Background: Redox of extracellular HMGB1 protein plays an important role in inflammation.
Results: The half-life of all-thiol HMGB1 was ∼17 min in serum and saliva and significantly longer in cancer cell culture medium and was modulated by exogenous ligands (e.g. heparin).
Conclusion: The extracellular environment dictates HMGB1 oxidation kinetics.
Significance: Our approach permits investigating protein oxidation in situ.
Keywords: Disulfide, Kinetics, Nuclear Magnetic Resonance, Oxidation-Reduction, Redox, In Situ
Abstract
Some extracellular proteins are initially secreted in reduced forms via a non-canonical pathway bypassing the endoplasmic reticulum and become oxidized in the extracellular space. One such protein is HMGB1 (high-mobility group box 1). Extracellular HMGB1 has different redox states that play distinct roles in inflammation. Using a unique NMR-based approach, we have investigated the kinetics of HMGB1 oxidation and the half-lives of all-thiol and disulfide HMGB1 species in serum, saliva, and cell culture medium. In this approach, salt-free lyophilized 15N-labeled all-thiol HMGB1 was dissolved in actual extracellular fluids, and the oxidation and clearance kinetics were monitored in situ by recording a series of heteronuclear 1H-15N correlation spectra. We found that the half-life depends significantly on the extracellular environment. For example, the half-life of all-thiol HMGB1 ranged from ∼17 min (in human serum and saliva) to 3 h (in prostate cancer cell culture medium). Furthermore, the binding of ligands (glycyrrhizin and heparin) to HMGB1 significantly modulated the oxidation kinetics. Thus, the balance between the roles of all-thiol and disulfide HMGB1 proteins depends significantly on the extracellular environment and can also be artificially modulated by ligands. This is important because extracellular HMGB1 has been suggested as a therapeutic target for inflammatory diseases and cancer. Our work demonstrates that the in situ protein NMR approach is powerful for investigating the behavior of proteins in actual extracellular fluids containing an enormous number of different molecules.
Introduction
Disulfide bonds between cysteine residues are very common in extracellular proteins. In the canonical pathway, disulfide bonds are formed via oxidation in the endoplasmic reticulum before secretion to the extracellular space (1, 2). However, there are many proteins that bypass the endoplasmic reticulum and have no disulfide bonds when released (3–5). Such proteins eventually become oxidized due to the oxidative nature of the extracellular environment. Various studies have shown that the functional roles of their reduced and oxidized forms are distinctly different (reviewed in Ref. 5). Therefore, the kinetics of conversion to the oxidized state must play an important role in dictating the functions of the two forms. Rapid disulfide bond formation (seconds to minutes) would suggest that the role for the reduced form is transient and that any function of the reduced form must be rapid and immediate. On the other hand, slow oxidation would suggest that the reduced form is the functionally relevant form, that disulfide formation regulates function, and/or that the function of the oxidized form, if important, emerges over longer time scales of hours. Therefore, the kinetics of the oxidation should be an important determinant of the balance between roles of reduced and oxidized states. This balance could be shifted in some diseases, including cancer (6, 7), where extracellular redox status is significantly modulated. Currently, there is a lack of knowledge of the kinetics of protein oxidation in the extracellular space. Analyzing the compositions of fluids is not enough to predict oxidation kinetics because a large number of chemical factors affect disulfide bond formation (8–12).
In this work, we studied the oxidation of HMGB1 (high-mobility group box 1) protein in extracellular fluids. This 25-kDa protein, which is normally located in nuclei, is passively released during cell injury and necrosis and actively secreted by some cell types via a non-canonical pathway that bypasses the endoplasmic reticulum (13, 14). Extracellular HMGB1 acts as a danger signal and inflammatory mediator via binding to cell surface receptors such as RAGE, TLR4 (Toll-like receptor 4), and CXCR4 (13–16). Extracellular HMGB1/receptor interactions promote inflammation and angiogenesis via activation of NF-κB (17, 18) and tumor proliferation via activation of p44/p42, p38, and SAPK/JNK MAPKs (19, 20). When released into the extracellular space, HMGB1 is initially in the reduced state (“all-thiol HMGB1”) (see Fig. 1A) but becomes oxidized due to the oxidative environment (5, 15, 21). HMGB1 has three cysteine residues. Cys-23 and Cys-45 in the A-domain are located in close proximity (see Fig. 1A). With a standard redox potential of −237 mV (21), these two cysteine residues can easily form a disulfide bond under relatively mild oxidative conditions (“disulfide HMGB1”). Cys-106 in the B-domain remains in the reduced state but can be sulfonated in the extracellular space when exposed to a large amount of reactive oxygen species from activated leukocytes (22). The three different redox states (i.e. all-thiol, disulfide, and sulfonated) of extracellular HMGB1 play distinct roles in inflammation (reviewed in Refs. 16 and 23). All-thiol HMGB1, but not disulfide HMGB1, is able to form a complex with CXCL12 for signaling via the CXCR4 receptor and exhibits chemoattractant activity (24). Only disulfide HMGB1 can interact with TLR4 and exhibits cytokine-inducing activity (24–26). Sulfonated HMGB1 is involved in the resolution of inflammation (22). Therefore, knowledge of the lifetimes of the different species is essential to better understand the relative roles of these different redox species.
FIGURE 1.

A, HMGB1 and its three cysteine residues. Cys-23 and Cys-45 form a disulfide bond under oxidative conditions. B, experimental scheme for the in situ protein NMR approach to investigate the behavior of HMGB1 in actual extracellular fluids. C, 1H-15N SOFAST-HMQC spectra recorded with eight scans at 37 °C for all-thiol HMGB1 (red) and disulfide HMGB1 (blue) immediately after dissolving in human serum. D, 1H-15N SOFAST-HMQC spectra of all-thiol HMGB1 (red) and disulfide HMGB1 (blue) immediately after dissolving in 20 mm HEPES-NaOH (pH 7.4), 120 mm NaCl, and 5% D2O. 135 μm 15N-labeled HMGB1 was used for both the serum and buffer samples. Line shapes of NMR signals from HMGB1 in serum were substantially broader than those in buffer presumably due to the molecular crowding environment of the serum. E, SDS-PAGE of the serum samples used for the NMR experiments. 3 μl of serum before and after dissolving 135 μm 15N-labeled HMGB1 was loaded. The arrow indicates the position of the HMGB1 band. The strongest bands are albumin. F, changes in the 1H-15N SOFAST-HMQC spectra recorded at 37 °C for all-thiol HMGB1 in human serum. Due to oxidation, signals from all-thiol HMGB1 became weaker, whereas those from disulfide HMGB1 became stronger.
