Abstract
Modern high-throughput HLA and KIR typing technologies are generating a wealth of immunogenomic data with the potential to revolutionize the fields of histocompatibility and immune-related disease association and population genetic research, much as SNP-based approaches have revolutionized association research. The Strengthening the Reporting of Genetic Association studies (STREGA) statement provides community-based data reporting and analysis standards for genomic disease-association studies, identifying specific areas in which adoption of reporting guidelines can improve the consistent interpretation of genetic studies. While aspects of STREGA can be applied to immunogenomic studies, HLA and KIR research requires additional consideration, as the high levels of polymorphism associated with immunogenomic data pose unique methodological and computational challenges to the synthesis of information across datasets. Here, we outline the principle challenges to consistency in immunogenomic studies, and propose that an immunogenomic-specific analog to the STREGA statement, a Strengthening the Reporting of Immunogenomic Studies (STREIS) statement, be developed as part of the 16th International HLA and Immunogenetics Workshop. We propose that STREIS extends at least four of the 22 elements of the STREGA statement to specifically address issues pertinent to immunogenomic data: HLA and KIR nomenclature, data-validation, ambiguity resolution, and the analysis of highly polymorphic genetic systems. As with the STREGA guidelines, the intent behind STREIS is not to dictate the design of immunogenomic studies, but to ensure consistent and transparent reporting of research, facilitating the synthesis of HLA and KIR data across studies.
Keywords: HLA, KIR, data management, data analysis, standards, reporting guidelines, STREIS, STREGA, STROBE
The Goal: Consistency in Immunogenomic Studies Through Community Data Reporting Standards
A consensus is emerging within the genomics community regarding the need for community data-reporting and analysis standards in genetic disease association studies. For example, the Human Genome Epidemiology Network (HuGENet) was established in 1998 by the Office of Public Health Genomics to advance the synthesis and interpretation of population data on human genetic variation in disease association. The publications of the ‘Strengthening the Reporting of Observational Studies in Epidemiology’ (STROBE) and ‘Strengthening the Reporting of Genetic Association studies’ (STREGA) statements(1, 2) represent further advances in these efforts, enumerating specific areas in which adoption of community-based reporting guidelines can improve the consistent interpretation of genetic studies, particularly for genome-wide association studies (GWAS) and meta-analyses of GWAS data. Published studies suggest that implementation of such data-reporting standards for genetic association studies could greatly enhance the utility of individual association studies for the larger community through increased transparency(3-7).
The STROBE and STREGA statements represent significant progress toward the widespread adoption of such reporting standards in the field of genetic epidemiology, and can be applied to gene-based population and evolutionary studies as well. Many of the data-reporting issues described in these statements are pertinent to immunogenetic studies. However, these statements pertain primarily to large cohorts and single nucleotide polymorphism (SNP) based studies. The high level of polymorphism associated with the HLA and KIR loci, the variety of HLA and KIR genotyping systems in use, the complexities of transplantation studies, and the unique role played by the MHC region in predisposition to disease require specific consideration for the development of reporting standards and recommendations that go beyond those defined in the STROBE and STREGA statements.
The HLA and KIR gene regions play pivotal roles in adaptive and innate immunity. Considered together, they constitute the core of the so-called immunogenome, the collection of all immune-related genes(8). As the application of highly automated, high-throughput genotyping systems capable of identifying individual HLA and KIR alleles becomes more accessible, the integrated analysis of these immunogenomic data will become central for transplantation, disease association, and population studies. The HLA community has pioneered the development of community-based standards and guidelines in histocompatibility research and clinical settings, but has not yet achieved a comparable level of standardization for basic immunogenetic studies of populations and diseases. The STROBE/STREGA statement is an important template that can be expanded upon to meet the needs of the immunogenetics and immunogenomics communities. Here, we outline the principle challenges to consistency in immunogenomic studies, in support of the development of an immunogenomic-specific analog to the STREGA statement, STREIS (STrengthening the REporting of Immunogenomic Studies). The proposed STREIS statement presented in Table 1 extends the principles of STREGA statement to immunogenomic studies and, was developed by the immunogenomics data-analysis working group (IDAWG).
Table 1.
Proposed STREIS Reporting Recommendations, Extended from STREGA Statement
| Item1 | Item Number1 | Strobe Guideline1 | STREGA Guideline1 | Extension for Immunogenomic Studies (STREIS) |
|---|---|---|---|---|
| Methods | ||||
| Variables | 7 | (a) Clearly define all outcomes, exposures, predictors, potential cofounders, and effect modifiers. Give diagnostic criteria, if applicable. | (b) Clearly define genetic exposures (genetic variants) using a widely-used nomenclature system. Identify variables likely to be associated with population stratification (confounding by ethnic origin) |
(c)
Describe HLA alleles in accordance with WHO Nomenclature Committee for Factors of the HLA System. Identify the IMGT/HLA Database release number pertinent to the data.
(d) Describe KIR alleles in accordance with the IPD-KIR Database. Identify the IPD-KIR Database release number pertinent to the data. |
| Data sources/measurement | 8 | (a) For each variable of interest, give sources of data and details of methods of assessment (measurement). Describe comparability of assessment methods if there is more than one group. | (b) Describe laboratory methods, including source and storage of DNA, genotyping methods and platforms (including the allele calling algorithm used, and its version), error rates and call rates. State the laboratory/center where genotyping was done. Describe comparability of laboratory methods if there is more than one group. Specify whether genotypes were assigned using all of the data from the study simultaneously or in smaller batches. |
(c)
Provide access to the primary, ambiguous genotype data for each individual.
(d) Describe the system(s) used to store, manage, and validate genotype and allele data, and to prepare data for analysis. (e) Use objective terms, identifying the assessed features of each gene, to describe genotyping systems and genotyping results. Avoid using subjective terms (e.g. low-, intermediate-, high-, or allele-resolution), that that may change over time, to describe genotyping systems and results. (f) Document all methods applied to resolve ambiguity. (g) Define any codes used to represent ambiguities. (h) Describe any binning or combining of alleles into common categories that were performed. |
| Statistical Methods | 12 | (a) Describe all statistical methods, including those used to control for confounding. | State software version used and options (or settings) chosen. |
(b)
Discuss any modifications made to the data in order to have them comport to the expectations of a method for the purpose of analysis.
(c) Document any caveats associated with each analysis as they pertain to immunogenomic data. |
| Discussion | ||||
| Limitations | 19 | Discuss limitations of the study, taking into account sources of potential bias or imprecision. Discuss both direction and magnitude of any potential bias. |
(a)
Discuss the impact of any modifications made to the data for the purpose of analysis.
(b) Discuss any caveats associated with each analysis as they pertain to immunogenomic data. (c) Discuss any potential impact of ambiguity resolution on the results. |
|
Challenges to Consistency in Immunogenomic Data Management
The international nature of the HLA and KIR genotyping communities, and the diversity of research interests within each, have resulted in a proliferation of methods for determining, recording, encoding, storing, and exchanging immunogenomic data (referred to here as immunogenomic data-management). In general, the various data-management methods in use have been designed and implemented independent of each other, and are often difficult to interrelate. In addition, the specifics of the data-management approach applied by a particular investigator or clinical laboratory will often be informed by that group's research goals or clinical responsibilities; a data-management approach favored by one group may be unsuitable for another. Finally, the process of preparing immunogenomic data for analysis or reporting (sometimes referred to as data “cleaning”) often involves the application of assumptions that remain undisclosed, which may not be applicable to all studies or research groups, and of which the general community may not be aware. As a result, the initial challenge to consistency in immunogenomic studies is the inability to ascertain the equivalence of data between research groups.
