Abstract
Derivation of embryonic stem (ES)-cell lines from genetically non-permissive mouse strains, such as FVB/N, has been difficult, despite this strain offering advantages for mouse transgenesis for developmental studies. We earlier generated β-actin promoter-driven enhanced green fluorescent protein (EGFP)-transgenic FVB/N mice, expressing EGFP in all cells. Here, by optimizing culture system and using RESGROTM ES-cell culture medium, we successfully derived EGFP-transgenic ES-cell line, ‘GS-2’ line, from F1 hybrid blastocysts, from wild-type 129/SvJ female X EGFP-transgenic homozygous FVB/N male. The GS-2 ES-cell line exhibited all defining criteria of a typical ES-cell line, including normal colony morphology and karyotype (40,XY), high replication-expansion efficiency (passages: >100), expression of pluripotent markers (Oct-4, Nanog, Sox-2, SSEA-1 and others) and, embryoid body (EB) development and EB differentiation to ecto-/meso-/endo-dermal cell types, expressing nestin, BMP-4 and α-fetoprotein, respectively. GS-2 ES-cells formed (i) teratoma containing three germ lineage-derived cell types, (ii) chimeric blastocysts and fetuses, following their aggregation with wild-type 8-cell embryos, (iii) functional cardiac clusters and (iv) predominantly neural cell types when EBs were developed in KOSR-supplemented medium. Taken together, we derived a robust EGFP-transgenic GS-2 ES-cell line, from a non-permissive transgenic (FVB/N) mouse by a single cross to 129/SvJ wild-type mouse. The GS-2 ES-cell line exhibited full differentiation potential, in vitro/in vivo, providing enormous opportunity for stem cell research, including experimental cell transplantation studies.
Keywords: Embryonic stem cells, non-permissive mouse strain, transgenic ES-cell line, differentiation, cardiogenesis
Introduction
Cellular and molecular regulation of early mammalian development, including embryonic stem cell (ES-cell) differentiation, has been an area of intense research. To gain insights into these events, genetic manipulation approaches such as transgenic and knock-out technologies are being employed [1-3]. Among reporter genes used in transgenesis-aided gene function studies, the green fluorescent protein (GFP) or enhanced GFP (EGFP) offer enormous advantages [4], including cell lineage-specific expression analysis and tracking of fluorescing cells during embryonic differentiation [5-7]. We earlier generated EGFP-transgenic ‘green’ mice, for the first time, in FVB/N strain since this genetic back ground is suitable for transgenesis [8-11]. The EGFP-transgenic mouse is a potential source for green fluorescing cell types [9,10,12], including derivation of ES-cell line required for developmental studies [5-7].
Though, transgenic mouse lines from FVB/N strain are available [8,9], established ES-cell lines are not available from these lines, in view of their non-permissivity for ES-cell derivation [13]. There is no information on the derivation of a permanent robust EGFP-expressing ES-cell line from transgenic FVB/N mice. Such cell lines could potentially be useful for animal cloning strategy via nuclear transfer or tetraploid blastocyst complementation experiments [14,15]. Derivation of ES-cell lines from FVB/N mice, employing conventional protocols, was unsuccessful [16] and it appears that derivation of ES-cell lines is highly mouse strain-dependent [17,18]. Following the first mouse ES-cell line derivation from the 129/Sv strain [20], several ES-cell lines were derived from permissive genetic strains [17,18,21,22]. However, by using conventional approach, derivation of stable ES-cell lines from non-permissive strains such as FVB/N [13], CBA [13,18,23], ICR [19], DDK [24]. NOD [25] was not successful. Consequently, transgenic or knockout ES-cell lines, from such important ‘mutant’ mice have been very limited for developmental studies.
To derive ES-cell lines from such non-permissive strains, several approaches were employed which included use of hybrid blastocysts or diapause-blastocysts [18,19,24,26], improved ES-cell derivation culture systems/media [17,22,27,28] or use of cell signaling inhibitors [16,29]. These approaches often involve a high level of experimental sophistication, expertise and normally difficult to carryout in most laboratories. Therefore, to overcome non-permissiveness of FVB/N strain to derive ES-cell line, our study developed a new protocol-strategy to successfully derive EGFP-transgenic ES-cell line, using an optimized culture system, from F1 hybrid blastocysts of our EGFP-transgenic ‘green’ FVB/N mouse [9] crossbred with 129/SvJ wild-type mouse. To our knowledge, this study is the first to use such a strategy to derive a robust ES-cell line from FVB/N EGFP-transgenic mouse strain.
Materials and methods
Derivation of ES-cell lines from EGFP-transgenic mice
Derivation of ES-cell lines was attempted from F1 hybrid blastocysts of EGFP-transgenic FVB/N mice crossed with 129/SvJ mice. Procedures used were as described by Hogan and colleagues [9,10,30]. Procedures on animal resource and experiments were as per the national guidelines with approval of Institutional Animal Ethics Committee. Blastocysts were co-cultured on a monolayer of mitotically inactivated mouse (day 13.5, CD1) embryonic fibroblasts (MEFs) in four-well plates in standard ES-cell culture medium supplemented with 1000 IU/ml mouse LIF (Chemicon) or in RESGRO™ medium (Chemicon) for four days [28,30,31]. Cultured blastocysts were monitored for development through hatching, attachment and outgrowths. The inner cell mass (ICM) were picked individually, washed in DPBS and placed in a 25 μl drop of 0.05% trypsin-EDTA solution. Dispersed cells/clumps were replated in a fresh 4-well plate on mitomycin-c (mit-c) treated MEFs and allowed to attach and grow for four days. Proliferated ES-cell like colonies were picked, re-trypsinized and plated as above.
Maintenance of ES-cell lines
ES-cell colonies were allowed to grow in 35 mm dishes in 2.5 ml of RESGRO™ medium or standard ES-cell culture medium [30,31] either without or with KOSR supplementation. At every 48 h, ES-cell colonies were trypsinized and passaged at 1:4 to 1:6 ratios. One-sixth of cells were replated as above and the rest (2 million) were transferred to vials for cryopreservation [31].
