Abstract
Tight junction proteins, including claudins, are often dysregulated during carcinogenesis and tumor progression. Moreover, the claudin expression pattern usually varies between different tumor entities. We aimed to investigate claudin expression profiles of primary and metastatic liver malignancies. We analyzed claudin-1, -2, -3, -4, and -7 expression by quantitative immunohistochemistry and real-time RT-PCR, respectively. Twenty hepatocellular carcinomas (HCCs) and liver metastases of 20 colorectal adenocarcinomas (CRLMs) and 15 pancreatic adenocarcinomas (PLMs) were studied together with paired surrounding non-tumorous liver samples and 5 normal liver samples. Strong claudin-3 and -7 immunohistochemical positivities were detected in CRLM samples, each with significantly stronger staining when compared with HCC and PLM groups. Claudin-1 protein was found highly expressed in CRLM, in contrast to lower expression in PLM and HCC. CRLMs and PLMs also were strongly positive for claudin-4, while being virtually undetectable in HCC. Claudin-2 showed strong positivity in non-tumorous liver tissue, whereas significantly weaker positivity was observed in all tumors. Differences in mRNA expression were mostly similar to those found by immunohistochemistry. In conclusion, HCC and both CRLM and PLM display distinct claudin expression profiles, which might provide better understanding of the pathobiology of these lesions and might be used for differential diagnosis.
Keywords: liver metastasis, hepatocellular carcinoma, tight junction, claudins
The liver is one of the most common sites of metastasis and also gives rise to highly malignant primary cancers. Colorectal and pancreatic carcinomas are common sources of hepatic metastases, which are approximately 20 times more frequently found than the most common primary liver cancer, hepatocellular carcinoma (HCC) (Kadry et al. 2001). Given the high prevalence and mortality rates of these focal lesions, a better molecular characterization is of great importance, especially because these lesions might also represent differential diagnostic problems.
Recently, proteins of the 26-member human claudin family were identified as participants in numerous normal and neoplastic processes (Mineta et al. 2011). Claudins are integral membrane proteins that form the indispensable backbone of tight junctions (TJs) (Tsukita and Furuse 1999). They play a crucial role in the cell-cell connection, maintaining epithelial cell polarity and regulating paracellular permeability (Martin and Jiang 2009). Besides these classical functions, claudins are newly discovered participants in various intriguing molecular events—for example, claudin-1, -6, and -9 seem to act as co-receptors for hepatitis C virus entry (Meertens et al. 2008), whereas claudin-4 and -3 are target molecules of Clostridium perfringens enterotoxin (Sonoda et al. 1999). Moreover, certain claudins have been implicated to be involved in carcinogenesis and even in tumor progression and metastasis formation. For example, claudin-1 overexpression alone was sufficient to provide invasive features to human liver cells (Yoon et al. 2010), whereas in colorectal cancer, claudin-1 is a suggested target of the β-catenin/Tcf signaling cascade (Miwa et al. 2001). Low claudin-4 expression was clearly correlated with invasion and metastasis of pancreatic cancer cells (Michl et al. 2003), and claudin-7 downregulation has also been shown to increase cell growth and invasion in esophageal cancer (Lioni et al. 2007). Increased expression of claudin-10 in HCC was suggested to be associated with invasiveness, recurrence, and shorter survival (Ip et al. 2007; Huang et al. 2011).
It is also noteworthy that claudins are expressed in a tissue-specific manner. Moreover, unique claudin composition is also characteristic of cancerous tissues, which exhibit tumor type- and even differentiation-specific up- or downregulation of certain claudins (Soini 2005; Nemeth et al. 2009; Szabo et al. 2009; Szekely et al. 2011; Torzsok et al. 2011). These specific patterns can even be exploited as differential diagnostic (Soini 2005; Lódi et al. 2006) or prognostic tools (Szasz et al. 2011). It has also been shown that, in response to various stimuli, claudins can dynamically change (e.g., during the metastatic process), as cells must break down the adhesion barrier at the primary site and then reform junctions at the site of metastasis (Martin and Jiang 2009). This might explain why claudin expression can differ between the primary and metastatic lesion of the same tumor (Tabaries et al. 2011). Moreover, tight junction proteins are involved in signal transduction and regulatory pathways (Balda and Matter 2009); consequently, the loss of cell-cell contact does not necessarily mean general downregulation of all claudins: Certain claudins are downregulated, whereas others are overexpressed.
Although it is clear that claudins might be key factors in tumor progression and many of them are suggested modulators of invasion and metastasis, there is limited knowledge about the claudin phenotype of metastatic lesions, especially regarding human samples (Ueda et al. 2007; Merikallio et al. 2011). Claudin expression alterations in primary cancers are much better characterized, including those of the breast (Kominsky et al. 2003; Tokes et al. 2005), colon (de Oliveira et al. 2005; Dhawan et al. 2005), prostate (Long et al. 2001), and pancreas (Nichols et al. 2004; Borka et al. 2007). Our group has also previously defined tight junctional protein changes in primary and metastatic cancers of the liver. For example, Lódi et al. (2006) found significantly increased claudin-4 expression in biliary tract cancers, which could differentiate tumors of biliary and hepatocellular origin, whereas the study by Orbán et al. (2008) revealed no occludin and zonula occludens-1 expression in HCC and high expression in colorectal liver metastasis.
To our knowledge, no study is available regarding comparative characterization of claudin expression profiles in hepatocellular carcinoma and liver metastases of gastrointestinal tumors. Therefore, in the present study, we aimed to investigate claudin-1, -2, -3, -4, and -7 expression in the most common primary liver carcinoma—HCC—and the most common metastatic liver cancer—colorectal liver metastasis (CRLM). In addition, because of shared developmental roots and supposed shared features between biliary tract cancers and pancreatic ductal adenocarcinoma (Geller et al. 2008), pancreatic liver metastasis (PLM) was also included in our study.
Materials and Methods
Tissue Samples
Surgically removed tissue specimens of 20 HCC, 20 CRLM, and 15 PLM cases with corresponding surrounding non-tumorous liver tissues were enrolled in the present study with the permission of the Regional Ethical Committee of the Semmelweis University (#137/2008). Tissue samples were collected over an 8-year period and the diagnoses were reevaluated and confirmed by expert pathologists (ZS, AK, GL) and further supported by clinical data specific to the location of the primary tumor. The median age and female to male ratio in the patient groups were as follows: 65 years, 7:13 (HCC); 65 years, 7:13 (CRLM); and 58 years, 9:6 (PLM). Five normal liver (NL) samples were obtained from patients who died in accidents.
Histology and Immunohistochemistry
Tissue samples were fixed in 10% neutral-buffered formalin and embedded in paraffin (FFPE), then cut and routinely stained with hematoxylin-eosin to establish the diagnosis.
Immunohistochemical detection of claudin-1, -2, -3, -4, and -7 proteins was performed on 3- to 4-µm-thick FFPE sections. Reactions were carried out using the Ventana ES automatic immunostainer (Ventana Medical Systems; Tucson, AZ) with the streptavidin-biotin-peroxidase technique and 3,3′-diaminobenzidine tetrahydrochloride as the chromogen. Well-characterized primary antibodies against claudin-1, -2, -3, -4, and -7 (Zymed; San Francisco, CA) were diluted 1:100 (Soini 2005; Laurila et al. 2007; Moldvay et al. 2007; Torzsok et al. 2011). Monoclonal mouse antibodies against claudin-2 and -4 and polyclonal rabbit antibodies against claudin-1, -3, and -7 were used. For negative controls, the specific antibody was omitted and either the antibody diluent alone or isotype-matched IgG serum was used. Positive controls were normal skin epithelium (claudin-1), normal colon mucosa (claudin-2, -3, -4), and breast ductal cells (claudin-7), as recommended by the manufacturer.