Here, we discuss some of the key challenges to consistency in immunogenomic data-management.
Immunogenomic Genotype Datasets
Typical HLA datasets for population or disease association studies include genotypes for anywhere from several hundred to a few thousand individuals, determined at between four and nine of the HLA loci, and based on DNA sequence data for one or two exons. The nature of HLA polymorphism, and the limitations of even the most robust genotyping methods (see below), often results in extensive ambiguity for a given sample at one or more loci. That is, for a given HLA locus, each genotyping result for a given sample may include multiple ambiguous genotypes, any of which might include multiple ambiguous alleles. In extreme cases [e.g. HLA-A genotyping results generated using diploid sequence-based typing (SBT) methodology for exons 2 and 3], a genotype for a single individual may include up to 14 ambiguous genotypes, and ambiguous allele strings may be composed of up to 28 alleles. Bone marrow donor registry (BMDR) datasets include HLA genotypes for several million individuals, usually typed at five to six HLA loci using multiple methodologies. Recognizing that the long term storage of HLA data can be challenging in the face of ongoing allele discovery, bone marrow donor registries have established practices for storing primary sequence information for use in matching applications (9).
A KIR dataset will contain genotype data for up to fourteen KIR loci. These data are often a combination of presence/absence typing at some KIR loci, combined with allelic typing at other KIR loci. As with the HLA data, the allelic KIR data may be composed of ambiguous genotypes and alleles. While the issue of ambiguity within HLA and KIR data is well known in the immunogenetic research community, the lack of standard protocols for recording and resolving these ambiguities can lead to major inconsistencies between data sets. Additional inconsistency may be present due to temporal changes to HLA and KIR nomenclature and differences in the sets of alleles that can be distinguished by different typing methods. As a result, it is often difficult or impossible to equate and compare data sets generated by different groups using different genotyping techniques at different time points.
HLA Nomenclature
The WHO Nomenclature Committee for factors of the HLA system has developed a hierarchical nomenclature for the representation of the complex polymorphisms that comprise HLA alleles(9-15). This nomenclature incorporates considerable information about HLA polymorphism into an allele name, and its development parallels improvements in the scope and performance of HLA genotyping methods over the last two decades. In 1987, HLA allele names were defined as four-digit numbers that represented a unique protein sequence; the first two digits described each allele's serologic specificity and the last two digits identified successive protein variants within that specificity(9). In 1990, the HLA nomenclature was updated to include a fifth digit, representing synonymous nucleotide polymorphisms(16). In 2002, the nomenclature was expanded to include four pairs of digits representing serological, protein, synonymous and non-coding nucleotide polymorphisms(14). In 2010, the nomenclature was updated to accommodate more than 99 variants in each of these polymorphic domains by explicitly defining each domain as a colon-delimited field(15).
Since 1999, the sequence definitions for each HLA allele and the nomenclature rules governing their names have been integrated in the ImMunoGeneTics (IMGT)/HLA Database (17-19). New versions of this database, released every three months, reflect current HLA sequence diversity and nomenclature rules, and are associated with a specific release number (e.g. the July 2011 release is Version 3.5.0). Each release number provides an unambiguous reference to a specific set of HLA allele sequences named under a specific set of nomenclature rules. For example, except where noted, the HLA alleles discussed here are included in release 3.5.0.
With each refinement to the nomenclature, the names of some alleles were changed (e.g., in 2002 as part of IHGT/HLA Database release number 1.16, B*1522 was changed to B*3543 after DNA sequences outside of exons 2 and 3 became available(14)). In the most recent major nomenclature refinement (release number 3.0.0), many allele names were changed in non-obvious ways (e.g., DPB1*0502 was changed to *104:01 when the allele names at the DPB1 locus were reorganized in relation to the order in which each unique DPB1 protein sequence was identified(14)), and the locus identifier for HLA-C locus alleles changed from Cw* to C*.
Such nomenclature refinements can have ramifications for the interpretation of allele names. For example, the concept of serologic specificity as such does not hold for the DPB1 locus; with only two exceptions (DPB1*02:01 and *02:02, and *04:01 and *04:02) the first field of the DPB1 allele name alone is sufficient to identify a unique protein sequence.
These nomenclature changes may not always be applied in a consistent manner to preexisting data. Even simple allele name changes (e.g., from 01011 to 010101) become time consuming when they must be made to large datasets, and non-obvious changes (e.g., changing B*1522 to B*3543) may not be adopted at all. As a result, HLA data reflecting older nomenclature versions may continue to be analyzed, published, and circulated long after a nomenclature change has occurred. As the IMGT/HLA Database release number for HLA data is often not recorded explicitly or reported in published datasets, incompatible HLA data (e.g., a dataset including both the B*1522 and B*3543 alleles) may be included in the same analysis, especially when datasets are combined, or when new genotypes are added to an existing dataset.
HLA Genotyping Ambiguity
The HLA region is characterized by extensive and repeated gene duplication, recombination and gene conversion events that have resulted in a complex ‘patchwork’ of polymorphic sequence motifs(20). Long before the advent of copy number variation (CNV) studies(21), the structural variation of the MHC region was well established in the HLA-DRB gene family(22). The high homology among HLA genes and the extensive polymorphism in exons that encode the peptide binding domain (PBD) pose challenges for the identification of individual HLA alleles in stem cell transplantation, population and epidemiologic studies. HLA alleles are defined by the variants at a number (~50 – 100) of polymorphic amino-acid positions in a specific HLA gene. It is often cost-prohibitive to identify all of the polymorphisms that distinguish closely-related alleles, and due to the patchwork of polymorphic motifs it is sometimes impossible to distinguish between genotypes that present identical sets of heterozygous polymorphisms. As a result, while an individual has only two HLA alleles per locus, many HLA genotyping results can be ambiguous in that the results for a given sample may be consistent with more than two potential alleles at a given locus.
Currently, SBT systems that assess the sequence of exon 2 for class II loci, and exons 2 and 3 for class I loci represent the gold-standard for reporting genotypes with minimal ambiguity. In the context of a given IMGT/HLA Database release, genotyping systems that assess fewer polymorphisms will generate HLA genotype data with greater levels of ambiguity, while those SBT systems that assess additional exons may report less ambiguity.
In addition, the potential for ambiguity increases as innovation in typing methodology extends knowledge of polymorphism to more regions (exons, introns, etc.) of a given gene; a genotype that may have been unambiguous in the context of one IMGT/HLA Database release may be ambiguous in the context of later releases (e.g. the DRB1*14:01:01 and *14:54 alleles, discussed below) as the result of previously undetectable polymorphisms. Knowledge of new polymorphisms in previously unassessed gene regions is necessary for researchers to be able to determine equivalence between older and more recent typing data.
