Background: Neurogenin3 is essential for pancreatic endocrine differentiation, but the mechanisms regulating its expression are poorly understood.
Results: Neurogenin3 transcriptionally activates its own gene in a direct autoregulatory loop involving Foxa2.
Conclusion: Neurogenin3 and Foxa2 cooperate during endocrine differentiation.
Significance: Elucidating the mechanisms governing Neurogenin3 expression and function is crucial to understand pancreatic endocrine differentiation and devise cell replacement therapies in diabetes.
Keywords: Diabetes, Differentiation, Embryo, Pancreatic Islets, Transcription/Developmental Factors, Foxa Factors, Neurogenin3, Beta Cells, Endocrine
Abstract
The transcription factor Neurogenin3 functions as a master regulator of endocrine pancreas formation, and its deficiency leads to the development of diabetes in humans and mice. In the embryonic pancreas, Neurogenin3 is transiently expressed at high levels for a narrow time window to initiate endocrine differentiation in scattered progenitor cells. The mechanisms controlling these rapid and robust changes in Neurogenin3 expression are poorly understood. In this study, we characterize a Neurogenin3 positive autoregulatory loop whereby this factor may rapidly induce its own levels. We show that Neurogenin3 binds to a conserved upstream fragment of its own gene, inducing deposition of active chromatin marks and the activation of Neurog3 transcription. Additionally, we show that the broadly expressed endodermal forkhead factors Foxa1 and Foxa2 can cooperate synergistically to amplify Neurogenin3 autoregulation in vitro. However, only Foxa2 colocalizes with Neurogenin3 in pancreatic progenitors, thus indicating a primary role for this factor in regulating Neurogenin3 expression in vivo. Furthermore, in addition to decreasing Neurog3 autoregulation, inhibition of Foxa2 by RNA interference attenuates Neurogenin3-dependent activation of the endocrine developmental program in cultured duct mPAC cells. Hence, these data uncover the potential functional cooperation between the endocrine lineage-determining factor Neurogenin3 and the widespread endoderm progenitor factor Foxa2 in the implementation of the endocrine developmental program in the pancreas.
Introduction
The pancreas consists of exocrine tissue (acinar and duct cells) and endocrine cells (insulin (β), glucagon (α), somatostatin (δ), pancreatic polypeptide (PP), and ghrelin (ϵ) cells) that are organized in the islets of Langerhans. During embryonic development, differentiation of these distinct cell types is controlled by the ordered and coordinated activation and inactivation of many transcriptional regulators. Among them, the basic helix-loop-helix (bHLH)3 transcription factor Neurogenin3 (Neurog3) implements the endocrine differentiation program in multipotent pancreatic progenitor cells (MPCs). Mice deficient in Neurog3 are born with no pancreatic endocrine cells (1), and genetic lineage tracing studies have demonstrated that all islet endocrine cells derive from Neurog3 positive progenitors (2). The finding that deficiency in Neurog3 caused by mutations in the NEUROG3 gene underlies neonatal and child-onset diabetes confirms the importance of this transcription factor in islet cell development and function in humans (3–5). Moreover, Neurog3 is sufficient to drive endocrine differentiation in a variety of in vivo and in vitro cellular contexts (6–9), highlighting its potential as a tool to generate replacement β-cells from other cell types for treatment of diabetes.
Neurog3 is transiently expressed in scattered MPCs within the trunk region of the developing pancreas, it is progressively down-regulated as the endocrine program is initiated and remains expressed at low levels in some adult islet cells (6, 10). Despite its relevance for endocrine cell formation, the molecular mechanisms that control Neurog3 expression are poorly understood. The transcription factors HNF6/Onecut1, HNF1b/Tcf2, HNF3b/Foxa2, Sox9, Pdx1, and Glis3 have all been acknowledged to be upstream regulators of the Neurog3 gene (11–15). Conversely, in clear parallelism to neural development, the Notch signaling pathway negatively regulates Neurog3 expression through the transcriptional repressor Hes1 (16), implying that release from Hes-1-mediated repression is required for Neurog3 gene activation in MPCs. Consistent with this notion, loss of Hes1 in the developing foregut endoderm is sufficient to induce ectopic endocrine cell formation (17).