Morphometrical Analysis
From each slide, 10 non-overlapping photographs were taken (×60 objective, Olympus BX50 microscope; Olympus Corporation, Tokyo, Japan), and quantification of the immunohistochemical staining was performed using Leica QWin V3 morphometrical software (Leica Microsystems Imaging Solutions Ltd.; Cambridge, UK). Quantification was done by measuring the area of immunohistochemical positivity, which correlates with both the presence and strength of the staining (Szekely et al. 2011). Area positivity above 3% was considered strong, 1.5% to 3% was moderate, and below 1.5% was weak.
Real-Time RT-PCR Analysis
RNA isolation
Histology-guided manual dissection of FFPE samples (20 HCC, 20 CRLM, 20 PLM, 5 NL) was performed, and tumorous and non-tumorous samples were processed separately. Total RNA was isolated with the High Pure RNA Paraffin Kit (Roche; Indianapolis, IN) according to the manufacturer’s instructions. Proteinase K digestion time was 16 h for each sample.
Reverse transcription
In total, 2 µg total RNA was reverse transcribed in a 20-µl mix of the High Capacity RNA-to-cDNA Kit (Applied Biosystems [ABI]; Foster City, CA) according to the manufacturer’s protocol.
Real-time RT-PCR
Real-time PCR reactions for claudin-1, -2, -3, -4, and -7 and β-actin (reference gene) were carried out in duplicates, applying 100 ng cDNA template in a total volume of 25 µl SYBR Green PCR Master Mix (ABI; AB4309155). The primer sequences are listed in Table 1. Real-time PCR conditions were as follows: initial denaturation at 95C for 2 min, then 40 cycles at 95C for 20 sec, 60C for 30 sec, and 72C for 1 min, using the ABI Prism 7000 sequence detection system (ABI). Relative quantification of target gene expression was performed using the E = 2ΔCq (ΔCq = Cqreference – Cqtarget) method, with the average value of β-actin reactions as reference.
Table 1.
Primer Sequences (5′–3′) and Amplicon Sizes of Claudins and β-actin
| Gene | Forward | Reverse | Amplicon Size, bp |
|---|---|---|---|
| claudin-1 | GTG CGA TAT TTC TTC TTG CAG G | TTC GTA CCT GGC ATT GAC TGG | 113 |
| claudin-2 | CTC CCT GGC CTG CAT TAT CTC | ACC TGC TAC CGC CAC TCT GT | 91 |
| claudin-3 | CTG CTC TGC TGC TCG TGT CC | TTA GAC GTA GTC CTT GCG GTC GTA G | 129 |
| claudin-4 | GGC TGC TTT GCT GCA ACT GTC | GAG CCG TGG CAC CTT ACA CG | 108 |
| claudin-7 | CAT CGT GGC AGG TCT TGC C | GAT GGC AGG GCC AAA CTC ATA C | 118 |
| β-actin | CCT GGC ACC CAG CAC AAT | GGG CCG GAC TCG TCA TAC | 144 |
Statistical Analysis
Statistical analysis was carried out using STATISTICA software v8.0 (StatSoft; Tulsa, OK). The surrounding non-tumorous livers were treated as one group for statistics. To examine the differences between the studied groups, the non-parametric Kruskal-Wallis test was used. Differences were considered statistically significant when p<0.05.
Results
Immunohistochemistry and Morphometry
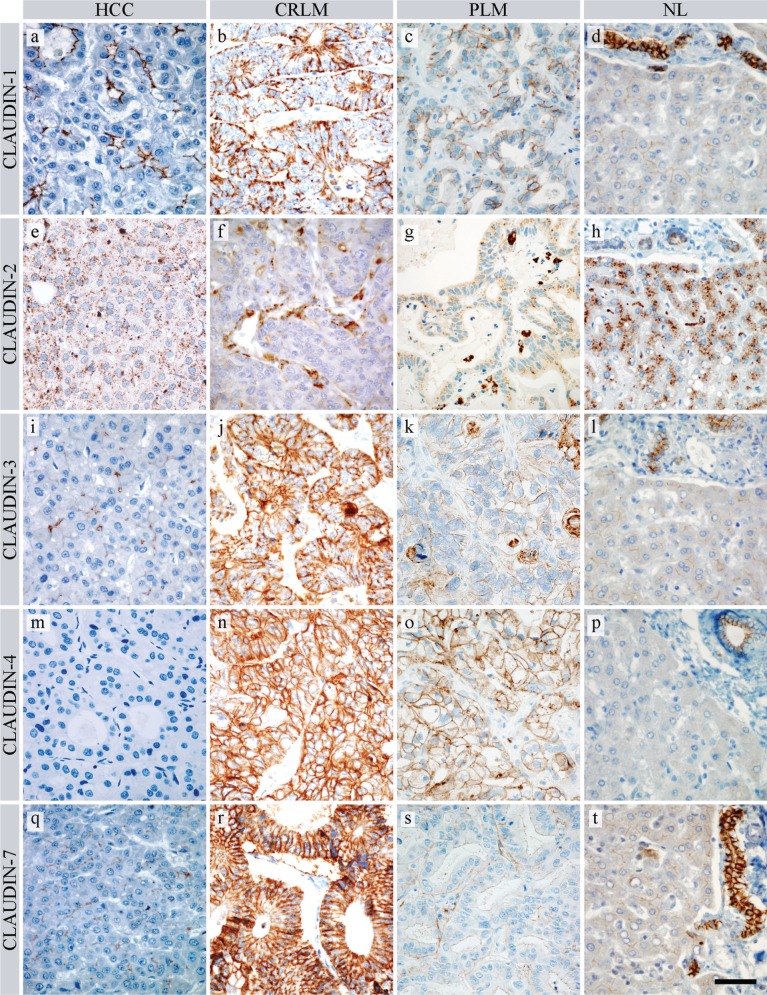
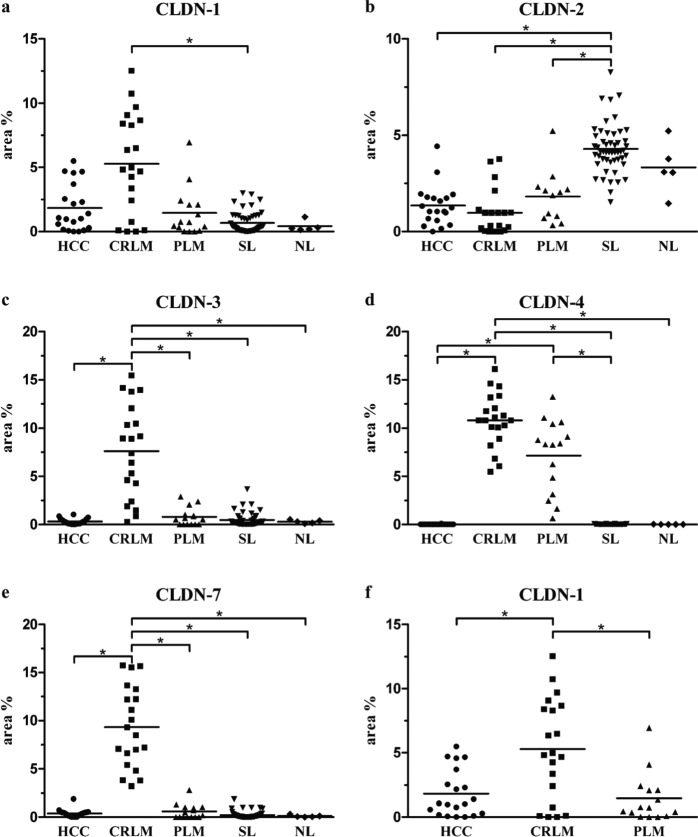
Membranous claudin-1 positivity was predominantly detected in an apical localization in HCC cells organized in trabecular/glandular/acinar structures (Fig. 1a), whereas an almost completely circumscribed positivity was revealed in tumor cells of CRLM and PLM (Fig. 1b,c). Normal and non-tumorous tissue surrounding liver bile ducts showed intense positivity, whereas hepatocytes revealed faint positivity, mainly in the apical position (Fig. 1d). Morphometrical analysis showed that the mean area percentage of claudin-1 staining was highest in the CRLM group (Table 2, Fig. 2a), although the difference between CRLM and the two other cancer groups became statistically significant only when the non-tumorous groups were excluded from the Kruskal-Wallis statistical comparison (Fig. 2f; CRLM vs HCC, p=0.032; CRLM vs PLM, p=0.008).
