Background: Tetraspanin-enriched microdomains (TEM) are ubiquitous specialized membrane platforms enriched in extracellular vesicles.
Results: Intracellular TEM interactome accounts for a great proportion of the exosome proteome. Selected CD81 ligands are depleted from exosomes in CD81-deficient cells.
Conclusion: Insertion into TEM may be necessary for protein inclusion into exosomes.
Significance: Exosome cargo selection remains largely unexplored. TEM may be specialized platforms to route exosome components.
Keywords: Exosomes, Membrane Structure, Proteomics, Tetraspanins, Vesicles
Abstract
Extracellular vesicles are emerging as a potent mechanism of intercellular communication because they can systemically exchange genetic and protein material between cells. Tetraspanin molecules are commonly used as protein markers of extracellular vesicles, although their role in the unexplored mechanisms of cargo selection into exosomes has not been addressed. For that purpose, we have characterized the intracellular tetraspanin-enriched microdomain (TEM) interactome by high throughput mass spectrometry, in both human lymphoblasts and their derived exosomes, revealing a clear pattern of interaction networks. Proteins interacting with TEM receptors cytoplasmic regions presented a considerable degree of overlap, although some highly specific CD81 tetraspanin ligands, such as Rac GTPase, were detected. Quantitative proteomics showed that TEM ligands account for a great proportion of the exosome proteome and that a selective repertoire of CD81-associated molecules, including Rac, is not correctly routed to exosomes in cells from CD81-deficient animals. Our data provide evidence that insertion into TEM may be necessary for protein inclusion into the exosome structure.
Introduction
Tetraspanin-enriched microdomains (TEM)3 are ubiquitous specialized membrane platforms (1) formed by the engagement of tetraspanins in molecular associations with lipids (2) and selected transmembrane proteins, mostly integrins, immunoglobulin-superfamily receptors, and metalloproteinases (1–3). In addition to the regulation of intracellular trafficking, insertion of receptors in TEM increases their local concentration at the membrane, which is crucial for adhesion processes during sperm-egg fusion, embryo development, immune system activation, and extravasation of immune or tumor cells (1). Different pathogens, such as hepatitis C, HIV and papilloma virus, or malaria-causing parasite, use TEM as entry gateways, intracellular reservoirs, or export routes (1).
Tetraspanin molecules are routinely used as protein markers of extracellular vesicles (EVs), such as exosomes or ectosomes, and regulate the internalization and recycling of associated receptors (4). Extracellular vesicles are emerging as a novel mechanism of intercellular communication (5), being involved in antigen presentation, antitumor immunity, and pathogen transmission (6). These vesicles carry genetic material, mRNA or microRNA (7–9), enabling their horizontal transfer between cells (7, 8, 10, 11) and acting as natural nanocarriers with a potential systemic distribution. However, little is known on how specific receptors or nucleic acids are routed to be exported into EVs. The expression in EVs of tetraspanins CD82 and Tspan8 confers them tumorigenic and angiogenic capacity (12, 13); however, the putative structural role of TEM in the unexplored process of cargo selection into exosomes has not been addressed.
Most studies on tetraspanins have emphasized their role as scaffolds of membrane microdomains, and some high throughput proteomic analyses of their associations at the membrane have been conducted (14). However, emerging evidence suggests that tetraspanins might also connect receptors to signaling cascades and the cytoskeleton. In this regard, the intracellular regions of tetraspanins are usually short, with no conserved features. The C-terminal regions of some tetraspanins have a net basic charge (15); others contain a PDZ-binding motif (16) or canonic internalization sequences (17, 18). Although it has been described that some tetraspanins associate with type-II phosphatidylinositol 4-kinase (19), activated PKC (20), 14-3-3 (21), syntenin-1 (16), or ERM proteins (ezrin, radixin, and moesin) (15), the identity of intracellular tetraspanin partners is still very incompletely defined.
In this work, we have used a collection of peptide ligands containing the intracellular C-terminal region of a representative set of tetraspanins and some tetraspanin-associated receptors and have performed a systematic characterization of intracellular TEM interactome in human lymphoblasts and their derived exosomes. Our results demonstrate that TEM ligands form a network of interactions and that this network accounts for a great proportion of the exosomal proteome, suggesting that TEM play a pivotal role in stabilizing the exosomal structure. Moreover, using CD81-deficient mice, we demonstrate by high throughput quantitative proteomics that depletion of this tetraspanin diminishes the concentration in exosomes of some of their associated partners in the network, thus providing evidence that TEM may play a role in defining the protein content of EVs.
EXPERIMENTAL PROCEDURES
Cell Cultures
Human primary lymphoblasts were obtained from peripheral blood lymphocytes of healthy donors by PHA/IL-2 stimulation. Mouse lymphoblasts were obtained from C57BL6 wt or CD81-KO animals (kindly provided by S. Levy, Stanford University). Splenocytes were stimulated with 2 μg/ml concanavalin A (Sigma) for 36 h and further cultured for 7 d.
Extracellular Vesicle Isolation and Quantification
For extracellular vesicle preparation, donor primary lymphoblast cells were cultured in RPMI 1640 supplemented with 10% FBS (depleted of bovine exosomes by overnight centrifugation at 100,000 × g). Cell supernatants were subjected to consecutive filtration and ultracentrifugation (100,000 × g) steps (6, 11), and the vesicles obtained were assessed by negative staining transmission electron microscopy. Extracellular vesicles were counted and sized by Nanoparticle Tracking Analysis (NTA) with Nanosight LM10 and NTA 2.3 Software (Nanosight, Wiltshire, UK).
Antibodies
Anti-ERM (90:3) has been described previously (15). Anti-filamin, anti-α-actinin, anti-nucleolin, and anti-β-actin antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), and anti-Rac1/2 and anti EF-1α were from Millipore (Billerica, MA).
Pulldown Assay
Biotinylated peptides with the following sequences were obtained from Ray Biotech, Inc. (Norcross, GA): CD147, biotin-SGSG-KRRKPDQTLDEDDPGAAPLKGSGHHMNDKDKNRQRNAT; CD69, MSSENCFVAENSSLHPESGQENDATSPHFSTRHEGSFQ-GSGSK-biotin; CD151, biotin-SGSG-YRSLKLEHY; CD81, biotin-SGSG-CCGIRNSSVY; EWI-2, biotin-SGSG-CCFMKRLRKR; ICAM-1, biotin-SGSG-RQRKIKKYRLQQAQKGTPMKPNTQATPP; VCAM-1, biotin-SGSG-RKANMKGSYSLVEAQKSKV; ICAM-3, biotin-REHQRSGSYHVREESTYLPLTSMQPTEAMGEEPSRAE; CD9, biotin-SGSG-CCAIRRNREMV; CCR7, biotin-KFRNDLFKLFKDLGCLSQEQLRQWSSCRHIRRSSMSVEAETTTTFSP; CXCR4, biotin-KFKTSAQHALTSVSRGSSLKILSKGKRGGHSSVSTESESSSFHSS. Each peptide (30 nmol) was coupled to 40 μl of streptavidin-Sepharose microbeads (GE Healthcare). Bound beads were incubated with total cell lysate or exosome extract for 60 min and washed three times in lysis buffer and 10 times in 50 mm ammonium bicarbonate, pH 8.8. Isolated proteins were then processed for digestion and peptide identification or Western blot.
Electron Microscopy
For electron microscopy, cell pellets were fixed in situ with 2% glutaraldehyde (Sigma) and 4% paraformaldehyde and embedded in epoxy resin (TAAB Laboratories). Ultrathin sections were counterstained with uranyl acetate and lead citrate and viewed under a Jeol JEM-1010 electron microscope. For negative staining of exosomes, ionized carbon and collodion-coated copper EM grids were floated on a sample drop, washed, and stained with 2% uranyl acetate (in double-distilled water) for 1 min.
Flow Cytometry
Cells were stained with the corresponding FITC-, phycoerythrin-, allophycocyanin-, and biotin-conjugated antibodies followed by streptavidin-allophycocyanin antibody. All antibodies used for flow cytometry were from BD Biosciences. Cells were analyzed using a FACSCanto II flow cytometer.
