Abstract
Background and aims
Drug-induced liver injury (DILI) is a serious adverse drug event that is suspected to have a heritable component. We carried out a genome-wide association study of 783 individuals of European ancestry who experienced DILI due to more than 200 implicated drugs.
Methods
DILI patients from the US-based Drug-Induced Liver Injury Network (n = 401) and three international registries (n = 382) were genotyped with the Illumina 1Mduo BeadChip and compared with population controls (n = 3001). Potential associations were tested in 307 independent Drug-Induced Liver Injury Network cases.
Results
After accounting for known major histocompatibility complex risk alleles for flucloxacillin-DILI and amoxicillin/clavulanate-DILI, there were no genome-wide significant associations, including in the major histocompatibility complex region. Stratification of DILI cases according to clinical phenotypes (injury type, latency, age of onset) also did not show significant associations. An analysis of hepatocellular DILI (n = 285) restricted to 193 single-nucleotide polymorphisms previously associated with autoimmune disease showed a trend association for rs7574865, in the vicinity of signal transducer and activator of transcription 4 (STAT4) (P = 4.5 × 10−4). This association was replicated in an independent cohort of 168 hepatocellular DILI cases (P = 0.011 and 1.5 × 10−5 for combined cohorts). No significant associations were found with stratification by other clinical or demographic variables.
Conclusion
Although not significant at the genome-wide level, the association between hepatocellular DILI and STAT4 is consistent with the emerging role of the immune system in DILI. However, the lack of genome-wide association study findings supports the idea that strong genetic determinants of DILI may be largely drug-specific or may reflect rare genetic variations, which were not assessed in our study.
Keywords: drug-induced liver injury, genome-wide association study, pharmacogenetics
Introduction
Idiosyncratic drug-induced liver injury (DILI) is a major cause of acute liver failure [1], but this statistic alone considerably underestimates the public health impact of DILI. For example, concern for DILI can limit patient access to drugs that might otherwise be beneficial. There are currently no tests available that can definitively establish the diagnosis of DILI or indicate which drug is responsible if a patient takes multiple medications. As a consequence, key drugs may be unnecessarily stopped, thereby creating a health risk to the patient. DILI is also the single, major adverse event that leads to termination of clinical drug development programs and to regulatory actions on approved drugs [2]. Concern for a DILI event during early clinical trials frequently leads to longer and larger clinical trials requested by the FDA, thus delaying patient access to important new medications and increasing the costs of bringing new drugs into the market. Improved understanding of underlying DILI mechanisms could lead to clinical tests that might be used to identify patients susceptible to DILI from a given drug, to confirm the diagnosis of DILI, and to help to identify the offending agent in a patient taking multiple drugs. This understanding should also lead to improved preclinical methodology to detect DILI potential in new drug candidates.
A strategy to identify the mechanisms underlying DILI has been genetic analysis of patients who have actually experienced DILI. The studies to date support the concept that DILI susceptibility results in part from genetic variation in at least four major classes of genes: drug metabolism including toxification and detoxification enzymes, drug and bile acid transporters, cellular stress response genes, and innate and adaptive immune response genes including the major histocompatibility complex (MHC) region genes [3,4].
We hypothesized that some DILI susceptibility factors are shared across multiple drugs for several reasons. Individual drug-metabolizing enzymes and drug transporters are often able to transport or metabolize many different drug substrates. Thus, individual DILI risk factors at the level of enzymes and transporters could incur a risk for multiple drugs. Similarly, cellular responses to stress are finite and risk factors in these pathways would likely not be drug-specific. Furthermore, previous genetic associations support the concept that specific genetic susceptibility variants may be shared by different drugs [3,4]. Finally, common risk factors for DILI are supported by the observation that in a cohort of Spanish DILI patients, nine had experienced more than one discrete episode of DILI due to different drugs [5], suggesting that some individuals may be particularly susceptible to DILI from multiple agents.
We carried out a genome-wide association study (GWAS) of DILI in 783 individuals of European ancestry who experienced DILI due to a large number of implicated drugs (> 200). Possible associations were then evaluated in a smaller replication cohort (n = 307). This report represents the largest investigation of DILI genetics to date.
Methods
Study participants
Cases (n = 783) were recruited from four separate studies [Drug-Induced Liver Injury Network (DILIN) [6], DILIGEN [7], EUDRAGENE [8], and the Spanish DILI Registry [9]]. A total of 565 cases were recruited from the DILIN network between August 2004 and April 2009 from eight DILIN clinical sites in the USA; 401 cases of European ancestry were included in the current study. The contribution of cases of European ancestry from the other networks was as follows: DILIGEN, 242 cases; EUDRAGENE, 89 cases; and Spanish DILI Registry, 51 cases. All participants provided written informed consent and each study was approved by the appropriate institutional review boards. Details of recruitment and the inclusion and exclusion criteria for these networks have been published previously [10]. For all cases in the DILIN, causality assessment was by expert consensus as described previously [6], whereas the Roussel-Uclaf causality assessment method (RUCAM) was used in the other three registries [11]. Notably, the cases studied here include 296 samples from DILI patients with injury attributed to flucloxacillin or amoxicillin/clavulanate that overlap almost completely with the samples used in previous reports [7,10]. These were included here to identify the common variants contributing to DILI susceptibility across different drugs, and should not be taken as replicating previous published genetic associations. The replication cohort consisted of 307 cases from the DILIN network. Genotyped controls (n = 655) from the Population Reference Sample (POPRES, n = 655) [12] and the 1958 British Birth Cohort (http://www.b58cgene.sgul.ac.uk) (n = 2346) were used for comparison. Replication studies utilized genotype data from the National Blood Service cohort (n = 2249). Genotype data for the 1958 British Birth Cohort and the National Blood Service cohort were provided by the Wellcome Trust Case–Control Consortium (http://www.wtccc.org.uk).
Secondary phenotypes
Cases were categorized as hepatocellular versus cholestatic versus mixed using the R-value [11]. Other secondary phenotypes included demographic or clinical strata (sex, age greater than or less than the median, time to onset greater than or less than the median, presence of eosinophilia, fever, or rash). Because experienced hepatologists enrolled each patient only after concluding that the patients probably had DILI, and because causality assessment protocols were not uniform across the different cohorts, our primary analyses were carried out on all cases where genotyping data were available. When potential associations were evident, post-hoc analyses were carried out by dichotomizing the sample into those with a causality score of at least probable (DILIN score of ‘probable’, ‘very likely’, or ‘definite’; RUCAM ≥ 6) versus those with causality less than probable (see Supplementary Tables S1 and S2, http://links.lww.com/FPC/A505).
Genotyping
DNA was prepared as described previously [10]. Genome-wide genotyping of the European DILI cases and POPRES controls was carried out by Expression Analysis Inc. (Durham, North Carolina, USA) and of the USA cases by the Center for Human Genome Variation, Duke University. Genotype data for the 1958 British Birth Cohort controls were provided by the Wellcome Trust Case–Control Consortium 2. All patients were genotyped using the Illumina Human1M or 1MDuo BeadChip (Illumina Inc., San Diego, California, USA), each containing greater than 1 000 000 markers. A total of 800 769 markers, 783 cases (401 DILIN, 242 DILIGEN, 89 EUDRAGENE, and 51 Spanish DILI Registry), and 3001 controls passed quality control carried out as described previously [13].