Reverse transcriptase (RT)-polymerase chain reaction (PCR) analysis
Total RNA was prepared from fresh or frozen ES-cells or from embryoid bodies (EBs) by using Tri reagent (Sigma). DNase-treated RNA samples were used for RT-PCR analysis [9] and for cDNA amplification, 100 ng of gene-specific primers (Table 1) were used.
Table 1.
Primers used for RT-PCR analysis*
| Sl. No. | Gene name | Forward and reverse primer sequences (5’→3’) | Amplicon size (bp) |
|---|---|---|---|
| 1. | Oct-4 | TCT TTC CAC CAG GCC CCC GGC TC | 224 |
| TGC GGG CGG ACA TGG GGA GAT CC | |||
| 2. | Nanog | AGG GTC TGC TAC TGA GAT GCT CTG | 364 |
| CAA CCA CTG GTT TTT CTG CCA CCG | |||
| 3. | Sox2 | TAG AGC TAG ACT CCG GGC GAT GA | 297 |
| TTG CCT TAA ACA AGA CCA CGA AA | |||
| 4. | Rex1 | AGC AGG ATC GCC TCA CTG | 189 |
| GGC CAC TTG TCT TTG CCG | |||
| 5. | Utf 1 | GGA TGT CCC GGT GAC TAC GTC TG | 344 |
| GGC GGA TCT GGT TAT CGA AGG GT | |||
| 6. | Esg1 | GAA GTC TGG TTC CTT GGC AGG ATG | 376 |
| ACT CGA TAC ACT GGC CTA GC | |||
| 7. | EGFP | CAC ATG AAG CAG CAC GAC TT | 267 |
| GAA GTT CAC CTT GAT GCC GT | |||
| 8. | β-Actin | TGA ACC CTA AGG CCA ACC GTG | 396 |
| GCT CAT AGC TCT TCT CCA GGG | |||
| 9. | Nestin | CCA AAG AGG TGT CCG ATC AT | 213 |
| TGA CAT CCT GGA CCT TGA CA | |||
| 10. | BMP-4 | CCT GGT AAC CGA ATG CTG AT | 260 |
| AGC CGG TAA AGA TCC CTC AT | |||
| 11. | α-Feto protein | TCC TCC TGC TAC ATT TCG CT | 199 |
| TTC TTC ATT GCA GCC AAC AC | |||
| 12. | NKx2.5 | CAG TGG AGC TGG ACA AAG CC | 217 |
| TAG CGA CGG TTC TGG AAC CA | |||
| 13. | GATA-4 | GCA GCA GCA GTG AAG AGA TG | 186 |
| GCG ATG TCT GAG TGA CAG GA | |||
| 14. | α-MHC | CTG CTG GAG AGG TTA TTC CTC G | 302 |
| GGA AGA GTG AGC GGC GCA TCA AGG |
Primers sets used are from published data and are exon-spanning ones.
Immunostaining of pluripotent markers expressed by ES-cells
ES-cells were fixed for 20 min at room temperature in 4% PFA in DPBS, washed with DPBS three times and then permeabilized for 10 min at room temperature with 0.1% Triton X-100 in DPBS. Washed cells were treated with blocking buffer (DPBS containing 1% BSA) for 30 min. at room temperature and incubated with primary antibodies in blocking buffer overnight in a humidified chamber at 40C. Primary antibodies included mouse monoclonal antibodies against OCT-4 (1:100; Santacruz) and SSEA-1 (1:20; gift from Dr. Peter Andrews, Univ. Sheffield, UK). Following washings with PBS, cells were exposed, for one hour at room temperature, to fluorescent Alexa Fluor 546-conjugated secondary anti-mouse IgG antibodies (1:100; Molecular Probes), Cells were washed with DPBS three times, counterstained with DAPI, mounted in prolong gold antifade (Invitrogen) and observed for immunolocalization using a confocal microscope [27].
Alkaline phosphatase (AP) assay
ES-cells were washed with DPBS, fixed for 10 min in 4% PFA in DPBS, again washed with DPBS then with 30 mM Tris-malate buffer (pH 9.0) and treated with the buffer containing napthol-AS-MX-phosphate (0.4 mg/ml), fast red TR salt (1 mg/ml) and 0.5 mM MgCl2 until the pink color developed. Cells were washed with DPBS and observed under the microscope.
Karyotype analysis
Karyotyping of ES-cells at passages 15 and 45 was performed by a standard G-banding of metaphase spreads using Giemsa staining procedure [31]. Approximately, ten metaphase spreads were counted, images acquired microscopically and the karyogram was prepared using the mouse karyogram software (Cytovision, Applied Imaging).
Chimeric embryo generation
Chimeric embryos were generated by aggregating zona-free, non-compact FVB/N 8-cell EGFP-transgenic embryos either with itself or with 15 ES-cells [30,31]. For aggregation, they were placed in 35 mm dishes in individual micro-wells, created by darning needle (BLS Ltd.). Embryos were cultured for 48 h in 1:1 mixture of M16 medium and ES-cell culture medium, overlaid with mineral oil. Embryo development was assessed and their confocal images were recorded. For embryo transfer, chimeric day-3 embryos were transferred, per-cervical, to day-2 pseudopregnant (CD-1)-mouse and their postimplantation development was assessed [9,30].
Teratoma formation and their histological analyses
About two million dispersed ES-cells, suspended in 250 μl of ES-cell medium without LIF, were injected subcutaneously into nude mice [31]. After six weeks, mice with well developed teratoma were euthanized. The tumor lump was surgically excised and was fixed in 10% buffered-formalin, processed for sectioning and H-E staining. They were assessed for differentiation and photographed [31].