Figure 1.
Claudin expression in HCC (hepatocellular carcinoma), CRLM (colorectal liver metastasis), PLM (pancreatic liver metastasis), and NL (normal liver). (a, b, c, d) Claudin-1 in HCC, CRLM, PLM, and NL, respectively. (e, f, g, h) Claudin-2 in HCC, CRLM, PLM, and NL, respectively. (i, j, k, l) Claudin-3 in HCC, CRLM, PLM, and NL, respectively. (m, n, o, p) Claudin-4 in HCC, CRLM, PLM, and NL, respectively. (q, r, s, t) Claudin-7 in HCC, CRLM, PLM, and NL, respectively. Each photograph was taken with the same magnification. Scale bar: 50 µm
Table 2.
Results of Morphometry with Statistical Comparison of Individual Groups
| Results of Morphometry |
||||||||
|---|---|---|---|---|---|---|---|---|
| Group | Area % | SEM | K-W Test, p Value | vs CRLM, p Value | vs PLM, p Value | vs SL, p Value | vs NL, p Value | |
| Claudin-1 | HCC | 1.83 | 0.41 | <0.001* | 0.443 | 1.000 | 0.390 | 1.000 |
| CRLM | 5.29 | 0.88 | 0.115 | <0.001* | 0.140 | |||
| PLM | 1.45 | 0.49 | 1.000 | 1.000 | ||||
| SL | 0.67 | 0.11 | 1.000 | |||||
| NL | 0.42 | 0.18 | ||||||
| Claudin-2 | HCC | 1.35 | 0.23 | <0.001* | 1.000 | 1.000 | <0.001* | 0.386 |
| CRLM | 0.98 | 0.27 | 1.000 | <0.001* | 0.085 | |||
| PLM | 1.82 | 0.39 | 0.001* | 1.000 | ||||
| SL | 4.29 | 0.18 | 1.000 | |||||
| NL | 3.33 | 0.61 | ||||||
| Claudin-3 | HCC | 0.31 | 0.07 | <0.001* | <0.001* | 1.000 | 1.000 | 1.000 |
| CRLM | 7.61 | 1.09 | <0.001* | <0.001* | 0.026* | |||
| PLM | 0.79 | 0.25 | 1.000 | 1.000 | ||||
| SL | 0.45 | 0.09 | 1.000 | |||||
| NL | 0.28 | 0.08 | ||||||
| Claudin-4 | HCC | 0.00 | 0.00 | <0.001* | <0.001* | <0.001* | 0.046* | 0.635 |
| CRLM | 10.82 | 0.63 | 1.000 | <0.001* | 0.029* | |||
| PLM | 7.16 | 0.98 | <0.001* | 0.212 | ||||
| SL | 0.01 | 0.00 | 1.000 | |||||
| NL | 0.01 | 0.00 | ||||||
| Claudin-7 | HCC | 0.38 | 0.09 | <0.001* | <0.001* | 1.000 | 0.443 | 1.000 |
| CRLM | 9.32 | 0.93 | <0.001* | <0.001* | 0.001* | |||
| PLM | 0.57 | 0.20 | 1.000 | 1.000 | ||||
| SL | 0.22 | 0.05 | 1.000 | |||||
| NL | 0.10 | 0.06 | ||||||
Mean and standard error values of morphometric analysis are listed. Statistical comparison of horizontal versus vertical groups was performed using the Kruskal-Wallis test. Significant differences (p<0.05) are marked by asterisks. CRLM, colorectal liver metastasis; HCC, hepatocellular carcinoma; K-W, Kruskal-Wallis; NL, normal liver; PLM, pancreatic liver metastasis; SEM, standard error of mean; SL, surrounding non-tumorous liver.
Figure 2.
Results of morphometric analysis. Stained area percentage of individual samples for each claudin (CLDN). Lines indicate the mean value of the groups. Asterisks mark significant differences (p<0.05). HCC (hepatocellular carcinoma), CRLM (colorectal liver metastasis), PLM (pancreatic liver metastasis), SL (surrounding non-tumorous liver), and NL (normal liver). Panel a shows the result of Kruskal-Wallis analysis, including non-tumorous groups, whereas panel f demonstrates the result of an analysis excluding non-tumorous groups
Claudin-2 immunostaining resulted in granular cytoplasmic positivity in both tumorous (Fig. 1e–g) and normal cells (Fig. 1h). A significant decrease in the stained area percentage was observed in all three tumor groups when compared with the surrounding non-tumorous liver tissue (Fig. 2b), where both bile ducts and hepatocytes stained intensely.
Claudin-3 gave a significantly stronger membranous reaction in CRLM (Fig. 1j) than in the other groups (Fig. 2c). HCC and non-tumorous liver cells exhibited weak and scattered positivity (Fig. 1i), along with stronger bile duct positivity (Fig. 1l). A weak and focal claudin-3 signal was also present in 10 of 15 PLM samples (Fig. 1k).
Claudin-4 showed definite strong membrane positivity in all CRLM and PLM samples (Fig. 1n,o) with no significant difference between these two groups (Fig. 2d). By contrast, this protein was virtually undetectable in HCC cells (Fig. 1m) and non-tumorous hepatocytes, but bile duct cells of non-tumorous and normal liver were positive (Fig. 1p).
Claudin-7 also showed significantly increased membranous positivity in CRLM (Fig. 1r), in comparison with the other groups (Fig. 2e). Of 15 PLM cases, only 9 showed faint and focal positivity (Fig. 1s), and only weak staining was present on normal hepatocytes and hepatocellular carcinoma cells (Fig. 1q,t).
In summary, HCC is associated with moderate claudin-1, weak claudin-2, -3, and -7, and no claudin-4 positivity. CRLM is characterized by strong claudin-1, -3, -4, and -7 and weak claudin-2 positivity, whereas strong claudin-4 and moderate or weak claudin-1, -2, -3, and -7 positivity are characteristic of PLM. Regarding the differentiation of HCC, claudin-1 positivity showed an increasing tendency to be expressed along with higher grades of HCC, which, however, did not prove to be significant. The other claudins did not reveal an association with grading (Suppl. Fig. 1.).