Recent studies have pointed out that attaining high Neurog3 levels is critical for endocrine cell commitment. Thus, low Neurog3-expressing MPCs can adopt alternative exocrine fates (18, 19) and Neurog3 haploinsufficiency results in decreased endocrine cell mass (18). Hence, activation mechanisms must operate to allow for rapid and substantial increases in Neurog3 expression in a narrow time window that is estimated to be <24 h (20, 19). One of the proposed mechanisms whereby Neurog3 may amplify its protein levels is through a positive feedback loop involving the Neurog3 target Myt1b, which activates Neurog3 gene transcription (21). Another mechanism used by transcription factors to control their protein levels is self-regulation. In this regard, exogenous Neurog3 has been shown to induce the endogenous mouse Neurog3 gene in pancreatic duct-like mPAC cells (8), thus revealing positive autoregulation as a potential mechanism that may contribute to the rapid accumulation of Neurog3 protein in endocrine progenitors. However, in apparent contradiction to results in mPAC cells, Neurog3 has also been shown to inhibit its own promoter in NIH3T3 fibroblasts (16). Given the transient nature of Neurog3 expression in endocrine progenitors, it is conceivable that positive and negative regulatory mechanisms function in a timely coordinated manner to ensure tight regulation of Neurog3 expression during pancreatic development.
Because of the essential role played by Neurog3 in the determination of endocrine cell fate in the pancreas, deciphering the molecular mechanisms that regulate its expression is important to fully understand how pancreatic endocrine cell differentiation is accomplished. Based on previously published studies indicating that Neurog3 is able to activate its own expression (8), here we sought to gain further insight into the mechanisms governing Neurog3 autoregulation. Using reporter luciferase and ChIP assays, we show that Neurog3 activates its own promoter by binding to a conserved upstream regulatory region. In addition, we demonstrate that the forkhead transcription factor Foxa2 synergizes with Neurog3 to autoactivate the Neurog3 gene. Importantly, we reveal that Foxa2 is not only instrumental for Neurog3 autoregulation but it is also required for the activation of other Neurog3 target genes, indicating that Neurog3 and Foxa2 functionally cooperate to switch on the endocrine differentiation program in the pancreas.
EXPERIMENTAL PROCEDURES
Mice
CD1 mice used in this study were maintained in a barrier facility according to protocols approved by the University of Barcelona Animal Welfare Committee. The morning of the appearance of a vaginal plug was considered as embryonic day (E) 0.5.
Luciferase and Expression Vectors
A 5-kb fragment of 5′-flanking sequence extending from −4864 nt to +88 nt (+1 is the transcription start site) of the mouse Neurog3 gene was amplified from mouse tail genomic DNA using primers A/B. The 5-kb fragment was then used as template to generate shorter promoter fragments by PCR using primers C/B (3kb, −3057 nt to +88 nt) and D/E (−3.2 kb cluster region, −3885 nt to −3217 nt), or by restriction digest with NheI-XhoI (0.9 kb, −932 nt to +88 nt). Primer sequences are listed in supplemental Tables. The 5, 3, and 1 kb fragments were cloned upstream from the firefly luciferase gene in the KpnI/XhoI sites of the pFOXluc vector, while the enhancer fragment was cloned upstream of the firefly luciferase gene in the KpnI-XhoI sites of the pGL3-promoter vector (Promega, Madison, WI). To generate mutations, we used the QuikChange II XL Site-directed mutagenesis kit (Stratagene, Santa Clara, CA) and the primers listed in supplemental Tables. The artificial E box-driven enhancer luciferase reporter constructs used to test E box preference of the Neurog3/E47 heterodimers were kindly provided by K. Kroll (Washington University). The Renilla luciferase reporter plasmid pRL-CMV (Promega) was used as transfection control in all luciferase assays.
The expression vectors for mouse Neurog3 (pCMV-TNT-Neurog3) and mouse E47 (pCMV-TNT-E47) were previously described (13, 22). Expression vectors encoding rat Foxa1 (pCMV-Bgal-Foxa1) and rat Foxa2 (pCMV-Bgal-Foxa2) were kindly provided by M. A. Navas (Universidad Complutense de Madrid, Spain). Expression vectors encoding mouse HNF6 (pCMV4-HNF6) and rat HNF1b (pRSV-HNF1b) were kindly provided by M. Gannon (Vanderbilt University, TN) and J. Ferrer (IDIBAPS, Barcelona, Spain) respectively.
Cell Culture and Adenoviral Infection
All cell lines (mPAC L20, αTC1.6, βTC3, NIH3T3, MIN6, PANC-1) were grown in Dulbecco's Modified Eagle's Medium-4.5 g/liter of glucose (Sigma-Aldrich) supplemented with 10% fetal bovine serum plus antibiotics. MIN6 cells were additionally supplemented with 2 mm l-glutamine and 50 μm β-mercaptoethanol.
