protocol (referred to as the standard protocol hereafter), hESCs progress through distinct stages (Fig. 2A). To mimic the interaction between pancreatic epithelium and pancreatic mesenchyme at ∼E11.5, the time point of gene array analysis, we tested the effects of the combined mesenchymal factors on primitive gut tube cells generated via the standard protocol (factors were added after day 6 of the standard protocol) (10). At that time, control samples received standard factors retinoic acid, sonic hedgehog inhibitor (3-Keto-N-[aminoethyl-aminocaproyl-dihydrocinnamoyl]-cyclopamine), and the bone morphogenetic protein antagonist Noggin (referred to as R/K/N hereafter) between days 7 and 9, while experimental samples were treated with R/K/N plus the 11 mesenchymal factors. Starting at day 7, cells aggregated into clusters under control and experimental conditions (Supplementary Fig. 2). Immunostaining for PDX1 and hepatic nuclear factor 6 (HNF6), two key pancreatic/foregut progenitor markers, were used to monitor the generation of pancreatic lineage at day 9 (21–23). Samples treated with mesenchymal factor displayed more PDX1-positive cells, and cultures expressed higher levels of PDX1 transcript than the control (Fig. 2B and C).
FIG. 2.
Combined treatment with mesenchymal factors affects pancreatic progenitor induction and endocrine differentiation. A: Schematic depicts differentiation procedure and genes used as markers for specific stages of hESC differentiation. ES/iPS, embryonic stem cells or induced pluripotent stem cells; DE, definitive endoderm; PG, primitive gut, PF, posterior foregut; β-cell, pancreatic β-cells. F7–9, cells treated with CF from day 7 to 9; F10–12, cells treated with CF from day 10 to 12. B: Immunostaining of PDX1 and HNF6 on day 9 shows induction of pancreatic progenitors. C: Q-PCR illustrates day 9 PDX1 expression level in samples with or without combined mesenchymal factors (n = 3 independent experiments). The error bars represent the SD for all graphs. Statistical analysis was performed using the Student t test. *P < 0.05. D: Immunostaining of insulin and glucagon shows differentiation of endocrine cells in control and samples treated with combined factors at different time points. E: Q-PCR analysis of day 15 INS expression levels in samples treated with or without combined mesenchymal factors (n = 3 independent experiments). E (inset): Day 15 relative INS expression in F7–9 samples. B: Scale bars = 200 μm. D: Scale bars = 100 μm.
To test the effects of mesenchymal factors on the continued differentiation of PDX1-positive cells into hormone-expressing cells, combined factors (CF) were added between days 10 and 12, immediately after the induction of PDX1. On day 15, samples were stained with insulin, glucagon, or PDX1 to identify the extent to which pancreatic endocrine cells had formed. In control samples, insulin- and glucagon-expressing cells were seen throughout the culture. Samples treated with CF between days 10 and 12 displayed a considerably lower number of hormone-expressing cells compared with the control samples (Fig. 2D). Also, despite significant increases in PDX1 expression at early stages, samples treated with mesenchymal factors between days 7 and 9 were almost completely devoid of insulin- or glucagon-positive cells (Fig. 2D). Quantitative (Q)-PCR analysis confirmed the dramatic inhibition of insulin (INS) expression in CF-treated samples (both between days 7 and 9 and days 10 and 12), with the most severe inhibition observed when cells were treated from day 7 to 9 (Fig. 2E). At day 15, a large population of PDX1-positive, insulin-negative cells was found in samples treated with CF from day 7 to 9 (data not shown). No excess amount of exocrine markers was detected by Q-PCR in the CF-treated samples (Supplementary Fig. 3). These results suggest that candidate factors identified from E11.5 pancreas mesenchyme promote the induction of the pancreatic and foregut progenitors but prohibit the further differentiation and maturation of endocrine cells. Furthermore, the effects of the mesenchymal factors seem to be stage-specific, with the most robust phenotypes observed when cells are treated from day 7 to 9.
Effects of individual mesenchymal factors on pancreatic progenitor determination.
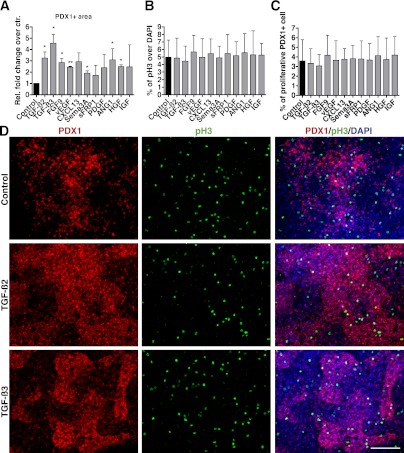
Because the combined factors dramatically enhanced the induction of pancreatic/foregut progenitors, we decided to test the activities of individual factors on the induction of PDX1-positive cells. Previous results with CF condition indicated that the most effective window for promoting pancreatic progenitors was between days 7 and 9. When single mesenchymal factors were applied alone (without R/K/N) from day 7 to 9, few PDX1-positive cells were detected on day 9 (Supplementary Fig. 4). This suggests that the mesenchymal factors tested individually do not effectively induce the formation of pancreatic progenitors. Considering that in the standard protocol R/K/N treatment is sufficient to induce PDX1 expression (4,10), we examined whether introducing single mesenchymal factors in combination with R/K/N could further promote PDX1 expression. Immunostaining performed at day 9 detected an increase in PDX1-positive cells in samples treated with R/K/N plus certain mesenchymal factors compared with R/K/N alone (Fig. 3A and D). Of note, treatment with individual factors did not dramatically affect the proliferative capacity of the entire cellular pool or the PDX1-positive cells (Fig. 3B and C). Thus, our results indicate that mesenchymal factors identified from the embryonic pancreas mesenchyme promote the formation of pancreatic progenitors in conjunction with R/K/N, likely through direct conversion of primitive gut cells into PDX1-expressing pancreatic progenitors instead of the expansion of existing PDX1-positive cells.