Real-Time RT-PCR Analysis
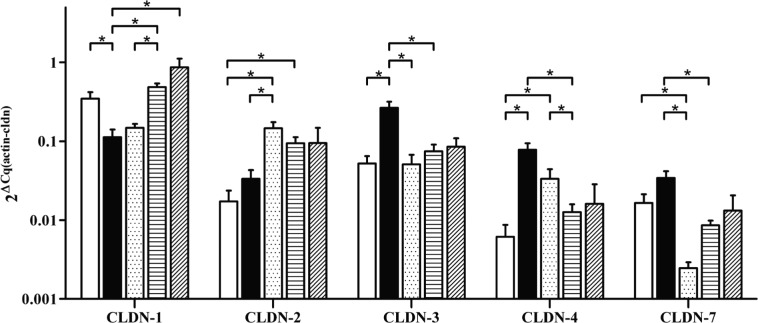
The differences in claudin expression at the protein level were mostly mimicked by differences in relative mRNA expression (β-actin was used as reference) (Table 3, Fig. 3.). Claudin-3 and -4 mRNA levels were in great concordance with morphometry, because their expression in CRLM were significantly higher than in non-tumorous surrounding liver (SL) and HCC. A significant difference was also present between CRLM and PLM regarding claudin-3. Furthermore, claudin-7 mRNA in CRLM was found to be significantly higher than in PLM and SL, although levels in HCC were comparable to levels in CRLM. Also coincident with morphometrical results, claudin-2 levels in HCC and CRLM were lower than in SL, with the difference reaching statistical significance for HCC, although this downregulation could not be observed in PLM. A striking difference was observed between protein and mRNA expression for claudin-1, which, instead of the expected upregulation, showed significant downregulation in CRLM when compared with HCC and SL; PLM also showed lower mRNA levels than HCC and SL.
Table 3.
Results of Real-time RT-PCR Analysis with Statistical Comparison of Individual Groups
| Results of Real-Time RT-PCR |
||||||||
|---|---|---|---|---|---|---|---|---|
| Group | 2ΔCq(actin – cldn) | SEM | K-W Test, p Value | vs CRLM, p Value | vs PLM, p Value | vs SL, p Value | vs NL, p Value | |
| Claudin-1 | HCC | 0.35 | 0.07 | <0.001* | 0.012* | 0.537 | 1.000 | 1.000 |
| CRLM | 0.11 | 0.03 | 1.000 | <0.001* | 0.004* | |||
| PLM | 0.15 | 0.02 | 0.006* | 0.062 | ||||
| SL | 0.49 | 0.05 | 1.000 | |||||
| NL | 0.86 | 0.25 | ||||||
| Claudin-2 | HCC | 0.02 | 0.01 | <0.001* | 0.927 | <0.001* | <0.001* | 0.461 |
| CRLM | 0.03 | 0.01 | 0.001* | 0.208 | 1.000 | |||
| PLM | 0.15 | 0.03 | 0.128 | 0.791 | ||||
| SL | 0.09 | 0.02 | 1.000 | |||||
| NL | 0.09 | 0.05 | ||||||
| Claudin-3 | HCC | 0.05 | 0.01 | <0.001* | <0.001* | 1.000 | 1.000 | 1.000 |
| CRLM | 0.27 | 0.05 | 0.001* | <0.001* | 1.000 | |||
| PLM | 0.05 | 0.02 | 1.000 | 1.000 | ||||
| SL | 0.07 | 0.02 | 1.000 | |||||
| NL | 0.09 | 0.02 | ||||||
| Claudin-4 | HCC | 0.01 | 0.00 | <0.001* | <0.001* | 0.003* | 1.000 | 1.000 |
| CRLM | 0.08 | 0.02 | 1.000 | <0.001* | 0.229 | |||
| PLM | 0.03 | 0.01 | 0.024* | 1.000 | ||||
| SL | 0.01 | 0.00 | 1.000 | |||||
| NL | 0.02 | 0.01 | ||||||
| Claudin-7 | HCC | 0.02 | 0.00 | <0.001* | 0.111 | 0.011* | 1.000 | 1.000 |
| CRLM | 0.03 | 0.01 | <0.001* | <0.001* | 0.679 | |||
| PLM | 0.00 | 0.00 | 0.051* | 0.454 | ||||
| SL | 0.01 | 0.00 | 1.000 | |||||
| NL | 0.01 | 0.01 | ||||||
Mean and standard error values of real-time RT-PCR analysis are listed. Statistical comparison of horizontal versus vertical groups was performed using the Kruskal-Wallis test. Significant differences (p<0.05) are marked by asterisks. CRLM, colorectal liver metastasis; HCC, hepatocellular carcinoma; K-W, Kruskal-Wallis; NL, normal liver; PLM, pancreatic liver metastasis; SEM, standard error of mean; SL, surrounding non-tumorous liver.
Figure 3.
Differences in claudin (CLDN) mRNA expression relative to β-actin. Asterisks mark significant differences (p<0.05). ( ) HCC (hepatocellular carcinoma), (
) HCC (hepatocellular carcinoma), ( ) CRLM (colorectal liver metastasis), (
) CRLM (colorectal liver metastasis), ( ) PLM (pancreatic liver metastasis), (
) PLM (pancreatic liver metastasis), ( ) SL (surrounding non-tumorous liver), (
) SL (surrounding non-tumorous liver), ( ) NL (normal liver)
) NL (normal liver)
mRNA expression did not differ significantly with respect to the different grades of HCC (Suppl. Fig. 2.).
Discussion
Claudins are the main constituents of tight junctions, and several lines of evidence propose that they contribute to the neoplastic processes. The altered expression of certain claudins can influence cancer progression indirectly by impairing tight junction functions or directly by modulating neoplastic signaling pathways. Furthermore, unique claudin profiles of different cancers are recognized to carry diagnostic, prognostic, and even therapeutic potential (Wang et al. 2011; Soini 2012).
Primary hepatocellular carcinoma and colorectal and pancreatic carcinomas are among the five most lethal carcinomas, and the latter two exhibit frequent association with liver metastases (Centeno 2006). Given that 90% of cancer-related deaths are due to metastases (Christofori 2006), of which hepatic metastases represent a great percentage, identification of metastasis-related molecular patterns is just as important as that of primary cancers. Thus, molecular profiling is crucial for a better understanding of these liver lesions and may contribute to a better therapeutic approach to these tumors in the future. Moreover, these profiles carry great differential diagnostic value.
To our knowledge, this is the first report to address comparative claudin characterization of primary and metastatic neoplasms of the liver. In the present study, distinct claudin expression profiles of HCC, CRLM, and PLM were identified: HCC moderately expressed claudin-1, weakly expressed claudin-2, -3, and -7, and lacked claudin-4. CRLM was characterized by strong claudin-1, -3, -4, and -7 and moderate claudin-2 positivities, whereas PLM showed strong claudin-4, moderate claudin-2, and weak claudin-1, -3, and -7 stainings.