Statistical analysis
Population structure of the combined case and control sample was estimated by principal components analysis using a modification of EIGENSTRAT [14] as described previously [13]. Tests for association between the single-nucleotide polymorphism (SNP) genotype and DILI were carried out using logistic regression in PLINK [15] using the top 14 principal components emerging from the EIGENSTRAT analyses as covariates in the model. Where applicable, the top-associated SNPs in the MHC region for flucloxacillin-DILI (rs2395029) and amoxicillin/clavulanate-DILI (rs2523822, rs3135388) that were present in the quality control filtered dataset were also included as covariates to determine whether any risk for all-drug DILI may be explained by other variants after correcting for these known DILI risk alleles. Association test results were annotated and visualized using the WGAviewer software [16]. We also carried out hypothesis-guided exploratory analysis of genes involved in drug absorption, distribution, metabolism, and excretion (ADME) [17], or non-HLA genes known to be generally involved in autoimmune diseases [18], as described previously [10].
Replication genotyping was carried out using TaqMan SNP Genotyping Assays (Life Technologies, Carlsbad, California, USA). Replication testing was carried out using the χ2 or Fisher’s exact test of allele frequency differences between 307 European-ancestry DILI cases enrolled in the DILIN network not included in the GWAS and 2587 samples of European descent from the National Blood Service. Combined P-values for the discovery and replication sets were calculated using Stouffer’s weighted Z method. Power estimates were performed using the PGA software [19].
Results
Characteristics of the study population
The DILI case samples included a discovery cohort of 783 patients of primarily European ancestry, all of which were genotyped on the Illumina 1M or 1Mduo BeadChip; a replication cohort consisting of 307 US patients who were self-identified White, non-Hispanic, was available from the DILIN for follow-up of variants of interest emerging from the GWAS. The demographic and clinical characteristics of both cohorts are shown in Table 1. The representation of implicated drugs (Table 2) was comparable between cohorts, with the exception of flucloxacillin, which was absent from the US-based (DILIN) replication cohort, and amoxicillin/clavulanate.
Table 1.
Clinical characteristics of the study population in the initial discovery sample used for genome-wide association study (n=783) and in the replication cohort (n=307)
|
N (%) or median (range) |
||
|---|---|---|
| Variables | Discovery sample | Replication sample |
| Sex | ||
| Male | 329 (42.0) | 136 (44.3) |
| Female | 454 (58.0) | 171 (55.7) |
| Age of onset (years) | 57.5 (1.8–98) | 54.2 (5.8–88) |
| Time to onset (days) | 24 (0–7045) | 39 (0–11463) |
| Injury type | ||
| Cholestatic | 187 (23.9) | 90 (29.3) |
| Mixed | 164 (20.9) | 64 (20.8) |
| Hepatocellular | 256 (32.7) | 139 (45.3) |
| Unknown/unclassified | 176 (22.5) | 14 (4.6) |
| Severity | ||
| Mild | 105 (13.4) | 50 (16.3) |
| Moderate | 80 (10.2) | 62 (20.2) |
| Moderate (hospitalized) | 124 (15.8) | 48 (15.6) |
| Severe | 47 (6.0) | 32 (10.4) |
| Fatal | 20 (2.6) | 16 (5.2) |
| Unknown/unclassified | 407 (52.0) | 99 (32.2) |
| Causality score | ||
| ‘Unlikely’ or ‘possible’ | 83 (10.6) | 35 (11.4) |
| ‘Probable’, ‘very likely’, ‘definite’ | 622 (79.4) | 173 (56.4) |
| Unknown/unclassified | 78 (10.0) | 99 (32.2) |
| Immunoallergic features | ||
| Eosinophilia | 46 (5.9) | NA |
| Fever | 161 (20.6) | NA |
| Rash | 125 (16.0) | NA |
| Any of above | 256 (32.7) | NA |
Table 2.
The most common drugs implicated in drug-induced liver injury cases (drugs with n ≥ 5) in both the discovery sample and in the individuals available in the Drug-Induced Liver Injury Network replication cohort
|
N (%) |
||
|---|---|---|
| Drugs | Discovery sample |
Replication sample |
| Amoxicillin/clavulanate | 208 (26.6) | 42 (13.7) |
| Flucloxacillin | 75 (9.6) | 0 (0.0) |
| Diclofenac | 30 (3.8) | 4 (1.3) |
| Nitrofurantoin | 24 (3.1) | 15 (4.9) |
| Isoniazid | 17 (2.2) | 7 (2.3) |
| Sulfamethoxazole/trimethoprim | 14 (1.8) | 14 (4.6) |
| Valproic acid | 13 (1.7) | 2 (0.6) |
| Nimesulide | 12 (1.5) | 0 (0.0) |
| Minocycline | 10 (1.3) | 17 (5.5) |
| Celecoxib | 8 (1.0) | 1 (0.3) |
| Flucloxacillin and amoxicillin/ clavulanate |
8 (1.0) | 0 (0.0) |
| Ciprofloxacin | 7 (0.9) | 9 (2.9) |
| Phenytoin | 7 (0.9) | 1 (0.3) |
| Azathioprine | 6 (0.8) | 5 (1.6) |
| Duloxetine | 6 (0.8) | 0 (0.0) |
| Telithromycin | 6 (0.8) | 0 (0.0) |
| Atorvastatin | 5 (0.6) | 3 (1.0) |
| Simvastatin | 5 (0.6) | 4 (1.3) |
| All other drugs and combinations | 322 (41.1) | 183 (59.6) |
Genome-wide association study results: all drug-induced liver injury cases
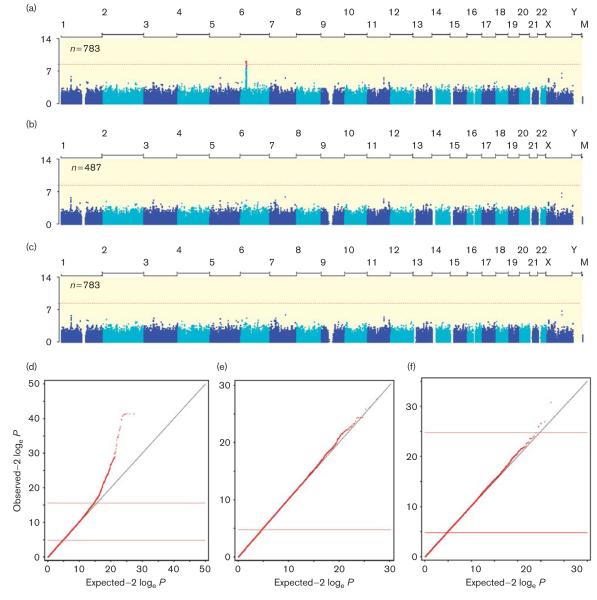
Principal components analysis showed that the 783 DILI cases and 3001 controls were well matched ancestrally (Supplementary Fig. S1, http://links.lww.com/FPC/A505). When all case patients were included in the analysis, we observed a large number of SNPs in the MHC region on chromosome 6p with genome-wide significance (Fig. 1a and d). The MHC region has been shown previously to contain high-impact risk alleles for DILI due to flucloxacillin and amoxicillin/clavulanate, and in fact, these MHC associations for flucloxacillin and amoxicillin/clavulanate have been reported previously using DNA samples from individuals overlapping significantly with the participants in the current study [7,10]. These cases were included here to improve power to detect novel genetic associations (anywhere in the genome) with DILI risk across all drugs. Given that these samples represent a large fraction of the case sample in the GWAS (Table 2) and may thus obscure other MHC associations with DILI due to other drugs, we also carried out a GWAS utilizing only DILI cases not explained by these two drugs. In the analysis of 487 DILI cases not attributed to flucloxacillin or amoxicillin/clavulanate, the observed P-value distribution genome-wide was not different from that expected under the null hypothesis, and no single SNP in the MHC region or elsewhere exceeded the typical threshold for genome-wide statistical significance (P < 10−8), (Fig. 1b and e). Similarly, when all drugs were considered, but the association tests were carried out conditional on the known MHC DILI risk SNPs for flucloxacillin and amoxicillin/clavulanate [7,10], no genome-wide significant associations were detected (Fig. 1c and f, Table 3).