FIG. 3.
Effect of single candidate factors on induction of pancreatic progenitors. A: Single mesenchymal factors tested in combination with R/K/N. Quantification of PDX1 expression in samples treated with single mesenchymal factors compared with control. Control condition is set to “1.” Ten independent fields were used for counting. Statistical analysis was performed using the Student t test. *P < 0.05 and **P < 0.01. B: Percentage of proliferating cells in control and experimental samples treated with single mesenchymal factors. Total number of cells counted for each condition is ≥14,000. C: Percentage of proliferating pancreatic progenitor cells in control and experimental samples treated with single mesenchymal factors. Total number of cells counted for each condition is ≥14,000. D: Immunostaining with PDX1 and pH3 indicates induction of progenitor cells under control, TGF-β2, and TGF-β3 conditions. D: Scale bar = 200 μm in all images.
TGF-β signaling improves PDX1 expression but inhibits endocrine cell differentiation.
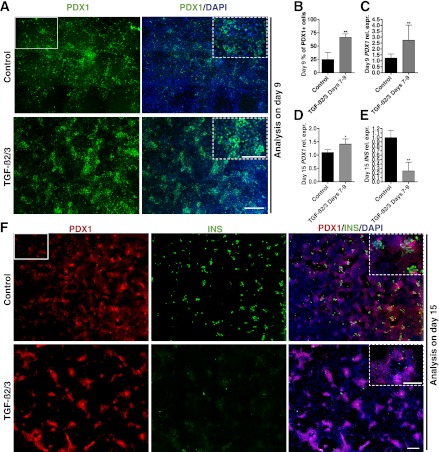
Among the 11 tested factors, TGF-β2 and TGF-β3 were more potent than the other mesenchymal factors, showing a 3.2- and 4.6-fold increase in PDX1-positive cells compared with the controls, respectively (Fig. 3A and D). TGF-β signaling is known to regulate multiple cell functions, including proliferation and differentiation in adult tissue and also during embryogenesis (24). When added together between days 7 and 9 in addition to R/K/N, TGF-β2 and 3 (referred to as TGF-β2/3 hereafter) induced a significant increase in PDX1-positive cells over the standard condition (analysis at day 9; Fig. 4A and B). Q-PCR analysis also revealed a significant increase in PDX1 gene expression on day 9 and day 15 in TGF-β2/3–treated samples (Fig. 4C and D). However, as observed under conditions when all factors are combined, TGF-β2/3 treatment from day 7 to 9 dramatically decreased INS expression levels and the number of insulin-positive cells on day 15 (Fig. 4E and F). At this point, most of the cells in TGF-β2/3– treated samples remain PDX1-positive but insulin-negative (Fig. 4F insets), suggesting the existence of a large population of pancreatic progenitors. Therefore, TGF-β signaling enhances the induction of pancreatic progenitors but subsequently blocks endocrine cell differentiation.
FIG. 4.
TGF-β signaling is important for efficient induction of pancreatic progenitors but prohibits endocrine differentiation. A: Immunostaining of PDX1 reveals induction of pancreatic progenitor cells in control and TGF-β2/3–treated samples. Each image is a composite of nine individual images at original magnification ×10 (representative ×10 image is shown as white rectangle) to cover a representative area of the culture (higher magnification pictures is marked by dotted lines). B: Quantification of PDX1-positive cells in day 9 samples with and without TGF-β2/3 treatment. Total number of cells counted for each condition is ≥10,000. C: Q-PCR showing day 9 PDX1 expression levels in control and TGF-β2/3-treated samples (n = 2 independent experiments). D: Q-PCR analysis of day 15 PDX1 expression levels in control and TGF-β2/3-treated samples (n = 3 independent experiments). E: Q-PCR analysis of day 15 INS expression levels in control and TGF-β2/3–treated samples (n = 3 independent experiments). F: Immunostaining of PDX1 and insulin in day 15 samples shows induction of endocrine cells in control but not in TGF-β2/3–treated samples. Each image is a composite of 20 individual original magnification ×10 images to cover a representative area of the culture (representative ×10 image is shown as white rectangle). Insets show higher magnification view. B–E: Statistical analysis was performed using the Student t test. *P < 0.05; **P < 0.01. A–F: Scale bars = 400 μm for all images. A and F insets: Scale bars = 200 μm.
Precise activation of TGF-β signaling improves endocrine cell maturation.
Because activation of the TGF-β pathway from day 7 to 9 resulted in a severe reduction of endocrine lineages, we next tried to regulate TGF-β signaling in a more precise manner by performing time course experiments between days 7 and 9. Q-PCR results on day 15 samples showed that addition of TGF-β2/3 for only 1 day (day 7), increased INS expression by twofold (Fig. 5A); 2-day TGF-β2/3 treatment (day 7–8) showed slightly lower but not significantly different INS expression levels, whereas applying TGF-β2/3 for 3 days (day 7 to 9) significantly decreased INS level compared with control. On the basis of these results, we decided to focus on a short, 1-day only (day 7) activation of TGF-β signaling for subsequent experiments. Cells were treated with R/K/N plus TGF-β2/3 for 24 h on day 7 and then continued to differentiate for 2 more days with R/K/N. On day 9, samples were stained for PDX1 and HNF6. TGF-β2/3–treated samples displayed more PDX1 and HNF6 double-positive cells (Fig. 5B), indicating efficient induction of pancreatic/foregut progenitors. Q-PCR also confirmed that 1-day TGF-β2/3 treatment enhanced PDX1 expression compared with control cells (Supplementary Fig. 5).