The role of certain claudins in tumorigenesis and tumor progression, particularly in metastasis formation, is largely undefined and sometimes appears contradictory. Whether overexpression of certain claudins is a cellular mechanism to compensate for the impaired tight junction function (Hadj-Rabia et al. 2004; Cheung et al. 2012) or is deeply involved in neoplastic signaling pathways is not yet clearly understood. However, tight junction strand disorganization and increased paracellular leakage were observed in colorectal tumor tissues despite upregulation of several claudins (de Oliveira et al. 2005), indicating that overexpression is not effective in maintaining tight junction function and thus may have other effects on cellular homeostasis.
Claudin-1 is one of the best characterized molecules of the claudin protein family, and its direct involvement in tumor progression and metastasis seems to be unequivocal, although there are some discrepancies as to its exact role in these processes. In colorectal carcinoma, overexpression and knockdown experiments support a role for claudin-1 as a promoter of invasion and metastasis, and a positive correlation between claudin-1 expression and tumor progression and metastasis has been established (Dhawan et al. 2005). In line with this, frequent upregulation of claudin-1 in human colorectal cancer tissues has been observed in several studies (de Oliveira et al. 2005; Grone et al. 2007; Kinugasa et al. 2007; Oshima et al. 2008; Huo et al. 2009). On the other hand, low claudin-1 expression seemed to correlate with poor differentiation, poor prognosis (Resnick et al. 2005), and lymph node metastasis in colorectal cancer (Ersoz et al. 2011), whereas, in lung adenocarcinoma, claudin-1 overexpression has been shown to suppress cancer invasion and metastasis (Chao et al. 2009). Interestingly, similarly controversial results can be found regarding HCC, in which reduced claudin-1 expression was shown to closely correlate with tumor dedifferentiation and poor prognosis (Higashi et al. 2007). Yet, HCC cell line experiments supported a role for claudin-1 as a potent inducer of cancer cell invasion (Yoon et al. 2010). In the present study, definite claudin-1 positivity was found in both primary HCC and metastatic colorectal and pancreatic carcinomas, underlining the involvement of this protein in hepatocarcinogenesis and metastasis formation. Our data on the increasing expression of claudin-1 with higher grades of HCC are also supported by a recent publication by Suh et al. (2012), which suggests a role for claudin-1 in epithelial-mesenchymal transition in hepatocarcinogenesis.
It has been shown that claudin-2 expression diminishes during breast cancer progression at the primary site (Kim et al. 2008), but the expression is regained in liver-metastasizing breast cancer cells, thus playing a promoter role in the formation of liver metastases (Tabaries et al. 2011). In our sample set, we found weak or moderate claudin-2 positivity in liver metastases of colorectal and pancreatic cancers. Obvious claudin-2 expression has already been described in 25.3% of colorectal (Aung et al. 2006) and 45% of pancreatic adenocarcinomas (Borka et al. 2007). Thus, claudin-2 may not only participate in the carcinogenesis of these tumors but may also contribute to a metastatic phenotype. Interestingly, we found a significant downregulation of claudin-2 at both protein and mRNA levels in HCC when compared with normal and surrounding liver tissues. Intriguingly, claudin-2 was shown to be positively regulated by the hepatocyte nuclear factor–1α (HNF-1α) (Sakaguchi et al. 2002), a factor with reduced expression in HCC (Zeng et al. 2011), and was also shown to be required for proper hepatic polarization (Son et al. 2009), which is frequently lost in tumors. Taken together, these results seem to support a tumor-suppressive role for claudin-2 in hepatocellular carcinogenesis.
Since the discovery of claudin-3 and -4 as receptors for the cytotoxic C. perfringens enterotoxin (Sonoda et al. 1999), much attention has been given to these claudin isoforms. Claudin-4, for example, soon became a candidate for claudin-based antitumor therapies (Suzuki et al. 2009) and the target of novel therapeutic approaches (Neesse et al. 2012). Claudin-4 expression was proposed to be a typical feature of adenocarcinomas (Michl et al. 2003), and frequent upregulation was described in several tumors, among others in colorectal carcinoma (Oshima et al. 2008), ovarian carcinoma (Rangel et al. 2003), biliary tract cancer (Nemeth et al. 2009), and pancreatic carcinoma (Borka et al. 2007). On the other hand, impaired claudin-4 expression seemed to be associated with less differentiated and invasive phenotypes, suggesting its inhibitory effect on invasion and metastasis of both pancreatic (Michl et al. 2003) and colorectal carcinomas (Ueda et al. 2007). Interestingly, when metastases of these tumors were investigated, decreased expression was documented in colorectal liver metastases (Ueda et al. 2007; Georges et al. 2012), but maintained claudin-4 positivity was reported in pancreatic liver metastases (Nichols et al. 2004). In our present study, both PLM and CRLM strongly expressed claudin-4, which underlines its role in the pathogenesis of these lesions. Considering HCC, no claudin-4 positivity was detected, which is in line with our previous findings (Lódi et al. 2006).
In our sample set, a faint claudin-3 signal was found in HCCs along with strong claudin-3 positivity in CRLM samples, and high levels already have been described in primary colorectal carcinoma (Oshima et al. 2008; Chen et al. 2010). An interesting finding of the present study was the presence of claudin-3 positivity in PLM samples. This can be important because no claudin-3 staining was detected in primary pancreatic cancer (Borka et al. 2007). Furthermore, intrahepatic bile duct cancers (IBDCs) were also characterized by the scarcity or absence of this claudin (Nemeth et al. 2009). Thus, claudin-3 expression might contribute to the metastatic progression of pancreatic carcinoma and could also be used to differentiate between IBDC and PLM.
Tumorigenic roles of claudin-7 in colorectal carcinoma cell lines have previously been described (Darido et al. 2008), and claudin-7 was proposed to exert its roles in complex with other proteins (Kuhn et al. 2007). Moreover, the frequent expression of claudin-7 and its associated molecules were detected in both primary colorectal carcinoma samples and liver metastases. In concordance with these results, we also found strong claudin-7 positivity in CRLM samples, but reduced claudin-7 expression was described to correlate with metastasis formation in esophageal (Usami et al. 2006) and breast (Sauer et al. 2005) cancers. Interestingly, early reduction of claudin-7 mRNA levels was also described during colorectal carcinogenesis (Bornholdt et al. 2011), whereas reexpression of this molecule was frequently found in lymph node metastases (Nakayama et al. 2008); this might also explain its positivity in our liver metastases samples. Furthermore, HCC showed weak claudin-7 positivity, with no significant change in comparison to non-tumorous liver. This might be in parallel with the results of a transgenic mouse HCC model, in which only small tumors showed enhanced claudin-7 expression (Borlak et al. 2005). When comparing our findings on PLM with the results described in primary pancreatic carcinoma and IBDC, interesting differences were observable. Claudin-7 positivity was described in all primary pancreatic adenocarcinomas (Borka et al. 2007) whereas, in PLMs, 6 cases were negative, and only 9 of 15 cases were faintly positive. In this regard, PLM appears to be more similar to bile duct cancers, which also exhibit low claudin-7 expression (Nemeth et al. 2009).