Allele ambiguity results when the polymorphisms that distinguish alleles fall outside of the regions assessed by the genotyping system. For example, the nucleotide sequence of DRB1*14:01:01 differs from *14:54 allele at cDNA nucleotide position 421 in exon 3. If a DRB1 typing system does not assess exon 3, these two alleles cannot be distinguished, and an ambiguous allele, DRB1*14:01:01/14:54, will be reported. In some cases, ambiguous alleles can consist of large numbers of possible alleles. In an extreme example, twenty-eight HLA-A alleles (A*02:01:01:01, *02:01:01:02L, *02:01:01:03, *02:01:08, *02:01:11, *02:01:14Q, *02:01:15, *02:01:21, *02:01:48, *02:01:50, *02:09, *02:43N, *02:66, *02:75, *02:83N, *02:89, *02:97:01, *02:97:02, *02:132, *02:134, *02:140, *02:241, *02:252, *02:256, *02:266, *02:291, *02:294 and *02:305N) share the same nucleotide sequence over HLA-A exons 2 and 3, and cannot be distinguished by genotyping systems that assess only these exons.
Genotype ambiguity results from an inability to establish chromosomal phase between identified polymorphisms. For example, the DRB1*04:01:01+DRB1*13:01:01, DRB1*04:01:01+DRB1*13:117, DRB1*04:13+DRB1*13:02:01, DRB1*04:14+DRB1*14:21, DRB1*04:35+DRB1*13:40 and DRB1*04:38+DRB1*13:20 genotypes cannot be distinguished by comparing DRB1 exon 2 nucleotide sequences, and a DRB1 SBT genotyping system that assesses only exon 2 will report the set of these six individual genotypes as a single ambiguous genotype. SBT genotyping systems that assess class I exons 2 and 3, or class II exon 2 sequences can report ambiguous genotypes composed of up to 14 or 15 individual genotypes (e.g. for HLA-A, HLA-B and DRB1).
The number of characters used to represent an ambiguous allele is often reduced by representing the string of alleles as a specific code. For example, “G groups” include all class II alleles that share the same exon 2 nucleotide sequence and class I alleles that share the same exon 2 and 3 sequence, and “P groups” include all alleles that encode the same PBD. The number of individual genotypes in the ambiguous genotypes discussed above includes ambiguous alleles represented as G groups. When these G grouped alleles are considered separately, the number of individual genotypes can increase dramatically (e.g. from 14 to 153 individual HLA-A genotypes). The DRB1*14:01:01/14:54 ambiguity can be represented as the DRB1*14:01:01G “G group”, and the more ambiguous DRB1*14:01:01/14:01:02/14:01:03/14:54 allele can be represented as the DRB1*04:01P “P group”. The National Marrow Donor Program (NMDP) uses an allele code system that assigns specific alphabetic strings to specific ambiguities that include only peptide-level allele-names. Using this system, the ambiguous DRB1*14:01:01/14:01:02/14:01:03/14:54 allele can be recorded as the DRB1*14:BCAD NMDP allele code. Some research groups implement their own coding systems for these purposes.
Ambiguous genotypes can also be represented in a variety of ways. In some cases, all of the alleles in a particular allele group are combined into a single allele code, so that all of the individual genotypes are “collapsed” into only two allele codes. For example, using NMDP allele codes, the ambiguous DRB1*04:02:01+DRB1*11:20, DRB1*04:14+DRB1*11:16 genotype could be represented as the DRB1*04:BK+DRB1*11:YR genotype. This practice of collapsing ambiguous genotypes to a pair of allele codes actually increases the ambiguity of a typing, because not all genotypic combinations implicit in a given allele code are possible in a single typing result; the information that excludes specific allele combinations is lost in the collapse. In the above example, in addition to the two reported possible results for the ambiguous genotype, the single collapsed genotype also includes two additional genotypes which were excluded in the original typing (i.e., DRB1*04:02:01+DRB1*11:16 and DRB1*04:14+DRB1*11:20).
The presence of allelic and genotypic ambiguity has resulted in the use of the term “typing resolution” to describe the extent of ambiguity in genotyping methodologies and genotyping results . For example, so-called “allele resolution” or “high resolution” typing methods generate “allele resolution” or “high resolution” genotyping results characterized by lower levels of ambiguity than so-called “intermediate resolution” or “low resolution” results generated by “intermediate resolution” or “low resolution” methods.
However, there are no universally recognized definitions for the various terms used to describe typing resolution. Hurley et al (23) discuss low, intermediate, and high level typing in terms of the number of alternative alleles included in a typing (i.e., the extent of ambiguity) and the number of allele name fields that can be distinguished (e.g. A*01:01:01:01 vs. A*01:01), and acknowledge that it is difficult to precisely define these terms. Alternatively, the 2010 European Federation for Immunogenetics (EFI) Standards for Providers of External Proficiency Testing defines high resolution typing as assessing, at a minimum, differences in exons 2 and 3 for class I loci and exon 2 for class II loci(24). The American Society for Histocompatibility and Immunogenetics (ASHI) Harmonization of Histocompatibility Typing Terms Working Group has recently developed definitions for low, high, and allelic resolution molecular typing results (25). This working group defines a high resolution typing result as a set of alleles that share the same protein sequence for the PBD and that are expressed on the cell surface (e.g. excluding null alleles), and an allelic typing result as being consistent with a single allele in the context of a specific WHO HLA nomenclature report. While the both the EFI and ASHI definitions of high resolution pertain to PBD polymorphisms, the distinctions between them are significant, and a high resolution typing as defined by EFI may involve less ambiguity than a high resolution typing as defined by ASHI. For example, DRB1*14:01:01/14:54 is a high resolution result under the EFI definition, while DRB1*14:01:01/14:01:02/14:01:03/14:54 is a high resolution result under the ASHI definition.