FIG. 5.
Short TGF-β2/3 treatment improves endocrine precursor induction as well as endocrine cell differentiation. A: Q-PCR analysis of day 15 INS expression levels in control and experimental samples treated with TGF-β2/3 for different durations (n = 3 independent experiments). B: Immunostaining of PDX1 and HNF6 shows induction of pancreatic progenitor cells in samples treated with TGF-β2/3 for 1 day compared with control. Insets depict higher magnification view. C: Q-PCR analysis of day 12 NGN3 expression levels in control and TGF-β2/3-treated samples (n = 3 independent experiments). D: Immunostaining of insulin and glucagon in day 15 samples indicates induction of endocrine cells in control and experimental samples treated with TGF-β2/3 for 1 day. Statistical analysis in panels A and C was performed using the Student t test. **P < 0.01. B and D: Scale bars = 200 μm for all images. B: Scale bar = 100 μm for all insets. D: Scale bar = 50 μm for all insets.
Proliferation and differentiation are two tightly regulated events during normal pancreas development. After expansion, pancreatic progenitors reduce proliferation rates and transiently turn on the endocrine master regulator neurogenin-3 (NGN3) (25,26). After 1 day of TGF-β2/3 treatment, we detected dramatically higher levels of NGN3 compared with the control cells (Fig. 5C). However, prolonged treatment with TGF-β2/3 for 3 days resulted in significantly lower NGN3 expression on day 9 (Fig. 5C). These results indicate that transient activation of TGF-β signaling facilitates pancreatic progenitor differentiation toward endocrine precursors at day 9 of in vitro differentiation. The increased expression of NGN3 in TGF-β2/3–treated samples resulted in a greater number of endocrine cells, including single- and double-hormone–positive cells compared with the samples treated with the standard protocol (Fig. 5D).
Inhibition of TGF-β signaling after progenitor induction further potentiates the differentiation of endocrine cells.
Taken together, our data demonstrate that prolonged activation of the TGF-β pathway prevented the generation of endocrine precursors. We then decided to test whether inhibiting TGF-β signaling by applying a TGF-β receptor (activin-like kinase 5) antagonist, 2-(3-(6-Methylpyridin-2-yl)-1H-pyrazol-4-yl)-1,5-naphthyridine (ALK5 inhibitor II) would promote the development of the endocrine lineage (27). Addition of the ALK5 inhibitor from day 7 to 9 led to decreased total cell numbers as well as the number of PDX1-positive cells on day 9 (data not shown). When the ALK5 inhibitor was added from day 10 onwards, significantly higher levels of NGN3 were observed on day 12 by Q-PCR, indicating the progression of pancreatic progenitor cells into endocrine precursors (Fig. 6A). To test whether addition of the ALK5 inhibitor could reverse the adverse effects of prolonged TGF-β activation at earlier time points, we performed time course experiments with TGF-β2/3 treatment at day 7, day 7–8, or day 7–9, followed by inhibition of this pathway via application of the ALK5 inhibitor from day 10 to 15. Similar to the earlier time course experiments (Fig. 5A), treatment with the ALK5 inhibitor after transient activation of TGF-β signaling on day 7 gave the most robust increase in INS expression over the standard protocol (Fig. 6B). In contrast, prolonged TGF-β activation resulted in smaller augmentation in INS expression (Fig. 6B).