The above-described findings rather suggest a complex and well-regulated system of claudin expression that is distinct in primary and metastatic liver tumors. Downregulation of certain claudins has been shown in advanced stages of colorectal carcinomas, but we found strong claudin positivity in CRLM. Several factors may attribute to this change (e.g., biological selection of tumor cells in tumor progression and microenvironmental influence) (Georges et al. 2012). It has also been shown that dedifferentiated colorectal adenocarcinomas undergo redifferentiation in liver metastases (Kaihara et al. 2003), which might also mean recovery of the claudin phenotype at the more differentiated stages. In the present study, a good correlation between protein and mRNA expression was found, with two interesting exceptions regarding HCC. HCC showed lower claudin-1 and -7 protein positivity than did CRLM, whereas mRNA levels were higher or comparable. This raises the possibility of a posttranscriptional regulation of claudin expression that might be mediated by microRNAs (Kwak et al. 2010). Rapid protein turnover might also be responsible for the discrepancy.
Taken together, our findings support the role of altered claudin expression in both hepatocarcinogenesis and metastasis formation of colorectal and pancreatic cancers. We found that HCC, CRLM, and PLM differentially express claudin-1, -2, -3, -4, and -7, which might even be exploited in the differential diagnosis of these tumor entities. Strong positivity of claudin-1, -3, -4, and -7 is characteristic of CRLM; HCC is associated with claudin-1 staining and a lack of claudin-4; and PLM shows strong claudin-4 and weak claudin-1 positivity. Furthermore, possible claudin-targeting or claudin-anchored therapies could be aimed at not only treating primary liver tumors, but patients with metastatic liver disease could also benefit from such therapeutic approaches. In conclusion, primary hepatocellular carcinoma and metastatic colorectal and pancreatic carcinomas display distinct claudin expression profiles. This provides further understanding of their pathobiology and might aid in the differential diagnosis of focal liver lesions. The observed expression changes clearly implicate these claudins in the process of carcinogenesis and metastasis formation.
Supplementary Material
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research and/or authorship of this article: This work was supported by grants from the Hungarian Scientific Research Found (OTKA) K101435 and the National Development Agency of Hungary (TÁMOP-4.2.1/B-09/1/KMR-2010-0001).
Supplementary material for this article is available on the Journal of Histochemistry & Cytochemistry Web site at http://jhc.sagepub.com/supplemental.
References
- Aung PP, Mitani Y, Sanada Y, Nakayama H, Matsusaki K, Yasui W. 2006. Differential expression of claudin-2 in normal human tissues and gastrointestinal carcinomas. Virchows Arch. 448:428–434 [DOI] [PubMed] [Google Scholar]
- Balda MS, Matter K. 2009. Tight junctions and the regulation of gene expression. Biochim Biophys Acta. 1788:761–767 [DOI] [PubMed] [Google Scholar]
- Borka K, Kaliszky P, Szabo E, Lotz G, Kupcsulik P, Schaff Z, Kiss A. 2007. Claudin expression in pancreatic endocrine tumors as compared with ductal adenocarcinomas. Virchows Arch. 450:549–557 [DOI] [PubMed] [Google Scholar]
- Borlak J, Meier T, Halter R, Spanel R, Spanel-Borowski K. 2005. Epidermal growth factor–induced hepatocellular carcinoma: gene expression profiles in precursor lesions, early stage and solitary tumours. Oncogene. 24:1809–1819 [DOI] [PubMed] [Google Scholar]
- Bornholdt J, Friis S, Godiksen S, Poulsen SS, Santoni-Rugiu E, Bisgaard HC, Lothe IM, Ikdahl T, Tveit KM, Johnson E, et al. 2011. The level of claudin-7 is reduced as an early event in colorectal carcinogenesis. BMC Cancer. 11:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centeno BA. 2006. Pathology of liver metastases. Cancer Control. 13:13–26 [DOI] [PubMed] [Google Scholar]
- Chao YC, Pan SH, Yang SC, Yu SL, Che TF, Lin CW, Tsai MS, Chang GC, Wu CH, Wu YY, et al. 2009. Claudin-1 is a metastasis suppressor and correlates with clinical outcome in lung adenocarcinoma. Am J Respir Crit Care Med. 179:123–133 [DOI] [PubMed] [Google Scholar]
- Chen JS, Chen KT, Fan CW, Han CL, Chen YJ, Yu JS, Chang YS, Chien CW, Wu CP, Hung RP, et al. 2010. Comparison of membrane fraction proteomic profiles of normal and cancerous human colorectal tissues with gel-assisted digestion and iTRAQ labeling mass spectrometry. FEBS J. 277:3028–3038 [DOI] [PubMed] [Google Scholar]
- Cheung ID, Bagnat M, Ma TP, Datta A, Evason K, Moore JC, Lawson ND, Mostov KE, Moens CB, Stainier DY. 2012. Regulation of intrahepatic biliary duct morphogenesis by Claudin 15–like b. Dev Biol. 361:68–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofori G. 2006. New signals from the invasive front. Nature. 441:444–450 [DOI] [PubMed] [Google Scholar]
- Darido C, Buchert M, Pannequin J, Bastide P, Zalzali H, Mantamadiotis T, Bourgaux JF, Garambois V, Jay P, Blache P, et al. 2008. Defective claudin-7 regulation by Tcf-4 and Sox-9 disrupts the polarity and increases the tumorigenicity of colorectal cancer cells. Cancer Res. 68:4258–4268 [DOI] [PubMed] [Google Scholar]
- de Oliveira SS, de Oliveira IM, De Souza W, Morgado-Diaz JA. 2005. Claudins upregulation in human colorectal cancer. FEBS Lett. 579:6179–6185 [DOI] [PubMed] [Google Scholar]
- Dhawan P, Singh AB, Deane NG, No Y, Shiou SR, Schmidt C, Neff J, Washington MK, Beauchamp RD. 2005. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J Clin Invest. 115:1765–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ersoz S, Mungan S, Cobanoglu U, Turgutalp H, Ozoran Y. 2011. Prognostic importance of Claudin-1 and Claudin-4 expression in colon carcinomas. Pathol Res Pract. 207:285–289 [DOI] [PubMed] [Google Scholar]
- Geller SA, Dhall D, Alsabeh R. 2008. Application of immunohistochemistry to liver and gastrointestinal neoplasms: liver, stomach, colon, and pancreas. Arch Pathol Lab Med. 132:490–499 [DOI] [PubMed] [Google Scholar]
- Georges R, Bergmann F, Hamdi H, Zepp M, Eyol E, Hielscher T, Berger MR, Adwan H. 2012. Sequential biphasic changes in claudin1 and claudin4 expression are correlated to colorectal cancer progression and liver metastasis. J Cell Mol Med. 16:260–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grone J, Weber B, Staub E, Heinze M, Klaman I, Pilarsky C, Hermann K, Castanos-Velez E, Ropcke S, Mann B, et al. 2007. Differential expression of genes encoding tight junction proteins in colorectal cancer: frequent dysregulation of claudin-1, -8 and -12. Int J Colorectal Dis. 22:651–659 [DOI] [PubMed] [Google Scholar]