Differentiation of ES-cells via EB development
Development of EBs from ES-cells were achieved by hanging drop method [32], using ES-cell medium containing 20% heat-inactivated FBS without LIF. EBs were cultured for 48 h, washed and suspended in the same medium in 60 mm bacteriological dishes (Greiner) for additional 3 days. On day 5, EBs were plated on 0.1% gelatin-coated 60 mm tissue culture dishes or 24-well plates and cultured for additional 25 days. Their proliferation, outgrowth and differentiation patterns were monitored daily. RT-PCR analysis of EB-derived differentiated cell types was carried out.
Assessment of functionality of beating cardiac patches
Wells containing EB-outgrowths with beating cardiac patches were placed on the temperature-controlled stage-top of the Olympus inverted microscope (IX-70). Cardiac beatings were videorecorded in real-time mode using CCD-camera (JVC) [32] for 3-5 min. Grabbed videographs were converted to digital mode and the specified recordings were described in Results section.
Results
Derivation of ES-cell line from EGFP-transgenic FVB/N mouse
We attempted deriving ES-cell line from FVB/N (GU3) X 129/SvJ hybrid blastocysts using standard culture conditions. We obtained 69% primary ES-cell like colonies and the rate of colonies, past 3-8 passages, achieved was superior to those obtained with the wild-type or transgenic FVB/N blastocysts (Table 2). We then optimized culture conditions, using RESGROTM medium (Chemicon) in order to achieve deriving ES-cell line. In this condition, cultured hybrid embryos hatched, attached and produced predominant ICMs as shown in Figure 1A-1C. Interestingly, they showed negligible trophoblast (TB) cells (Figure 1C). We derived three primary ES-cell like colonies which propagated past-five passages. One of them proliferated robustly and produced efficient ES-cell colonies (Figure 1D-1F) and the other two putative ES-cell likecolonies were frozen for future characterization. ES-cell colonies were initially sub-cultured, up to 10 passages, in RESGROTM medium and then in standard ES-cell culture medium. Continuous sub-cultures of ES-cells were performed up to 100 passages and cryopreserved. Thus, we successfully established the EGFP-expressing ES-cell line and we designated this as ‘GS-2’ ES-cell line.
Table 2.
Summary on derivation efficiencies of ES-like cells from FVB/N wild-type and EGFP-transgenic blastocysts or from FVB/N GU-3X129/SvJ F1 hybrid blastocysts.
| Steps in ES-cells derivation | Values | ||
|---|---|---|---|
|
| |||
| Wild Type | Transgenic | GU3X129/SvJ | |
| No. of embryo donors* | 14 | 14 | 05 |
| No. of blastocysts recovered (Avg./donor) | 185 (13) | 48 (12) | 19 (3.8) |
| No. of blastocysts cultured (% of recovered) | 159 (85.9%) | 39 (81.3%) | 16 (84.2%) |
| No. of blastocysts hatched (% of cultured) | 140 (88.1%) | 34 (87.2%) | 16 (100%) |
| No. of blastocyst attached (% of hatched) | 128 (91.4%) | 27 (79.5%) | 16 (100%) |
| No. of ICM picked, trypsinized and plated (% of attached) | 119 (92.9%) | 24 (88.9%) | 13 (81.3% ) |
| No. of primary colonies (% of ICM picked) | 102 (85.7%) | 16 (66.7%) | 9 (69.2% ) |
| No. of colonies, past 3-8 passages (% of primary colonies) | 18 (17.6%) | 4 (25.0%) | 5 (55.6% ) |
Wild-type and transgenic FVB/N mice were super-ovulated with PMSG/hCG, while 129/SvJ female (mated with FVB/N-GU3 male) donors were un-stimulated.
Figure 1.
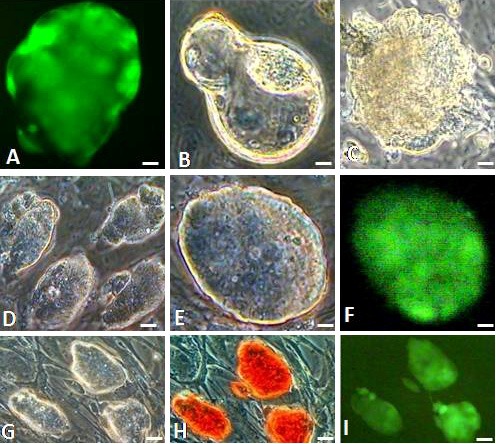
Morphological assessment of GS-2 ES-Cells. (A) FVB/N (GU3) X 129/SvJ F1 hybrid green fluorescent blastocyst, (B) hatching (24 h) and (C) attached (72 h) blastocysts. (D-I) Images of GS-2 ES-cell colonies in phase contrast (D, E, G), fluorescence (F, I) and AP-staining (H). Magnification bars. A-C: 10 μm; D, G-I: 25 μm; E, F: 10 μm.
Characterization of GS-2 ES-cell line
The GS-2 ES-cell colonies robustly proliferated and individual colonies had distinct boundaries, containing a few hundred cells as shown in Figure 1D-1F. Cells appeared to have high ratios of nucleus to cytoplasm with prominent nucleoli and the colony morphology was similar to that of R1 or D3 mouse ES-cell lines, cultured parallely. The GS-2 ES-cell line had a high replicative efficiency and did not show any replication crisis or loss of EGFP expression during passages. The GS-2 ES-cell line exhibited excellent propagation efficiency. When sub-cultured at 1:4 or 1:8 or 1:16 passage ratios, they showed increases in cell counts and colony forming efficiency as shown in Figure 2 and Table 3. Moreover, the GS-2 ES-cells were also highly adaptable to ES-cell maintenance culture media tested and other culture media such as ES-cell medium supplemented with only 15% FBS or with 15% KOSR (devoid of FBS).
Figure 2.