FIG. 6.
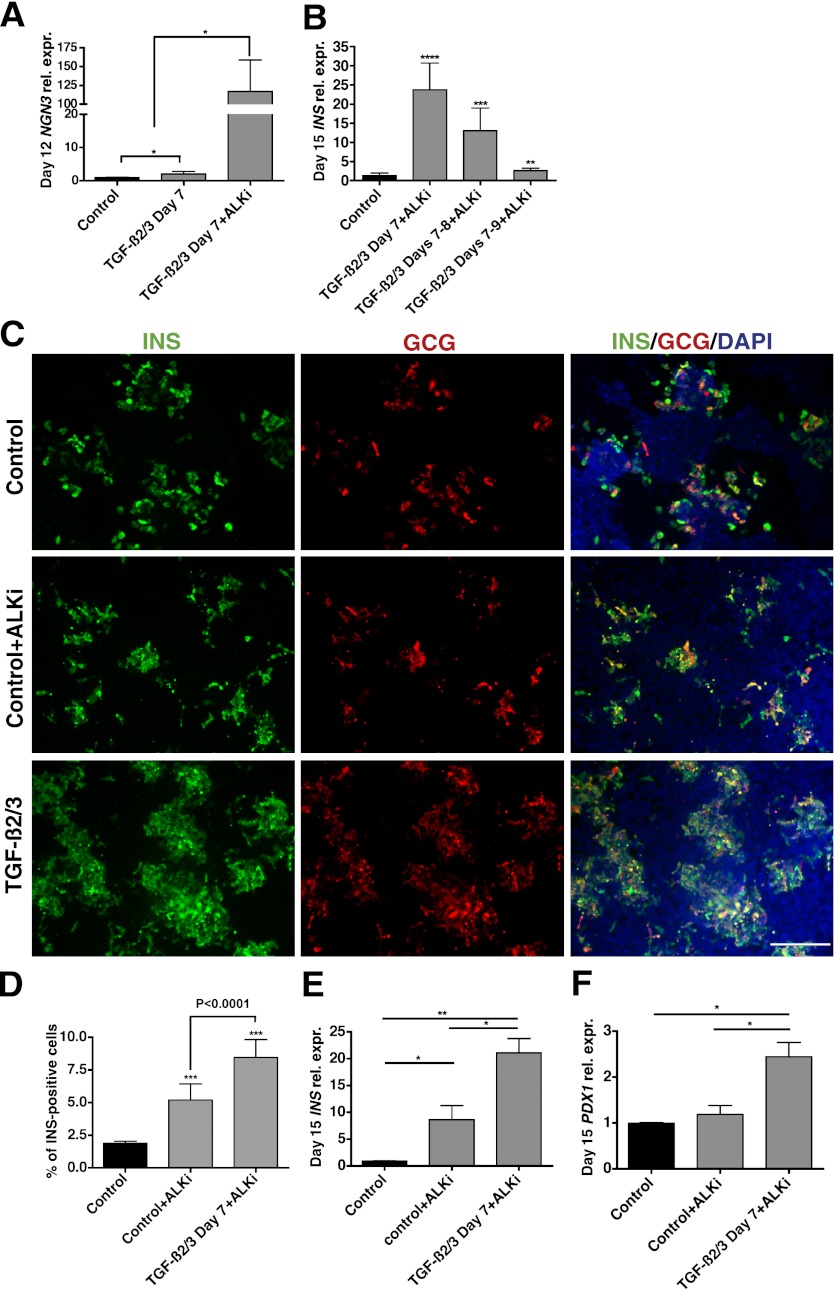
Inhibition of TGF-β pathway further improves endocrine cell differentiation.ic>: Q-PCR analysis is shown for day 12 NGN3 expression levels in control, TGF-β2/3–treated, and TGF-β2/3+ALK5 inhibitor samples (n = 3 independent experiments). B: Q-PCR analysis of day 15 INS expression levels in samples treated with TGF-β2/3 for different lengths, followed by ALK5 inhibitor from day 10 to 15 (n = 3 independent experiments). C: Immunostaining of insulin and glucagon in day 15 samples showing induction of endocrine cells in control and experimental samples treated with ALK5 inhibitor alone (day 10–15) or TGF-β2/3 (day 7) plus ALK5 inhibitor (day 10–15). D: Percentage of insulin-positive cells on day 15 in control samples, samples treated with ALK5 inhibitor alone, and samples treated with TGF-β2/3 plus ALK5 inhibitor (n = 3 independent experiments). Q-PCR analysis of day 15 INS (E) and PDX1 (F) expression levels in control, samples treated with ALK5 inhibitor alone, and with TGF-β2/3 plus ALK5 inhibitor (n = 3 independent experiments). A, B, and D–F: Statistical analysis was performed using the Student t test. *P < 0.05, * *P < 0.01, * * *P < 0.001, and * * * *P < 0.0001. C: Scale bar = 200 μm for all images.
To confirm that TGF-β activation is necessary before inhibition of the pathway, we differentiated hESCs according to the standard protocol until day 9 and then applied the ALK5 inhibitor to block the TGF-β pathway from day 10 to 12. Confirming prior studies (27–29), we observed higher numbers of insulin- and glucagon-positive cells as well as elevated INS expression level under these conditions compared with the standard condition (Fig. 6C–E). However, short activation of TGF-β2/3 at day 7, followed by ALK5 inhibition from day 10 to 15, resulted in the most significant increase in the number of endocrine cells. Detailed quantitative analysis revealed that the modified protocol typically generated 2.3 million cells at day 15 from a starting population of 1 million undifferentiated cells, among which an average of 8.5% and as many as 15% are insulin-expressing cells (Fig. 6C and D and Supplemental Fig. 6). Q-PCR analysis revealed an almost threefold increase in INS gene expression in TGF-β2/3–ALK5 inhibitor samples over those treated with the standard protocol plus ALK5 inhibitor alone (Fig. 6E). Samples receiving TGF-β2/3 and ALK5 inhibitor also showed a significantly higher level of PDX1 expression compared with control samples and samples treated only with the ALK5 inhibitor (Fig. 6F). Similar results were obtained when the modified protocol was tested on MEL1 INSGFP/w hESCs (19), with an average of 8.7% cells being insulin-positive on day 15 (Supplementary Fig. 7). Thus, our data demonstrate that finely tuned activation of TGF-β signaling, followed by pathway inhibition, increases formation of pancreas progenitors and insulin-producing endocrine cells.
Characterization of hESC-derived insulin-producing cells.