Quantitative RT-PCR
For quantitative RT-PCR, total cell mRNA was isolated with RNeasy Plus mini kit (Qiagen) and subjected to RT-PCR with High Capacity cDNA reverse transcription kit (Applied Biosystems) followed by quantitative PCR with Power SYBR Green PCR master mix (Applied Biosystems). mRNAs were normalized to that of β-actin, YWHAZ, and B2M housekeeping genes.
In-gel Protein Digestion
Proteins were in-gel digested using a previously described protocol (22). Briefly, 75 μl of beads were suspended in 25 μl of sample buffer and loaded in 2.8-cm-wide wells of an SDS-PAGE gel. The run was stopped as soon as the front entered 3 mm into the resolving gel. The protein band was visualized by Coomassie Blue staining, excised, and digested overnight at 37 °C with 60 ng/μl trypsin at 5:1 protein:trypsin (w/w) ratio in 50 mm ammonium bicarbonate, pH 8.8, containing 10% acetonitrile and 0.01% 5-cyclohexyl-1-pentyl-β-d-maltoside. The resulting tryptic peptides were extracted by a 1-h incubation in 12 mm ammonium bicarbonate, pH 8.8. TFA was added to a final concentration of 1%, and the peptides were finally desalted onto C18 Oasis cartridges, dried down, and analyzed separately by RP-HPLC-LIT for protein identification or subjected to 18O/16O labeling and isoelectric focusing fractionation for protein quantification.
Peptide 18O Labeling and Isoelectric Focusing Fractionation
Dried peptides from wild-type mice were labeled with 16O and peptides from CD81 knock-out mice were labeled with 18O as described previously (22). Labeling with 16O or 18O (95%; Isotec, Miamisburg, OH) was performed by incubation in 500 mm sodium citrate, pH 6, 20% acetonitrile, at a 1:200 (v/w) immobilized trypsin/protein ratio. After labeling, trypsin was removed by filtration (Wizard minicolumns, Promega) and by adding the irreversible trypsin inhibitor 2Nα-p-tosyl-l-lysine chloromethyl ketone at 1 mm for 1 h at 37 °C. The paired labeled samples were diluted to 2% acetonitrile, pH adjusted to 3 with 1 m ammonium formate, desalted onto C18 Oasis cartridges, and dried down. The peptide pools were taken up in focusing buffer (5% glycerol and 2% IPG buffer, pH 3–10, GE Healthcare), loaded into 24 wells over a 24-cm-long Immobiline DryStrip gel, pH 3–10 (GE Healthcare), and separated by isoelectric focusing on a 3100 OFFGEL fractionator (Agilent, Santa Clara, CA). The recovered fractions were desalted, using OMIX C18 tips, with 50% acetonitrile in 5 mm ammonium formate, pH 3, and peptides were dried down prior to RP-HPLC-LIT.
Mass Spectrometry and Data Analysis
Isolated peptides were analyzed by LC-MS/MS using a Surveyor LC system coupled to a linear ion trap mass LTQ spectrometer (Thermo Fisher) as described previously (23). For peptide identification, the LTQ was operated in a data-dependent MS/MS mode using the 15 most intense precursors detected in a survey scan from 400 to 1,600 m/z, whereas for quantification of 16O/18O labeled peptides, the LTQ was operated in data-dependent ZoomScan and MS/MS switching mode (23, 24). Protein identification was carried out as described previously (23) using the SEQUEST algorithm (Bioworks 3.2 package, Thermo Finnigan). The MS/MS raw files were searched against the Human Swiss-Prot database (UniProt release 14.0, 19,929 sequence entries for human) supplemented with porcine trypsin and bacterial streptavidin. SEQUEST results were validated using the probability ratio method (25), and false discovery rates were calculated using the refined method (26). Peptide and scan counting was performed assuming as positive events those with an FDR equal to or lower than 5%. Peptide quantification from ZoomScan spectra and calculation of labeling efficiencies for all identified peptides with an FDR lower than 5% were performed as described (27, 28).
Statistical Analysis of Proteomics Data
The median number of peptides obtained in the pulldowns from all the baits and from the negative control was used as a reference for each protein, and the standard deviation was estimated by the median absolute deviation method. A protein interaction was considered to be specific in a given experiment when the number of peptides identified with a bait increased with respect to the median with a statistical significance of p < 0.05. In experiments with fewer than four baits, statistical significance was calculated using the negative control as reference. To detect interactions with low abundance proteins, we also considered as specific those proteins for which one peptide was detected with only one bait. Specific interactions were considered significant when found in at least half of the experiments performed with the bait. Interactions in exosome pulldown experiments were validated by using one-tailed paired Student's t test, comparing results from either CD81 or EWI-2 pulldown experiments with those from the controls, and applying a statistical significance of p < 0.05. Dataset normalization was performed by taking the maximum number of unique peptides identified with each bait as reference. Quantitative data from O18 labeling were analyzed on the basis of a random effects hierarchical model implemented in the QuiXoT software platform (28). Details of the statistical model and the algorithm can be found in previous publications (22, 28).
Systems Biology Analysis of Interacting Proteins
Interactomes were analyzed with Ingenuity Pathway Analysis (IPA; Ingenuity Systems, Mountain View, CA).
RESULTS
Proteomic Characterization of Intracellular Tetraspanin-enriched Microdomain Interactome
Our fist aim was to define the full interactome of tetraspanin-based membrane platforms in a high throughput approach. We performed a series of pulldown assays using as baits biotinylated peptides spanning the C-terminal regions of tetraspanins CD9, CD81, and CD151 and of the tetraspanin-associated immunoglobulin superfamily receptors ICAM-1, VCAM-1 (29) and EWI-2 (30, 31) (Fig. 1A), which were bound to streptavidin-coated Sepharose beads. As controls, we used uncoated beads as well as beads coated with biotinylated peptides spanning the intracellular sequences of immunoglobulin superfamily members CD147 and ICAM-3, the C-lectin CD69, and the chemokine receptors CXCR4 and CCR7.
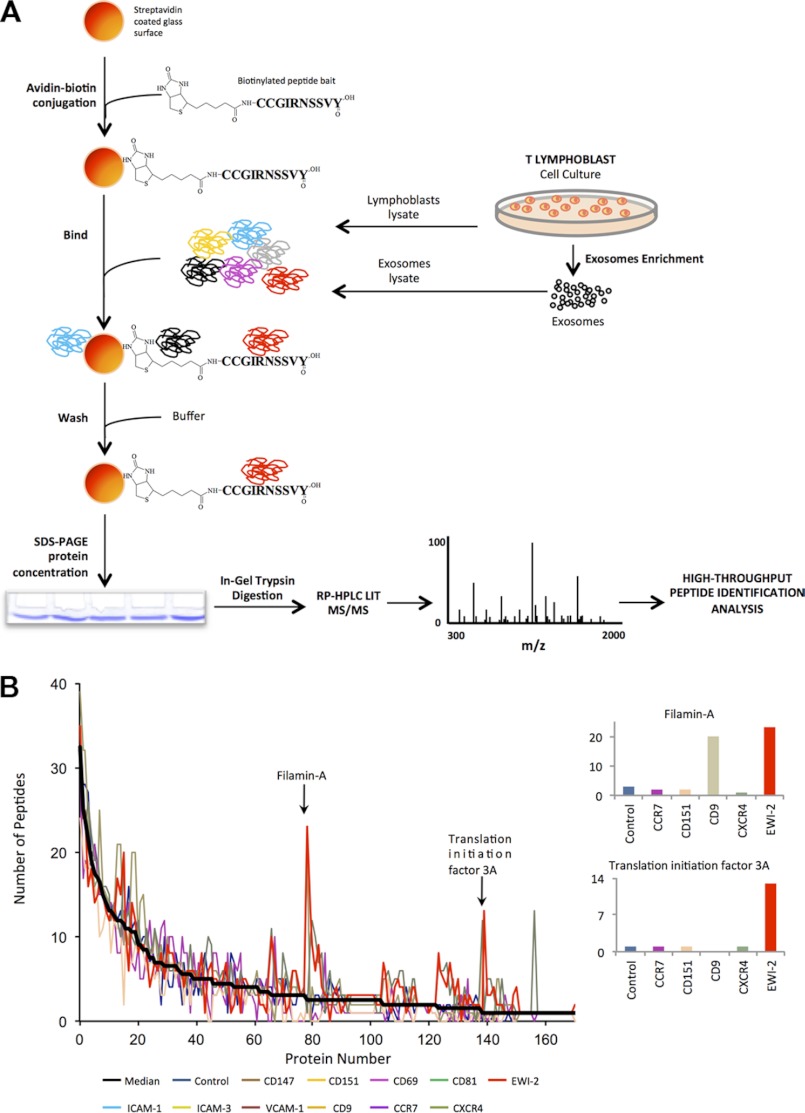
FIGURE 1.