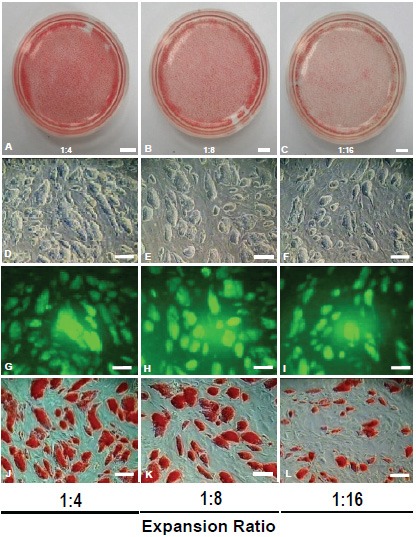
Colony forming efficiency of GS-2 ES-cell line. (A-C) AP-stained ES-cell colonies (p40) with expansion ratios at 1:4 (A), 1:8 (B) and 1:16 (C). (D-F) ES-cell colony morphology formed at above ratios, (G-I) their respective fluorescence and (J-L) AP-stained colonies. Magnification bars. A-C: 150 μm; D-L: 50 μm.
Table 3.
Colony forming efficiency of GS-2 ES-cells
| Expansion Ratio | Input cells | Cell counts* (48 hrs.) | No. of colonies/dish |
|---|---|---|---|
| 1:4 | 0.6 X 106 | 2.97 X 106 | ~42,268 |
| 1:8 | 0.3 X 106 | 2.04 X 106 | ~39785 |
| 1:16 | 0.15 X106 | 1.67 X 106 | ~28,382 |
Cell counts shown are approximate numbers based on visual countings of alkaline phosphatase stained cells from the grabbed images of the colonies.
The GS-2 ES-cell line expressed key pluripotent marker genes i.e., Oct-4, Nanog, Sox-2, Rex-1, UTF-1 and Esg1 as shown in Figure 3I. In addition to expressing EGFP (Figure 1F, 1I), they also expressed AP activity (Figure 1H), OCT-4 and SSEA-1 (Figure 3II). The OCT-4 staining was specifically in the nucleus with a clear colocalization with the DAPI-stained DNA as shown in Figure 3IIA-C. In contrast, the SSEA-1 immunostaining was exclusively in the ES-cell’s plasma membrane (Figure 3IID-F). Moreover, the GS-2 ES-cell line was from a male embryo, showing a normal karyotype (40, XY) as shown in Figure 3III.
I.
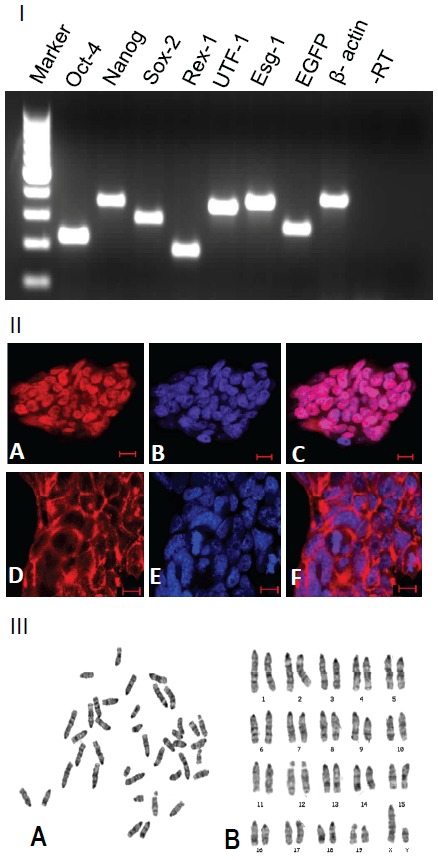
RT-PCR analysis of GS-2 ES-cell line. Lanes (left to right): 100 bp DNA ladder marker, Oct-4 (224 bp), Nanog (364 bp), Sox2 (297 bp), Rex1 (189 bp), UTF1 (344 bp), Esg1 (376 bp), EGFP (267 bp) and β-actin (396 bp); last two lanes: -RT (for β-actin amplification primer set) and no template controls, respectively. (II) Immunostaining of OCT-4 (A), DAPI-nuclear staining (B) and their colocalization in the nucleus (C); immunostaining of SSEA-1 (D) DAPI-nuclear staining (E) and their distinct localizations in the plasma membrane and nucleus (F). Magnification bars. A-F: 10 μm. (III) Karyotype analysis of GS-2 ES-cells (p15). (A) Metaphase chromosome spread (B) Karyogram with 40, XY. Similar observation made with p45 GS-2 ES-cells.
Differentiation of GS-2 ES-cells
GS-2 ES-cells showed excellent differentiation potential both in vitro and in vivo (Figure 4). In vitro, we routinely generated hundreds of EBs from GS-2 ES-cells. Developed EBs were uniformly spherical shaped and expressed EGFP as shown in Figure 4IA, F. As shown in Figure 4, EBs efficiently attached and exhibited extensive proliferation of differentiated cells such as ectodermally-derived neuronal cells (I, B, G), mesodermally-derived beating cardiac patches (I, C, H) and innervated- skeletal tissues (I, D, I) and endodermal-like cells (I, E, J). As seen in Videos 1, 2a and 2b, cardiac patches show vigorous beatings. When EBs were generated in KOSR-supplemented medium, we obtained robust differentiation to predominantly neural progenitors with neuronal cells and, cardiac cells were not detected (Figures 4IA and 5IA). As shown in Figure 5, GS-2 ES-cells spontaneously differentiated to osteoblast-like cells (I, B) and endoderm-like cells (I, C). Besides, they formed skeletal muscle tubes showing twitchingtype contractions (Video-3). Beyond day 20, EBoutgrowths spontaneously showed massively contracting innervated smooth muscle-like tissues as shown in Video-4.
I.