In this study, using a unique NMR-based approach (schematically depicted in Fig. 1B), we investigated the kinetics of HMGB1 oxidation and the half-lives of all-thiol and disulfide HMGB1 proteins in actual extracellular fluids: serum, saliva, and cell culture medium. We also studied how oxidation kinetics are affected by exogenous ligands such as heparin, which mimics cell surface heparan sulfate.
EXPERIMENTAL PROCEDURES
Human Serum
The serum sample for this study was prepared from a blood sample obtained from a consenting healthy male (71 years old) by our approved Institutional Review Board protocol. The blood was centrifuged at 1000 × g for 5 min at 20 °C. The supernatant was transferred into BD Vacutainer red-top tubes (non-heparinized) and allowed to clot by leaving it undisturbed for 30 min at room temperature. The clot was removed by centrifugation at 200 × g for 5 min at room temperature. Without disturbing the coagulated pellet, the resultant supernatant (i.e. serum) was carefully transferred into 2-ml polypropylene microtubes and stored at −80 °C until used.
Human Saliva
Human saliva was sampled from a healthy male (42 years old). Before sampling, the mouth was washed with 18-megohm water, and the saliva was collected 10 min later. The saliva was centrifuged at 2000 × g for 10 min at room temperature, and the supernatant was immediately used for the NMR experiment.
Extracellular Fluids of PC-3M Cell Culture
Human prostate cancer PC-3M cells (27) were cultured at 37 °C in RPMI 1640 medium (Invitrogen) with 10% FBS (Atlanta Biologicals). Cells were trypsinized for seeding. To remove trypsin, the cells were centrifuged at 1000 rpm for 5 min at room temperature, and the cell pellet was washed once with phosphate-buffered saline (Invitrogen) and then suspended with RPMI 1640 medium plus 10% FBS. Three different titrating amounts of the cell suspension were used for seeding to obtain cultures with three different cell confluences at the same time. Culture supernatants were collected after the cells were cultured for 48 h at 37 °C under 5% CO2. As a control, a blank (without cells) RPMI 1640 medium plus 10% FBS was also incubated and collected at the same time. Each fluid was filtered with a 0.22-μm filter and kept at −80 °C until used.
HMGB1 Ligands
Glycyrrhizin was purchased from Nacalai USA, Inc. Heparin octasaccharide was purchased from Iduron. These materials were dissolved in 18-megohm water and lyophilized before use for in situ protein NMR experiments (see Fig. 1B).
Preparation of HMGB1
Full-length HMGB1 protein was expressed in Escherichia coli strain Rosetta 2(DE3) cells harboring a pET-11d-derived plasmid with the human HMGB1 gene inserted between the NcoI and BamHI sites. For preparation of 15N- or 13C/15N-labeled proteins, [15N]ammonium chloride and [13C]glucose were used as the sole sources of nitrogen and carbon, respectively, in the E. coli culture medium. Protein expression was induced with 0.4 mm isopropyl β-d-thiogalactopyranoside, and the E. coli culture was continued at 18 °C for 16 h. Harvested cells were suspended in buffer containing 20 mm sodium phosphate (pH 6.0), 1 mm EDTA, 100 mm NaCl, 2 mm DTT, and 5% glycerol and disrupted at 4 °C by sonication. The supernatant of the cell lysate was loaded onto an SP cation exchange column equilibrated with 20 mm sodium phosphate (pH 6.0) and 100 mm NaCl and eluted with a gradient of 100–2000 mm NaCl. Fractions containing HMGB1 were concentrated and passed through an S-100 column equilibrated with buffer containing 20 mm Tris-HCl (pH 8.0), 1 mm EDTA, and 200 mm NaCl. After size exclusion chromatography, HMGB1 was further purified by RESOURCE Q anion exchange chromatography with a gradient of 0–1500 mm NaCl in 50 mm Tris-HCl (pH 7.5). No reducing regent was used in the column chromatography procedures except in the sonication buffer. Complete disulfide bond formation in HMGB1 was confirmed by NMR (21).
Preparation of Salt-free Powders of 15N-Labeled All-thiol HMGB1
To break the disulfide bond, 5 mm DTT was added to a solution of 15N-labeled disulfide HMGB1, and the mixture was kept overnight at 4 °C. Using an Amicon Ultra-15 unit, the solvent was exchanged with water containing 1 mm β-mercaptoethanol. Because of the lower solubility of HMGB1 in the absence of salt, the process of the solvent exchange was carried out at protein concentrations lower than 200 μm. The final solution was divided into aliquots (each containing 68 nmol of HMGB1), frozen at −80 °C, and lyophilized. β-Mercaptoethanol was removed during the lyophilization process, and all-thiol HMGB1 remained as a powder. The obtained powder was stored at −20 °C until used.
Preparation of Salt-free Powders of 15N-Labeled Disulfide HMGB1
Salt-free powders of 15N-labeled disulfide HMGB1 were prepared in the same way as for all-thiol HMGB1 except that the samples were not treated with either DTT or β-mercaptoethanol. Disulfide HMGB1 in water was lyophilized as described above.
In Situ NMR Experiments to Measure the Half-lives of All-thiol and Disulfide HMGB1 Species in Extracellular Fluids
For the kinetic measurements, the salt-free powder of 15N-labeled all-thiol HMGB1 was dissolved in 500 μl of extracellular fluid (i.e. serum, saliva, or cell culture medium) plus 1% D2O (for the NMR lock). The solution was immediately sealed in a 5-mm NMR tube with ambient oxygen pressure, and a series of 1H-15N correlation spectra were recorded at 37 °C using Bruker AVANCE III spectrometers equipped with cryogenic probes (1H frequencies, 800 and 600 MHz). In all experiments except those involving the cell culture medium, band-selective optimized flip-angle short-transient heteronuclear multiple quantum coherence (SOFAST-HMQC)2 spectra (28, 29) were recorded with a recycle delay of 0.1 s. For the cell culture medium, in which HMGB1 oxidation is relatively slow, the kinetics were measured with a series of sensitivity-enhanced 1H-15N transverse relaxation optimized spectroscopy spectra (30, 31) recorded with a recycle delay of 0.8 or 1.0 s. Values of pH for the NMR samples were confirmed to be ∼7.4. Kinetic rate constants were determined via nonlinear least-squares fitting with MATLAB software (MathWorks). Other details of the calculations are given in the figure legends.