Scheme of the protocol followed for the high throughput analysis of the interactome of the tetraspanin-enriched microdomains. A, biotinylated, synthetic peptides containing the intracellular regions (C-terminal in all cases except for CD69) of tetraspanins CD9, CD81(depicted in the figure), and CD151, their associated receptors (EWI-2, ICAM-1, and VCAM-1), or different control receptors (CD147, CD69, ICAM-3, CCR7, and CXCR4) were bound to streptavidin-Sepharose beads and incubated with protein extracts from whole cell lysates or exosomes from human primary lymphoblasts. Beads were washed and directly subjected to concentrating SDS-PAGE. Protein bands were trypsin-digested, and the resulting peptides were identified by liquid chromatography in tandem with linear ion trap mass spectrometry. B, peptide counting analysis reveals specific protein interactions with peptide baits. The left plot represents the distribution of the number of peptides identified per each protein in a representative pulldown assay. On the right, the number of peptides from the proteins is indicated by the arrows, which show specific interactions with one or more baits are shown.
To identify interacting partners, the beads were incubated with total cell lysates of human primary lymphoblasts. Bead-bound proteins were buffer-washed, and after SDS addition, they were subjected to concentrating SDS-PAGE, according to a previously optimized in-gel trypsin digestion protocol whose reproducibility has been previously tested using stable isotope labeling approaches (22); the resulting peptides were then identified by high throughput mass spectrometry (Fig. 1A). Interaction specificity of a protein with a bait was attested by statistical comparison with the results obtained with all other baits and naked beads. The background distribution of nonspecific interactions was constructed by ordering proteins by the median of the number of peptides identified in all pulldowns and was remarkably similar for the different baits used in the same pulldown experiment so that specific interactions clearly stood out from the background distribution (Fig. 1B) and could be detected by a simple statistical test, as explained under “Experimental Procedures.” To sort out interactions that showed a biological reproducibility, a second specificity filter was imposed to the data by comparing the results obtained from different experimental replicas and performing a second statistical analysis. In total, 53 pulldown experiments, each one followed by high throughput mass spectrometry analysis, were performed using lysates from primary lymphoblasts from seven different human donors (supplemental Table S1). A total of 4,918 unique peptides, belonging to 1,957 different proteins, were identified from all the pulldown experiments (supplemental Data File 1). The majority of specific interactions were consistently observed in biological replicates; however, to increase the sensitivity and detect those interactions with low abundance proteins, we considered as specific interactions those that were detected in at least half of the experimental replicas. Finally, and as a further validation step, some selected interactions were confirmed by Western blot analysis (Fig. 2).
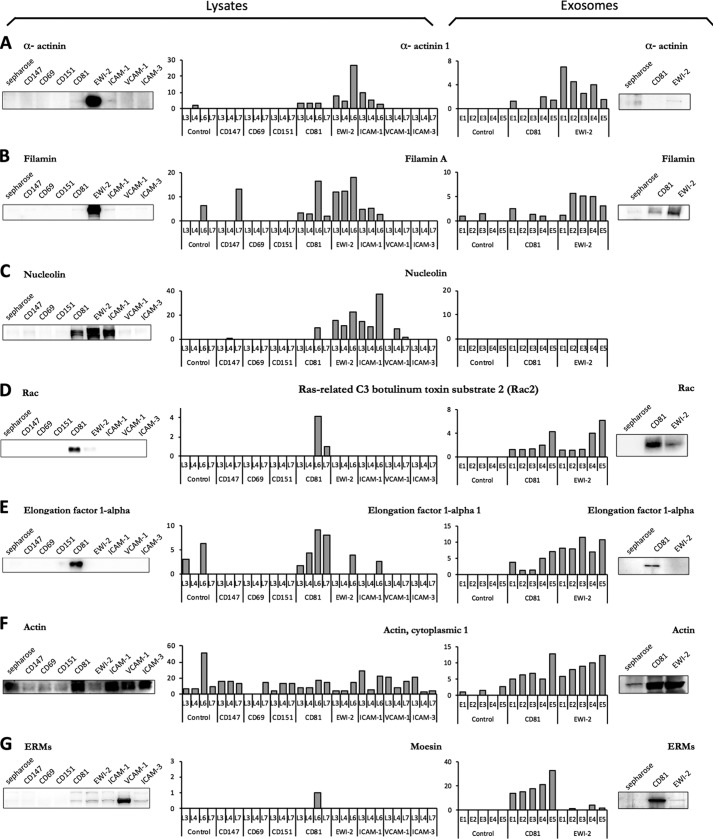
FIGURE 2.
Specificity of interactions between TEM receptors and intracellular proteins in cell lysates and exosomes of human primary lymphoblasts. Charts show proteomic analysis (high throughput mass spectrometry) of pulldown assays in whole cell lysates (left) or exosome extracts (right) of primary human lymphoblasts. Data correspond to interaction with α-actinin (A), filamin (B), nucleolin (C), Rac (D), elongation factor 1-α (E), actin (F), and ERMs (G). Western blotting of equivalent pulldowns in whole cell lysates and exosomes confirms these interactions.
A total of 240 protein partners showed specific interaction with at least one bait. From these, a very limited number of interactions were observed with baits CD147, CD69, CD151, VCAM-1, and ICAM-3, indicating that the specificity controls were stringent enough to sort out nonspecific interactions. In clear contrast, EWI-2, ICAM-1, and CD81 produced a large number of interactions judged to be specific according to the same criteria. There were protein interactions, such as those with actin, that were detected with a large number of peptides, but with most of the peptide baits, and hence did not pass the specificity filters (Fig. 2F). In contrast, some of the specifically interacting proteins, such as α-actinin, filamin, and nucleolin (Fig. 2, A–C, left panels), were also detected with a large number of peptides; however, they were only detected in the pulldowns with CD81, ICAM-1, and EWI-2 baits and barely detected with any of the other baits. Interestingly, ICAM-1, EWI-2, and CD81 shared a large proportion of binding patterns (supplemental Table S2 and Fig. 3B). Nevertheless, some of the interactions were specific of only one of these baits; this is the case for Ras-related proteins such as the GTPase Rac (Fig. 2D) and elongation factor 1-α (EF1-α), which were only detected with CD81 (Fig. 2E). Finally, when interactions were analyzed with IPA software, a clear network of interconnected interactions was revealed that linked most of the detected partners of the three TEM components. The proteins in the network formed five main clusters, including actin-related proteins, tubulins, ribosomal and nucleic acid-related proteins, and histones (Fig. 3A), which were not detected with any of the other 8 baits.
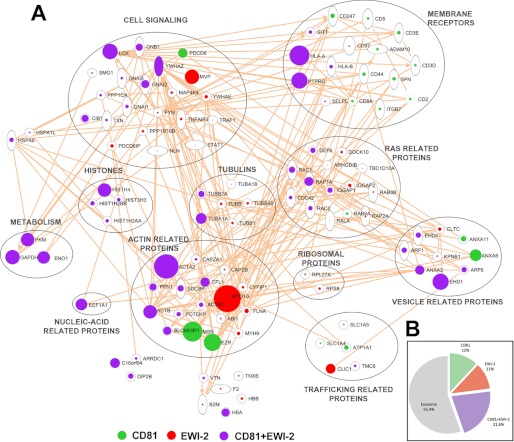
FIGURE 3.