Fig. 1.
Manhattan and Quantile–quantile plots of association study results including (a, d) or excluding (b, e) drug-induced liver injury (DILI) cases due to amoxicillin/clavulanate or flucloxacillin, for which strong genetic risk variants in the MHC region have been identified previously [7]. The genome-wide association study was also carried out including all cases, but conditioned on the known major histocompatibility complex single-nucleotide polymorphisms associated with flucloxacillin-induced or amoxicillin/clavulanate-induced DILI (c, f). The Manhattan plot shows the location along the genome on the x-axis and the –log10 P-value along the x-axis. A P-value less than 10−8 was considered significant. The quantile–quantile plot shows in rank order the observed P-values versus those expected under the null hypothesis, showing adequate correction for population structure (genomic inflation factor, λ = 1.02) and significant deviation from the null for the top-associated SNPs only in the analysis including amoxicillin/clavulanate and flucloxacillin cases.
Table 3.
The top-associated variants in all drug-induced liver injury casesa for all single-nucleotide polymorphisms and selected subsets of single-nucleotide polymorphisms associated with absorption, distribution, metabolism, and excretion and autoimmune genes
| SNP Sets | SNP | P-value | Chromosome:coordinate | Function | Nearest gene |
|---|---|---|---|---|---|
| Genome-wide | rs35709459 | 1.99E – 07 | X:91516677 | Intronic | PCDH11X |
| rs35925943 | 1.38E – 06 | X:91516339 | Intronic | PCDH11X | |
| rs11767067 | 1.64E – 06 | 7:93022840 | Intergenic | CALCR | |
| rs1187997 | 2.18E – 06 | 1:58455538 | Intronic | DAB1 | |
| rs656437 | 4.24E – 06 | 11:95770378 | Intronic | MAML2 | |
| ADME subset | rs3740065 | 0.0001 | 10:101605693 | Intronic | ABCC2 |
| rs2756113 | 0.0008 | 10:101569371 | Intronic | ABCC2 | |
| rs2804397 | 0.0011 | 10:101559415 | Intronic | ABCC2 | |
| rs8187710 | 0.0013 | 10:101611294 | Missense (C1515Y) | ABCC2 | |
| rs11816708 | 0.0013 | 10:101615015 | Downstream | ABCC2 | |
| Autoimmune subset | rs2476601 | 2.26E – 05 | 1:114377568 | Missense (R620W) | PTPN22 |
| rs6679677 | 4.18E – 05 | 1:114303808 | Upstream | RSBN1 | |
| rs7202877 | 0.0002 | 16:75247245 | Intergenic | CTRB1 | |
| rs1464510 | 0.003 | 3:188112554 | Intronic | LPP | |
| rs12928822 | 0.0033 | 16:11403893 | Upstream | MIR548H2 |
ADME, absorption, distribution, metabolism, and excretion; SNP, single-nucleotide polymorphism.
Analysis conditioned on rs2395029, rs2523822, and rs3135388, known to be highly associated with drug-induced liver injury due to flucloxacillin or amoxicillin/clavulanate.
As the MHC region has been implicated previously in DILI risk through GWAS, we determined whether the MHC region may be enriched for SNPs associated with DILI risk after the exclusion of the flucloxacillin and amoxicillin/clavulanate cases. Among the 487 non-flucloxacillin, non-amoxicillin/clavulanate-related DILI cases, we observed a lower P-value distribution among SNPs in the extended MHC region (chr6: 26 000 000–35 500 000) compared with all other autosomal SNPs (Supplementary Fig. S4A, S4B, http://links.lww.com/FPC/A505). To account for linkage disequilibrium prevalent in the MHC region, we randomly permuted the phenotype–genotype relationship among all individuals and repeated the association tests in the MHC region 1000 times. Among 1000 permutations, the observed median P-value in the observed (unpermuted) data (0.381) was in the lower 1.5 percentile of the median P-values in the MHC region (empirical P = 0.015, Supplementary Fig. S4C, http://links.lww.com/FPC/A505).
Drug metabolism and transporter single-nucleotide polymorphism analysis
Because drug-metabolizing enzymes and transporters have been assumed to play critical roles in mechanisms underlying DILI, we tested whether any SNPs in the vicinity of 130 genes involved in ADME [17] showed a greater degree of association with DILI risk than expected by chance. We found that several SNPs in the region of ABCC2 (encoding the multidrug resistance-associated protein MRP2) were the most associated SNPs in the ADME category (Table 3). The P-value corresponding to the highest associated SNP (rs3740065; P = 2.6 × 10−5) approached the specified threshold for significance after multiple test correction (P < 10−5). In addition, post-hoc analysis showed that the association with this SNP became stronger as the analysis was restricted to individuals with time to onset less than 30 days and causality scores of ‘probable’ or greater (RUCAM ICC ≥ 6 or DILIN causality of ‘probable’, ‘very likely’, or ‘definite’) (P = 2.5 × 10−6) (Supplementary Table S4, http://links.lww.com/FPC/A505). However, in a replication cohort of 304 independently genotyped DILI cases and 2249 controls from the National Blood Service cohort, there was no evidence of an association between rs3740065 and DILI risk (P = 0.49) (Table 4). Of note, no significant association with SNPs in the vicinity of the bile salt excretory protein gene (ABCB11) or manganese superoxide dismutase (SOD2) genes was found, although these have been implicated previously in DILI mechanisms [3,4].
Table 4.