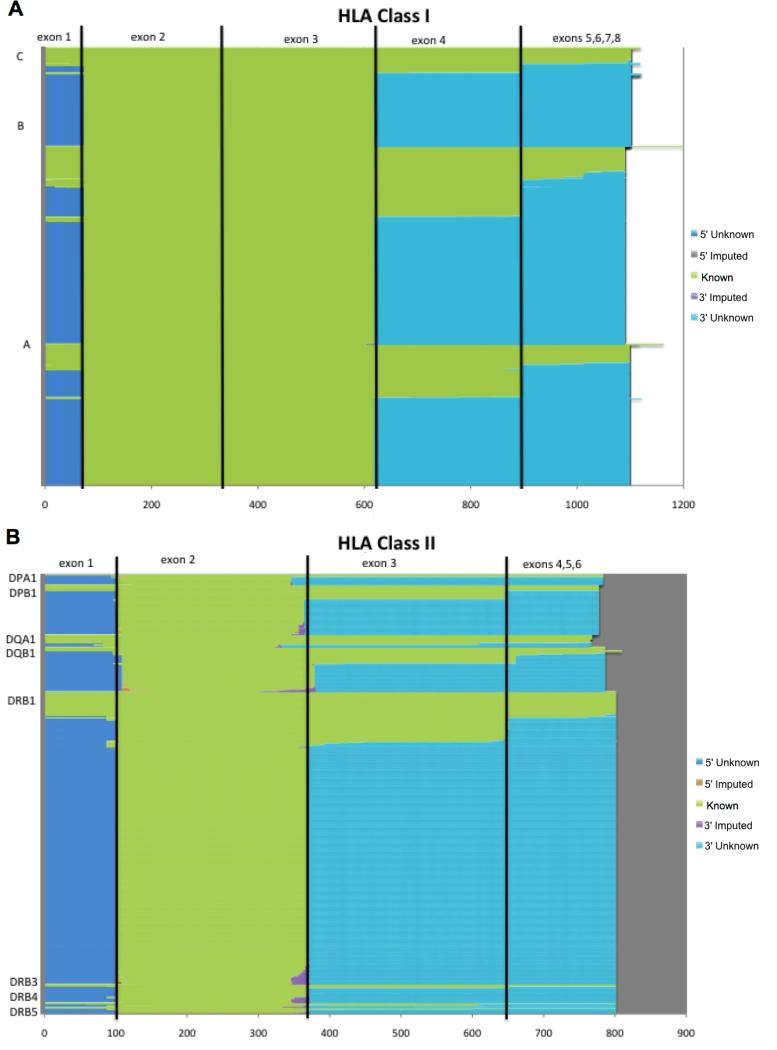
In addition to being poorly-defined and often conflicting, these typing resolution terms have the potential change over time as new alleles are identified. As sequences from additional exons and introns, as well as upstream and downstream regions, are incorporated into the IMGT/HLA Database, a typing result described as “allele resolution” in the context of one IMGT/HLA Database release may become a “high resolution” type with a subsequent release, and an “intermediate resolution” type with later releases. Figure 1 illustrates the extent to which the exon sequences for current classical HLA allele alleles are known, unknown and imputed. The number of distinct alleles will only increase as exon sequences that are currently unknown are determined. Similarly, knowledge about the extent to which an allele is or is not expressed may change as well. The confusion that arises from the lack of standardized terminology results in difficulty for researchers and clinicians in the immunogenetics field in their ability to synthesize and equate HLA genotyping results across typing methodologies. These difficulties are likely even more pronounced for investigators from other disciplines, who may be less familiar with this issue. One approach for addressing this problem of typing resolution terminology is to use objective descriptions that identify the regions of the gene that were assessed by the typing system used. Where a “high resolution” typing result may someday become an “intermediate resolution” result, an “exons 2 and 3” typing result or a “PBD resolution” typing result will not change.
Figure 1. Known, Unknown and Imputed Exon Sequences for Classical HLA Genes.
Sequence categories for all HLA-A, B, C, DPA1, DPB1, DQA1, DQB1, DRB1, DRB3, DRB4, and DRB5 alleles in IMGT/HLA Database release 3.2.0 are color coded. Known sequences are shown in green, unknown sequences that are 5′ of the peptide binding domain (PBD) encoding exons (exons 2 and 3 for class I, and exon 2 for class II alleles) are shown in blue, imputed sequences that are 5′ of the PBD exons are shown in red, imputed sequences that are 3′ of the PBD exons are shown in purple, and unknown sequences that are 3′ of the PBD exons are shown in aqua. The horizontal scale indicates the distance in nucleotide bases from the start codon.
A. Sequence categories for 4268 class I alleles over exons 1-8.
B. Sequence categories for 1248 class II alleles over exons 1-6.
Resolving Ambiguity
Most population genetic and epidemiologic analytical methods, and the tools that implement them, require genetic data that reflect the diploid genetic state of two alleles per locus per individual. Therefore, in the case of HLA genotype data, allele and genotype ambiguities must be “resolved” in order to make diploid allele assignments. There are currently no standards for making these allelic assignments, or even reporting how they were accomplished; they are based on the individual investigator's accumulated and empirical knowledge of the data under study. This knowledge often derives from the availability of allele-frequency distributions in different ethnic groups, established patterns of linkage disequilibrium (LD) between alleles(resulting in haplotypes) (26), and the categorization of individual alleles and haplotypes as being either common (likely to be observed) or rare (unlikely to be observed) (27, 28). However, in the absence of a consistent approach for applying this knowledge, different allelic assignments may be made by different investigators considering the same data. Furthermore, the lack of reporting standards for immunogenetic data means that the HLA literature often contains no reference to the methods used for ambiguity resolution, or even to the application of these methods in a given study. While most researchers in the field are aware that some degree of ambiguity resolution necessarily occurred prior to publication of a ‘clean’ (completely diploid and unambiguous) dataset, the precise methods applied will remain a mystery. More problematically, investigators less familiar with the HLA system may be unaware that any ambiguity resolution methods were applied prior to publication of the data.
The categorization of alleles into common and rare categories is facilitated by the availability of HLA data for large numbers of populations worldwide. In 2007, Cano et al.(28), defined the 2285 HLA-A, -C, -B, -DRB1, -DQA1, -DQB1 and -DPB1 alleles in IMGT/HLA database release 2.15 as being either “common” (with allele frequencies > 0.001 in reference populations), “well-documented” (observed in at least three unrelated individuals), or “rare” (frequencies or observations too low to be included in the other categories), and identified 693 alleles (30%) as being common and well-documented (CWD) and 70% as being rare. The Rare Alleles Project of the Allele Frequencies Net Database (AFND) (27) has built on these CWD definitions by integrating allele-count data from the AFND and NMDP database with genotyping reports from 53 participating laboratories. This expanded reference dataset can be queried to report the number of times any of the 5827 classical HLA alleles in IMGT/HLA database release 3.3.0 have been observed(29), and reveals that of the 5799 observed HLA-A, -C, -B, -DRB1, -DQA1, -DQB1 and -DPB1 alleles, 2958 (63%) are rare (observed less than 3 times), and are unlikely to be present in most datasets. However, current typing methods do not distinguish common from rare alleles. As a result, much of the ambiguity present in HLA genotypes is due to rare alleles, and much of the effort of modern sequence-based genotyping methods is the detection or exclusion of rare alleles.
Furthermore, the categorization of an allele as common or rare may change as typing methodologies improve. For example, many specimens that were originally genotyped as having the DRB1*14:01:01 allele have recently been recognized as having *14:54 alleles; in some cases, 88-100% of DRB1*14:01:01 alleles have been reassigned as *14:54 alleles(30, 31). As the number of exons that can be assessed in each gene by modern HLA genotyping systems increases, it seems likely that such inversions in the perceived frequencies of well-known and more-recently identified alleles will become more common (e.g., the A*11:02:01 and A*11:53 alleles may prove to be a second example where this occurs), requiring the reassessment of much previously published data, with the potential for reassignment of alleles.
It seems likely that there will always be variation in the ability of different typing systems to detect and discriminate between closely related alleles. As long as the approaches used by individual investigators to make any necessary allele assignments and report genotype data continue to be applied in the absence of a reporting standard, the reported genotype for a given specimen may vary between different laboratories, and between studies carried out at different times. Inconsistency in the detailing and disclosure of the manner in which HLA genotyping data have been generated and managed undermines the capacity of researchers to determine whether significant (or non-significant) findings for HLA data are consistent across multiple studies; such inconsistency also greatly diminishes the ability to replicate those studies and pool data resources.