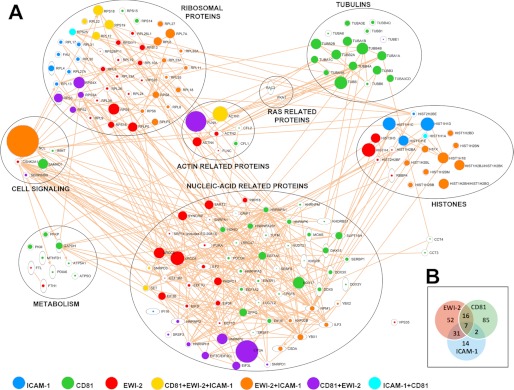
Protein interaction networks for ligands of CD81, EWI-2, and ICAM-1. A, intracellular ligands of CD81, EWI-2, or ICAM-1 were manually clustered into groups according to their function using the IPA analysis program. Symbol size is proportional to the number of peptides identified for each protein in the pulldown assays. B, Venn diagram showing the degree of overlap between the sets of proteins interacting with CD81, EWI-2, and ICAM-1 baits.
Tetraspanin Interactome in Exosomes
The interactions of TEM components with ribosomal, nucleic acid-binding, and processing proteins may have functional consequences during adhesion and spreading (32, 33). They may also be relevant for the sorting of mRNA and microRNA to extracellular vesicles, including exosomes, for which tetraspanin proteins are commonly employed as molecular markers (34). To explore the role of TEM in EVs, we isolated them from primary human lymphoblast cultures. Electronic microscopy of negatively stained samples revealed a prevalence of cup-shaped vesicles with the typical size of exosomes (not shown). High throughput mass spectrometry protein identification of our samples revealed the presence of 489 proteins in at least three out of five different human donors. This list is the most detailed characterization to date of the protein composition of primary human T-lymphoblast-derived exosomes (supplemental Table S3 and supplemental Data File 2), with 74% of the identified peptides belonging to proteins found in ExoCarta database, confirming the purity of our samples. EVs were enriched in tetraspanins or previously identified tetraspanin-associated molecules such as MHC isoforms, CD4, CD8, ADAM-10, CD44, CD98, integrins, ERMs, and syntenin (1) (supplemental Table S3). These data have been also incorporated into the Vesiclepedia database (59).
We then performed an exhaustive analysis of the exosomal TEM interactome, using the same approach employed for total cell lysates of human primary lymphoblasts. For these analyses, we chose CD81 and EWI-2 C-terminal peptide baits, which produced the most representative results in cell lysates, and also uncoated beads as negative controls. We performed a total of 15 pulldown experiments using EVs from lymphoblasts from five different human donors. A total of 172 proteins were found that interacted specifically with CD81, EWI-2, or both (supplemental Table S4 and S5, Fig. 2, right panels, and supplemental Data File 3). Consistently, a large number of interacting partners previously identified in total cell lysates were also detected in exosome extracts, including α-actinin, filamin, histones, tubulins, ribosomes, and Ras-related proteins. IPA analysis of the exosomal interactome revealed again a coherent network (Fig. 4A), containing the same clusters detected in cell lysates. Some of these clusters, such as those containing actin- and Ras-related and cell signaling proteins, were now detected with a larger number of proteins due to the increased sensitivity attained in a less complex proteome. Interactions with proteins highly enriched in exosomes such as ERMs could now be detected (Figs. 2G and 4A). In addition, new interaction clusters were now revealed that were not detected before; these included membrane receptors and trafficking- and vesicle-related proteins. These categories included tetraspanin-associated transmembrane receptors such as CD44, ADAM-10, CD8, HLA, and integrins, suggesting that intracellular cross-linking of proteins in TEM might reinforce their associations.
FIGURE 4.
Protein interaction networks for CD81 and EWI-2 ligands identified in human primary lymphoblast-derived exosomes. A, intracellular ligands of CD81 and EWI-2 were manually clustered into groups according to their function using the IPA analysis program. Symbol size is proportional to the number of peptides identified for each protein in the pulldown assays. B, proportions of proteins in exosomes that were identified as ligands of EWI-2 and CD81 in exosomes. The proportions were calculated on the basis of the number of peptides identified per protein in total exosome lysates.
Role of Tetraspanin-enriched Microdomains in the Protein Composition of Exosomes
To analyze the relative proportion of the tetraspanin network in the protein composition of exosomes, we analyzed how many of the protein components of the interactomes of EWI-2 and CD81 were detected in the total exosomal proteome. Assuming that the protein amount is roughly proportional to the number of peptides identified by mass spectrometry (35), we found that 45% of the protein content of exosomes was formed by members of the interactome of these TEM components (Fig. 4B). In contrast, only three out of the 753 proteins annotated in the Human MitoCarta database coincided with those found in pulldowns with CD81 or EWI-2 baits with either total cell lysates or exosome extracts (not shown). These findings support the hypothesis that the incorporation of receptors or soluble proteins into TEM might route them to exosome compartmentalization.
To directly explore the potential role of TEM in exosomal cargo selection, we analyzed the effect of depleting tetraspanin CD81 on the protein composition of exosomes. For this purpose, we made a comparative study of exosomes secreted from lymphoblast cultures obtained from wild-type and CD81-deficient mice. The use of a KO mouse model to study CD81-dependent protein cargo in exosomes allowed us a complete depletion of tetraspanin CD81 in primary cells, which cannot be attained with siRNA in primary T-lymphoblasts of human origin. Moreover, thanks to the high proteome coverage we obtained in both models, we could confirm that exosome cargo similarity is higher than 80% (supplemental Data Files 2 and 4).
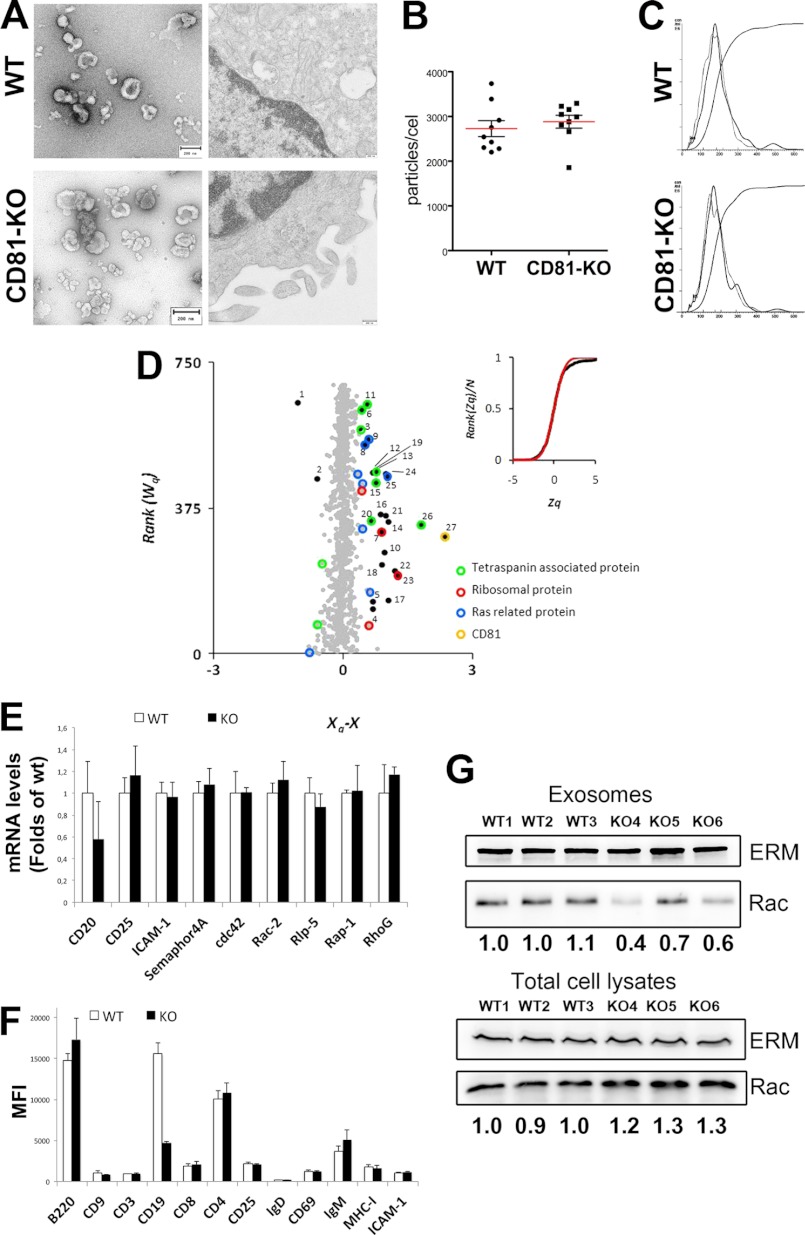