NMR Resonance Assignment
Because the spectra of HMGB1 samples dissolved in extracellular fluids and in buffers are very similar, resonances of HMGB1 in extracellular fluids were assigned using the NMR spectra recorded for HMGB1 in a buffer. 1H/13C/15N resonances for oxidized and reduced HMGB1 proteins were assigned with three-dimensional HNCA, HN(CO)CA, HNCO, HN(CA)CO, HNCACB, CBCA(CO)NH, C(CO)NH, 15N-edited NOESY-heteronuclear single quantum coherence; and F1-15N/F2-15N-edited HMQC-NOESY-heteronuclear single quantum coherence spectra (32) recorded at 25 °C on 0.7 mm 13C/15N-labeled proteins in buffer containing 20 mm Tris-HCl (pH 7.5), 120 mm NaCl, and 5% D2O. For all-thiol HMGB1 in the buffer, 5 mm DTT was also added, and the NMR tube was sealed in the presence of argon gas. Resonance assignment was also aided by previous assignment data for HMGB1 under different conditions (21, 33–35). NMR data were processed and analyzed using the NMRPipe (36) and NMRView (37) programs.
In this study, we use the residue numbering scheme from the initial methionine in the gene (i.e. Met-1–Gly-2–Lys-3 … ) because this has become the accepted norm, although the numbering scheme from the actual N-terminal glycine (i.e. Gly-l–Lys-2 … ) of HMGB1 has been used in many previous articles (including ours).
RESULTS
NMR of All-thiol and Disulfide HMGB1 Proteins Dissolved in Serum
We prepared salt-free lyophilized powders of 15N-labeled all-thiol HMGB1 and HMGB1 proteins and dissolved them in human serum to characterize the actual redox kinetics of extracellular HMGB1 by NMR. Fig. 1C shows NMR spectra recorded at 37 °C for all-thiol HMGB1 and disulfide HMGB1 in human serum. The spectra were similar to spectra recorded for all-thiol HMGB1 and disulfide HMGB1 in buffer containing 20 mm HEPES-NaOH (pH 7.4), 120 mm NaCl, and 5% D2O (Fig. 1D). This suggests that the HMGB1 structures in serum and in buffer are identical, although serum is a molecular crowding environment with numerous proteins as shown by the SDS-PAGE data (Fig. 1E). Our NMR data also indicate that the proteins from the salt-free lyophilized powders were properly folded when dissolved in extracellular fluids. As demonstrated in our previous study on the A-domain of HMGB1 (21), residues in close proximity to Cys-23 or Cys-45 exhibited large 1H/15N chemical shift differences between all-thiol HMGB1 and disulfide HMGB1. Due to rapid oxidation, however, the spectra recorded for the sample of all-thiol HMGB1 in serum gradually changed and finally became identical to the spectra of disulfide HMGB1 (Fig. 1F). We used this change in NMR spectra to investigate the oxidation kinetics of HMGB1 in serum.
Kinetics of HMGB1 Oxidation in Serum
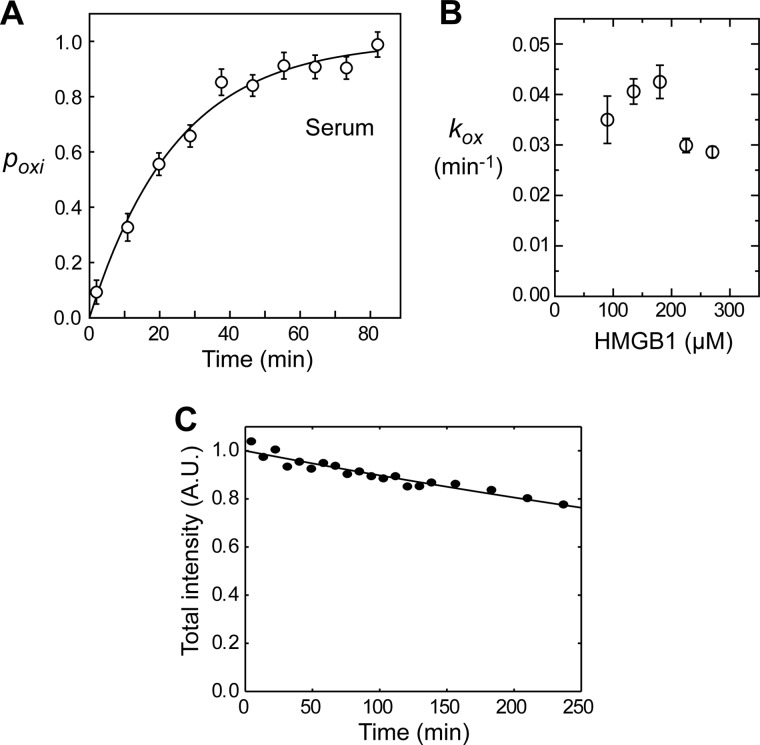
We recorded a series of 1H-15N SOFAST-HMQC spectra immediately after dissolving 15N-labeled all-thiol HMGB1 in serum. SOFAST-HMQC allows acquisition of high-resolution 1H-15N correlation spectra in a few minutes and is therefore ideally suited for measuring real-time kinetics (28, 29). NMR signals from disulfide HMGB1 gradually became stronger and predominant, whereas signals from all-thiol HMGB1 became weaker and eventually disappeared in 50 min (Fig. 1F). This clearly reflects the oxidation process of all-thiol HMGB1 in serum. Using the SOFAST-HMQC spectra, we obtained the time course data of the fraction of oxidation (Fig. 2A). By nonlinear least-squares fitting with the experimental data, we determined the apparent pseudo-first-order rate constant (kox) for HMGB1 oxidation. The half-life (t½) of all-thiol HMGB1, which was calculated as kox−1 ln 2, was determined to be 17 ± 1 min (Table 1). We also measured kox at different concentrations of HMGB1 (Fig. 2B). kox was slightly smaller at concentrations higher than 200 μm. This is probably because the overall concentration of the oxidants that oxidized HMGB1 was not high enough to make the process completely pseudo-first-order. Because the in vivo concentration of extracellular HMGB1 is lower than micromolar, HMGB1 oxidation in vivo should occur in a completely pseudo-first-order manner, and the half-life in vivo should be only slightly shorter than what we observed in situ.