Top-associated variant and replication test statistics for each of the clinical strata and drug-specific or drug class-specific analyses
| Discovery cohort |
Replication cohort |
Combined |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Phenotypes | SNP set | SNP | Nearest gene | Na | P-value | OR (95%CI) | Nb | P-value | OR (95%CI) | N | P-value |
| All DILI | ADME | rs3740065 | ABCC2 | 783 | 2.6×10−5 | 1.45 (1.22–1.72) | 304 | 0.49 | 0.90 (0.68–1.20) | 1087 | 1.9×10−3 |
| Cholestatic | AI and GWAS | rs2476601 | PTNP22 | 187 | 5.4×10−7 | 2.20 (1.62–3.00) | 105 | 0.26 | 0.74 (0.44–1.24) | 292 | 8.0×10−4 |
| Hepatocellular | AI | rs7574865 | STAT4 | 256 | 4.5×10−4 | 1.45 (1.18–1.79) | 168 | 0.011 | 1.39 (1.08–1.78) | 424 | 1.5×10−5 |
| Diclofenacc | GWAS | rs17036170 | PPARG | 30 | 1.0×10−8 | 11.3 (4.9–25.9) | 26 | 0.088 | 3.59 (0.83–15.6) | 56 | 2.0×10−8 |
| Fluoroquinolones | ADME | rs17862876 | UGT1A | 12 | 3.4×10−5 | 11.6 (3.6–37.4) | 10 | 1.00 | NA | 22 | 8.0×10−4 |
| NSAIDs | GWAS | rs9376256 | IL22RA2 | 68 | 6.7×10−8 | 3.42 (2.19–5.34) | 12 | 0.42 | 0.44 (0.06–3.23) | 80 | 1.0×10−4 |
| Statins | AI | rs7574865 | STAT4 | 27 | 4.9×10−5 | 3.35 (1.87–6.00) | 16 | 0.58 | 0.77 (0.31–1.91) | 43 | 3.5×10−3 |
ADME, ADME SNP-restricted analyses; AI, autoimmune disease SNP-restricted; GWAS, column ‘SNP set’ indicates whether the target SNP was brought forward for replication on the basis of the results of the genome-wide analysis.
ADME, absorption, distribution, metabolism, and excretion; DILI, drug-induced liver injury; GWAS, genome-wide association study; OR, odds ratio; SNP, single-nucleotide polymorphism; STAT4, signal transducer and activator of transcription 4.
N indicates number of case samples compared with 3001 population controls from the Population Reference Sample and 1958 British Birth Cohort.
N indicates number of case samples compared with 2249 population controls from the National Blood Service cohort.
Diclofenac-DILI samples for replication were genotyped specifically for rs17036170 and were not included in omnibus or injury-type-specific tests.
Autoimmune single-nucleotide polymorphism analysis
We next tested whether any of the 193 SNPs associated previously with autoimmune diseases might have a stronger association with DILI risk than expected by chance. This analysis (conditioned on genotypes at the known MHC risk alleles for DILI due to flucloxacillin and amoxicillin/clavulanate) showed rs2476601 in the protein tyrosine phosphatase nonreceptor type 22 (PTPN22) gene to be the top-associated autoimmune SNP (Table 3). This association appeared to be driven solely by the cases classified as having a cholestatic injury type (see the section below).
Genome-wide association study results: clinical strata and drug-specific or class-specific analyses
The results of the stratified analyses are shown in Supplementary Figs S5–S38 (http://links.lww.com/FPC/A505) and Supplementary Tables S5–S38 (http://links.lww.com/FPC/A505). Among the clinically defined strata, the most notable results were from the autoimmune-associated SNP sets, which showed experiment-wide significance (i.e. after correction for multiple testing of 193 autoimmune-associated SNPs) for an association between rs2476601, a nonsynonymous coding SNP in the PTPN22 gene, and cholestatic injury (P = 5.4 × 10−7), and rs7574865 in the vicinity of signal transducer and activator of transcription 4 (STAT4), which showed an almost experiment-wide significant (P = 4.5 10−4) association with hepatocellular injury (Table 4). Although the association between rs2476601 and cholestatic injury was almost genome-wide significant and would have been declared significant within the autoimmune-restricted SNP analysis, this association was not replicated in the independent cohort of 105 cholestatic DILI cases (P = 0.26, with the trend in the opposite direction of that observed in the discovery cohort). The lack of replication could not be attributed to the lower representation of amoxicillin/clavulanate and the absence of flucloxacillin cases in the replication cohort as the other cholestatic DILI cases [n = 77, odds ratio (OR) 2.32 (1.45–3.70), P = 4.3 × 10−4] contributed to an equivalent degree to the initial apparent association as did the flucloxacillin and amoxicillin/clavulanate cases [n = 110, OR 2.14 (1.46–3.18), P = 1.2 × 10−4]. The association between rs7574865 located near the STAT4 gene and hepatocellular injury was found to replicate in an independent set of 168 cases, although the estimated effect size and strength of association was reduced compared with the discovery cohort (Table 4).
No other associations approaching the specified levels of significance were observed, including examinations by time to onset, age of onset, presence of extrahepatic signs, or severity of injury.
Among the drug-specific or class-specific tests, the most notable results in the GWAS analyses were found for diclofenac (rs17036170, upstream of the PPARG gene, P = 1.07 × 10−8) and NSAIDs (rs9376256, an intergenic SNP in the region of IL20RA and IL22RA2, P = 6.7 × 10−8). In the ADME-restricted analysis of fluoroquinolone DILI cases, rs17862876 in the region of the UDP-glycosyltransferase-encoding UGT1A gene showed almost experiment-wide significance (3.4 × 10−5). In the autoimmune-restricted analysis of DILI due to statins, the rs7574865 polymorphism near STAT4 reached experiment-wide significance (P = 4.9 × 10−5). However, for all drug and drug class-specific associations, attempts to replicate the observed findings in independent cohorts were unsuccessful (or ambiguous in the case of diclofenac/PPARG) (Table 4). The lack of replication was not explained by differences in the allele frequency between discovery and replication control cohorts (P ≥ 0.1 for the difference in frequency between control datasets for all SNPs, except rs17862876, which showed a slightly lower risk allele frequency in the replication controls (P = 0.018) and thus would be biased toward association in the same direction as in the discovery set). Neither could it be explained by platform differences in genotyping: among 401 patients of European ancestry from the DILIN cohort genotyped using both the Illumina 1Mduo BeadChip and single-site TaqMan assays, there was perfect concordance in genotype calls across platforms. No associations approaching significance were observed in other drug or drug class cohorts.
Power calculations
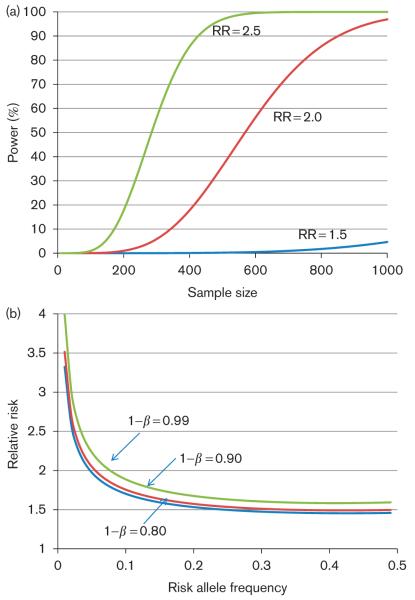
As no genome-wide significant associations were found in our analysis, we then calculated what magnitude of effect [in terms of relative risk (RR)] from truly associated common SNPs would likely have been missed in our analyses. As shown in Fig. 2a, the current sample size (793) provides sufficient power (> 80%) to detect SNPs conferring RR more than 2, and almost complete power to detect the effects of SNPs carrying RR greater than 2.5, even for a relatively low-frequency SNP [minor allele frequency (MAF) = 0.05, typically among the lowest frequency SNPs included in GWAS] (Fig. 2a). For the majority of the common SNP frequency spectrum (i.e. MAF > 10%), the current study was estimated to have almost complete power to detect the effects of SNPs carrying RR greater than 1.5 (Fig. 2b). However, analyses of less than 200 cases, which were typical in our analyses of clinical strata, would have fairly low power to detect RR less than 3, and the drug-specific or class-specific analyses utilizing case samples totaling 30 or fewer would generally be powered only to detect common variants with fairly large effects (RR > 5) (Table 5). Further, we estimated the influence of the use of unselected population controls relative to drug-exposed controls on statistical power to detect a genetic association. As suggested previously [20], for rare diagnoses such as DILI, the low rate of expected misclassification of control samples has a negligible effect on power (Supplementary Table S39, http://links.lww.com/FPC/A505).