Consistently, with the highly redundant nature of TEM, depletion of CD81 did not produce an appreciable effect in the size or amount of released exosomes, as revealed by both electron microscopy (Fig. 5A) and NTA analyses (Fig. 5, B and C). We could readily find multivesicular bodies in both WT and CD81-deficient lymphoblasts (Fig. 5A). We then performed a high throughput quantitative proteomic analysis using the 18O-stable isotope labeling technique (22). The protein extracts from exosomes from WT and CD81-deficient animals were digested, and the peptides were labeled with either 16O or 18O, mixed, fractionated, and subjected to mass spectrometry quantification. We were able to quantify a total of 1,945 peptides corresponding to 692 proteins (supplemental Data File 4). Most proteins were similarly abundant in both genotypes (Fig. 5D, gray dots), and consistently, the distribution of protein log2 ratios closely followed the null hypothesis trend described by the statistical model (Fig. 5B, inset). These results indicated that the vast majority of proteins were present in the same proportion in the two exosome preparations. However, a small proportion of proteins, 25 out of the 692 quantified proteins, was significantly less abundant in the CD81-deficient EV preparation (Fig. 5D, numbered dots, Table 1, and supplemental Data File 4). The affected molecules included CD81-associated transmembrane molecules, such as components of the B-cell receptor complex, CD20, ICAM-1, and HLA isotypes; ion transport channels; and ribosomal proteins (Table 1). Notably, there was a marked reduction in the group of Ras-related proteins (Fig. 5D, colored dots). To rule out the possibility that changes in exosome composition just reflected changes in total protein expression in the cell, we analyzed the expression by RT-PCR (Fig. 5E) or flow cytometry (Fig. 5F) of a number of molecules shown to be reduced in exosomes from CD81 KO lymphoblasts. We analyzed by RT-PCR the mRNA levels of nine different molecules, including membrane receptors (CD20, CD25, ICAM-1, and semaphorin 4A) and GTPases (Cdc42, Rac-2, Rlp5, Rap-1, and RhoG). From all these molecules, only CD20 expression levels were found to be reduced in CD81-deficient cells (Fig. 5D). Moreover, when we analyzed by flow cytometry the actual protein content of the cells, we could not detect any reduction in CD25, ICAM-1, CD4, IgM, IgD, or MHC-I mean fluorescence intensity (Fig. 5F). As controls, we confirmed previous data that reported no changes in CD4, CD8, and B220 but a drastic decrease on CD19 surface levels (36–38)(Fig. 5F). The activation markers CD69 or tetraspanin CD9 were also unaffected (Fig. 5F).
FIGURE 5.
Lack of CD81 alters the protein composition of exosomes. A, exosomes (left panels) were enriched from lymphoblast culture media as described under “Experimental Procedures” and negatively stained. Cells (right panels) were pelleted and fixed with 2% glutaraldehyde and 4% paraformaldehyde and embedded into epoxy resin. Ultrathin sections were viewed in a Jeol JEM-1010 electron microscope after counterstaining with uranyl acetate and lead citrate. A multivesicular body is shown. B, exosome counts in the supernatants of 7-day cultures of mouse blasts from WT or KO animals by NTA. Data represent the mean ± S.E. of individual measurements. C, NTA profile of exosome samples derived from the supernatants of 7-day cultures of mouse blasts from WT or CD81 KO animals. Two different profiles are overlaid for each sample. D, quantitative proteomics comparison of the compositions of pooled exosome extracts from three WT or three CD81 knock-out mouse lymphoblast cultures by stable isotope labeling with 16O (wild type) or 18O (CD81 KO). The plot shows the distribution of log2 ratios of individual proteins, corrected for the grand mean, and ranked by their statistical weight, which is the inverse of the variance and measures the accuracy of the quantification. Black dots indicate proteins with statistically significant abundance changes with respect to the bulk of nonchanging proteins (gray dots). The quantified proteins are listed in supplemental Data File 4. The numbers indicate the proteins as listed in Table 1. The inset plot shows the cumulative frequency distribution of the standardized variable Zq (black line), which expresses the quantitative data in terms of units of variance, showing the agreement with the expected null hypothesis distribution (red line) and the deviation due to proteins changing their abundance. Quantified proteins from the same ontological categories show a similar pattern of abundance changes. Colored points indicate ribosomal proteins, Ras-related proteins, and membrane proteins associated with tetraspanins with a Zq ≥ |2.4|. E, quantitative RT-PCR analyses of mRNA of selected molecules in 7-day cultures of mouse blasts from three WT animals or three CD81 KO mice (mean ± S.D.) F, flow cytometry analyses of selected receptors in 7-day cultures of mouse blasts from three WT animals or three CD81 KO mice. Data represent the mean fluorescent intensity (MFI) (mean ± S.D.) of the positive cells for each marker. G, Western blot analysis of Rac content in exosomes and total cell lysates from independent lymphoblast cultures from wild-type (WT1–3) or CD81 knock-out (KO4–6) mice. Numbers represent densitometric values of Rac signal corrected with ERM loading and normalized to WT1 sample.
TABLE 1.
Protein abundance changes between exosomes from CD81 knock-out mice as compared with exosomes obtained from wild type animals
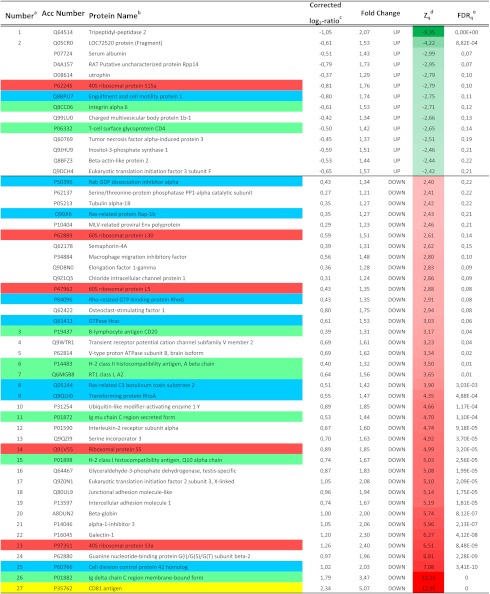
a Proteins showing statistically significant abundance changes are numbered as in Fig. 5B.
b Proteins belonging to some selected ontological categories are colored using the same set of colors as in Fig. 5B.
c Log2 ratios were corrected by subtracting the grand mean value.
d Standardized normal values were obtained by dividing protein log2 ratios by their variance. Only proteins with |Zq| ≥ 2.4 are listed. Data are colored according to the color scales on the bottom.
e False discovery rate of quantification at the protein level.
To confirm our proteomic results, we performed a semiquantitative Western blot analysis in both exosome lysates and total cell extracts obtained from animals belonging to the two genotypes on the CD81-specific interactor Rac. Our results revealed that overall cellular Rac levels were unaltered (Fig. 5G); however, the proportion of Rac in EVs released by CD81-deficient cells was lower than in EVs from the control cells (Fig. 5G), confirming the results obtained by quantitative proteomics. Taken together, these results indicate that despite the redundancy among tetraspanins and the overlap in the interactomes of CD81 and its partners EWI-2 or ICAM-1, the genetic deletion of CD81 specifically impairs the inclusion into EVs of a selective repertoire of CD81-exclusive partners. These findings suggest that TEM may act as compartmentalizing platforms of receptors and signaling proteins for their selection toward exosomes.