FIGURE 2.
Kinetics of oxidation and clearance of HMGB1 in human serum. A, oxidation kinetics of HMGB1 in human serum measured by in situ protein NMR. The vertical axis represents the fraction of oxidation (poxi) measured as (1/N)ΣiNID,i/(IA,i + ID,i), where IA and ID are signals intensities for all-thiol and disulfide HMGB1 proteins, respectively, and i is indices for analyzed residues. Intensities were measured for NMR signals from residues that exhibit relatively strong, well isolated signals for both all-thiol and disulfide states (i.e. those from Thr-22, Lys-29, His-31, Thr-51, and Gly-58 backbone amide groups and the Gln-21 side chain NH2 group). Experimental pox data were fitted to 1 − exp(−kox(t − t0)), where kox represents a pseudo-first-order rate constant for the oxidation of all-thiol HMGB1, and t0 represents a time shift due to the difference between the effective times and the mid-times for recording NMR spectra. This shift was applied to the horizontal axis of the time course data shown. B, kinetic rate constants (kox) for HMGB1 at different concentrations (90, 135, 180, 225, and 270 μm) in human serum. C, decrease in the total intensity of NMR signals from amide groups whose 1H/15N chemical shifts were unaffected by oxidation. SOFAST-HMQC spectra recorded for 135 μm 15N-labeled HMGB1 in human serum were analyzed. The curve represents the best fit to aexp(−kclt), where kcl is a pseudo-first-order rate constant for HMGB1 clearance. The vertical axis represents the total intensity divided by a. A.U., arbitrary unit.
TABLE 1.
Half-lives of all-thiol HMGB1 and disulfide HMGB1 in extracellular fluids measured by in situ NMR
135 μm 15N-labeled all-thiol HMGB1 was used for each experiment.
| Extracellular fluids | Oxidation t½a | Clearance t½b |
|---|---|---|
| min | min | |
| Human serum | 17 ± 1 | 642 ± 49 |
| Human saliva | 18 ± 2 | 65 ± 6 |
| RPMI 1640 medium + 10% FBSc | 25 ± 3 | 464 ± 41 |
| PC-3M cell culture medium | ||
| 30% cell confluencyd | 27 ± 3 | 480 ± 48 |
| 60% cell confluencye | 84 ± 3 | >1500f |
| 95% cell confluencyg | 201 ± 6 | >1500f |
| PC-3Mg + heparinh | 43 ± 2 | >1500f |
| PC-3Mg + glycyrrhizini | 289 ± 9 | >1500f |
a Determined as kox−1 ln 2. This corresponds to the half-life of all-thiol HMGB1.
b Determined as kcl−1 ln 2. This corresponds to the half-life of disulfide HMGB1.
c Control for PC-3M data. No cells were cultured (see “Experimental Procedures”).
d Extracellular fluid at a cell confluency of 30%.
e Extracellular fluid at a cell confluency of 60%.
f Too slow to determine.
g Extracellular fluid at a cell confluency of 95%.
h 2.5 mm heparin octasaccharide dissolved together with HMGB1.
i 2.0 mm glycyrrhizin dissolved together with HMGB1.
Clearance of HMGB1 in Serum
Our NMR data also provide kinetic information on the clearance of HMGB1 due to protease activities in extracellular fluids. Fig. 2C shows the time course of the loss in total intensity of NMR signals whose positions (i.e. 1H/15N chemical shifts) were unaffected by the oxidation. The HMGB1 band in SDS-PAGE became weaker over hours as well, which indicates that the decrease in NMR signal intensities is caused by a decrease in the total amount of HMGB1. Apparent kinetic rate constants for HMGB1 clearance (kcl) were determined via monoexponential fitting with the time course data shown in Fig. 2C. The half-life of disulfide HMGB1 in serum was calculated as kcl−1 ln 2 to be 642 ± 49 min (Table 1). Interestingly, neither 1H-15N spectra nor SDS-PAGE showed degradation products of HMGB1. These results imply that, once cleavage occurs, HMGB1 molecules become highly susceptible to complete digestion by extracellular proteases to an amino acid level.
Kinetics of HMGB1 Oxidation and Clearance in Saliva
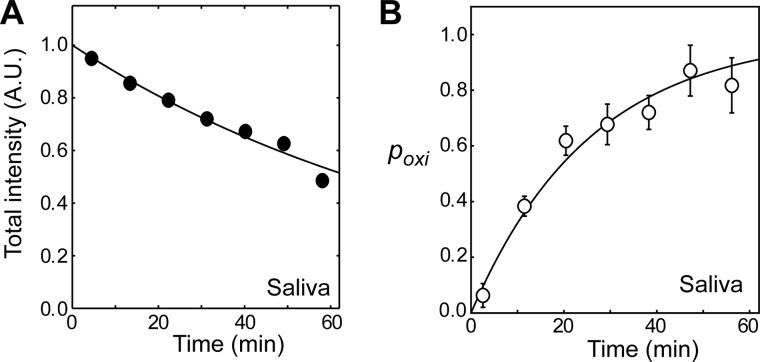
Salivary gland cells are known to produce extracellular HMGB1 (38). To examine whether or not the molecular behavior of HMGB1 in saliva is similar to that in serum, we also investigated oxidation and clearance of HMGB1 in saliva using the in situ protein NMR approach. Interestingly, HMGB1 clearance in saliva was found to be 10-fold faster than that in serum (Fig. 3A), whereas HMGB1 oxidation in saliva was as fast as that in serum (Fig. 3B). Thus, the function of different redox forms of extracellular HMGB1 is very likely dependent on the nature of the extracellular fluid.
FIGURE 3.