- Hadj-Rabia S, Baala L, Vabres P, Hamel-Teillac D, Jacquemin E, Fabre M, Lyonnet S, De Prost Y, Munnich A, Hadchouel M, et al. 2004. Claudin-1 gene mutations in neonatal sclerosing cholangitis associated with ichthyosis: a tight junction disease. Gastroenterology. 127:1386–1390 [DOI] [PubMed] [Google Scholar]
- Higashi Y, Suzuki S, Sakaguchi T, Nakamura T, Baba S, Reinecker HC, Nakamura S, Konno H. 2007. Loss of claudin-1 expression correlates with malignancy of hepatocellular carcinoma. J Surg Res. 139:68-76 [DOI] [PubMed] [Google Scholar]
- Huang GW, Ding X, Chen SL, Zeng L. 2011. Expression of claudin 10 protein in hepatocellular carcinoma: impact on survival. J Cancer Res Clin Oncol. 137:1213–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Q, Kinugasa T, Wang L, Huang J, Zhao J, Shibaguchi H, Kuroki M, Tanaka T, Yamashita Y, Nabeshima K, et al. 2009. Claudin-1 protein is a major factor involved in the tumorigenesis of colorectal cancer. Anticancer Res. 29:851–857 [PubMed] [Google Scholar]
- Ip YC, Cheung ST, Lee YT, Ho JC, Fan ST. 2007. Inhibition of hepatocellular carcinoma invasion by suppression of claudin-10 in HLE cells. Mol Cancer Ther. 6:2858–2867 [DOI] [PubMed] [Google Scholar]
- Kadry Z, Malekkiani N, Clavien PA. 2001. Treatment of primary and secondary liver malignancy. Swiss Med Wkly. 131:338–345 [DOI] [PubMed] [Google Scholar]
- Kaihara T, Kawamata H, Imura J, Fujii S, Kitajima K, Omotehara F, Maeda N, Nakamura T, Fujimori T. 2003. Redifferentiation and ZO-1 reexpression in liver-metastasized colorectal cancer: possible association with epidermal growth factor receptor–induced tyrosine phosphorylation of ZO-1. Cancer Sci. 94:166–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Huh JH, Lee S, Kang H, Kim GI, An HJ. 2008. Down-regulation of claudin-2 in breast carcinomas is associated with advanced disease. Histopathology. 53:48–55 [DOI] [PubMed] [Google Scholar]
- Kinugasa T, Huo Q, Higashi D, Shibaguchi H, Kuroki M, Tanaka T, Futami K, Yamashita Y, Hachimine K, Maekawa S, et al. 2007. Selective up-regulation of claudin-1 and claudin-2 in colorectal cancer. Anticancer Res. 27:3729–3734 [PubMed] [Google Scholar]
- Kominsky SL, Argani P, Korz D, Evron E, Raman V, Garrett E, Rein A, Sauter G, Kallioniemi OP, Sukumar S. 2003. Loss of the tight junction protein claudin-7 correlates with histological grade in both ductal carcinoma in situ and invasive ductal carcinoma of the breast. Oncogene. 22:2021–2033 [DOI] [PubMed] [Google Scholar]
- Kuhn S, Koch M, Nubel T, Ladwein M, Antolovic D, Klingbeil P, Hildebrand D, Moldenhauer G, Langbein L, Franke WW, et al. 2007. A complex of EpCAM, claudin-7, CD44 variant isoforms, and tetraspanins promotes colorectal cancer progression. Mol Cancer Res. 5:553–567 [DOI] [PubMed] [Google Scholar]
- Kwak PB, Iwasaki S, Tomari Y. 2010. The microRNA pathway and cancer. Cancer Sci. 101:2309–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurila JJ, Karttunen T, Koivukangas V, Laurila PA, Syrjala H, Saarnio J, Soini Y, Ala-Kokko TI. 2007. Tight junction proteins in gallbladder epithelium: different expression in acute acalculous and calculous cholecystitis. J Histochem Cytochem. 55:567–573 [DOI] [PubMed] [Google Scholar]
- Lioni M, Brafford P, Andl C, Rustgi A, El-Deiry W, Herlyn M, Smalley KS. 2007. Dysregulation of claudin-7 leads to loss of E-cadherin expression and the increased invasion of esophageal squamous cell carcinoma cells. Am J Pathol. 170:709–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lódi C, Szabo E, Holczbauer A, Batmunkh E, Szijarto A, Kupcsulik P, Kovalszky I, Paku S, Illyes G, Kiss A, et al. 2006. Claudin-4 differentiates biliary tract cancers from hepatocellular carcinomas. Mod Pathol. 19:460–469 [DOI] [PubMed] [Google Scholar]
- Long H, Crean CD, Lee WH, Cummings OW, Gabig TG. 2001. Expression of Clostridium perfringens enterotoxin receptors claudin-3 and claudin-4 in prostate cancer epithelium. Cancer Res. 61:7878–7881 [PubMed] [Google Scholar]
- Martin TA, Jiang WG. 2009. Loss of tight junction barrier function and its role in cancer metastasis. Biochim Biophys Acta. 1788:872–891 [DOI] [PubMed] [Google Scholar]
- Meertens L, Bertaux C, Cukierman L, Cormier E, Lavillette D, Cosset FL, Dragic T. 2008. The tight junction proteins claudin-1, -6, and -9 are entry cofactors for hepatitis C virus. J Virol. 82:3555–3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merikallio H, Kaarteenaho R, Paakko P, Lehtonen S, Hirvikoski P, Makitaro R, Harju T, Soini Y. 2011. Zeb1 and twist are more commonly expressed in metastatic than primary lung tumours and show inverse associations with claudins. J Clin Pathol. 64:136–140 [DOI] [PubMed] [Google Scholar]
- Michl P, Barth C, Buchholz M, Lerch MM, Rolke M, Holzmann KH, Menke A, Fensterer H, Giehl K, Lohr M, et al. 2003. Claudin-4 expression decreases invasiveness and metastatic potential of pancreatic cancer. Cancer Res. 63:6265–6271 [PubMed] [Google Scholar]
- Mineta K, Yamamoto Y, Yamazaki Y, Tanaka H, Tada Y, Saito K, Tamura A, Igarashi M, Endo T, Takeuchi K, et al. 2011. Predicted expansion of the claudin multigene family. FEBS Lett. 585:606–612 [DOI] [PubMed] [Google Scholar]
- Miwa N, Furuse M, Tsukita S, Niikawa N, Nakamura Y, Furukawa Y. 2001. Involvement of claudin-1 in the beta-catenin/Tcf signaling pathway and its frequent upregulation in human colorectal cancers. Oncol Res. 12:469–476 [DOI] [PubMed] [Google Scholar]
- Moldvay J, Jackel M, Paska C, Soltesz I, Schaff Z, Kiss A. 2007. Distinct claudin expression profile in histologic subtypes of lung cancer. Lung Cancer. 57:159–167 [DOI] [PubMed] [Google Scholar]
- Nakayama F, Semba S, Usami Y, Chiba H, Sawada N, Yokozaki H. 2008. Hypermethylation-modulated downregulation of claudin-7 expression promotes the progression of colorectal carcinoma. Pathobiology. 75:177–185 [DOI] [PubMed] [Google Scholar]
- Neesse A, Griesmann H, Gress TM, Michl P. 2012. Claudin-4 as therapeutic target in cancer. Arch Biochem Biophys. 524: 64–70 [DOI] [PubMed] [Google Scholar]
- Nemeth Z, Szasz AM, Tatrai P, Nemeth J, Gyorffy H, Somoracz A, Szijarto A, Kupcsulik P, Kiss A, Schaff Z. 2009. Claudin-1, -2, -3, -4, -7, -8, and -10 protein expression in biliary tract cancers. J Histochem Cytochem. 57:113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols LS, Ashfaq R, Iacobuzio-Donahue CA. 2004. Claudin 4 protein expression in primary and metastatic pancreatic cancer: support for use as a therapeutic target. Am J Clin Pathol. 121:226–230 [DOI] [PubMed] [Google Scholar]