DISCUSSION
In this work, we have set up a method to undertake a systematic exploration of the intracellular connections of tetraspanins and tetraspanin-associated receptors. The method is based on a robust in-gel digestion protocol that was previously demonstrated to produce highly reproducible results in terms of peptide identification as judged by statistical modeling by using stable isotope labeling approaches (22). This reproducibility was essential to discern specific interactions from the bulk of nonspecific ones. We used as baits the cytoplasmic tails from a collection of tetraspanins, as well as a series of related proteins, which were bound to streptavidin-Sepharose beads. This approach has been previously validated; the relative performance of this method to identify interacting molecules was seen to be better than that obtained using as baits GST fusion proteins and reproduced by direct immunoprecipitation of endogenous molecules (15). Basic stretches that function as ERM-binding sites are found in the C-terminal regions of several tetraspanin-associated receptors (15, 39) and CD81 (15). Therefore, to rule out that the associations found in our screen are merely charge-driven electrostatic interactions or that they take place by indirect binding via ERM proteins, we included as negative controls C-terminal peptides from ICAM-3, an ERM-binding receptor (40) not reported to associate with tetraspanins, and from other related non-ERM-binding proteins. Indeed, we were able to confirm the association of ERM proteins with the cytoplasmic regions of VCAM-1, and to a lesser extent, EWI-2, ICAM-1, ICAM-3, and CD81. Note that despite the large number of peptide baits tested in this work, our method only produced a significant number of specific interactions with three of the partners used, which were clearly detected over the nonspecific background produced by the rest of the baits. Some of the interactions were further validated by Western blotting. Although our results have been obtained using an in vitro assay, several lines of evidence indicate that these interactions are specific. Firstly, a large proportion of the observed interactions were consistently reproduced in both cell lysates and exosomal preparations. Secondly, they could be reproduced and validated in several experiments performed using extracts prepared from different human donors. Thirdly, the observed interactions formed a consistent network in the two models used. In addition, there is evidence that at least some of the observed interactions are biologically relevant; thus, depletion of one of the tetraspanins in a mouse model produced a highly selective decrease in the proportion of some of the detected interacting partners in exosomes. Besides, some of the interactions identified in this work have been previously demonstrated to take place in a more biological context by coimmunoprecipitation of the endogenous molecules. Such is the case for ICAM-1/filamin (41); ICAM-1/actinin (42); CD81/actinin (43); EWI-2/actinin (44); or CD81/Rac. (45). Some of these interactions have been proven to be direct using purified systems, such as CD81/ERMs, EWI-2/ERMs (15), and CD81/Rac (45). However, we should note here that the experimental design used in this work does not allow determining whether all these interactions involve direct protein-protein contacts or are mediated through intermediate partners. Hence, the strong binding to adaptor molecules α-actinin and filamin, known to colocalize with elongation factors and polysomes at the branch points of actin filaments (46), might mediate the interaction of ICAM-1, EWI-2, and CD81 with nucleic acid-binding proteins, which are detected in our analyses with a lower number of peptides, being in any case specific and therefore potentially relevant.
Tetraspanin-enriched microdomains play a role in many biological phenomena that involve adhesion to the extracellular matrix or to other cells. Insertion into these specialized membrane domains ensures a correct level of aggregation or clustering of the associated adhesion receptors, necessary for coordinated cell adhesion and migration (1). However, mounting evidence suggests that tetraspanins may also connect these receptors with specific signaling pathways or link them to the underlying cortical cytoskeleton. Nevertheless, the interactions taking place in their cytoplasmic domains are very poorly characterized, so an important result in this work corresponds to the detailed map of TEM intracellular interactome.
Our data provide firm evidence for the role of tetraspanin-enriched microdomain-dependent compartmentalization of intracellular components. The connection of tetraspanin-enriched microdomains with RNA-binding proteins has potential relevance in the process of cell spreading and migration. Nascent adhesions incorporate RNA and RNA-binding proteins in a structure called the spreading initiation center (33). Moreover, ribosomes are recruited to focal adhesion sites by integrin ligation and mechanical tension (32). Adhesion to the extracellular matrix regulates translation (47), and some translation initiation factors and RNA-binding proteins have been shown to interact with integrins (48–50). In addition, the connection with RNA and DNA-binding proteins might be important for the incorporation of mRNA and microRNA into released vesicles such as exosomes (6, 11, 51).
Multivesicular bodies and exosomes are well known to be highly enriched in tetraspanins (52, 53). We have therefore decided to expand our analyses to a cellular model, i.e. exosomes secreted by human primary T-lymphoblasts, with a high biological relevance. The intracellular interactomes of CD81 and EWI-2 were found to contain a large number of interacting partners previously identified in cell lysates. In addition, exosome lysates provided a richer source of potential tetraspanin-interacting ligands, and the exosomal interactome contained new clusters of proteins. These included ERM proteins, TEM-associated transmembrane receptors, and integrins, suggesting the existence of an intracellular network of proteins stabilized through TEM. Remarkably, the tetraspanin interactome accounted for almost half of the protein composition of exosomes, suggesting that TEM may play a regulatory role in exosome structure by targeting a selected repertoire of receptors to exosomes. This issue is further supported by the decrease in abundance of a selected set of CD81-binding partners found in exosomes from CD81-deficient animals.
The role of tetraspanins in exosome biogenesis has been addressed only tangentially. Defective exosome release was reported in CD9 knock-out dendritic cells (13), based on Western blot measurements of flotillin. Although the exosomal abundance of CD81 is normally higher than that of CD9, we detected no gross defects in exosome release or multivesicular body morphology in CD81-deficient lymphocytes. This lack of gross alterations in the number or size of exosome is consistent with the TEM structure defined in our proteomic analysis, which is based in a very redundant network of protein-protein interactions, so that a given intracellular molecule may bind to different transmembrane components of TEM. Although we did detect reduced flotillin expression, CD81-deficient exosomes did not show a reduction in total protein content or in the expression of other abundant proteins in exosomes such as ERMs. In other studies, tetraspanin CD63 has been shown to be fundamental to the biogenesis of lysosome-related organelles (54) and to bind to syntenin (16), a PDZ protein necessary for intraluminal vesicle formation (55). However, in these studies, flotillin-enriched vesicles were found to be different from those containing CD63 (55).
Regarding the selection of cargo for packaging into exosomes, targeting of CD9P-1 to exosomes occurs, at least partially, even after silencing of its direct tetraspanin partners CD9 and CD81 (56). In contrast, detection of E-cadherin and β-catenin in the exosomal fraction depends on their association with tetraspanin CD82 (13). Our own data suggest that genetic deletion of CD81 impairs the inclusion of a selective repertoire of molecules into exosomes. This repertoire includes some transmembrane CD81 partners, such as MHC molecules, B-cell receptor, and ICAM-1. Moreover, lack of CD81 diminished the proportion of Rac present in exosomes, and insertion of Rac into tetraspanin-enriched microdomains was very specifically directed by its association with CD81 and was not observed with other tetraspanins or EWI-2.
Although to date the physiological functions of exosomes are still mostly unknown, some of the phenotypic characteristics exhibited by CD81 KO mice may be attributable to both cell and exosome dysfunction (i.e. female infertility (57, 58) or T-cell-dependent B-cell response (36, 38)). Future research will need to explore this intriguing possibility.
In conclusion, our results suggest that TEM act as specialized platforms for the compartmentalization of receptors and signaling proteins in the plasma membrane and that this compartmentalization aids the selection of receptors and intracellular components to be sorted toward exosomes. Tetraspanin-driven insertion into exosomes therefore may have potential in the design of genetic therapies to route a desired agent to these natural nanocarriers.
Acknowledgments
We thank Dr. M. Rejas for help with electron microscopy, Dr. S. Levy for CD81-KO mice, and S. Bartlett for English editing.
This work was supported by Grants PI080794 and PI11/01645 from the Instituto de Salud Carlos III (to M. Y.-M.); Grants ERC-2011-AdG 294340-GENTRIS, and SAF2011-25834 from the Ministerio de Ciencia e Innovación (MCINN) and INDISNET S2011/BMD-2332 from the Comunidad de Madrid (CAM) (to F. S. M); Grants BIO2009-07990 from the MCINN and BIO/0194/2006 from the CAM (to J. V.); and Grant RECAVA RD06/0014 from the Fondo de Investigaciones Sanitarias (MCINN) (to J. V. and F. S. M.).

This article contains supplemental Tables S1–S5 and Data Files 1–4.