Fig. 2.
Power estimates for the identification of drug-induced liver injury risk variants given available sample sizes. (a) Power versus sample size over a range of relative risk, assuming a minor allele frequency of 0.05, sample size: n = 783 cases versus n = 3000 controls, population prevalence of 1 : 10 000, and significance threshold P < 10−8. (b) Detectable relative risk (RR) versus risk allele frequency over a range of power (1 – β), assuming r2 = 1 between the marker and the causal risk allele, and sample sizes and significance threshold as above.
Table 5.
Detectable effect size for common variants (minor allele frequency = 10%) as a function of the number of case samples versus 3000 controls
| Sample size | Detectable relative risk at 80% power |
|---|---|
| 783 | 1.7 |
| 200 | 2.3 |
| 100 | 2.9 |
| 30 | 5.1 |
| 10 | 10.8 |
Available sample sizes for stratified analyses include those on the order of 100–200 for injury type and other clinical strata, which should be able to detect moderate effects of common variants (genotype relative risk on the order of 2–3). Some drug-specific analyses were carried out using sample sizes in the range of 10–30 individuals, which have low power to detect moderate effects but are well powered to detect very large effects (genotype relative risks of 5–10 or greater). These estimates assume a minor allele frequency of 0.10, a control sample of 3000, a population prevalence of drug-induced liver injury 1 : 10 000, and a significance threshold P<10−8.
Discussion
If only a limited number of mechanisms were responsible for idiosyncratic DILI events, preclinical tests could reliably screen and limit drug candidates for DILI liability, and in principle, a ‘DILI chip’ could be used to screen patient populations to identify susceptible individuals and to potentially aid in the diagnosis of DILI. We therefore hoped to find strong and unambiguous associations between common variants and all-cause DILI at the genome-wide significance threshold. However, once the previously described MHC associations with flucloxacillin and amoxicillin/clavulanate-DILI were excluded, no significant genome-wide associations were found in our large cohort of DILI cases attributed to diverse drugs. Because the proposed mechanisms of DILI involve drug metabolism and disposition and immune response, we next restricted our analysis to SNPs in the vicinity of ADME genes and those associated previously with susceptibility to autoimmune diseases. None of the ADME SNP associations suggested in the test cohort were confirmed in the validation cohort.
For the autoimmune analysis only, the association between hepatocellular DILI and an SNP near the STAT4 gene was also observed in the validation cohort; however, the effect appeared to be attenuated in the validation sample. STAT4 is involved in inflammation through the regulation of several cytokines and has been implicated in T-cell maturation [21]. To our knowledge, this is the first reported association of an innate immune gene with DILI across a large number of implicated drugs, and supports a potential role of innate immunity in DILI. However, this association did not meet the typical rigorous threshold for significance in a genome-wide study. The reduced effect size observed in the validation cohort suggests the possibility of a ‘winner’s curse’ effect, but is also consistent with a chance association: given the seven SNPs carried forward for validation testing, it is not unlikely that one of them would continue to show an association in the validation cohort simply due to chance. We therefore cannot conclude that this is a true association, but we believe further study of the possible relationship between STAT4 variation and hepatocellular DILI is warranted.
Although our sample sizes for each implicated drug were generally small, we found some interesting trends toward associations with specific drugs. For example, an SNP upstream of the PPARG gene was associated with diclofenac DILI at almost the genome-wide significance level and with an OR of 30 (Table 4). Diclofenac has been shown previously to interact with the PPARγ receptor at pharmacologic concentrations of the drug [22], and PPARγ activation has been shown to influence inflammation, specifically by diclofenac [23,24]. Although this strong association was not replicated in our second cohort, the trend was in the same direction, the replication cohort was small, and the association in the combined cohorts remained almost genome-wide significant. Given the biological plausibility of this association, additional studies are warranted. For statin cases, an association with a STAT4 SNP reached specified significance for the autoimmune-restricted analysis but also did not replicate in our second smaller cohort. Given the large population receiving statin medications and the current controversy about the DILI risk that statins pose [25], this observation also requires additional studies.
It seems unlikely that the lack of replication of tentative associations indicates differences in the characteristics of the discovery and replication samples. The larger proportion of amoxicillin/clavulanate and flucloxacillin-DILI cases in the discovery cohort did not appear to account for the failure to replicate, and other characteristics such as severity of injury, causality assessment, or patient characteristics appeared to be comparable between cohorts. Genotype calls between the GWAS chip and the targeted TaqMan assays were also perfectly concordant for all samples examined. The most likely reason for failure to replicate associations of interest (Table 4) therefore may be that these were simply false associations. This is consistent with the fact none of the variants carried forward to replication achieved genome-wide significance in the discovery cohort, and most of the variants selected on the basis of the ADME-restricted or autoimmune-restricted analysis were only approaching or barely surpassing a more lenient threshold for significance.
Recently reported GWAS have identified common alleles in the MHC region as risk factors for DILI [7,10,26,27,28]. If an adaptive immune attack on the liver is the final common event that causes significant liver injury, it is possible that only patients carrying certain MHC alleles are at risk of DILI from a specific drug, that is, ‘upstream’ events (e.g. hepatocyte accumulation of critical reactive metabolites or bile acids) may be necessary but not sufficient to produce DILI. We therefore carried out an exploratory analysis of the association signal in the MHC region compared with autosomal regions outside of the MHC after excluding DILI cases due to flucloxacillin or amoxicillin/clavulanate. Although the P-value distribution was lower in the MHC region compared with elsewhere (Supplementary Fig. S4, http://links.lww.com/FPC/A505), after permutation testing, the enrichment of the association signal in the MHC region only approached significance (empirical P = 0.015). The observed trend toward enrichment could be due to the extremely high degree of linkage disequilibrium among SNPs in the MHC region and/or an artifact of uncorrected population structure, as low-frequency subpopulation-specific variants in the MHC may not be accounted for by principal components analysis based largely on common variants outside of the MHC. Our analysis therefore does not support the concept of common MHC variation as a general risk factor for DILI, but does not exclude drug-specific or drug subset-specific effects.