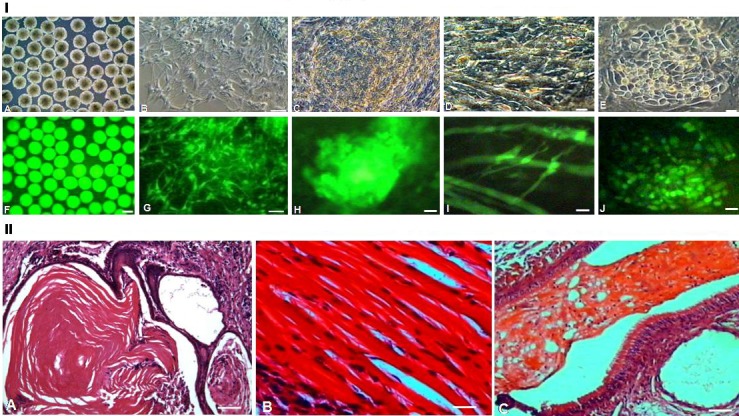
Differentiation of GS-2 ES-cells. (A, F) GS-2 ES-cells-derived EBs. (B-E & G-J) Differentiated cell types: neuronal cells (B, G), cardiac cluster (C, H), innervated myotubes (D, I) and endoderm-like cells (E, J). A-E, phase contrast and F-J, respective fluorescence images. Magnification bars. A, F: 50 μm; B, G: 25 μm; C-E & H-J: 10 μm. (II) H-E stained teratoma sections showing ectodermally-derived epidermis (A), mesodermally-derived skeletal muscle (B) and endodermally-derived respiratory ciliated epithelium (C). Magnification bars. A-C: 25 μm.
I.
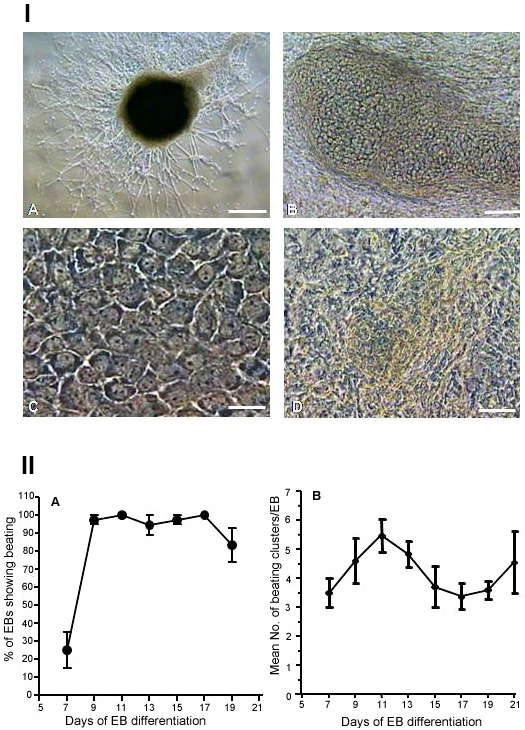
EB-derived differentiation of GS-2 ES-cells. (A) Neural cell types (KOSR-medium). (B) Osteoblast-like cells. (C) Endodermal-like cells. (D) Functional cardiac patch. Magnification bars. A: 50 μm; B: 25 μm; C, D: 10 μm. (II). Profiles of cardiac differentiation of GS-2 ES-cells. (A) Percentage of EBs showing cardiac beatings. (B) Mean number of beating clusters per EB.
As described above, we observed spontaneous cardiac differentiation with vigorously beating clusters beginning from day 7 through day 19 of EB development (Videos-1,-2). These were observed without any addition of cardiogenic inducers. We measured cardiac differentiation efficiency of GS-2 ES-cells as shown in Figure 5II. More than 80% of EBs consistently showed beating clusters with 3-6 mean number of clusters per EB and rates of beating clusters and their frequencies increased from day 7, reaching maximum by day 11 (Figure 5II). Cardiac clusters showed beatings at 60-70 (day 7) up to 80-120 (day 10) beats/min.
As seen in Figure 6, RT-PCR analysis reveal that lineage-specific cell differentiation markers were expressed for the three germ layer-derived cell types. These included nestin for ectoderm lineage, markers such as BMP-4, Nkx-2.5, α-MHC and GATA-4 for mesoderm/cardiac lineage and, α-fetoprotein (AFP) for endoderm lineage. While the expression of stemness gene markers such as Oct-4, Nanog, Sox-2 and Rex-1 decreased as the differentiation of EB proceeded from day 2 through day 13, the expression of differentiated cell types-associated gene markers increased during the same period (data not shown).
Figure 6.
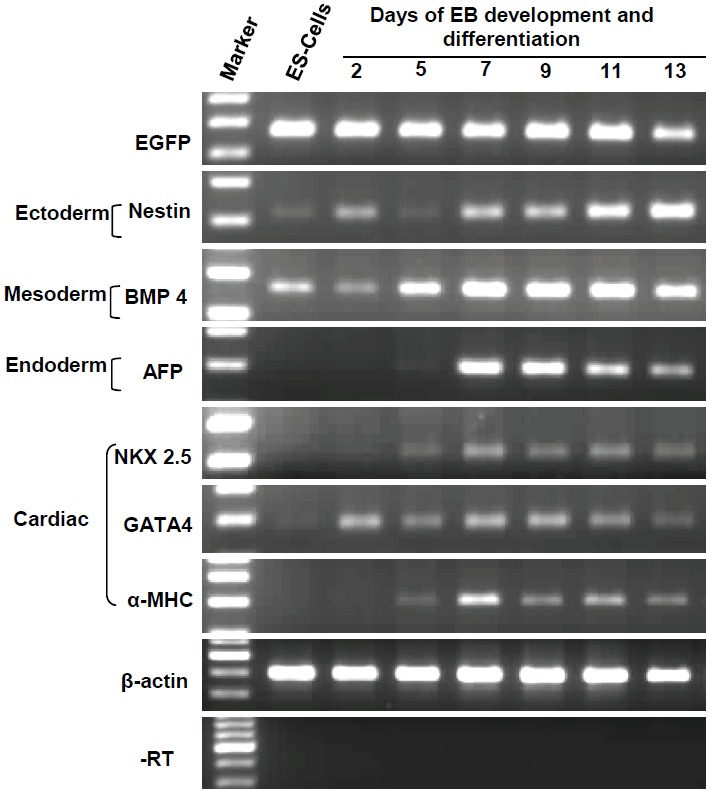
RT-PCR analysis of EBs and their outgrowths during differentiation. Rows (top to bottom): EGFP (267 bp), nestin (213 bp), BMP-4 (260 bp), AFP (199 bp), Nkx 2.5 (217 bp), GATA-4 (186 bp), αMHC (302 bp), β-actin (396 bp) and -RT control (for β-actin amplification primer set). Left two lanes: 100 bp DNA ladder-marker and amplicons from GS-2 ES-cells-derived cDNA.