- TEM
- tetraspanin-enriched microdomain(s)
- EV
- extracellular vesicle
- ERM
- ezrin-radixin-moesin
- IPA
- Ingenuity Pathway Analysis
- NTA
- Nanoparticle Tracking Analysis
- RP-HPLC-LIT
- reverse phase-HPLC-linear ion trap
- FDR
- false discovery rate.
REFERENCES
- 1. Yáñez-Mó M., Barreiro O., Gordon-Alonso M., Sala-Valdés M., Sánchez-Madrid F. (2009) Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol. 19, 434–446 [DOI] [PubMed] [Google Scholar]
- 2. Charrin S., le Naour F., Silvie O., Milhiet P. E., Boucheix C., Rubinstein E. (2009) Lateral organization of membrane proteins: tetraspanins spin their web. Biochem. J. 420, 133–154 [DOI] [PubMed] [Google Scholar]
- 3. Yáñez-Mó M., Gutiérrez-López M. D., Cabañas C. (2011) Functional interplay between tetraspanins and proteases. Cell. Mol. Life Sci. 68, 3323–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berditchevski F., Odintsova E. (2007) Tetraspanins as regulators of protein trafficking. Traffic 8, 89–96 [DOI] [PubMed] [Google Scholar]
- 5. Mittelbrunn M., Sánchez-Madrid F. (2012) Intercellular communication: diverse structures for exchange of genetic information. Nat. Rev. Mol. Cell Biol. 13, 328–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Théry C., Ostrowski M., Segura E. (2009) Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 9, 581–593 [DOI] [PubMed] [Google Scholar]
- 7. Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J. J., Lötvall J. O. (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 [DOI] [PubMed] [Google Scholar]
- 8. Skog J., Würdinger T., van Rijn S., Meijer D. H., Gainche L., Sena-Esteves M., Curry W. T., Jr., Carter B. S., Krichevsky A. M., Breakefield X. O. (2008) Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Balaj L., Lessard R., Dai L., Cho Y. J., Pomeroy S. L., Breakefield X. O., Skog J. (2011) Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pegtel D. M., Cosmopoulos K., Thorley-Lawson D. A., van Eijndhoven M. A., Hopmans E. S., Lindenberg J. L., de Gruijl T. D., Würdinger T., Middeldorp J. M. (2010) Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. U.S.A. 107, 6328–6333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mittelbrunn M., Gutiérrez-Vázquez C., Villarroya-Beltri C., González S., Sánchez-Cabo F., González M. Á., Bernad A., Sánchez-Madrid F. (2011) Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2, 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nazarenko I., Rana S., Baumann A., McAlear J., Hellwig A., Trendelenburg M., Lochnit G., Preissner K. T., Zöller M. (2010) Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. 70, 1668–1678 [DOI] [PubMed] [Google Scholar]
- 13. Chairoungdua A., Smith D. L., Pochard P., Hull M., Caplan M. J. (2010) Exosome release of β-catenin: a novel mechanism that antagonizes Wnt signaling. J. Cell Biol. 190, 1079–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Le Naour F., André M., Boucheix C., Rubinstein E. (2006) Membrane microdomains and proteomics: lessons from tetraspanin microdomains and comparison with lipid rafts. Proteomics 6, 6447–6454 [DOI] [PubMed] [Google Scholar]
- 15. Sala-Valdés M., Ursa A., Charrin S., Rubinstein E., Hemler M. E., Sánchez-Madrid F., Yáñez-Mó M. (2006) EWI-2 and EWI-F link the tetraspanin web to the actin cytoskeleton through their direct association with ezrin-radixin-moesin proteins. J. Biol. Chem. 281, 19665–19675 [DOI] [PubMed] [Google Scholar]
- 16. Latysheva N., Muratov G., Rajesh S., Padgett M., Hotchin N. A., Overduin M., Berditchevski F. (2006) Syntenin-1 is a new component of tetraspanin-enriched microdomains: mechanisms and consequences of the interaction of syntenin-1 with CD63. Mol. Cell. Biol. 26, 7707–7718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu L., He B., Liu W. M., Zhou D., Cox J. V., Zhang X. A. (2007) Tetraspanin CD151 promotes cell migration by regulating integrin trafficking. J. Biol. Chem. 282, 31631–31642 [DOI] [PubMed] [Google Scholar]
- 18. Stipp C. S., Kolesnikova T. V., Hemler M. E. (2003) Functional domains in tetraspanin proteins. Trends Biochem. Sci. 28, 106–112 [DOI] [PubMed] [Google Scholar]
- 19. Berditchevski F., Tolias K. F., Wong K., Carpenter C. L., Hemler M. E. (1997) A novel link between integrins, transmembrane-4 superfamily proteins (CD63 and CD81), and phosphatidylinositol 4-kinase. J. Biol. Chem. 272, 2595–2598 [DOI] [PubMed] [Google Scholar]
- 20. Zhang X. A., Bontrager A. L., Hemler M. E. (2001) Transmembrane-4 superfamily proteins associate with activated protein kinase C (PKC) and link PKC to specific β1 integrins. J. Biol. Chem. 276, 25005–25013 [DOI] [PubMed] [Google Scholar]
- 21. Clark K. L., Oelke A., Johnson M. E., Eilert K. D., Simpson P. C., Todd S. C. (2004) CD81 associates with 14-3-3 in a redox-regulated palmitoylation-dependent manner. J. Biol. Chem. 279, 19401–19406 [DOI] [PubMed] [Google Scholar]
- 22. Bonzon-Kulichenko E., Pérez-Hernández D., Núñez E., Martínez-Acedo P., Navarro P., Trevisan-Herraz M., Ramos Mdel C., Sierra S., Martínez-Martínez S., Ruiz-Meana M., Miró-Casas E., García-Dorado D., Redondo J. M., Burgos J. S., Vázquez J. (2011) A robust method for quantitative high-throughput analysis of proteomes by 18O labeling. Mol. Cell. Proteomics 10, M110.003335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. López-Ferrer D., Martínez-Bartolomé S., Villar M., Campillos M., Martín-Maroto F., Vázquez J. (2004) Statistical model for large-scale peptide identification in databases from tandem mass spectra using SEQUEST. Anal. Chem. 76, 6853–6860 [DOI] [PubMed] [Google Scholar]
- 24. Jorge I., Casas E. M., Villar M., Ortega-Pérez I., López-Ferrer D., Martínez-Ruiz A., Carrera M., Marina A., Martínez P., Serrano H., Cañas B., Were F., Gallardo J. M., Lamas S., Redondo J. M., García-Dorado D., Vázquez J. (2007) High-sensitivity analysis of specific peptides in complex samples by selected MS/MS ion monitoring and linear ion trap mass spectrometry: application to biological studies. J. Mass Spectrom. 42, 1391–1403 [DOI] [PubMed] [Google Scholar]
- 25. Martínez-Bartolomé S., Navarro P., Martín-Maroto F., López-Ferrer D., Ramos-Fernández A., Villar M., García-Ruiz J. P., Vázquez J. (2008) Properties of average score distributions of SEQUEST: the probability ratio method. Mol. Cell. Proteomics 7, 1135–1145 [DOI] [PubMed] [Google Scholar]
- 26. Navarro P., Vázquez J. (2009) A refined method to calculate false discovery rates for peptide identification using decoy databases. J. Proteome Res. 8, 1792–1796 [DOI] [PubMed] [Google Scholar]
- 27. Ramos-Fernández A., López-Ferrer D., Vázquez J. (2007) Improved method for differential expression proteomics using trypsin-catalyzed 18O labeling with a correction for labeling efficiency. Mol. Cell. Proteomics 6, 1274–1286 [DOI] [PubMed] [Google Scholar]
- 28. Jorge I., Navarro P., Martínez-Acedo P., Núñez E., Serrano H., Alfranca A., Redondo J. M., Vázquez J. (2009) Statistical model to analyze quantitative proteomics data obtained by 18O/16O labeling and linear ion trap mass spectrometry: application to the study of vascular endothelial growth factor-induced angiogenesis in endothelial cells. Mol. Cell. Proteomics 8, 1130–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barreiro O., Yáñez-Mó M., Sala-Valdés M., Gutiérrez-López M. D., Ovalle S., Higginbottom A., Monk P. N., Cabañas C., Sánchez-Madrid F. (2005) Endothelial tetraspanin microdomains regulate leukocyte firm adhesion during extravasation. Blood 105, 2852–2861 [DOI] [PubMed] [Google Scholar]