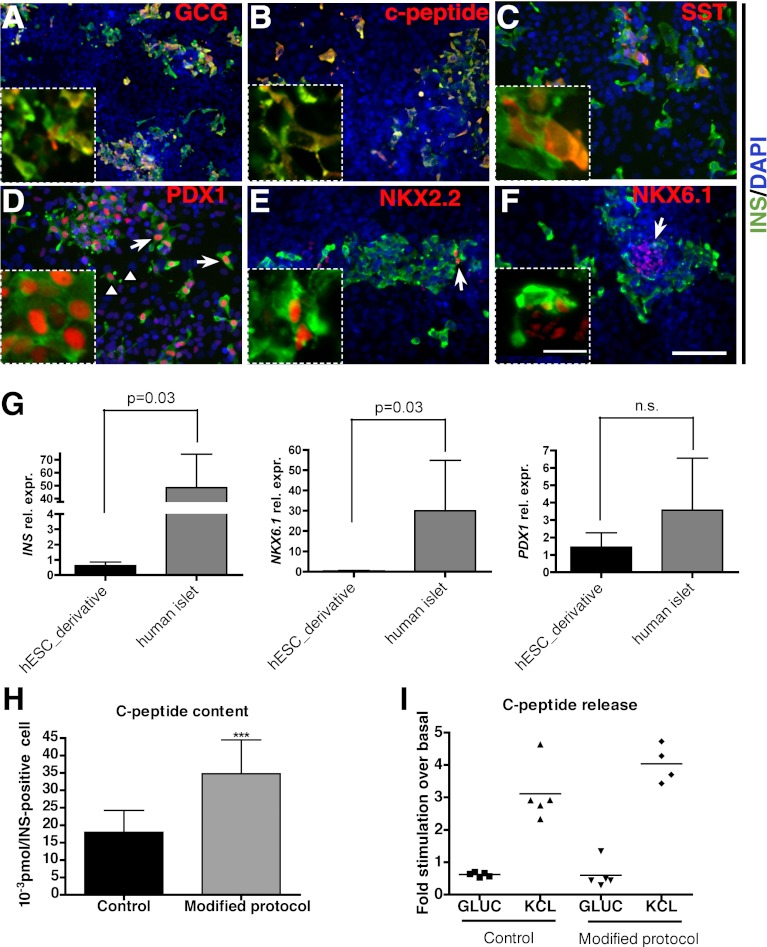
To further characterize the insulin-producing cells generated in vitro with our modified differentiation protocol (TGF-β2/3 on day 7, followed by ALK5 inhibition between day 10 and 15), we performed immunostaining with multiple markers. Approximately 40% of insulin-expressing cells were insulin-, glucagon double-positive cells, as previously reported (4) and likely resemble immature β-cells (Fig. 7A and data not shown). Most of the insulin-positive cells were double-labeled with C-peptide, indicating that these cells can properly process proinsulin (Fig. 7B). We also observed a small percentage of somatostatin-positive cells, with or without insulin staining (Fig. 7C). More important, most of the insulin-positive cells expressed PDX1 at high levels compared with the insulin-negative, PDX1-positive cells, likely representing pancreatic/foregut progenitors (Fig. 7D, arrows vs. arrowheads). Immunostaining with two essential NK homeodomain factors found in islet β-cells, NKX2.2 and NKX6.1 (30,31), revealed that a subset of insulin-expressing cells were positive for these markers (Fig. 7E and F). These staining results indicate that the insulin-producing cells generated in vitro possess many but not all characteristics of “authentic” pancreatic β-cells. When compared with adult human islets using Q-PCR, hESC-derived tissue showed lower expression of INS and NKX6.1 but a comparable level of PDX1 (Fig. 7G).
FIG. 7.
Characterization of insulin-producing cells generated from hESCs. Immunostaining with insulin and glucagon (A); C-peptide (B); somatostatin (C); PDX1 (D); NKX2.2 (E); and NKX6.1 (F). D: The arrows mark insulin- and PDX1 double-positive cells; arrowheads mark PDX1-positive, insulin-negative cells. E and F: Arrows mark insulin-producing cells coexpressing NKX2.2 or NKX6.1, respectively. Insets show higher magnification view. G: Q-PCR analysis comparing INS, PDX1, and NKX6.1 expression levels between hESC-derived cultures and adult human islets. H: Corrected C-peptide content in control and samples treated with TGF-β2/3 for 1 day plus ALK5 inhibitor from day 10 to 15. Total C-peptide content is divided by the percentage of insulin-positive cells in culture. I: C-peptide release assay demonstrates cells respond efficiently to KCL stimulation but not to high glucose. Statistical analysis in panels G and H was performed using the Student t test. ***P < 0.001. C–F: Scale bar = 100 μm. C–F (insets): Scale bar = 25 μm. A and B: Scale bar = 200 μm. A and B (insets): Scale bar = 50 μm.
We furthermore examined the C-peptide content in the insulin-expressing cells. After 15 days of differentiation, the insulin-producing cells generated under modified conditions had significantly higher C-peptide content compared with control cells (Fig. 7H).
To further examine the physiological function of the insulin-expressing cells generated in vitro, we monitored C-peptide release upon stimulation. Insulin-producing cells from the control and modified protocols can both be directly depolarized by chemicals, such as potassium chloride (KCL), which promote insulin secretion without glucose stimulation, and showed a fourfold increase in C-peptide release compared with basal conditions (Fig. 7I). In contrast, in only one of five times did we detect slight C-peptide induction upon d-glucose stimulation over basal condition in samples treated with the modified protocol (Fig. 7I). Our results therefore suggest that the insulin-producing cells generated in vitro may have immature glucose-sensing and -transporting machinery compared with the bona fide pancreatic β-cells. This agrees with the recent finding that hESC-generated insulin-producing cells have less efficient Ca2+ influx upon glucose stimulation (32).
Finally we examined whether hESC-derived pancreatic endoderm can further mature in vivo. Four months after transplantation, pancreatic progenitors generated with TGF-β2/3 treatment on D7 matured to single hormone-positive, insulin-expressing cells with high levels of PDX1 and NKX6.1, as well as CK19-positive ductal cells and rare amylase-positive acinar cells (Supplementary Fig. 8), indicating their full differentiation potential toward pancreatic lineages.