We also obtained an excellent in vivo evidence for pluripotency of the GS-2 ES-cells by obtaining well-formed teratoma in nude mouse. The formed teratoma (2 X 3 cm) contained three germ layer derived tissues and their differentiated progeny cells. As shown in Figure 4, they revealed ectodermally-derived epidermis (II, A), mesodermally-derived skeletal muscle (II, B) and endodermally-derived respiratory ciliatedepithelium (II, C).
GS-2 ES-cells contribute to embryonic chimera
The GS-2 ES-cells readily formed chimeric blastocysts (17/21; 80%) when they were aggregated with non-compact wild-type FVB/N 8-cell embryos. As shown in Figure 7, following blastocyst differentiation, GS-2 ES-cells were localized exclusively in the ICM. The chimeric blastocyst generation efficiency was dependent on the stoichiometry of ES-cells used for aggregation with non-compact 8-cell embryos and the culture medium used. Following transfer of chimeric blastocysts into uteri of pseudo-pregnant CD-1 recipient mice, developing chimeric fetal inclusions, with green-fluorescence in situ, were observed in the transferred uterine horns (data not shown).
Figure 7.
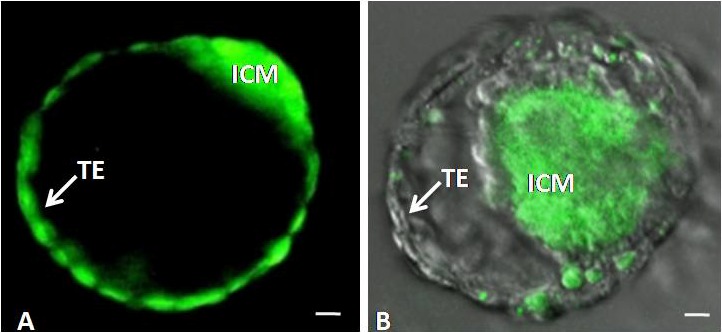
Chimeric blastocyst derived by aggregation of either two EGFP-transgenic 8-cell embryos (A) or wild-type 8-cell embryo and GS-2 ES-cells (B). Abbreviations used: ICM, inner cell mass. TE, trophectoderm. Magnification bars. A, B: 10 μm.
Discussion
Using a two-pronged approach, we successfully derived a robust, constitutively EGFP-transgene expressing permanent GS-2 ES-cell line from the GU3-EGFP-transgenic ‘green’ FVB/N mouse strain [9,11,13]. This involved the use of FVB/N X 129/SvJ hybrid EGFP-expressing blastocysts and an optimized culture system using the RES-GRO™ medium [28]. The newly derived transgenic GS-2 ES-cell line exhibited all defining criteria of a typical-authentic mouse ES-cell line with excellent capability to differentiate to multilineage cell types, both in vitro and in vivo, including chimeric embryogenesis. The GS-2 ES-cell line is the first of its kind derived from the EGFP-transgenic mice in the FVB/N genetic background, the strain known to be most refractory for ES-cell derivation [16]. We believe that the GS-2 ES-cell line is superior to many GFP-expressing ES-cell lines generated by GFP-expression vector-mediated transfection methods, which are variable, unstable and unsuitable so far as the sustenance of fluorescence in long-term (in vivo) studies are concerned.
It appears that the genetic constitution of inbred mouse strains have a strong bearing over successful derivation of an ES-cell line [17,18]. In this regard, the genomic constitution of 129/SvJ strain is highly conducive for ES-cell derivation with high efficiency [18,21]. By crossbreeding approach, the ES-cell derivation enabling property of the 129/SvJ mouse strain could faithfully be transferred to non-permissive genetic strain of mice such as FVB/N. Our observation is consistent with the report on the derivation of NOD/129 hybrid ES-cells [25]. It could be speculated that in the 129/SvJ mouse, vis-à-vis. other strains of mice, the (epi)genetic regulatory mechanisms controlling the desirable level of expression of pluripotency-conferring genes, such as Oct-4, Nanog, Sox-2 is highly conducive for ES-cell derivation and proliferation of ES-cells [13,18]. Our improvised strategy to derive ES-cells from mixed-strains of mice could be useful to carry out immunological and stem cell biology-related mouse experiments that are critically genotype-dependent.
Interestingly, we observed that the RESGRO™ medium selectively supported development and proliferation of blastocysts with excellent ICMs and negligible TBs, post-attachment (see results), unlike the observations made when blastocysts were cultured under conventional ES-cell culture conditions. It appears that the rabbit LIF-transduced rabbit fibroblasts culture conditioned-medium, i.e., RESGRO™ medium [22], is highly supportive of ICM proliferation, while inhibiting TB proliferation. This ES-cell derivation enabling property of the RESGRO™ medium coupled with the use of genetically permissive 129/SvJ female as embryo donor, following cross-breeding with relatively non-permissive male mice (GU3-EGFP transgenic FVB/N male) and timed handling of proliferating blastocyst and picking of ICMs in culture have led to our successful establishment of GS-2 ES-cell line.