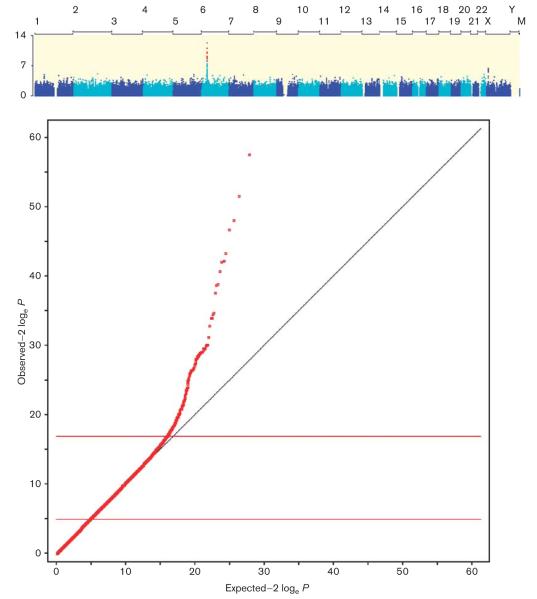
On the basis of the power calculations carried out (Fig. 2), we estimate that common variants conferring the risk of a moderate effect size (i.e. RR ≥ 1.5) to all-cause DILI should have been discovered in our study. It is of note that in our initial GWAS analysis of all DILI cases, the MHC signals driven solely by the flucloxacillin and amoxicillin/clavulanate cases were appreciable at the genome-wide significance level (Fig. 1), although these cases account for only 38% of the cohort. As a further example of the ability to detect strong associations despite gross misclassification of phenotype, an analysis that included DILI cases due to flucloxacillin and all other drugs except amoxicillin/clavulanate identified the rs2395209 association (due to flucloxacillin) at the genome-wide significance level, despite the fact that flucloxacillin made up only 13.4% of the total cases (Fig. 3). Individuals carrying the HLA-B*5701 allele have about an 80-fold increased risk of developing DILI when treated with flucloxacillin [7]. Hence, if any combination of individuals comprising a similarly small fraction of our cohort had a common risk factor with comparable effect, we would have detected it in our GWAS analysis. The fact that no other risk variants were detected in the genome-wide analyses is therefore an important negative finding for identifying common SNPs with a very strong effect on DILI risk.
Fig. 3.
Manhattan (a) and Quantile–quantile (b) plots of genome-wide association study results for the analysis of all drug-induced liver injury (DILI) cases including flucloxacillin, but excluding amoxicillin/clavulanate-induced DILI cases. Despite the fact that flucloxacillin-DILI cases make up only 13.4% of the total case sample (75/560), there remains clear evidence of an HLA-B*5701 (rs2395029) association with DILI risk in the combined sample, suggesting that misclassification or heterogeneity of cases is unlikely to obscure the true DILI risk variants of a very large effect (the HLA-B*5701 allele confers an 80-fold increased risk of DILI from flucloxacillin [7]).
Our negative GWAS findings may suggest that future efforts should focus on collecting and analyzing DILI cases due to single drugs. However, collecting sufficient cases will be a challenge; although the HLA-B*5701 association with flucloxacillin-DILI (OR~80) would have been detected by GWAS with as few as 12 cases, our data suggest that such strong associations with common variants are unusual. With amoxicillin/clavulanate-DILI, more than 50 cases would be required to demonstrate an HLA association by GWAS (OR~3) [10]. Test and replication cohorts of this size may simply not be feasible for most drugs implicated in the existing registries. If there in fact exist common genetic variants that underlie susceptibility to DILI, it may still be feasible to identify them with GWAS if a sufficient number of DILI cases due to diverse drugs could be ‘clustered’ by a common mechanism or surrogates for mechanism. For example, it may be possible to use chemoinformatic approaches or biological effects (e.g. induction of STAT4 in cell culture or transport by MRP2) as surrogates for mechanisms to create subgroupings for GWAS.
Our use of population controls deserves comment. As DILI is such a rare event (occurring on average in fewer than one in 10 000 patients), the expected rate of misclassification of control individuals using unselected population controls is very low and should not appreciably impact statistical power to detect a genetic association ([20], Supplementary Table S39, http://links.lww.com/FPC/A505). However, this study design does not allow us to directly test for the effects of environmental risk factors for DILI, such as age, sex, or diagnosis/indication for the precipitant drug(s), among other factors. In particular, polypharmacy (i.e. patients taking a large number of concomitant drugs, even if only one is truly hepatotoxic) has been suggested as a risk factor for DILI [29], perhaps due to competition for, metabolism, or active transport out of hepatocytes or enzyme induction.
Only common genetic variants (predominantly variants with allele frequencies exceeding 5%) were genotyped in our patients. For an event frequency of less than one in 10 000, as is typical for DILI, it is not possible for common variants individually to have highly specific associations with susceptibility. Analytical approaches that could detect the synergistic effects of multiple SNPs are fraught with statistical (multiple comparison) and computational hurdles and such investigations would probably need to be directed by solid data on biological plausibility. It should also be acknowledged that effects not captured by GWAS or genome sequencing are likely to also contribute to DILI risk, including epigenetic or environmental effects as described above. Nonetheless, the availability of the DILI gene banks and the rapidly evolving genomic technology available currently make genetic investigation of DILI a high-priority area of research. Weak but detectable genetic contributions to risk should show important mechanistic pathways whose epigenetic or environmental influences could then be studied specifically.
Conclusion
GWAS of the largest DILI cohort assembled to date was unable to identify any major genetic risk factors associated with DILI. Restricted analyses showed a replicated association between all-cause hepatocellular injury and STAT4, which should be further confirmed. Although the lack of significant GWAS findings does not rule out a highly polygenic model of many variants with a small effect on DILI risk, our data support the idea that very strong associations may be largely drug-specific or reflect rare genetic variations that may only be shown through ongoing advancements in next-generation sequencing. It also seems likely that the maximal yield from genetic interrogation will require parallel experimental approaches to define subgroups of drugs likely to have common mechanisms of DILI or to identify specific biological pathways implicated in DILI.
Supplementary Material
Acknowledgements
The authors thank the DILIGEN, EUDRAGENE, Spanish DILI, iSAEC, and DILIN collaborators for their help with case recruitment.
The authors acknowledge use of genotype data from the British 1958 Birth Cohort DNA collection and UK National Blood Service control cohorts. This study makes use of data generated by the Wellcome Trust Case–Control Consortium. A full list of the investigators who contributed to the generation of the data is available from http://www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under award 076113 and 085475.
The DILIN network is structured as a U01 cooperative agreement with funds provided by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) under grants: 2U01-DK065176-06 (Duke), 2U01-DK065201-06 (UNC), 2U01-DK065184-06 (Michigan), 2U01-DK065211-06 (Indiana), 5U01DK065193-04 (UConn), 5U01-DK065238-08 (UCSF/CPMC). Additional funding is provided by CTSA grants: UL1 RR025761 (Indiana), UL1 RR025747 (UNC), and UL1 UL1 RR024986 (UMich).