KIR Genotyping
The many technical challenges to the analysis of KIR data derive from the structural variation of the region. KIR haplotypes include 7-12 polymorphic genes; the minimal KIR haplotype (haplotype A), contains 7 expressed KIR genes and two pseudogenes. KIR A haplotypes include only one activating gene (KIR2DS4), and KIR B haplotypes generally include more than one activating gene(32, 33). Like HLA, KIR alleles are highly polymorphic (http://www.ebi.ac.uk/cgi-bin/ipd/kir); in addition to heterogeneity in gene content and allelic variation, KIR cell-surface expression is variable, and may be allele-specific. Furthermore, the extensive KIR locus- and haplotype-level polymorphisms include locus-level CNVs. For example, the locus KIR2DS3/S5 can be found in the centromeric or telomeric regions of KIR haplotypes or in both portions(34). Additional copy number diversity has been detected primarily through a limited number of family studies(35); while its extent is unclear, the issue of locus copy number will be clarified with the advent of improved allele-level typing. For example, allowing specific KIR2DS3/S5 alleles to be assigned to a centromeric or telomeric KIR2DS3/S5 locus.
The extensive sequence homology among the KIR loci has resulted in a variety of KIR typing methods that generate KIR data at a range of levels. The level of typing currently reported and analyzed in the literature is predominantly the presence or absence of KIR loci (locus-level typing). Most laboratories use 3-5 different molecular techniques [e.g., multiple PCRs, cloning and/or sequence specific oligonucleotide probe (SSOP), sequence-specific priming (SSP) and sequencing assays] to accomplish allele-level typing. Even with these efforts, allele-level KIR genotyping discrepancy rates between laboratories can be very high; the large number of exons that must be typed results in extensive ambiguity.
Continuing new allele discovery has also presented difficulties for the allele-level typing of KIR. In a preliminary study of allele-level KIR typing performed by the NMDP, greater than seventy new alleles were discovered in the typing of 435 samples. Finally, allele and genotype ambiguities also pertain to allele-level KIR typing, and as with HLA genotyping ambiguities, KIR genotyping ambiguities are sometimes resolved to individual allelic assignments through the use of observed allele-frequency distributions in different populations (e.g., using the KIR Allele/Gene Frequency section of the AFND(27)). Compounding these issues, nomenclature inconsistencies (e.g., distinct ‘loci’ that segregate as alleles, or duplicate loci on the centromeric and telomeric ends of the cluster) confound consistent interpretation of results by the KIR community, and can introduce analytical biases. For example, KIR2DL2 and KIR2DL3 are the major allelic groups of one locus; these allelic classes were previously thought to be separate loci, and are still often treated as such in the literature and public databases.
As with the IMGT/HLA Database for HLA, the Immuno Polymorphism Database (IPD) – KIR Database is a central repository for KIR sequence and nomenclature information(36) and each IPD – KIR Database release number provides an unambiguous reference to a specific set of KIR allele sequences named under a specific set of nomenclature rules.
Challenges to the Consistent Application of Analytical Methods for Immunogenomic Data
The methods routinely used for the analysis of immunogenomic data were originally developed and implemented for data characterized by low numbers of variants at small numbers of loci and levels of polymorphism that were characteristic of molecular and serological immunological data at the time. More-recently developed genomic analysis methods and applications (e.g., PLINK(37)) have been developed specifically for use with SNP data generated through GWAS and CNV data derived from SNPs. These data differ significantly from immunogenomic data in that they are minimally polymorphic across many hundreds of thousands of loci, LD between all but the most proximate loci is negligible and, for the most part, the variants have no known function(38). As a result, these recently developed methods cannot easily be applied to the analysis of HLA and KIR data.
Because modern immunogenomic data are characterized by high levels of polymorphism across large numbers of functionally related loci with relatively high LD, the dilemma facing the modern immunogenomic investigator is that neither modern genomic analytical methods nor historically-employed genetic methods are ideally suited for the analysis of their data. For immunogenomic investigators, the primary obstacle to analytical consistency is the documentation of the data processing that must be done in order to perform analyses. This is especially true when using methods that were developed for use with less polymorphic data.
In this section, we provide a brief overview of some of the challenges to consistency in immunogenomic data-analyses. Additional details can be found in these references ((39-44)).
Heterogeneity in Data Management
In any study, the primary data being analyzed are assumed to be valid and reliable. However, in addition to the specific issues regarding data ambiguity and nomenclature detailed above, the complex structure of HLA allele names renders these data prone to specific types of errors; manual data entry and modification of allele names can result in transcription errors, and commonly used spreadsheet applications routinely introduce a variety of errors (e.g., 01010101 might be presented as 1010101 or 1,010,101, and 01:60 may be presented as 0.0833333333333333), as they were not designed for HLA data-management. Before any data analysis can be undertaken, internal quality control (QC) measures (e.g., validating allele-names against a specific nomenclature release) must be applied to ensure the integrity of the primary data.
For the purposes of comparing and combining different study results and larger scale meta-analyses, there must be appropriate binning (equating allele assignments made using different data-management methods) of alleles if the same set of alleles was not detectable in all populations, or if the data were generated at different times (i.e., under different nomenclature versions) or by different groups(45). For example, the DRB1*14:01:01 and DRB1*14:54 alleles in two datasets would be binned into a common allele-category for meta-analysis if it was not clear that DRB1*14:54 had been excluded in the typing of DRB1*14:01:01. Failure to account for this methodological heterogeneity can lead to errors in association studies, haplotype estimates, and genetic distance measures, and spurious results can ensue when datasets are combined without careful consideration of this heterogeneity.
Most statisticians, even those familiar with HLA or KIR nomenclature, do not know the specifics of the typing systems used, and will not always recognize when appropriate binning of data is necessary. For example, HLA data available from many public databases, as well as online material supplementing published studies, are being studied by groups unfamiliar with HLA. These meta-analytical studies are most prone to potential errors in the absence of a standardized approach for documenting the management of HLA genotype data.
Low-Frequency Alleles
Low-frequency alleles (generally with frequencies less than one to two percent, depending on the size of the dataset) typify highly polymorphic immunogenomic data, and present challenges to the interpretation of haplotype estimates, measures of LD and association testing in case/control studies. Failure to properly account for low-frequency alleles can lead to spurious or meaningless results. For tests with explicit distributional assumptions (e.g., χ2 tests), low-frequency alleles and genotypes affect the validity of asymptotic approximations for the test statistic(40). While the impact of low-frequency alleles may not be problematic for analyses of individual SNPs or other loci with low levels of polymorphism where constraints on minor allele frequencies to be included in analyses are imposed, this issue must be addressed for polymorphic loci such as HLA, KIR, and SNP haplotypes.