DISCUSSION
Seminal work performed in the 1960s demonstrated crucial roles for the pancreatic mesenchyme in regulating epithelial cell development and differentiation (18,33). Although the filter pores used in the classical mesenchyme and epithelial coculture studies were large enough to allow cell–cell contact (34), subsequent work demonstrated that soluble factors produced by the mesenchyme regulate specific processes within the forming pancreas epithelium. Elegant work using transgenic animals identified FGF10 as one of these mesenchyme-produced factors that directly influence the developing pancreatic endoderm (35). Other mesenchyme-derived factors, such as activin, follistatin, retinoic acid, and TGF-β1, have also been shown to regulate differentiation of the pancreatic epithelium (36–39). However, until recently, the lack of suitable transgenic mouse models allowing for specific manipulation of pancreatic mesenchyme has prevented detailed studies regarding the requirement for specific mesenchymal factors (40).
Here, we set out to directly translate gene expression analysis performed on embryonic tissues to hESC to β-cell differentiation efforts. We used a precise way of isolating pancreatic mesenchyme from the epithelium using LCM, thus allowing for a direct comparison of gene expression in different tissues. Most of the 11 factors selected showed positive effects on the induction of pancreatic/foregut progenitors (Fig. 3A). From this list, we focused on two ligands from the TGF-β family, TGF-β2 and TGF-β3. TGF-βs are multifunctional cytokines that affect cell growth, differentiation, migration, and survival (24). Canonical TGF-β signaling is mediated through transmembrane serine/threonine kinase receptors. Upon ligand activation, TGF-β receptor type II (TBR II) phosphorylates and forms heterodimer with TBR type I (TBR I), also called ALK5, to initiate downstream signaling cascades via SMAD proteins (24,41). During normal pancreas development, TGF-β receptors localize to the early pancreatic epithelium as well as the mesenchyme (39). Inhibition of the TGF-β pathway using a dominant-negative receptor leads to increased endocrine differentiation and abnormal exocrine development (42,43). Although our in vitro differentiation data using TGF-β inhibition confirmed findings from these previous studies, more importantly, our analysis at earlier time points (day 7 to 9, which is approximately equivalent to E9.5 to E11.5 of mouse embryogenesis) revealed that addition of TGF-β ligands increases the number of progenitor cells and thus supports expansion of the pancreas epithelium. Our data also suggest that the TGF-β pathway is dynamically regulated to control the balance between progenitor specification and endocrine differentiation. A short activation of the pathway leads to increased expression of NGN3, whereas prolonged treatment with TGF-β ligands impairs endocrine development by keeping cells in the progenitor stage. Thus, TGF-β signaling needs to be precisely regulated to allow generation of a certain number of progenitor cells before differentiation proceeds to the next stage. A gradual shift of TGF-β receptor expression from the uncommitted epithelium to exocrine cells may indicate how this transient activation of TGF-β signaling in certain cell populations is achieved in vivo (39). Importantly, these data also point to dynamic changes in gene expression in mesenchyme throughout pancreatic development because early signals need to be downregulated to allow cell differentiation. However, it is also possible that over time, pancreatic epithelium becomes insensitive to TGF-β signaling through upregulation of inhibitory SMADs.
During mid-to-late gestation, TGF-β signaling is believed to play a role in preventing the recruitment of ductal and periductal cells into the endocrine lineage (43). In our hESC differentiation system, inhibition of the TGF-β pathway using a TGFR-I receptor antagonist significantly increased the number of insulin-producing cells, a phenomenon reminiscent of the increased accumulation of periductal endocrine cell phenotype observed in dominant-negative TBR-II receptor in mice (43).
Recently, several groups reported that inhibition of the TGF-β pathway promotes the differentiation of hESCs into hormone-producing cells in vitro (27–29). Nostro et al. (29) and Inman et al. (44) showed increased endocrine differentiation by inhibiting TGF-β/activin/nodal signaling with a small molecule ALK4/5/7 inhibitor SB431542. Afrikanova et al. (28) further reported that inhibition of Src and focal adhesion kinase promotes endocrine specification through inhibition of the TGF-βR/SMAD pathway and that addition of an ALK5 inhibitor during the PDX1 induction stage caused progenitors to exit the cell cycle. Our work supports these findings and provides novel information regarding the necessity to precisely tune the level of TGF-β signaling in a temporal manner. Activating TGF-β signaling during the pancreatic progenitor stage before inhibiting the pathway at the endocrine differentiation stage increases the number of PDX1-positive progenitor cells and induces a higher number of insulin-positive cells. Thus, our combined brief activation of the pathway, followed by inhibition of TGF-β signaling, more closely mimics the in vivo scenario in which TGF-β promotes the generation of pancreatic progenitors during earlier pancreatic differentiation but inhibits endocrine specification thereafter (43).
Interestingly, persistent TGF-β activation during the PDX1 induction phase leads to an increased number of PDX1-positive, insulin-negative cells at a later differentiation stage. Notably, we did not detect significant increases in pancreatic exocrine and duct or CDX2-marked intestinal cells under these conditions. Prolonged TGF-β inhibition after exogenous TGF-β stimulation results in a much higher number of insulin-producing cells, indicating the increased number of PDX1-positive cells represent pancreatic progenitors capable of undergoing further endocrine differentiation.