- Orbán E, Szabo E, Lotz G, Kupcsulik P, Paska C, Schaff Z, Kiss A. 2008. Different expression of occludin and ZO-1 in primary and metastatic liver tumors. Pathol Oncol Res. 14:299–306 [DOI] [PubMed] [Google Scholar]
- Oshima T, Kunisaki C, Yoshihara K, Yamada R, Yamamoto N, Sato T, Makino H, Yamagishi S, Nagano Y, Fujii S, et al. 2008. Reduced expression of the claudin-7 gene correlates with venous invasion and liver metastasis in colorectal cancer. Oncol Rep. 19:953–959 [PubMed] [Google Scholar]
- Rangel LB, Agarwal R, D’Souza T, Pizer ES, Alo PL, Lancaster WD, Gregoire L, Schwartz DR, Cho KR, Morin PJ. 2003. Tight junction proteins claudin-3 and claudin-4 are frequently overexpressed in ovarian cancer but not in ovarian cystadenomas. Clin Cancer Res. 9:2567–2575 [PubMed] [Google Scholar]
- Resnick MB, Konkin T, Routhier J, Sabo E, Pricolo VE. 2005. Claudin-1 is a strong prognostic indicator in stage II colonic cancer: a tissue microarray study. Mod Pathol. 18:511–518 [DOI] [PubMed] [Google Scholar]
- Sakaguchi T, Gu X, Golden HM, Suh E, Rhoads DB, Reinecker HC. 2002. Cloning of the human claudin-2 5′-flanking region revealed a TATA-less promoter with conserved binding sites in mouse and human for caudal-related homeodomain proteins and hepatocyte nuclear factor–1alpha. J Biol Chem. 277:21361–21370 [DOI] [PubMed] [Google Scholar]
- Sauer T, Pedersen MK, Ebeltoft K, Naess O. 2005. Reduced expression of Claudin-7 in fine needle aspirates from breast carcinomas correlate with grading and metastatic disease. Cytopathology. 16:193–198 [DOI] [PubMed] [Google Scholar]
- Soini Y. 2005. Expression of claudins 1, 2, 3, 4, 5 and 7 in various types of tumours. Histopathology. 46:551–560 [DOI] [PubMed] [Google Scholar]
- Soini Y. 2012. Tight junctions in lung cancer and lung metastasis: a review. Int J Clin Exp Pathol. 5:126–136 [PMC free article] [PubMed] [Google Scholar]
- Son S, Kojima T, Decaens C, Yamaguchi H, Ito T, Imamura M, Murata M, Tanaka S, Chiba H, Hirata K, et al. 2009. Knockdown of tight junction protein claudin-2 prevents bile canalicular formation in WIF-B9 cells. Histochem Cell Biol. 131:411–424 [DOI] [PubMed] [Google Scholar]
- Sonoda N, Furuse M, Sasaki H, Yonemura S, Katahira J, Horiguchi Y, Tsukita S. 1999. Clostridium perfringens enterotoxin fragment removes specific claudins from tight junction strands: evidence for direct involvement of claudins in tight junction barrier. J Cell Biol. 147:195–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh Y, Yoon CH, Kim RK, Lim EJ, Oh YS, Hwang SG, An S, Yoon G, Gye MC, Yi JM, et al. 2012. Claudin-1 induces epithelial-mesenchymal transition through activation of the c-Abl-ERK signaling pathway in human liver cells [published online November 19, 2012]. Oncogene. 10.1038/onc.2012.505 [DOI] [Google Scholar]
- Suzuki M, Kato-Nakano M, Kawamoto S, Furuya A, Abe Y, Misaka H, Kimoto N, Nakamura K, Ohta S, Ando H. 2009. Therapeutic antitumor efficacy of monoclonal antibody against Claudin-4 for pancreatic and ovarian cancers. Cancer Sci. 100:1623–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo I, Kiss A, Schaff Z, Sobel G. 2009. Claudins as diagnostic and prognostic markers in gynecological cancer. Histol Histopathol. 24:1607–1615 [DOI] [PubMed] [Google Scholar]
- Szasz AM, Nemeth Z, Gyorffy B, Micsinai M, Krenacs T, Baranyai Z, Harsanyi L, Kiss A, Schaff Z, Tokes AM, et al. 2011. Identification of a claudin-4 and E-cadherin score to predict prognosis in breast cancer. Cancer Sci. 102:2248–2254 [DOI] [PubMed] [Google Scholar]
- Szekely E, Torzsok P, Riesz P, Korompay A, Fintha A, Szekely T, Lotz G, Nyirady P, Romics I, Timar J, et al. 2011. Expression of claudins and their prognostic significance in noninvasive urothelial neoplasms of the human urinary bladder. J Histochem Cytochem. 59:932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabaries S, Dong Z, Annis MG, Omeroglu A, Pepin F, Ouellet V, Russo C, Hassanain M, Metrakos P, Diaz Z, et al. 2011. Claudin-2 is selectively enriched in and promotes the formation of breast cancer liver metastases through engagement of integrin complexes. Oncogene. 30:1318–1328 [DOI] [PubMed] [Google Scholar]
- Tokes AM, Kulka J, Paku S, Szik A, Paska C, Novak PK, Szilak L, Kiss A, Bogi K, Schaff Z. 2005. Claudin-1, -3 and -4 proteins and mRNA expression in benign and malignant breast lesions: a research study. Breast Cancer Res. 7:R296–R305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torzsok P, Riesz P, Kenessey I, Szekely E, Somoracz A, Nyirady P, Romics I, Schaff Z, Lotz G, Kiss A. 2011. Claudins and ki-67: potential markers to differentiate low- and high-grade transitional cell carcinomas of the urinary bladder. J Histochem Cytochem. 59:1022–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita S, Furuse M. 1999. Occludin and claudins in tight-junction strands: leading or supporting players? Trends Cell Biol. 9:268–273 [DOI] [PubMed] [Google Scholar]
- Ueda J, Semba S, Chiba H, Sawada N, Seo Y, Kasuga M, Yokozaki H. 2007. Heterogeneous expression of claudin-4 in human colorectal cancer: decreased claudin-4 expression at the invasive front correlates cancer invasion and metastasis. Pathobiology. 74:32–41 [DOI] [PubMed] [Google Scholar]
- Usami Y, Chiba H, Nakayama F, Ueda J, Matsuda Y, Sawada N, Komori T, Ito A, Yokozaki H. 2006. Reduced expression of claudin-7 correlates with invasion and metastasis in squamous cell carcinoma of the esophagus. Hum Pathol. 37:569–577 [DOI] [PubMed] [Google Scholar]
- Wang X, Tully O, Ngo B, Zitin M, Mullin JM. 2011. Epithelial tight junctional changes in colorectal cancer tissues. Sci World J. 11:826–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon CH, Kim MJ, Park MJ, Park IC, Hwang SG, An S, Choi YH, Yoon G, Lee SJ. 2010. Claudin-1 acts through c-Abl-protein kinase Cdelta (PKCdelta) signaling and has a causal role in the acquisition of invasive capacity in human liver cells. J Biol Chem. 285:226–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Lin Y, Yin C, Zhang X, Ning BF, Zhang Q, Zhang JP, Qiu L, Qin XR, Chen YX, et al. 2011. Recombinant adenovirus carrying the hepatocyte nuclear factor-1alpha gene inhibits hepatocellular carcinoma xenograft growth in mice. Hepatology. 54:2036–2047 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.