Kinetics of HMGB1 clearance (A) and oxidation (B) in human saliva measured by in situ protein NMR. The clearance of HMGB1 in saliva was 10-fold faster than that in serum (see Table 1). A.U., arbitrary unit.
Kinetics of HMGB1 Oxidation and Clearance in Extracellular Fluids from Cell Culture
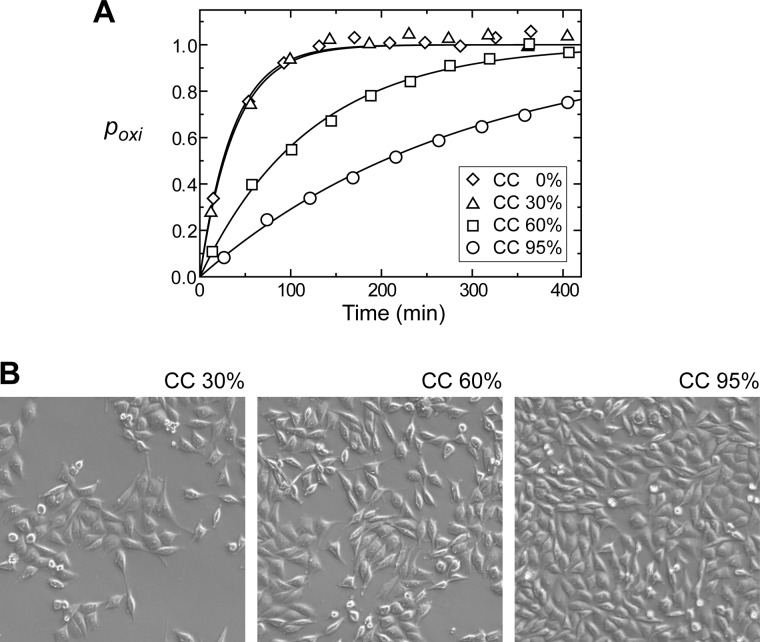
Using the in situ protein NMR approach, we also investigated the kinetics of HMGB1 oxidation and clearance in extracellular fluids from the culture of the prostate cancer cell line PC-3M (27). Extracellular fluids at different cell confluences (0, 30, 60, and 95%) were analyzed. Interestingly, the oxidation kinetics were found to strongly depend on the cell confluency. HMGB1 oxidation was slower at a higher confluency (Fig. 4). At the highest confluency, the half-life of all-thiol HMGB1 was longer than 3 h (Table 1), suggesting a more hypoxic environment in the extracellular fluid. In such a hypoxic environment (e.g. in cancer), slow oxidation can suppress the role of disulfide HMGB1.
FIGURE 4.
A, oxidation kinetics of HMGB1 in extracellular fluids from cultures of the prostate cancer cell line PC-3M studied by in situ protein NMR. Oxidation of HMGB1 was slower at higher cell confluency (CC). B, images of the cultures when the extracellular fluids were sampled. The total time of incubation at 37 °C was identical for all of the cell culture samples (see “Experimental Procedures”). Different cell confluences were achieved by using different amounts of seeding.
Impact of Exogenous Ligands on the Oxidation of HMGB1
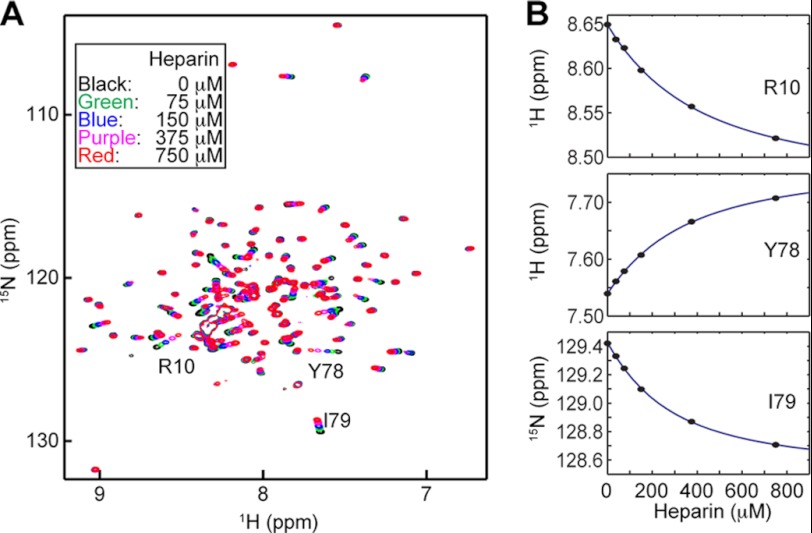
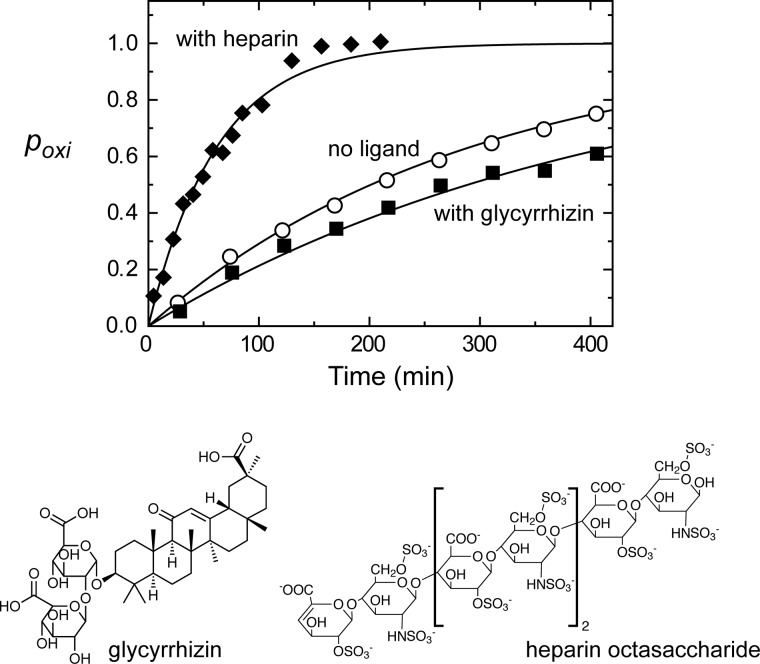
Since blockade of extracellular HMGB1 signaling was shown to be effective for suppressing tumor growth and metastases (20), a large number of strategies to block HMGB1 signaling have been proposed for therapeutics of cancer and inflammatory diseases (e.g. reviewed in Refs. 14, 15, and 39–43). Compounds that directly bind to HMGB1 were discovered previously. Among them are glycyrrhizin and heparin (44–46). Using extracellular fluids from PC-3M cell culture (95% confluency), we investigated the impact of these compounds on the oxidation and degradation of HMGB1. The dissociation constant (Kd) for the HMGB1-glycyrrhizin complex was reported to be 156 ± 3 μm (45). By NMR-based titration, we determined the Kd for the HMGB1-heparin octasaccharide complex to be 259 ± 72 μm (Fig. 5). For these ligands, we compared the kinetics of HMGB1 oxidation for free and ligand-bound HMGB1 in PC-3M extracellular fluids. In these experiments, the concentrations of glycyrrhizin and heparin were 2.0 and 2.5 mm, respectively, at which >90% of HMGB1 should be bound. The oxidation time course data for these samples are shown in Fig. 6, and the half-lives determined from the data are shown in Table 1. Although both glycyrrhizin and heparin are inactive in terms of thiol/disulfide redox chemistry, the binding of these compounds significantly affected the oxidation of HMGB1. The binding of heparin was found to promote the oxidation by 4-fold. It is well known that HMGB1 undergoes conformational equilibrium between the open and closed states via intramolecular interactions between the C-terminal acidic tail and A/B-domains (34, 35). Because heparin is also highly negatively charged, HMGB1/heparin interactions may weaken intramolecular interactions between the A-domain and the acidic tail and increase the exposure of the pair of the Cys-23/Cys-45 thiol groups, thereby enhancing their oxidation. On the contrary, glycyrrhizin binding to HMGB1 was found to slow HMGB1 oxidation by ∼1.4-fold (Fig. 6). According to the NMR-based structure model of the HMGB1-glycyrrhizin complex of Mollica et al. (45), glycyrrhizin bound to HMGB1 covers the regions of the Cys-23 and Cys-45 side chains, and so glycyrrhizin may block oxidant accessibility to the Cys-23/Cys-45 thiol groups. A computational study suggested that the HMGB1/glycyrrhizin interactions could be non-directional and dynamic (44), which may weaken the protection of the thiol pair from oxidants. Our present data suggest that non-redox-active ligands can regulate HMGB1 oxidation.
FIGURE 5.