- 30. Charrin S., Le Naour F., Labas V., Billard M., Le Caer J. P., Emile J. F., Petit M. A., Boucheix C., Rubinstein E. (2003) EWI-2 is a new component of the tetraspanin web in hepatocytes and lymphoid cells. Biochem. J. 373, 409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stipp C. S., Kolesnikova T. V., Hemler M. E. (2001) EWI-2 is a major CD9 and CD81 partner and member of a novel Ig protein subfamily. J. Biol. Chem. 276, 40545–40554 [DOI] [PubMed] [Google Scholar]
- 32. Chicurel M. E., Singer R. H., Meyer C. J., Ingber D. E. (1998) Integrin binding and mechanical tension induce movement of mRNA and ribosomes to focal adhesions. Nature 392, 730–733 [DOI] [PubMed] [Google Scholar]
- 33. de Hoog C. L., Foster L. J., Mann M. (2004) RNA and RNA binding proteins participate in early stages of cell spreading through spreading initiation centers. Cell 117, 649–662 [DOI] [PubMed] [Google Scholar]
- 34. Zöller M. (2009) Tetraspanins: push and pull in suppressing and promoting metastasis. Nat. Rev. Cancer 9, 40–55 [DOI] [PubMed] [Google Scholar]
- 35. Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., Mann M. (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics 4, 1265–1272 [DOI] [PubMed] [Google Scholar]
- 36. Maecker H. T., Levy S. (1997) Normal lymphocyte development but delayed humoral immune response in CD81-null mice. J. Exp. Med. 185, 1505–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tsitsikov E. N., Gutierrez-Ramos J. C., Geha R. S. (1997) Impaired CD19 expression and signaling, enhanced antibody response to type II T independent antigen, and reduction of B-1 cells in CD81-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 94, 10844–10849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Miyazaki T., Müller U., Campbell K. S. (1997) Normal development but differentially altered proliferative responses of lymphocytes in mice lacking CD81. EMBO J. 16, 4217–4225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Barreiro O., Yanez-Mo M., Serrador J. M., Montoya M. C., Vicente-Manzanares M., Tejedor R., Furthmayr H., Sanchez-Madrid F. (2002) Dynamic interaction of VCAM-1 and ICAM-1 with moesin and ezrin in a novel endothelial docking structure for adherent leukocytes. J. Cell Biol. 157, 1233–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Serrador J. M., Alonso-Lebrero J. L., del Pozo M. A., Furthmayr H., Schwartz-Albiez R., Calvo J., Lozano F., Sánchez-Madrid F. (1997) Moesin interacts with the cytoplasmic region of intercellular adhesion molecule-3 and is redistributed to the uropod of T lymphocytes during cell polarization. J. Cell Biol. 138, 1409–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kanters E., van Rijssel J., Hensbergen P. J., Hondius D., Mul F. P., Deelder A. M., Sonnenberg A., van Buul J. D., Hordijk P. L. (2008) Filamin B mediates ICAM-1-driven leukocyte transendothelial migration. J. Biol. Chem. 283, 31830–31839 [DOI] [PubMed] [Google Scholar]
- 42. Carpén O., Pallai P., Staunton D. E., Springer T. A. (1992) Association of intercellular adhesion molecule-1 (ICAM-1) with actin-containing cytoskeleton and α-actinin. J. Cell Biol. 118, 1223–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mazzocca A., Liotta F., Carloni V. (2008) Tetraspanin CD81-regulated cell motility plays a critical role in intrahepatic metastasis of hepatocellular carcinoma. Gastroenterology 135, 244–256 [DOI] [PubMed] [Google Scholar]
- 44. Gordón-Alonso M., Sala-Valdés M., Rocha-Perugini V., Pérez-Hernández D., López-Martín S., Ursa A., Alvarez S., Kolesnikova T. V., Vázquez J., Sánchez-Madrid F., Yáñez-Mó M. (2012) EWI-2 association with α-actinin regulates T cell immune synapses and HIV viral infection. J. Immunol. 189, 689–700 [DOI] [PubMed] [Google Scholar]
- 45. Tejera E., Rocha-Perugini V., López-Martín S., Pérez-Hernández D., Bachir A. I., Horwitz A. R., Vázquez J., Sánchez-Madrid F., Yáñez-Mo M. (2013) CD81 regulates cell migration through its association with Rac GTPase. Mol. Biol. Cell 24, 261–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bassell G. J., Powers C. M., Taneja K. L., Singer R. H. (1994) Single mRNAs visualized by ultrastructural in situ hybridization are principally localized at actin filament intersections in fibroblasts. J. Cell Biol. 126, 863–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gorrini C., Loreni F., Gandin V., Sala L. A., Sonenberg N., Marchisio P. C., Biffo S. (2005) Fibronectin controls cap-dependent translation through β1 integrin and eukaryotic initiation factors 4 and 2 coordinated pathways. Proc. Natl. Acad. Sci. U.S.A. 102, 9200–9205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sanvito F., Piatti S., Villa A., Bossi M., Lucchini G., Marchisio P. C., Biffo S. (1999) The β4 integrin interactor p27BBP/eIF6 is an essential nuclear matrix protein involved in 60S ribosomal subunit assembly. J. Cell Biol. 144, 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schiller H. B., Friedel C. C., Boulegue C., Fässler R. (2011) Quantitative proteomics of the integrin adhesome show a myosin II-dependent recruitment of LIM domain proteins. EMBO Rep. 12, 259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Humphries J. D., Byron A., Bass M. D., Craig S. E., Pinney J. W., Knight D., Humphries M. J. (2009) Proteomic analysis of integrin-associated complexes identifies RCC2 as a dual regulator of Rac1 and Arf6. Sci. Signal. 2, ra51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Simons M., Raposo G. (2009) Exosomes–vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 21, 575–581 [DOI] [PubMed] [Google Scholar]
- 52. Escola J. M., Kleijmeer M. J., Stoorvogel W., Griffith J. M., Yoshie O., Geuze H. J. (1998) Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 273, 20121–20127 [DOI] [PubMed] [Google Scholar]
- 53. Théry C., Boussac M., Véron P., Ricciardi-Castagnoli P., Raposo G., Garin J., Amigorena S. (2001) Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol. 166, 7309–7318 [DOI] [PubMed] [Google Scholar]
- 54. van Niel G., Charrin S., Simoes S., Romao M., Rochin L., Saftig P., Marks M. S., Rubinstein E., Raposo G. (2011) The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev. Cell 21, 708–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Baietti M. F., Zhang Z., Mortier E., Melchior A., Degeest G., Geeraerts A., Ivarsson Y., Depoortere F., Coomans C., Vermeiren E., Zimmermann P., David G. (2012) Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 14, 677–685 [DOI] [PubMed] [Google Scholar]
- 56. Abache T., Le Naour F., Planchon S., Harper F., Boucheix C., Rubinstein E. (2007) The transferrin receptor and the tetraspanin web molecules CD9, CD81, and CD9P-1 are differentially sorted into exosomes after TPA treatment of K562 cells. J. Cell. Biochem. 102, 650–664 [DOI] [PubMed] [Google Scholar]
- 57. Miyado K., Yoshida K., Yamagata K., Sakakibara K., Okabe M., Wang X., Miyamoto K., Akutsu H., Kondo T., Takahashi Y., Ban T., Ito C., Toshimori K., Nakamura A., Ito M., Miyado M., Mekada E., Umezawa A. (2008) The fusing ability of sperm is bestowed by CD9-containing vesicles released from eggs in mice. Proc. Natl. Acad. Sci. U.S.A. 105, 12921–12926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rubinstein E., Ziyyat A., Prenant M., Wrobel E., Wolf J. P., Levy S., Le Naour F., Boucheix C. (2006) Reduced fertility of female mice lacking CD81. Dev. Biol. 290, 351–358 [DOI] [PubMed] [Google Scholar]
- 59. Kalra H., Simpson R. J., Ji H., Aikawa E., Altevogt P., Askenase P., Bond V. C., Borràs F. E., Breakefield X., Budnik V., et al. (2012) Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 10, e1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]