The DILIGEN sample collection was partly funded by the UK Department of Health (Pharmacogenetics Initiative ref PHGX10A), UK NIHR funding to the Nottingham Digestive Diseases Centre, and the iSAEC. The EUDRAGENE collaboration has received support from the EC 5th Framework program (QLRI-CT-2002-02757) and the iSAEC. M.M. has received funding from a UK NIHR postdoctoral award and is an investigator for the FP7 EU-ADR (ICT-215847) and ARITMO (HEALTH-241679) projects on drug safety. The Spanish DILI Registry is partly funded by the Spanish Medicine Agency and has received support from the iSAEC. CIBERehd is funded by Instituto de Salud Carlos III.
The genome-wide genotyping of DILIN cases was funded by the NIDDK under grant 2U01-DK065176-06. Genome-wide genotyping of the DILIGEN, Dundee, EUDRAGENE, Spanish DILI Registry and GSK POPRES controls was funded by iSAEC, with support from Abbott, Daiichi-Sankyo, GSK, J & J, Novartis, Pfizer, Roche, Sanofi-Aventis, Takeda, the Wellcome Trust and Wyeth.
Footnotes
T.J.U., Y.S., and A.F. were responsible for data management; T.J.U. and Y.S. performed the data analysis; T.J.U., A.S., N.C., R.J.F., J.R., J.S., D.B.G., and P.B.W. coordinated the genetic studies for DILIN; D.G. provided bioinformatic support; K.V.S. was responsible for generation and quality control of the genotype data; A.K.D. and G.P.A. recruited and phenotyped patients for DILIGEN; M.I.L. recruited and phenotyped patients for the Spanish DILI Registry; M.M. recruited and phenotyped patients for EUDRAGENE; A.S., N.C., R.J.F., H.B., T.J.D., W.M.L., V.J.N., J.A.T., and P.B.W. recruited and phenotyped patients for DILIN; Y.S., A.S., N.C., R.J.F., A.K.D., M.I.L., M.R.N., M.M., and T.J.D. provided comments on the research design and contributed to the manuscript; T.J.U., D.B.G., and P.B.W. designed the study; T.J.U., and P.B.W. wrote the manuscript.
DILIGEN investigators: Ann K. Daly (PI), Newcastle University; Guruprasad P. Aithal, Nottingham Digestive Diseases Centre, NIHR Biomedical Research Unit, Nottingham; John F. Dillon, Dundee University; Munir Pirmohamed, University of Liverpool; Christopher P. Day, Newcastle University; Peter T. Donaldson, Newcastle University, William Bernal, Kings College, London; B. Kevin Park, University of Liverpool; Saye Khoo, University of Liverpool, Ian Gilmore, University of Liverpool.
Other DILIGEN collaborators: Research nurses – J. Henderson (Newcastle University), C. Davies (Nottingham Digestive Diseases Centre), K. Hawkins, A. Hanson, J. Evely (University of Liverpool). Other contributors to case recruitment – H. Hussaini (Truro), P. Mills (Glasgow), W. Griffiths (Addenbrooks Hospital, Cambridge), J. Collier (John Radcliffe Infirmary, Oxford), A. Brind (North Staffordshire), N. Fisher (Dudley), J. Shearman (South Warwick), E. Elias (Birmingham), A. Grant (Leicester Royal Infirmary), A. Austin (Derby), F. Gordon (Bristol), M. Cramp (Plymouth), S. Saksena (North Durham), H.J. McMurtry (Chorley), N. Thompson (Freeman Hospital, Newcastle), R. Williams and M. Morgan (Royal Free and University College Medical School), E.M. Phillips (Hexham), M. Patel (Merthyr Tydfil), H. Mitchison (Sunderland), J.G. Kingham (Singleton Hospital, Swansea), D. Das (Stepping Hill Hospital, Stockport), J. Collier (John Radcliffe Infirmary, Oxford), S. Hellier (Worcester), M. Groome (Ninewells Hospital, Dundee), M. Miller (Ninewells Hospital, Dundee).
EUDRAGENE Investigators: Paul McKeigue (co PI), University of Edinburgh, Scotland; Bruno Stricker, Erasmus MC, NL (Data Analysis Committee); Qun-Ying Yue, Medical Products Agency, Uppsala, Sweden (Data Analysis Committee); Mia Wadelius, Uppsala Clinical Research Centre University Hospital, Sweden; Erik Eliasson, Karolinska Institutet, Sweden; Pär Hallberg, Uppsala Clinical Research Centre University Hospital, Sweden; Håkan Melhus, Uppsala Clinical Research Centre University Hospital, Sweden; Alfonso Carvajal, Universidad de Valladolid, Spain; Luisa Ibáñez, Fundació Institut Català de Farmacologia, Hospital Universitari Vall d’ Hebron, Universitat Autònoma, Barcelona, Spain; Emmanuelle Bondon-Guitton, Université de Toulouse, France; Maryse Lapeyre-Mestre, Université de Toulouse, France Jean-Louis Montastruc, Université de Toulouse, France; Dr Anita Conforti, University Hospital, Verona, Italy; Professor Giampaolo Velo, University Hospital, Verona, Italy; Michel Eichelbaum, Dr Maragrete Fischer-Bosch Institute Clinical Pharmacology, Stuttgart.
Other contributors to case recruitment: Inés Salado, Universidad de Valladolid, Spain; Maria Sainz, Universidad de Valladolid, Spain; Lourdes Vendrell, Fundació Institut Català de Farmacologia; Dr Francesca Succurro, University Hospital, Verona; Dr Marco Smerghetto, University Hospital, Verona; Ramazan Buyukcelik, Erasmus MC, NL; Pascal Arp, Erasmus MC, NL.
Pharmacovigilance centers involved in case ascertainment (France, Spain, Italy, UK): L’Association Française des Centres Régionaux de Pharmacovigilance (CRPV), Spanish Pharmacovigilance Centres (Vallodolid, Madrid, Murcia, Badalona, Galicia, Asturias, Vall d’Hebron, Pais Vasco, Asistencial de Avila, Agencia Espanola de Medicamentos y Products Sanitarios), Participating Italian Pharmacovigilance Centres (Veneto region), MHRA, UK.
Spanish DILI Registry collaborators: Investigators: R.J. Andrade (PI), M.I. Lucena, C. Stephens, E. Ulzurrun, Y. Borraz (Málaga University), F. Ruiz-Cabello, M.A. Lopez-Nevot (H. Virgen de las Nieves, Granada). Contributors to case recruitment: M. Romero-Gomez (H. Valme, Sevilla), M.C. Fernández, G. Peláez, M. Casado (H. Torrecárdenas, Almería), J.M. Navarro (H. Costa del Sol, Málaga), C. Guarner, G. Soriano, E.M. Roman (H. San Pau, Barcelona), T. Muñoz-Yague, J.A. Solís-Herruzo (H. 12 de Octubre, Madrid), A. Castiella, E.M. Zapata (H. de Mendaro, Guipuzcuo), M. Moreno (H. la Laguna, Tenerife), J.L. Calleja, J. de la Revilla (H. Puerta de Hierro, Madrid), O. Lo Lacono (H. del Tajo, Madrid).
Conflicts of interest There are no conflicts of interest.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Website (www.pharmacogeneticsandgenomics.com).