Haplotype Estimation
Estimated haplotypes and haplotype frequencies play a central role in most genetic studies. Haplotype-level analyses are important to studies of the etiology of human disease, selective forces acting on populations, and optimal sizes for bone-marrow donor registries (BMDRs). Associations between markers and disease loci that are not evident with a single-marker locus may be identified in multi-locus marker analyses using estimated haplotype frequencies (HFs). However, the diversity and complexity of immunogenomic data pose challenges for haplotype estimation. For both KIR and HLA, the frequency of the alleles, the sample size of the dataset, the various levels of missing information, and the various levels of LD influence the accuracy of the estimation(42, 44). In addition, the high levels of allele diversity at these loci may result in more haplotype parameters being estimated than the number of actual phenotypes observed.
Linkage Disequilibrium
Extensive LD has been documented between loci across the 1MB KIR and 3.6MB MHC regions. This LD requires pairwise computation of association measures between as many as fourteen (e.g., in the KIR complex) highly polymorphic loci(39, 42, 43). Standard pair-wise measures of LD have multi-allelic extensions for use with highly polymorphic data, however these measures may not always capture important aspects of associations due to implicit assumptions of symmetry. For example, Wn, a multi-allelic extension of the r2 LD correlation measure, is always symmetric with respect to two loci; however, the number of alleles reported at each locus can differ considerably leading to asymmetries. Likewise, the other commonly used measure of overall LD, D’, is highly sensitive to the presence of rare alleles, a major feature of immunogenomic loci. It is therefore important to explore results from a variety of LD measures and not to over interpret any specific LD measure for a locus pair with highly asymmetric numbers of alleles.
Association Studies
The characteristic features of immunogenomic data require careful consideration in disease association studies, Low-frequency alleles pose a challenge in traditional association (case/control) tests in disease studies that use the χ2 test to compare cases and controls(39, 41). The χ2 test has been the standard approach at the overall locus-level, but this test can lead to false acceptance or rejection of the null hypothesis when the expected genotype counts in a contingency table are small (aka, “sparse cells”); the χ2 test is inappropriate if any expected count is substantially less than 1, or if more than 20% of the cells in a contingency table have expected counts less than five. For example, it is not unusual for 30 or more alleles to be observed at the HLA loci, with a wide range of frequencies, resulting in many sparse cells. The standard approach in these cases is to create combined classes of low-frequency alleles. However, there are alternatives to this approach; genotypes containing low-frequency alleles may be entirely excluded from analysis, or genotypes can be analyzed hierarchically, beginning with the most common alleles and creating combined categories of these alleles with successively less common alleles. Nonparametric resampling and exact tests may be alternatives in some cases, although their application can also be problematic for high dimensional tables with sparse cells. There are currently no standard recommendations for the application of these procedures. In addition, the large number of alleles at immunogenomic loci requires specific corrections for multiple comparisons; where these corrections depend on the number of markers tested in GWAS, they may also depend on the number of alleles tested at each marker in immunogenomic association studies.
The issue of potential population stratification is also important to consider in association studies for immunogenetic data, since both selection and demography have shaped allele frequency distributions within specific ethnic and geographic groups. If samples are not collected with scrupulous attention to homogeneity of ancestry background, investigators run the risk of misinterpreting genetic differences between cases and controls. In these cases, heterogeneity between cases and controls due to allele frequency differences related to population stratification may be mistaken for association with a particular locus(46). Genome wide markers can be helpful to detect such stratification issues.
Hardy-Weinberg Testing
The Hardy-Weinberg (HW) principle provides a useful model for primary QC verification of the integrity of genotype data, as genotyping errors may result in both individual genotype deviations and overall deviations from HW equilibrium (HWE). Confidence in the accuracy of HW testing is therefore crucial for confidence in subsequent analyses, as many analytical methods (e.g., haplotype estimation) are predicated on an assumption of HW equilibrium in the data set. HW testing can be particularly challenging for highly-polymorphic datasets(40, 44). As with association studies, the χ2 test has been the standard for testing fit to HWEP. However, while the minimum number for an observed genotype must be 1, the minimum number of expected genotypes under HWEP can be much less than one; this can result in what appear to be significant deviations from HWEP where a genotype with a fractional expected count is observed only once. Three approaches can be taken to increase the accuracy of the test: (1) low-frequency alleles can be combined in a unified class for the χ2 test to be effective; (2) an exact test enumerating all possible tables of all possible genotypes consistent with observed allele frequencies can be undertaken, or (3) approximations to such complete enumerations can be made via resampling.
Phylogenetic Analysis
Phylogenetic trees (or dendrograms) generated using allele and haplotype frequencies have been used to investigate population relationships and test hypotheses regarding human history and migration. However, many of the applications commonly employed to carry out these analyses do not have the capacity to consider the extent of sequence differences between alleles, and as a result, alleles that are related by descent will be evaluated in the same manner as alleles that are un-related. For example, while the DRB1*08:07 allele differs from the *08:02:01 allele by a non-synonymous nucleotide replacement in codon 56, and is thought to be derived from *08:02:01, these alleles are considered to be as distinct from each other as each is from the DRB1*01:01:01 in most frequency-based phylogenetic analyses, and any information about the relatedness of these alleles that might be derived from their sequences is ignored. The binning of allele names below the exon level may be an acceptable approach for low-frequency alleles that are related by descent, but is not appropriate for more common alleles.
Summary
In conclusion, the large number of alleles per locus (often > 50) and high haplotype diversity (often > 1000) at the HLA and KIR systems present specific, persistent challenges in immunogenomics research. At the same time, these features are the very reason why immunogenomic loci should be treated as model systems for genomic research; if these immunogenomic challenges can be met and overcome, the analytical challenges of less complex genomic regions will seem mundane. Over the past thirty years, the immunogenomics community has seen an exponential increase in the number of recognized HLA alleles, leading to regular nomenclature revisions. This phenomenon now extends to the KIR genes(47). Furthermore, in addition to the continual discovery of new alleles, heterogeneity of typing resolution, typing techniques and allele nomenclatures are common to both the MHC and KIR regions. In addition, KIR and HLA data are very sensitive to ethnic background diversity. The potential for population sub-structure is particularly relevant for immunogenomic data, as both MHC and KIR genes carry signatures of both the selective and demographic histories of human populations. These issues are exacerbated in BMDRs where sample sizes for specific research questions are often very large (>100,000). Taken together, these features support the idea that the “MHC continues to provide new insights and remains in the vanguard of contemporary research in human genomics”(48).
While the conclusions of genetic data analyses are inextricably linked to the criteria used for data generation and management, a consistent and integrated data management and analysis approach is currently absent from immunogenomics research, limiting the ability to pool data resources and accurately interpret results across studies. The goal of the immunogenomics data-analysis working group (IDAWG) is to develop consensus-based community data standards for the HLA and KIR gene systems. The integration of standards for both immunogenetic systems will allow for consistent, reproducible, and easily combined analyses for each system, and will facilitate the analysis of KIR and HLA interactions.
The first step in achieving this approach is to build on the principles outlined in the STREGA statement and develop a set of community-based documentation guidelines intended to strengthen the reporting of immunogenomic studies. Consistent reporting of the manner in which the data in immunogenomic studies are managed and analyzed will facilitate the reproducibility of studies, enhance data-sharing and meta-analyses and make immunogenomic research more accessible to the larger genomics community. While the guidelines described in the STROBE and STREGA statements need to be applied to immunogenomic studies, a STREIS statement is also needed to extend these guidelines as described in Table 1.