For adenoviral infection experiments, cells were seeded onto 6-well plates or 10-cm plates and treated 1 day later (or when reaching 70–80% confluence) with adenoviruses at a multiplicity of infection (moi) of 40 for 2–5 h at 37 °C in culture medium. Then, virus-containing media was replaced, and cells were cultured for an additional 44–48 h period. Generation and amplification of recombinant adenoviruses encoding human NEUROG3 and β-galactosidase was previously described (8). The adenovirus encoding the HA-tagged version of mouse Neurog3 used in ChIP and co-immunoprecipitation assays was kindly provided by G. Gradwohl (IGBMC, INSERM, University of Strasbourg, France).
Transient Transfections and Luciferase Assays
For luciferase assays, 1.2–1.5 × 104 mPAC or NIH3T3 cells were plated onto 96-well culture tissue plates 1 day before transfection. Transient transfections were performed using Metafectene (mPAC) from Biontex Laboratories GmbH (Martinsried, Germany) or Transfast (NIH3T3) from Promega according to the manufacturer's instructions. The amount of DNA used per well was 125 ng of firefly luciferase reporter vector, 2.5 ng of pRL.CMV and 10 ng of any cotransfected transcription factor cDNA. Empty expression vector was added when necessary to keep the amount of DNA equal in all wells. Cells were harvested 48 h after transfection and luciferase activity was analyzed using the Dual-Luciferase Reporter Assay System (Promega) and a Veritas microplate luminometer (Promega). Luciferase readings were normalized to activities of the internal control vector pRL.CMV.
For siRNA transfections, 2 × 105 mPAC cells were seeded onto 12-well plates and simultaneously transfected with 100 pmols of SMART pool mouse Foxa2 siRNA or a siControl (Dharmacon, Lafayette, CO) using Metafectene®SI (Biontex) following the manufacturer's instructions. The following day, cells were infected with recombinant adenoviruses as detailed above.
RNA Isolation and RT-PCR
Total RNA was prepared from cell lines and embryonic mouse pancreas using the RNeasy kit (Qiagen, Hilden, Germany). RNA samples were treated with DNase to remove contaminating genomic DNA. First-strand cDNA was prepared using 1–2 μg of total RNA, Superscript III enzyme (Invitrogen, Carlsbad, CA) and random hexamer primers (Invitrogen) in a total volume of 20 μl. Reverse transcription reaction was carried for 90 min at 50 °C and an additional 10 min at 55 °C. 1/20 or 1/40 of the transcribed cDNA was used as template for conventional RT-PCR or real time PCR, respectively. Real time PCR was performed on an ABI Prism 7900 sequence detection system using SybrGreen reagents (Express Greener, Invitrogen). PCR primer sequences are provided in the supplemental Tables.
Chromatin IP (ChIP) Assays
mPAC cells or E15.5 pancreata were fixed in 1% formaldehyde for 10–15 min and cross-linking was quenched by addition of 0.125 mm glycine. Histone ChIPs were performed as described elsewhere (22). ChIP against transcription factors were performed using the EpiQuik kit (Epigentek, Farmingdale, NY) following the manufacturer's instructions. Antibodies used were: rabbit anti-dimethylated and tri-methylated H3K4 (Millipore, Billerica, MA), goat anti-Foxa2 (HNF3β/M20) (Santa Cruz Biotechnology, Santa Cruz, CA), mouse anti-HA clone HA-7 (Sigma) and normal mouse and rabbit IgG as controls (Sigma). Immunoprecipitated DNA was assayed by real time PCR to test for the precipitation of specific promoter fragments with primers listed in the supplemental Tables. Real time PCR was performed on an ABI Prism 7900 sequence detection system using SybrGreen reagents (Express Greener, Invitrogen). Percentage of input was calculated as follows: 2−(Cp antibody-Cp input) − 2−(Cp IgG-Cp input).
Co-immunoprecipitation and Immunoblot Analysis
For co-immunoprecipitation assays, mPAC cells were lysed in coIP buffer (Tris-HCl 20 mm pH 7.5, NaCl 100 mm, EDTA 1 mm, Igepal CA-630 1%, NaF 5 mm, 10% protease inhibitor mixture from Sigma). Lysates were incubated for 15 min at 4 °C and cellular debris was removed by centrifugation at 10,000 × g for 15 min at 4 °C. Cellular lysates (500 μg) were then immunoprecipitated with 5 μg of anti-Foxa2 (Santa Cruz Biotechnology) or mouse IgG (Sigma) overnight at 4 °C. Immunoconjugates were recovered using protein G-coupled magnetic beads (Millipore) and, after 3 washes with coIP buffer, they were eluted by boiling in SDS-Laemli buffer. For preparing whole cell extracts, mPAC cells were lysed in triple detergent lysis buffer (Tris-HCl 50 mm, NaCl 150 mm, 0.1% SDS, 1% Nonidet P-40, and 0.5% sodium deoxycholate). Lysates were incubated for 20 min at 4 °C and cellular debris was removed by centrifugation at 10,000 × g for 15 min at 4 °C.