HMGB1/heparin interactions observed by NMR. A, 1H-15N transverse relaxation optimized spectroscopy spectra recorded at 37 °C for 135 μm all-thiol HMGB1 in the presence of heparin octasaccharide at different concentrations. The samples were dissolved in 20 mm Tris-HCl (pH 7.5), 120 mm NaCl, and 5 mm DTT. B, determination of the dissociation constant (Kd) from 1H and 15N chemical shifts as a function of heparin concentration. The titration data, together with the best fit curves, are shown for three residues whose signals are indicated in A. Fourteen signals from residues showing large chemical shift perturbation were analyzed, from which the Kd was determined to be 259 ± 72 μm.
FIGURE 6.
Impact of ligand binding on HMGB1 oxidation in extracellular fluids from PC-3M cell culture (95% cell confluency). Time courses of oxidation obtained from in situ protein NMR data are plotted. ○, data for the sample without ligands; ♦ and ■, data for the samples containing heparin octasaccharide (2.5 mm) and glycyrrhizin (2 mm), respectively.
DISCUSSION
Relevance to Inflammation
All-thiol HMGB1 is considered to recruit leukocytes in inflammation (16, 23, 24). Venereau et al. (24) found that this role is associated with the binding of the all-thiol HMGB1-CXCL12 heterodimer to CXCR4 on leukocytes. In this study, we have shown that the half-life of all-thiol HMGB1 is only ∼17 min in serum. Our data suggest that the half-life can become even shorter via interactions with heparan sulfate, which is structurally analogous to heparin and highly abundant on the cell surface. To recruit leukocytes, the binding of the all-thiol HMGB1-CXCL12 heterodimer to CXCR4 should occur rapidly before the HMGB1 molecule is oxidized. Heterodimer formation with CXCL12 may protect HMGB1 from oxidants. This is possible, as we found that the binding of ligands significantly modulates the kinetics of HMGB1 oxidation. Because of the short half-life of all-thiol HMGB1, most of the extracellular HMGB1 molecules at the injury site should be disulfide HMGB1 when leukocytes arrive as a result of the chemoattractant activity of all-thiol HMGB1. Disulfide HMGB1, but not all-thiol HMGB1, is capable of activating the leukocytes and triggering the release of proinflammatory cytokines/chemokines (16, 23, 24, 26). In this sense, the short half-life of all-thiol HMGB1 in serum seems advantageous for the timely switching of HMGB1 function in inflammation.
Variations in the Half-lives of HMGB1 in Different Extracellular Fluids
We found large variations in the half-lives of HMGB1 in different extracellular fluids. For example, the half-life of all-thiol HMGB1 in PC-3M cell culture medium (95% confluency) was as long as ∼3 h, whereas those in serum and saliva were ∼17 min. Because all experiments were conducted under ambient oxygen pressure, the kinetic variation in HMGB1 oxidation can be largely due to different levels of redox factors intrinsically present in individual fluids. Our data also indicate that the half-lives of disulfide HMGB1 in serum and saliva differ by 10-fold, most likely due to different degrees of protease activity. These data suggest that the balance between the roles of all-thiol HMGB1 and disulfide HMGB1 depends significantly on the extracellular environment.
Potential Application of in Situ Protein NMR
This work has demonstrated that in situ protein NMR can provide important insights into the molecular behavior of a particular protein in actual extracellular fluids. The NMR data for 15N-labeled HMGB1 in human serum provided information specific to the behavior of HMGB1, although serum is a molecular crowding fluid with an enormous number of different proteins (60–80 mg/ml) and metabolites. Requiring only 0.50 ml of fluid, in situ protein NMR can potentially be used to investigate the molecular behaviors of extracellular proteins in various clinical specimens (e.g. serum, saliva, mucus, etc.). This approach is in contrast to NMR metabolomics, which focuses exclusively on the metabolite compositions of clinical specimens. Because of the orthogonal information they can provide, the combined use of NMR metabolomics and in situ protein NMR might allow more robust detection of abnormalities associated with diseases.