It is a basic tenet in scientific research that the laboratory methods employed to generate data must be described in sufficient detail to enable reproduction of the reported results. With the advent of genotyping methods that generate enormous quantities of complex data that are stored and transmitted electronically, and are publically available, the specific and detailed reporting of data management and analysis methods is equally necessary. Immunogenomic typing and analysis results that do not meet these minimum standards for documentation should be treated as invalid, and should be recognized as such during the peer-review process. Strict adherence to these guidelines by the editorial staff and expert reviewers for the major journals in the field would allow these standards to be implemented consistently. Furthermore, journals publishing histocompatibility, immunogenetics, and immunogenomics studies should consider the joint data archiving policy adopted by journals in the fields of ecology and evolution, which requires that published data be deposited in public digital archives (49).
The time has come for the consistent application of these STREIS principles to immunogenomic data management and analysis. We hope that this commentary will initiate a community-wide discussion of this issue that will lead to a consensus standard as recently promoted by Nature Genetics (50). The IDAWG has initiated a 16th International HLA and Immunogenetics Workshop project for the development of this STREIS statement, and the Histocompatibility and Immunogenetics community is invited to participate. More information can be found online at www.immunogenomics.org.
Acknowledgements
This work was supported by National Institutes of Health (NIH) grants U01AI067068 (JAH, SJM) and U19 AI067152 (PAG) awarded by the National Institute of Allergy and Infectious Diseases (NIAID) and by NIH/NIAID contract AI40076 (RMS and GT). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health. Australian governments fully fund the Australian Red Cross Blood Service for the provision of blood products and services to the Australian community.
REFERENCES
- 1.von Elm E, Altman DG, Egger M, Pocock SJ, Gotzsche PC, Vandenbroucke JP. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Epidemiology. 2007;18:800–4. doi: 10.1097/EDE.0b013e3181577654. [DOI] [PubMed] [Google Scholar]
- 2.Little J, Higgins JP, Ioannidis JP, et al. Strengthening the reporting of genetic association studies (STREGA): an extension of the STROBE Statement. Hum Genet. 2009;125:131–51. doi: 10.1007/s00439-008-0592-7. [DOI] [PubMed] [Google Scholar]
- 3.Khoury MJ, Little J, Higgins J, Ioannidis JP, Gwinn M. Reporting of systematic reviews: the challenge of genetic association studies. PLoS Med. 2007;4:e211. doi: 10.1371/journal.pmed.0040211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khoury MJ, Little J, Gwinn M, Ioannidis JP. On the synthesis and interpretation of consistent but weak gene-disease associations in the era of genome-wide association studies. Int J Epidemiol. 2007;36:439–45. doi: 10.1093/ije/dyl253. [DOI] [PubMed] [Google Scholar]
- 5.Khoury MJ, McBride CM, Schully SD, et al. The Scientific Foundation for personal genomics: recommendations from a National Institutes of Health-Centers for Disease Control and Prevention multidisciplinary workshop. Genet Med. 2009;11:559–67. doi: 10.1097/GIM.0b013e3181b13a6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yesupriya A, Evangelou E, Kavvoura FK, et al. Reporting of human genome epidemiology (HuGE) association studies: an empirical assessment. BMC Med Res Methodol. 2008;8:31. doi: 10.1186/1471-2288-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yesupriya A, Yu W, Clyne M, Gwinn M, Khoury MJ. The continued need to synthesize the results of genetic associations across multiple studies. Genet Med. 2008;10:633–5. doi: 10.1097/gim.0b013e3181815360. [DOI] [PubMed] [Google Scholar]
- 8.Miretti MM, Beck S. Immunogenomics: molecular hide and seek. Human genomics. 2006;2:244–51. doi: 10.1186/1479-7364-2-4-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.WHO Nomenclature Committee for Factors of the HLA System Nomenclature for factors of the HLA system, 1987. Tissue Antigens. 1988;32:177–87. doi: 10.1111/j.1399-0039.1988.tb01655.x. [DOI] [PubMed] [Google Scholar]
- 10.WHO Nomenclature Committee for Factors of the HLA System Nomenclature for factors of the HL-a system. Bull World Health Organ. 1968;39:483–6. [PMC free article] [PubMed] [Google Scholar]
- 11.WHO Nomenclature Committee for Factors of the HLA System Nomenclature for factors of the HLA system. Bull World Health Organ. 1975;52:261–5. [PMC free article] [PubMed] [Google Scholar]
- 12.WHO Nomenclature Committee for Factors of the HLA System Nomenclature for factors of the HLA system--1977. Tissue Antigens. 1978;11:81–6. doi: 10.1111/j.1399-0039.1978.tb01231.x. [DOI] [PubMed] [Google Scholar]
- 13.Bodmer JG, Marsh SG, Albert ED, et al. Nomenclature for factors of the HLA system, 1991. WHO Nomenclature Committee for factors of the HLA system. Tissue Antigens. 1992;39:161–73. doi: 10.1111/j.1399-0039.1992.tb01932.x. [DOI] [PubMed] [Google Scholar]
- 14.Marsh SG, Albert ED, Bodmer WF, et al. Nomenclature for factors of the HLA system, 2002. Tissue Antigens. 2002;60:407–64. doi: 10.1034/j.1399-0039.2002.600509.x. [DOI] [PubMed] [Google Scholar]
- 15.Marsh SG, Albert ED, Bodmer WF, et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens. 75:291–455. doi: 10.1111/j.1399-0039.2010.01466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bodmer JG, Marsh SG, Albert ED, et al. Nomenclature for factors of the HLA system, 1990. Tissue Antigens. 1991;37:97–104. doi: 10.1111/j.1399-0039.1991.tb01853.x. [DOI] [PubMed] [Google Scholar]
- 17.Bodmer JG, Marsh SG, Albert ED, et al. Nomenclature for factors of the HLA System, 1998. Hum Immunol. 1999;60:361–95. doi: 10.1016/s0198-8859(99)00042-7. [DOI] [PubMed] [Google Scholar]
- 18.Robinson J, Waller MJ, Parham P, Bodmer JG, Marsh SG. IMGT/HLA Database--a sequence database for the human major histocompatibility complex. Nucleic Acids Res. 2001;29:210–3. doi: 10.1093/nar/29.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson J, Mistry K, McWilliam H, Lopez R, Parham P, Marsh SG. The IMGT/HLA database. Nucleic Acids Res. 2011;39:D1171–6. doi: 10.1093/nar/gkq998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer D, Mack S. ENCYCLOPEDIA OF LIFE SCIENCES. John Wiley & Sons, Ltd; Chichester: 2008. Major Histocompatibility Complex (MHC) Genes: Polymorphism. [Google Scholar]
- 21.Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–51. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 22.Klein J, Figueroa F. Evolution of the major histocompatibility complex. Crit Rev Immunol. 1986;6:295–386. [PubMed] [Google Scholar]
- 23.Hurley CK, Setterholm M, Lau M, et al. Hematopoietic stem cell donor registry strategies for assigning search determinants and matching relationships. Bone Marrow Transplant. 2004;33:443–50. doi: 10.1038/sj.bmt.1704365. [DOI] [PubMed] [Google Scholar]
- 24.European Federation for Immunogenetics [April 5 2011];Standards for PROVIDERS of External Proficiency Testing (EPT) schemes – Version 5.0. Available at: http://www.efiweb.eu/fileadmin/user_upload/EPT/EFI_EPT_Standards_for_Providers_v_5_0.pdf.