50 μg of whole cell extracts or immunoprecipitates were separated by PAGE-SDS electrophoresis, transferred to a Polyscreen PVDF membrane (Perkin Elmer, Waltham, MA) and incubated overnight at 4 °C with goat anti-mouse Foxa2 (1:1000, Santa Cruz Biotechnology), mouse anti-HA (1:1000, Sigma) or beta actin (1:1000, Sigma). Blots were visualized with ECL Reagent (Pierce Biotechnology) using a LAS4000 Lumi-Imager (Fuji Photo Film, Valhalla, NY). Protein spots were evaluated with Image J software.
Immunofluorescence and Immunocytochemistry
Mouse embryos were dissected in phosphate-buffered saline (PBS) and their intestinal tracts, including stomach, pancreas, and spleen, were fixed in 4% paraformaldehyde (PFA) for 3 to 6 h. After three washes in PBS, tissues were subsequently dehydrated, embedded in paraffin wax, and sectioned at 3 μm. For immunofluorescence, a standard immunodetection protocol was followed. Briefly, tissues were rehydrated and subjected to heat-mediated antigen retrieval in citrate buffer in a pressure cooker for 10 min. After a blocking step in 3% donkey serum/0.2% Triton X-100 tissue sections were incubated with the following primary antibodies: goat anti-Foxa2/HNF3β 1:100, rabbit anti-Sox9 1:200, rabbit anti-mucin-1 1:100 (Santa Cruz Biotechnology), mouse anti-Neurogenin3 1:500, mouse anti-Nkx6.1 1:500 (Developmental Studies Hybridoma Bank, Iowa City, IA), guinea pig anti-Pdx1 1:500 (Abcam, Cambridge, UK), guinea pig anti-insulin 1:500 (DAKO, Glostrup, Denmark), rabbit anti-chromogranin A 1:200 (Thermo Scientific). The secondary antibodies used were donkey anti-goat Cy2, donkey anti-mouse Cy3, donkey anti-rabbit Cy2 Cy3, or Alexa643, and donkey anti-guinea pig Cy3 (Jackson ImmunoResearch, Suffolk, UK).
For immunocytochemistry, mPAC and MIN6 cells were seeded onto coverslips and transduced with the β-galactosidase and NEUROG3 adenoviruses as described above. Cells were fixed 44–48 h after infection in 4% PFA for 20 min and permeabilized in PBS with 0.2–0.5% Triton X-100. After a blocking step of 1 h in 3% normal donkey serum, cells were incubated overnight with an antibody against mouse Neurog3 at 1:500 dilution. The secondary antibody used was donkey anti-mouse Cy3. Before mounting onto slides, nuclei were stained for 3 min in a 1:500 dilution of Hoechst 33258 (Sigma). Fluorescent images were captured using a Leica DMI 6000B widefield microscope or a Leica TCS SPE confocal microscope.
For quantification of Neurog3 and Foxa2 levels, single confocal scans were analyzed using image J software. Briefly, Foxa2 and Neurog3 fluorescence intensities of >100 Neurog3 positive cells were quantified and normalized for each channel to the maximum intensity (100%) for each of the images. To assign cells a qualitative fluorescence intensity value (HIGH or LOW), cells displaying fluorescence levels over 60% were arbitrarily considered HIGH and vice versa, and plotted onto a bar chart. For statistics, linear regression analysis was performed using GraphPad Prism.
Statistical Analysis
Data are presented as means ± S.E. of the mean (S.E.). Statistical significance was tested using Student's t test.