References
- 1.Ostapowicz G, Fontana RJ, Schiodt FV, Larson A, Davern TJ, Han SH, et al. Results of a prospective study of acute liver failure at 17 tertiary care centers in the United States. Ann Intern Med. 2002;137:947–954. doi: 10.7326/0003-4819-137-12-200212170-00007. [DOI] [PubMed] [Google Scholar]
- 2.Watkins PB, Seligman PJ, Pears JS, Avigan MI, Senior JR. Using controlled clinical trials to learn more about acute drug-induced liver injury. Hepatology. 2008;48:1680–1689. doi: 10.1002/hep.22633. [DOI] [PubMed] [Google Scholar]
- 3.Andrade RJ, Robles M, Ulzurrun E, Lucena MI. Drug-induced liver injury: insights from genetic studies. Pharmacogenomics. 2009;10:1467–1487. doi: 10.2217/pgs.09.111. [DOI] [PubMed] [Google Scholar]
- 4.Daly AK. Drug-induced liver injury: past, present and future. Pharmacogenomics. 2010;11:607–611. doi: 10.2217/pgs.10.24. [DOI] [PubMed] [Google Scholar]
- 5.Lucena MI, Kaplowitz N, Hallal H, Castiella A, Garcia-Bengoechea M, Otazua P, et al. Recurrent drug-induced liver injury (DILI) with different drugs in the Spanish Registry. The dilemma of the relationship to autoimmune hepatitis. J Hepatol. 2011 doi: 10.1016/j.jhep.2010.12.041. [DOI] [PubMed] [Google Scholar]
- 6.Fontana RJ, Watkins PB, Bonkovsky HL, Chalasani N, Davern T, Serrano J, et al. Drug-Induced Liver Injury Network (DILIN) prospective study: rationale, design and conduct. Drug Saf. 2009;32:55–68. doi: 10.2165/00002018-200932010-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daly AK, Donaldson PT, Bhatnagar P, Shen Y, Péer I, Floratos A, et al. HLA-B*5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nat Genet. 2009;41:816–819. doi: 10.1038/ng.379. [DOI] [PubMed] [Google Scholar]
- 8.Molokhia M, McKeigue P. EUDRAGENE: European collaboration to establish a case-control DNA collection for studying the genetic basis of adverse drug reactions. Pharmacogenomics. 2006;7:633–638. doi: 10.2217/14622416.7.4.633. [DOI] [PubMed] [Google Scholar]
- 9.Andrade RJ, Lucena MI, Fernandez MC, Pelaez G, Pachkoria K, Garcia-Ruiz E, et al. Drug-induced liver injury: an analysis of 461 incidences submitted to the Spanish registry over a 10-year period. Gastroenterology. 2005;129:512–521. doi: 10.1016/j.gastro.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 10.Lucena MI, Molokhia M, Shen Y, Urban TJ, Aithal GP, Andrade RJ, et al. Susceptibility to amoxicillin-clavulanate-induced liver injury is influenced by multiple HLA class I and II alleles. Gastroenterology. 2011 doi: 10.1053/j.gastro.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danan G, Benichou C. Causality assessment of adverse reactions to drugs – I. a novel method based on the conclusions of international consensus meetings: application to drug-induced liver injuries. J Clin Epidemiol. 1993;46:1323–1330. doi: 10.1016/0895-4356(93)90101-6. [DOI] [PubMed] [Google Scholar]
- 12.Nelson MR, Bryc K, King KS, Indap A, Boyko AR, Novembre J, et al. The Population Reference Sample, POPRES: a resource for population, disease, and pharmacological genetics research. Am J Hum Genet. 2008;83:347–358. doi: 10.1016/j.ajhg.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 14.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 15.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ge D, Zhang K, Need AC, Martin O, Fellay J, Urban TJ, et al. WGAViewer: software for genomic annotation of whole genome association studies. Genome Res. 2008;18:640–643. doi: 10.1101/gr.071571.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahmadi KR, Weale ME, Xue ZY, Soranzo N, Yarnall DP, Briley JD, et al. A single-nucleotide polymorphism tagging set for human drug metabolism and transport. Nat Genet. 2005;37:84–89. doi: 10.1038/ng1488. [DOI] [PubMed] [Google Scholar]
- 18.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menashe I, Rosenberg PS, Chen BE. PGA: power calculator for case-control genetic association analyses. BMC Genet. 2008;9:36. doi: 10.1186/1471-2156-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edwards BJ, Haynes C, Levenstien MA, Finch SJ, Gordon D. Power and sample size calculations in the presence of phenotype errors for case/control genetic association studies. BMC Genet. 2005;6:18. doi: 10.1186/1471-2156-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korman BD, Kastner DL, Gregersen PK, Remmers EF. STAT4: genetics, mechanisms, and implications for autoimmunity. Curr Allergy Asthma Rep. 2008;8:398–403. doi: 10.1007/s11882-008-0077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adamson DJ, Frew D, Tatoud R, Wolf CR, Palmer CN. Diclofenac antagonizes peroxisome proliferator-activated receptor-gamma signaling. Mol Pharmacol. 2002;61:7–12. doi: 10.1124/mol.61.1.7. [DOI] [PubMed] [Google Scholar]
- 23.Kaur J, Sanyal SN. Modulation of inflammatory changes in early stages of colon cancer through activation of PPARgamma by diclofenac. Eur J Cancer Prev. 2010;19:319–327. doi: 10.1097/CEJ.0b013e32833ad385. [DOI] [PubMed] [Google Scholar]
- 24.Zieleniak A, Wojcik M, Wozniak LA. Structure and physiological functions of the human peroxisome proliferator-activated receptor gamma. Arch Immunol Ther Exp (Warsz) 2008;56:331–345. doi: 10.1007/s00005-008-0037-y. [DOI] [PubMed] [Google Scholar]
- 25.Bader T. Liver tests are irrelevant when prescribing statins. Lancet. 2010;376:1882–1883. doi: 10.1016/S0140-6736(10)62142-3. [DOI] [PubMed] [Google Scholar]
- 26.Singer JB, Lewitzky S, Leroy E, Yang F, Zhao X, Klickstein L, et al. A genome-wide study identifies HLA alleles associated with lumiracoxib-related liver injury. Nat Genet. 2010;42:711–714. doi: 10.1038/ng.632. [DOI] [PubMed] [Google Scholar]
- 27.Spraggs CF, Budde LR, Briley LP, Bing N, Cox CJ, King KS, et al. HLA-DQA1*02:01 Is a major risk factor for lapatinib-induced hepatotoxicity in women with advanced breast cancer. J Clin Oncol. 2011;29:667–673. doi: 10.1200/JCO.2010.31.3197. [DOI] [PubMed] [Google Scholar]
- 28.Kindmark A, Jawaid A, Harbron CG, Barratt BJ, Bengtsson OF, Andersson TB, et al. Genome-wide pharmacogenetic investigation of a hepatic adverse event without clinical signs of immunopathology suggests an underlying immune pathogenesis. Pharmacogenomics J. 2008;8:186–195. doi: 10.1038/sj.tpj.6500458. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki A, Yuen N, Walsh J, Papay J, Hunt CM, Diehl AM. Co-medications that modulate liver injury and repair influence clinical outcome of acetaminophen-associated liver injury. Clin Gastroenterol Hepatol. 2009;7:882–888. doi: 10.1016/j.cgh.2009.03.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.