We consistently observed a greater propensity of spontaneous cardiac lineage commitment and differentiation of GS-2 ES-cells vis-à-vis wild-type D3 ES-cells (data not shown). Additional striking feature is the almost predominant formation of neural progenitors and neuronal cell types with virtual absence of cardiac clusters when EBs were formed in KOSR medium. The superior property of proliferative and differentiation potential in general and cardiac and neural differentiation (in KOSR condition), in particular, exhibited by the GS-2 ES-cells, could be attributed to the hybrid vigor phenomenon arising from the 129/SvJ X FVB/N derived F1 blastocysts. This is consistent with the reported observation in mice [15].
Because the GS-2 ES-cell line could provide a limitless source of intrinsically green fluorescent-marked stem cells and their-derived lineagespecific progenitors/differentiated cells and because they exhibit greater propensity to differentiate to cardiac and neural lineages, we envisage that the GS-2 ES-cells could potentially be useful in stem cell differentiation biology. These include, (1) use of genetically modified GS-2 ES-cells-derived cell types in experimental cell transplantation studies to understand mechanisms of their structural-functional integration (homing) in a host tissue [6,7]. (2) Use of GS-2 ES-cells for gene-targeting (knock-in/knock-out) studies, for transfection of desired developmentally-regulated genes with different reporter systems and in cell-reconstitution experiments both in vitro and in vivo. (3) In view of their inherent hybrid vigor, the GS-2 ES-cells could potentially be used for improved animal cloning strategy via nuclear transfer or tetraploid blastocyst complementation approaches [14,15] with improved progeny generation rates, surviving through adulthood since most nuclear transfer and cloning experiments involving inbred ES-cells often result in high rates of neonatal mortality, leading to non-viable progeny [14,15].
In conclusion, we demonstrate successful derivation of a permanent EGFP-transgene-expressing ‘GS-2’ ES-cell line using an improved culture system from non-permissive FVB/N mouse strain. The robustly differentiating capability of the GS-2 ES cell line could be exploited for stem cell differentiation studies in vitro and in vivo. The two pronged ES-cell derivation strategy that we developed could be adapted to other difficult strains which could be of immense and versatile use not only in developmental and stem cell biology but also in immunological and oncological research.
Acknowledgements
The authors thank Drs. Jamie Thomson and Ruth Sullivan for interpretation of teratoma tissue sections; Drs. Jerry Schatten and Stacie Oliver for interpretation of mouse karyograms; Dr. Peter Andrews for providing anti SSEA-1 antibodies; Dr. UdayKumar Kolkundkar, Ms. Deepti Abbey and Mr. Sukesh Bhupathy for their technical help; Dr. Panicker M for providing 129/SvJ mice; Dr. Krishnamurthy HS for help in confocal imaging analysis; Ms. Padmavathi MS for help in the preparation of the manuscript. This work was supported by funds from DBT and ICMR, Govt. of India.
Abbreviations
- SSEA-1
stage-specific embryonic antigen
- BMP
bone morphogenic protein
- KOSR
knockout serum replacement
Video1
Video2a
Video2b
Video3
Video4
References
- 1.Palmiter RD, Brinster RL. Transgenic mice. Cell. 1985;41:343–345. doi: 10.1016/s0092-8674(85)80004-0. [DOI] [PubMed] [Google Scholar]
- 2.Capecchi MR. Generating mice with targeted mutations. Nat Med. 2001;7:1086–1090. doi: 10.1038/nm1001-1086. [DOI] [PubMed] [Google Scholar]
- 3.Guan C, Ye C, Yang Xgao J. A review of current large-scale mouse knockout efforts. Genesis. 2010;48:73–85. doi: 10.1002/dvg.20594. [DOI] [PubMed] [Google Scholar]
- 4.Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 5.Zernicka-Goetz M, Pines J. Use of green fluorescent protein in mouse embryos. Methods. 2001;24:55–60. doi: 10.1006/meth.2001.1157. [DOI] [PubMed] [Google Scholar]
- 6.Min J, Yang Y, Converso KL, Liu L, Uhang Q, Morgan JP, Xiao Y. Transplantation of embryonic stem cells improves cardiac function in postinfarcted rats. J Appl Physiol. 2002;92:288–296. doi: 10.1152/jappl.2002.92.1.288. [DOI] [PubMed] [Google Scholar]
- 7.Hedlund E, Pruszak J, Lardaro T, Ludwig W, Viñuela A, Kim KS, Isacson O. Embryonic stem cell-derived Pitx3-enhanced green fluorescent protein midbrain dopamine neurons survive enrichment by fluorescence-activated cell sorting and function in an animal model of Parkinson's disease. Stem Cells. 2008;26:1526–1536. doi: 10.1634/stemcells.2007-0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taketo M, Schroeder AC, Mobraaten LE, Bunning KB, Hanten G, Fox RR, Roderick TH, Stewart CL, Lilly F, Hansen CT, Overbeek PA. FVB/N: an inbred mouse strain preferable for transgenic analysis. Proc Nat Acad Sci USA. 1991;88:2065–2069. doi: 10.1073/pnas.88.6.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devgan V, Thomas M, Ullas KS, Rao MR, Seshagiri PB. Embryo culture-based generation of enhanced green fluorescent protein-transgenic mice. Biochem Biophys Res Commun. 2003;303:994–1001. doi: 10.1016/s0006-291x(03)00483-2. [DOI] [PubMed] [Google Scholar]
- 10.Devgan V, Rao MR, Seshagiri PB. Impact of embryonic expression of enhanced green fluorescent protein on early mouse development. Biochem Biophys Res Commun. 2004;313:1030–1036. doi: 10.1016/j.bbrc.2003.11.184. [DOI] [PubMed] [Google Scholar]