Acknowledgments
We thank Carrie Maxwell and Yash Tarkunde for assistance in sample preparation, Tianzhi Wang for the maintenance of NMR equipment, and Lavanya Rajagopalan for editorial assistance.
This work was supported, in whole or in part, by National Institutes of Health Clinical and Translational Science Award 8UL1TR000071, Grant 3R01 CA97959 (to P. S.), and Grant 1P01 HL107152-01 (to K. R.). This work was also supported by the Institute for Translational Sciences at University of Texas Medical Branch and American Heart Association Grant 12BGIA8960032 (to J. I.).
- SOFAST-HMQC
- band-selective optimized flip-angle short-transient heteronuclear multiple quantum coherence.
REFERENCES
- 1. Ellgaard L. (2004) Catalysis of disulphide bond formation in the endoplasmic reticulum. Biochem. Soc. Trans. 32, 663–667 [DOI] [PubMed] [Google Scholar]
- 2. Ellgaard L., Helenius A. (2003) Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4, 181–191 [DOI] [PubMed] [Google Scholar]
- 3. Chimini G., Rubartelli A. (2005) Novel pathways of protein secretion. in Molecular Chaperones and Cell Signaling (Henderson B., Pockley A. G., eds) pp. 45–60, Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 4. Muesch A., Hartmann E., Rohde K., Rubartelli A., Sitia R., Rapoport T. A. (1990) A novel pathway for secretory proteins? Trends Biochem. Sci. 15, 86–88 [DOI] [PubMed] [Google Scholar]
- 5. Rubartelli A., Lotze M. T. (2007) Inside, outside, upside down: damage-associated molecular-pattern molecules (DAMPs) and redox. Trends Immunol. 28, 429–436 [DOI] [PubMed] [Google Scholar]
- 6. Chaiswing L., Oberley T. D. (2010) Extracellular/microenvironmental redox state. Antioxid. Redox Signal. 13, 449–465 [DOI] [PubMed] [Google Scholar]
- 7. Grek C. L., Tew K. D. (2010) Redox metabolism and malignancy. Curr. Opin. Pharmacol. 10, 362–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blanco R. A., Ziegler T. R., Carlson B. A., Cheng P. Y., Park Y., Cotsonis G. A., Accardi C. J., Jones D. P. (2007) Diurnal variation in glutathione and cysteine redox states in human plasma. Am. J. Clin. Nutr. 86, 1016–1023 [DOI] [PubMed] [Google Scholar]
- 9. Dröge W. (2002) The plasma redox state and ageing. Ageing Res. Rev. 1, 257–278 [DOI] [PubMed] [Google Scholar]
- 10. Hildebrandt W., Alexander S., Bärtsch P., Dröge W. (2002) Effect of N-acetylcysteine on the hypoxic ventilatory response and erythropoietin production: linkage between plasma thiol redox state and O2 chemosensitivity. Blood 99, 1552–1555 [DOI] [PubMed] [Google Scholar]
- 11. Hildebrandt W., Kinscherf R., Hauer K., Holm E., Dröge W. (2002) Plasma cystine concentration and redox state in aging and physical exercise. Mech. Ageing Dev. 123, 1269–1281 [DOI] [PubMed] [Google Scholar]
- 12. Schafer F. Q., Buettner G. R. (2001) Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic. Biol. Med. 30, 1191–1212 [DOI] [PubMed] [Google Scholar]
- 13. Bianchi M. E., Manfredi A. A. (2007) High-mobility group box 1 (HMGB1) protein at the crossroads between innate and adaptive immunity. Immunol. Rev. 220, 35–46 [DOI] [PubMed] [Google Scholar]
- 14. Lotze M. T., Tracey K. J. (2005) High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat. Rev. Immunol. 5, 331–342 [DOI] [PubMed] [Google Scholar]
- 15. Tang D., Kang R., Zeh H. J., 3rd, Lotze M. T. (2011) High-mobility group box 1, oxidative stress, and disease. Antioxid. Redox Signal. 14, 1315–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Venereau E., Schiraldi M., Uguccioni M., Bianchi M. E. (December 1, 2012) HMGB1 and leukocyte migration during trauma and sterile inflammation. Mol. Immunol. 10.1016/j.molimm.2012.10.037 [DOI] [PubMed] [Google Scholar]
- 17. Orlova V. V., Choi E. Y., Xie C., Chavakis E., Bierhaus A., Ihanus E., Ballantyne C. M., Gahmberg C. G., Bianchi M. E., Nawroth P. P., Chavakis T. (2007) A novel pathway of HMGB1-mediated inflammatory cell recruitment that requires Mac-1-integrin. EMBO J. 26, 1129–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Palumbo R., Galvez B. G., Pusterla T., De Marchis F., Cossu G., Marcu K. B., Bianchi M. E. (2007) Cells migrating to sites of tissue damage in response to the danger signal HMGB1 require NF-κB activation. J. Cell Biol. 179, 33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Riuzzi F., Sorci G., Donato R. (2006) The amphoterin (HMGB1)/receptor for advanced glycation end products (RAGE) pair modulates myoblast proliferation, apoptosis, adhesiveness, migration, and invasiveness. Functional inactivation of RAGE in L6 myoblasts results in tumor formation in vivo. J. Biol. Chem. 281, 8242–8253 [DOI] [PubMed] [Google Scholar]
- 20. Taguchi A., Blood D. C., del Toro G., Canet A., Lee D. C., Qu W., Tanji N., Lu Y., Lalla E., Fu C., Hofmann M. A., Kislinger T., Ingram M., Lu A., Tanaka H., Hori O., Ogawa S., Stern D. M., Schmidt A. M. (2000) Blockade of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature 405, 354–360 [DOI] [PubMed] [Google Scholar]