In summary, our study identified a set of mesenchymal factors that promote induction of pancreatic progenitors. When used individually, several factors, including TGF-β2, TGF-β3, FGF9, and VEGF, promote the expansion of pancreatic progenitors. In particular, the dynamic regulation of the TGF-β pathway is important for recapitulating the developmental events during progenitor expansion and endocrine specification. More detailed analysis using a combination of TGF-β ligands and some other beneficial factors identified from this study may help discover crosstalk between different signaling pathways. We anticipate that further investigation of mesenchymal factors expressed at later stages of development will help to optimize the late stages of hESC to β-cell differentiation that are currently not well recapitulated under cell culture conditions (45). In particular, it will be interesting to investigate whether the sequential TGF-β activation and inhibition also promotes differentiation of endodermal progenitor cells described recently by Gadue and colleagues (46) into fully differentiated β-cells.
Supplementary Material
ACKNOWLEDGMENTS
Work in the laboratory of M.H. was supported by the Juvenile Diabetes Research Foundation, the Brehm Coalition, and the Leona M. and Harry B. Helmsley Charitable Trust. T.G. was supported by a postdoctoral fellowship from the Juvenile Diabetes Research Foundation. L.L. was supported by a postdoctoral fellowship from the American Diabetes Association. M.H. has equity interest in ViaCyte Inc.
No other potential conflicts of interest relevant to this article were reported.
T.G. performed the studies and assisted in the study design and its interpretation and in writing the manuscript. L.L. assisted in executing the study and study interpretation. N.L. assisted in performing the studies. M.H. contributed to the study design, study interpretation, and to preparation of the manuscript. M.H. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
The authors thank Dr. Nissim Benvenisty, of The Hebrew University of Jerusalem, for providing human islets samples and Drs. Grace Wei, Holger Ross, and Sapna Puri, of the University of California, San Francisco, for discussion and thoughtful comments on the manuscript.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db12-0167/-/DC1.
L.L. is currently affiliated with the Department of Cell and Developmental Biology, Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel.
REFERENCES
- 1.Posselt AM, Szot GL, Frassetto LA, et al. Islet transplantation in type 1 diabetic patients using calcineurin inhibitor-free immunosuppressive protocols based on T-cell adhesion or costimulation blockade. Transplantation 2010;90:1595–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Posselt AM, Szot GL, Frassetto LA, Masharani U, Stock PG. Clinical islet transplantation at the University of California, San Francisco. Clin Transpl 2010:235–243 [PubMed] [Google Scholar]
- 3.Wiseman AC, Wainright JL, Sleeman E, et al. An analysis of the lack of donor pancreas utilization from younger adult organ donors. Transplantation 2010;90:475–480 [DOI] [PubMed] [Google Scholar]
- 4.D’Amour KA, Bang AG, Eliazer S, et al. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol 2006;24:1392–1401 [DOI] [PubMed] [Google Scholar]
- 5.Jiang J, Au M, Lu K, et al. Generation of insulin-producing islet-like clusters from human embryonic stem cells. Stem Cells 2007;25:1940–1953 [DOI] [PubMed] [Google Scholar]
- 6.Jiang W, Shi Y, Zhao D, et al. In vitro derivation of functional insulin-producing cells from human embryonic stem cells. Cell Res 2007;17:333–344 [DOI] [PubMed] [Google Scholar]
- 7.Chen S, Borowiak M, Fox JL, et al. A small molecule that directs differentiation of human ESCs into the pancreatic lineage. Nat Chem Biol 2009;5:258–265 [DOI] [PubMed] [Google Scholar]
- 8.Maehr R, Chen S, Snitow M, et al. Generation of pluripotent stem cells from patients with type 1 diabetes. Proc Natl Acad Sci U S A 2009;106:15768–15773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D’Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol 2005;23:1534–1541 [DOI] [PubMed] [Google Scholar]
- 10.Kroon E, Martinson LA, Kadoya K, et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol 2008;26:443–452 [DOI] [PubMed] [Google Scholar]
- 11.Wells JM, Melton DA. Vertebrate endoderm development. Annu Rev Cell Dev Biol 1999;15:393–410 [DOI] [PubMed] [Google Scholar]
- 12.Hebrok M, Kim SK, Melton DA. Notochord repression of endodermal Sonic hedgehog permits pancreas development. Genes Dev 1998;12:1705–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim SK, Hebrok M. Intercellular signals regulating pancreas development and function. Genes Dev 2001;15:111–127 [DOI] [PubMed] [Google Scholar]
- 14.Kim SK, Hebrok M, Melton DA. Notochord to endoderm signaling is required for pancreas development. Development 1997;124:4243–4252 [DOI] [PubMed] [Google Scholar]
- 15.Lammert E, Cleaver O, Melton D. Induction of pancreatic differentiation by signals from blood vessels. Science 2001;294:564–567 [DOI] [PubMed] [Google Scholar]
- 16.Gittes GK. Developmental biology of the pancreas: a comprehensive review. Dev Biol 2009;326:4–35 [DOI] [PubMed] [Google Scholar]
- 17.Scharfmann R. Control of early development of the pancreas in rodents and humans: implications of signals from the mesenchyme. Diabetologia 2000;43:1083–1092 [DOI] [PubMed] [Google Scholar]
- 18.Golosow N, Grobstein C. Epitheliomesenchymal interaction in pancreatic morphogenesis. Dev Biol 1962;4:242–255 [DOI] [PubMed] [Google Scholar]