- 11.Devgan V, Seshagiri PB. Successful development of viable blastocysts from enhanced green fluorescent protein transgene-microinjected mouse embryos: comparison of culture media. Mol Reprod Dev. 2003;65:269–277. doi: 10.1002/mrd.10306. [DOI] [PubMed] [Google Scholar]
- 12.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 13.Buehr M, Smith A. Genesis of embryonic stem cells. Philos Trans R Soc Lond B Biol Sci. 2003;358:1397–1402. doi: 10.1098/rstb.2003.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rideout WM, Wakayama T, Wutz A, Eggan K, Jackson-Grusby L, Dausman J, Yanagimachi R, Jaenisch R. Generation of mice from wild-type and targeted ES cells by nuclear cloning. Nat Genet. 2000;24:109–110. doi: 10.1038/72753. [DOI] [PubMed] [Google Scholar]
- 15.Eggan K, Akutsu H, Loring J, Jackson-Grusby L, Klemm M, Rideout WM, Yanagimachi R, Jaenisch R. Hybrid vigor, fetal overgrowth and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc Natl Acad Sci USA. 2001;98:6209–6214. doi: 10.1073/pnas.101118898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buehr M, Nichols J, Stenhouse F, Mountford P, Greenhalgh CJ, Kantachuvesiri S, Brooker G, Mullins J, Smith AG. Rapid Loss of Oct-4 and pluripotency in cultured rodent blastocysts and derivative cell lines. Biol Repro. 2003;68:222–229. doi: 10.1095/biolreprod.102.006197. [DOI] [PubMed] [Google Scholar]
- 17.Kawase E, Suemori H, Takahashi N, Okazaki K, Hashimoto K, Nakatsuji N. Strain difference in establishment of mouse embryonic stem (ES) cell lines. Int J Dev Biol. 1994;38:385–390. [PubMed] [Google Scholar]
- 18.Brook FA, Gardner RL. The origin and efficient derivation of embryonic stem cells in the mouse. Proc Natl Acad Sci USA. 1997;94:5709–5712. doi: 10.1073/pnas.94.11.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki O, Matsuda J, Takano K, Yamamoto Y, Asano T, Naiki M, Kusanagi M. Effect of genetic background on establishment of mouse embryonic stem cells. Exp Anim. 1999;48:213–216. doi: 10.1538/expanim.48.213. [DOI] [PubMed] [Google Scholar]
- 20.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 21.Auerbach W, Dunmore JH, Fairchild-Huntress V, Fang Q, Auerbach AB, Huszar D, Joyner AL. Establishment and chimera analysis of 129/SvEv- and C57BL/6-derived mouse embryonic stem cell lines. Biotechniques. 2000;29:1024–1028. doi: 10.2144/00295st04. [DOI] [PubMed] [Google Scholar]
- 22.Schoonjans L, Kreemers V, Danloy S, Moreadith RW, Laroche Y, Collen D. Improved generation of germline-competent embryonic stem cell lines from inbred mouse strains. Stem Cells. 2003;21:90–97. doi: 10.1634/stemcells.21-1-90. [DOI] [PubMed] [Google Scholar]
- 23.McWhir J, Schnieke AE, Ansell R, Wallace H, Colman A, Scott AR, Kind AJ. Selective ablation of differentiated cells permits isolation of embryonic stem cell lines from murine embryos with a non-permissive genetic background. Nat Genet. 1996;14:223–226. doi: 10.1038/ng1096-223. [DOI] [PubMed] [Google Scholar]
- 24.Kress C, Vandormael-Pournin S, Baldacci P, Cohen-Tannoudji M, Babinet C. Non-permissiveness for mouse embryonic stem (ES) cell derivation circumvented by a single backcross to 129/Sv strain: establishment of ES cell lines bearing the Omd conditional lethal mutation. Mamm Genome. 1998;9:998–1001. doi: 10.1007/s003359900914. [DOI] [PubMed] [Google Scholar]
- 25.Brook FA, Evans EP, Lord CJ, Lyons PA, Rainbow DB, Howlett SK, Wicker LS, Todd JA, Gardner RL. The derivation of highly germline-competent embryonic stem cells containing NOD-derived genome. Diabetes. 2003;52:205–208. doi: 10.2337/diabetes.52.1.205. [DOI] [PubMed] [Google Scholar]
- 26.Yagi T, Tokunaga T, Furuta Y, Nada S, Yoshida M, Tsukada T, Saga Y, Takeda N, Ikawa Y, Aizawa S. A novel ES-cell line, TT2, with high germline-differentiating potency. Anal Biochem. 1993;21:70–76. doi: 10.1006/abio.1993.1458. [DOI] [PubMed] [Google Scholar]
- 27.Bryja V, Bonilla S, Cajánek L, Parish CL, Schwartz CM, Luo Y, Rao MS, Arenas E. An efficient method for the derivation of mouse embryonic stem cells. Stem Cells. 2006;24:844–849. doi: 10.1634/stemcells.2005-0444. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Schoonjans L, Tielens S, Speleman F, Cornelissen M, De Sutter P, Dhont M, Van der Elst J. Culturing in vitro produced blastocysts in sequential media promotes ES-cell derivation. Mol Reprod Dev. 2006;73:1017–1021. doi: 10.1002/mrd.20512. [DOI] [PubMed] [Google Scholar]
- 29.Wray J, Kalkan T, Smith AG. The ground state of pluripotency. Biochem Soc Trans. 2010;38:1027–32. doi: 10.1042/BST0381027. [DOI] [PubMed] [Google Scholar]
- 30.Hogan B, Beddington R, Costantini F, Lacy E. A Laboratory Manual. Cold Spring Harbor, Planiview, NY: Cold Spring Harbor Laboratory press; 1994. Manipulating the mouse embryo; pp. 174–175. [Google Scholar]
- 31.Robertson EJ, editor. Teratocarcinomas and embryonic stem cells, A Practical Approach. Oxford: IRL Press; 1987. Embryo-derived stem cell lines; pp. 71–112. [Google Scholar]
- 32.Wobus AM, Guan K, Yang HT, Boheler KR. Embryonic stem cells as a model to study cardiac, skeletal muscle, and vascular smooth muscle cell differentiation. Methods Mol Biol. 2002;185:127–156. doi: 10.1385/1-59259-241-4:127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