- 21. Sahu D., Debnath P., Takayama Y., Iwahara J. (2008) Redox properties of the A-domain of the HMGB1 protein. FEBS Lett. 582, 3973–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kazama H., Ricci J. E., Herndon J. M., Hoppe G., Green D. R., Ferguson T. A. (2008) Induction of immunological tolerance by apoptotic cells requires caspase-dependent oxidation of high-mobility group box-1 protein. Immunity 29, 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tang D., Billiar T. R., Lotze M. T. (2012) A Janus tale of two active HMGB1 redox states. Mol. Med. 18, 1360–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Venereau E., Casalgrandi M., Schiraldi M., Antoine D. J., Cattaneo A., De Marchis F., Liu J., Antonelli A., Preti A., Raeli L., Shams S. S., Yang H., Varani L., Andersson U., Tracey K. J., Bachi A., Uguccioni M., Bianchi M. E. (2012) Mutually exclusive redox forms of HMGB1 promote cell recruitment or proinflammatory cytokine release. J. Exp. Med. 209, 1519–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu A., Fang H., Dirsch O., Jin H., Dahmen U. (2012) Oxidation of HMGB1 causes attenuation of its pro-inflammatory activity and occurs during liver ischemia and reperfusion. PLoS ONE 7, e35379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang H., Lundbäck P., Ottosson L., Erlandsson-Harris H., Venereau E., Bianchi M. E., Al-Abed Y., Andersson U., Tracey K. J., Antoine D. J. (2012) Redox modification of cysteine residues regulates the cytokine activity of HMGB1. Mol. Med. 18, 250–259 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27. Kozlowski J. M., Fidler I. J., Campbell D., Xu Z. L., Kaighn M. E., Hart I. R. (1984) Metastatic behavior of human tumor cell lines grown in the nude mouse. Cancer Res. 44, 3522–3529 [PubMed] [Google Scholar]
- 28. Schanda P., Brutscher B. (2005) Very fast two-dimensional NMR spectroscopy for real-time investigation of dynamic events in proteins on the time scale of seconds. J. Am. Chem. Soc. 127, 8014–8015 [DOI] [PubMed] [Google Scholar]
- 29. Schanda P., Kupce E., Brutscher B. (2005) SOFAST-HMQC experiments for recording two-dimensional heteronuclear correlation spectra of proteins within a few seconds. J. Biomol. NMR 33, 199–211 [DOI] [PubMed] [Google Scholar]
- 30. Pervushin K., Riek R., Wider G., Wüthrich K. (1997) Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. U.S.A. 94, 12366–12371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rance M., Loria J. P., Palmer A. G. (1999) Sensitivity improvement of transverse relaxation-optimized spectroscopy. J. Magn. Reson. 136, 92–101 [DOI] [PubMed] [Google Scholar]
- 32. Clore G. M., Gronenborn A. M. (1998) Determining the structures of large proteins and protein complexes by NMR. Trends Biotechnol. 16, 22–34 [DOI] [PubMed] [Google Scholar]
- 33. Knapp S., Müller S., Digilio G., Bonaldi T., Bianchi M. E., Musco G. (2004) The long acidic tail of high mobility group box 1 (HMGB1) protein forms an extended and flexible structure that interacts with specific residues within and between the HMG boxes. Biochemistry 43, 11992–11997 [DOI] [PubMed] [Google Scholar]
- 34. Stott K., Watson M., Howe F. S., Grossmann J. G., Thomas J. O. (2010) Tail-mediated collapse of HMGB1 is dynamic and occurs via differential binding of the acidic tail to the A and B domains. J. Mol. Biol. 403, 706–722 [DOI] [PubMed] [Google Scholar]
- 35. Watson M., Stott K., Thomas J. O. (2007) Mapping intramolecular interactions between domains in HMGB1 using a tail-truncation approach. J. Mol. Biol. 374, 1286–1297 [DOI] [PubMed] [Google Scholar]
- 36. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe–a multidimensional spectral processing system based on Unix pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 37. Johnson B. A., Blevins R. A. (1994) NMRView–a computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 38. Ek M., Popovic K., Harris H. E., Nauclér C. S., Wahren-Herlenius M. (2006) Increased extracellular levels of the novel proinflammatory cytokine high mobility group box chromosomal protein 1 in minor salivary glands of patients with Sjogren's syndrome. Arthritis Rheum. 54, 2289–2294 [DOI] [PubMed] [Google Scholar]
- 39. Ellerman J. E., Brown C. K., de Vera M., Zeh H. J., Billiar T., Rubartelli A., Lotze M. T. (2007) Masquerader: high mobility group box-1 and cancer. Clin. Cancer Res. 13, 2836–2848 [DOI] [PubMed] [Google Scholar]
- 40. Logsdon C. D., Fuentes M. K., Huang E. H., Arumugam T. (2007) RAGE and RAGE ligands in cancer. Curr. Mol. Med. 7, 777–789 [DOI] [PubMed] [Google Scholar]
- 41. Lotze M. T., DeMarco R. A. (2003) Dealing with death: HMGB1 as a novel target for cancer therapy. Curr. Opin. Investig. Drugs 4, 1405–1409 [PubMed] [Google Scholar]
- 42. Tang D., Kang R., Zeh H. J., 3rd, Lotze M. T. (2010) High-mobility group box 1 and cancer. Biochim. Biophys. Acta 1799, 131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ulloa L., Messmer D. (2006) High-mobility group box 1 (HMGB1) protein: friend and foe. Cytokine Growth Factor Rev. 17, 189–201 [DOI] [PubMed] [Google Scholar]
- 44. Mollica L., Curioni A., Andreoni W., Bianchi M. E., Musco G. (2008) The binding domain of the HMGB1 inhibitor carbenoxolone: theory and experiment. Chem. Phys. Lett. 456, 236–242 [Google Scholar]
- 45. Mollica L., De Marchis F., Spitaleri A., Dallacosta C., Pennacchini D., Zamai M., Agresti A., Trisciuoglio L., Musco G., Bianchi M. E. (2007) Glycyrrhizin binds to high-mobility group box 1 protein and inhibits its cytokine activities. Chem. Biol. 14, 431–441 [DOI] [PubMed] [Google Scholar]
- 46. Wake H., Mori S., Liu K., Takahashi H. K., Nishibori M. (2009) Histidine-rich glycoprotein inhibited high mobility group box 1 in complex with heparin-induced angiogenesis in Matrigel plug assay. Eur. J. Pharmacol. 623, 89–95 [DOI] [PubMed] [Google Scholar]