- 19.Micallef SJ, Li X, Schiesser JV, et al. INS(GFP/w) human embryonic stem cells facilitate isolation of in vitro derived insulin-producing cells. Diabetologia 2012;55:694–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szot GL, Koudria P, Bluestone JA. Transplantation of pancreatic islets into the kidney capsule of diabetic mice. J Vis Exp 2007:e404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 1994;371:606–609 [DOI] [PubMed] [Google Scholar]
- 22.Offield MF, Jetton TL, Labosky PA, et al. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development 1996;122:983–995 [DOI] [PubMed] [Google Scholar]
- 23.Jacquemin P, Durviaux SM, Jensen J, et al. Transcription factor hepatocyte nuclear factor 6 regulates pancreatic endocrine cell differentiation and controls expression of the proendocrine gene ngn3. Mol Cell Biol 2000;20:4445–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massagué J. TGF-beta signal transduction. Annu Rev Biochem 1998;67:753–791 [DOI] [PubMed] [Google Scholar]
- 25.Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development 2002;129:2447–2457 [DOI] [PubMed] [Google Scholar]
- 26.Gradwohl G, Dierich A, LeMeur M, Guillemot F. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci USA 2000;97:1607–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rezania A, Riedel MJ, Wideman RD, et al. Production of functional glucagon-secreting α-cells from human embryonic stem cells. Diabetes 2011;60:239–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Afrikanova I, Yebra M, Simpkinson M, Xu Y, Hayek A, Montgomery A. Inhibitors of Src and focal adhesion kinase promote endocrine specification: impact on the derivation of β-cells from human pluripotent stem cells. J Biol Chem 2011;286:36042–36052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nostro MC, Sarangi F, Ogawa S, et al. Stage-specific signaling through TGFβ family members and WNT regulates patterning and pancreatic specification of human pluripotent stem cells. Development 2011;138:861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sander M, Sussel L, Conners J, et al. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of beta-cell formation in the pancreas. Development 2000;127:5533–5540 [DOI] [PubMed] [Google Scholar]
- 31.Wang J, Elghazi L, Parker SE, et al. The concerted activities of Pax4 and Nkx2.2 are essential to initiate pancreatic beta-cell differentiation. Dev Biol 2004;266:178–189 [DOI] [PubMed] [Google Scholar]
- 32.Basford CL, Prentice KJ, Hardy AB, et al. The functional and molecular characterisation of human embryonic stem cell-derived insulin-positive cells compared with adult pancreatic beta cells. Diabetologia 2012;55:358–371 [DOI] [PubMed] [Google Scholar]
- 33.Grobstein C. Inductive epitheliomesenchymal interaction in cultured organ rudiments of the mouse. Science 1953;118:52–55 [DOI] [PubMed] [Google Scholar]
- 34.Kallman F, Grobstein C. Fine structure of differentiating mouse pancreatic exocrine cells in transfilter culture. J Cell Biol 1964;20:399–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhushan A, Itoh N, Kato S, et al. Fgf10 is essential for maintaining the proliferative capacity of epithelial progenitor cells during early pancreatic organogenesis. Development 2001;128:5109–5117 [DOI] [PubMed] [Google Scholar]
- 36.Maldonado TS, Kadison AS, Crisera CA, et al. Ontogeny of activin B and follistatin in developing embryonic mouse pancreas: implications for lineage selection. J Gastrointest Surg 2000;4:269–275 [DOI] [PubMed] [Google Scholar]
- 37.Miralles F, Czernichow P, Scharfmann R. Follistatin regulates the relative proportions of endocrine versus exocrine tissue during pancreatic development. Development 1998;125:1017–1024 [DOI] [PubMed] [Google Scholar]
- 38.Kobayashi H, Spilde TL, Bhatia AM, et al. Retinoid signaling controls mouse pancreatic exocrine lineage selection through epithelial-mesenchymal interactions. Gastroenterology 2002;123:1331–1340 [DOI] [PubMed] [Google Scholar]
- 39.Crisera CA, Rose MI, Connelly PR, et al. The ontogeny of TGF-beta1, -beta2, -beta3, and TGF-beta receptor-II expression in the pancreas: implications for regulation of growth and differentiation. J Pediatr Surg 1999;34:689–693; discussion 693–684 [DOI] [PubMed]
- 40.Landsman L, Nijagal A, Whitchurch TJ, et al. Pancreatic mesenchyme regulates epithelial organogenesis throughout development. PLoS Biol 2011;9:e1001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Derynck R, Feng XH. TGF-beta receptor signaling. Biochim Biophys Acta 1997;1333:F105–F150 [DOI] [PubMed] [Google Scholar]
- 42.Böttinger EP, Jakubczak JL, Haines DC, Bagnall K, Wakefield LM. Transgenic mice overexpressing a dominant-negative mutant type II transforming growth factor beta receptor show enhanced tumorigenesis in the mammary gland and lung in response to the carcinogen 7,12-dimethylbenz-[a]-anthracene. Cancer Res 1997;57:5564–5570 [PubMed] [Google Scholar]
- 43.Tulachan SS, Tei E, Hembree M, et al. TGF-beta isoform signaling regulates secondary transition and mesenchymal-induced endocrine development in the embryonic mouse pancreas. Dev Biol 2007;305:508–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inman GJ, Nicolás FJ, Callahan JF, et al. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol 2002;62:65–74 [DOI] [PubMed] [Google Scholar]
- 45.Guo T, Hebrok M. Stem cells to pancreatic beta-cells: new sources for diabetes cell therapy. Endocr Rev 2009;30:214–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng X, Ying L, Lu L, et al. Self-renewing endodermal progenitor lines generated from human pluripotent stem cells. Cell Stem Cell 2012;10:371–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.