- 25.Nunes E, Heslop H, Fernandez-Vina M, et al. Definitions of histocompatibility typing terms: harmonization of histocompatibility typing terms working group. Hum Immunol. 2011;72(12):1214–6. doi: 10.1016/j.humimm.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 26.Noble JA, Valdes AM, Varney MD, et al. HLA class I and genetic susceptibility to type 1 diabetes: results from the Type 1 Diabetes Genetics Consortium. Diabetes. 59:2972–9. doi: 10.2337/db10-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Middleton D, Menchaca L, Rood H, Komerofsky R. New allele frequency database: http://www.allelefrequencies.net. Tissue Antigens. 2003;61:403–7. doi: 10.1034/j.1399-0039.2003.00062.x. [DOI] [PubMed] [Google Scholar]
- 28.Cano P, Klitz W, Mack SJ, et al. Common and well-documented HLA alleles: report of the Ad-Hoc committee of the american society for histocompatiblity and immunogenetics. Hum Immunol. 2007;68:392–417. doi: 10.1016/j.humimm.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 29.Middleton D, Gonzalez F, Fernandez-Vina M, et al. A bioinformatics approach to ascertaining the rarity of HLA alleles. Tissue Antigens. 2009;74:480–5. doi: 10.1111/j.1399-0039.2009.01361.x. [DOI] [PubMed] [Google Scholar]
- 30.Furst D, Solgi G, Schrezenmeier H, Mytilineos J. The frequency of DRB1*1454 in South German Caucasians. Tissue Antigens. 76:57–9. doi: 10.1111/j.1399-0039.2010.01461.x. [DOI] [PubMed] [Google Scholar]
- 31.Yang KL, Chen MJ, Lee SK, Lin CC, Tsai MJ, Chiu HM, Jiang S, Chao YC, Chen SP, Lin S, Shyr MH, Lin PY. New allele name of some HLA-DRB1*1401: HLA-DRB1*1454. Int J Immunogenet. 2009;36:119–20. doi: 10.1111/j.1744-313X.2008.00826.x. [DOI] [PubMed] [Google Scholar]
- 32.Hsu KC, Chida S, Geraghty DE, Dupont B. The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polymorphism. Immunol Rev. 2002;190:40–52. doi: 10.1034/j.1600-065x.2002.19004.x. [DOI] [PubMed] [Google Scholar]
- 33.Martin AM, Kulski JK, Gaudieri S, et al. Comparative genomic analysis, diversity and evolution of two KIR haplotypes A and B. Gene. 2004;335:121–31. doi: 10.1016/j.gene.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 34.Vilches C, Pando MJ, Rajalingam R, Gardiner CM, Parham P. Discovery of two novel variants of KIR2DS5 reveals this gene to be a common component of human KIR ’B’ haplotypes. Tissue Antigens. 2000;56:453–6. doi: 10.1034/j.1399-0039.2000.560510.x. [DOI] [PubMed] [Google Scholar]
- 35.Middleton D, Meenagh A, Gourraud PA. KIR haplotype content at the allele level in 77 Northern Irish families. Immunogenetics. 2007;59:145–58. doi: 10.1007/s00251-006-0181-7. [DOI] [PubMed] [Google Scholar]
- 36.Robinson J, Mistry K, McWilliam H, Lopez R, Marsh SG. IPD--the Immuno Polymorphism Database. Nucleic Acids Res. 38:D863–9. doi: 10.1093/nar/gkp879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Ronaghi M, Chong SS, Lee CG. pfSNP: An integrated potentially functional SNP resource that facilitates hypotheses generation through knowledge syntheses. Hum Mutat. 2011;32:19–24. doi: 10.1002/humu.21331. [DOI] [PubMed] [Google Scholar]
- 39.Farewell VT, Dahlberg S. Some Statistical Methodology for the Analysis of HLA Data. Biometrics. 1984;40:547–60. [PubMed] [Google Scholar]
- 40.Thomson G, Maldonado-Torres H, Lancaster AK, Hollenbach JA, Barcellos LF, Mack SJ, Single RM. Hardy-Weinberg Proportions Methods Manual Version 0.1.2: Testing fit of genotype frequencies to Hardy-Weinberg proportions. 2009 Available online at: www.ImmPort.org.
- 41.Thomson G, Barcellos LF, Valdes AM. Searching for additional disease loci in a genomic region. Adv Genet. 2008;60:253–92. doi: 10.1016/S0065-2660(07)00411-7. [DOI] [PubMed] [Google Scholar]
- 42.Single RM, Martin MP, Meyer D, Gao X, Carrington M. Methods for assessing gene content diversity of KIR with examples from a global set of populations. Immunogenetics. 2008;60:711–25. doi: 10.1007/s00251-008-0331-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gourraud PA, Meenagh A, Cambon-Thomsen A, Middleton D. Linkage disequilibrium organization of the human KIR superlocus: implications for KIR data analyses. Immunogenetics. 2010;62:729–40. doi: 10.1007/s00251-010-0478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Single RM, Meyer D, Thomson G. Statistical Methods for Analysis of Population Genetic Data. In: Hansen JA, editor. Immunobiology of the Human MHC Proceedings of the 13th International Histocompatibility Workshop and Conference. IHWG Press; Seattle: 2007. [Google Scholar]
- 45.Mack SJ, Sanchez-Mazas A, Meyer D, Single RM, Tsai Y, Erlich HA. Methods used in the generation and preparation of data for analysis in the 13th International Histocompatibility Workshop. 13th International Histocompatibility Workshop Anthropology/Human Genetic Diversity Joint Report. In: Hansen JA, editor. Immunobiology of the Human MHC: Proceedings of the 13th International Histocompatibility Workshop and Conference. IHWG Press; Seattle,WA: 2007. [Google Scholar]
- 46.Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association Mapping in Structured Populations. The American Journal of Human Genetics. 2000;67:170–81. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson J, Waller MJ, Fail SC, Marsh SG. The IMGT/HLA and IPD databases. Hum Mutat. 2006;27:1192–9. doi: 10.1002/humu.20406. [DOI] [PubMed] [Google Scholar]
- 48.Vandiedonck C, Knight JC. The human Major Histocompatibility Complex as a paradigm in genomics research. Brief Funct Genomic Proteomic. 2009;8:379–94. doi: 10.1093/bfgp/elp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fairbairn DJ. The advent of mandatory data archiving. Evolution. 2011;65:1–2. doi: 10.1111/j.1558-5646.2010.01182.x. [DOI] [PubMed] [Google Scholar]
- 50.Discussing standards. Nat Genet. 2010;42:915. doi: 10.1038/ng1110-915. [DOI] [PubMed] [Google Scholar]