RESULTS
Neurog3 Activates Its Own Promoter
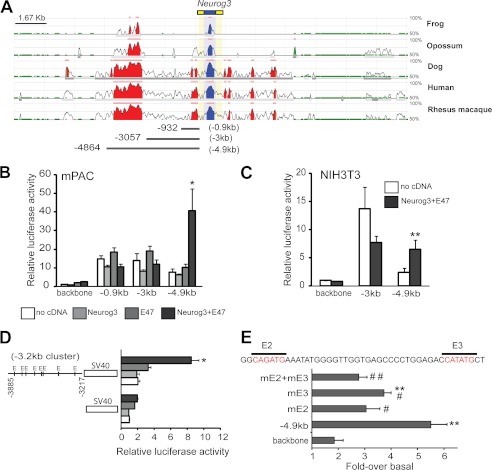
As a first step to locate cis-regulatory sequences potentially involved in Neurog3 autoregulation, we searched a 20 kb genomic region surrounding the mouse Neurog3 gene for evolutionarily conserved regions. We identified two areas that displayed extensive conservation throughout vertebrates: a ∼2 kb region located between −3 kb and −5 kb and a 0.3 kb region lying immediately upstream of the Neurog3 transcription initiation site (Fig. 1A).
FIGURE 1.
A, ECR browser visualization of 20 kb of the Neurog3 gene locus. The conservation profile of the mouse gene in comparison with the frog, opossum, dog, human, and rhesus macaque are shown. Coding exons are depicted in blue and yellow, blue corresponding to coding exons and yellow to UTRs. Conserved alignments are shown in blue if they overlap with a coding exon and in red if they correspond to intergenic regions. The pink bar on top of every layer provides an overview of the distribution of conserved regulatory elements. B and C, promoter fragments containing sequences extending from the indicated 5′-end to the +88 site of the mouse Neurog3 gene (as depicted in A) were ligated upstream of the firefly luciferase gene and transiently cotransfected with a CMV promoter-driven Renilla luciferase reporter plasmid and CMV-driven expression vectors encoding Neurog3 and/or E47 or an empty vector (no cDNA) into mPAC (B) and NIH3T3 (C) cells. Reporter gene activity is expressed relative to the activity of the promoter-less parental vector pFOXLuc (backbone) and the empty expression vector, set at 1. All data are presented as the mean ± S.E. from 4–8 independent experiments. D, fragment of the upstream Neurog3 conserved region (−3885 to −3217, or −3.2 kb cluster region) was ligated in front of the SV40 promoter in the pGL3-promoter luciferase reporter vector. mPAC cells were transiently transfected with this vector or the parental pGL3-promoter and expression vectors encoding Neurog3 and/or E47 or an empty vector as in B. Data shown represent the fold-activation relative to the activity of the parental reporter vector and the empty expression vector (set at 1) and are expressed as mean ± S.E. from 3 independent transfection experiments. E, mutational analysis of E boxes E2 and E3 present in the conserved −3.2 kb cluster region of the Neurog3 promoter. Constructs were derived from the −4.9 kb Neurog3 promoter fragment. Each vector was co-transfected with Neurog3 and E47 or an empty expression vector in mPAC cells. The activation of each construct by Neurog3+E47 is represented as fold-activation. Data are mean ± S.E. from 4–5 independent experiments. **, p < 0.01 compared with fold-response of the pFOXLuc backbone vector and #, p < 0.05, ##, p < 0.01 compared with fold-response of the −4.9 kb unmutated promoter construct.
To define sequences potentially involved in Neurog3 autoregulation, we focused on the 5-kb region located upstream of the Neurog3 TSS and cloned 5′ deletion fragments of a 5 kb mouse Neurog3 genomic construct, depicted in Fig. 1A, in front of the luciferase gene. We used the pancreatic cell line mPAC and NIH3T3 fibroblasts, where positive and negative Neurog3 self-regulatory loops have been previously described (8, 16), to test whether Neurog3 alone, or in conjunction with its dimeric partner E47, increased the transcriptional activity of these reporter vectors. We found that, in both cell types, Neurog3+E47 activated the 5kb Neurog3 promoter but were unable to do so when using the shorter promoter constructs containing 3 kb and 0.9 kb promoter fragments (Fig. 1, B and C). Altogether, our results demonstrate that Neurog3 can autoactivate its own expression and point to the distal conserved area (between −3 and −5 kb) as a required element in this autoregulatory loop. In agreement with these data, it is worth noting that sequences upstream from −2.6 kb have been shown to be necessary for high expression levels of a Neurog3 promoter-driven reporter gene construct in the embryonic pancreas of transgenic mice (12).
Detailed homology analyses revealed that, within the ∼2 kb distal conserved region, sequences from −3.1 to −3.9 kb displayed the highest conservation (92%) between the mouse and human genes. Moreover, we located 8 E boxes as putative Neurog3 binding sites in this highly similar fragment, all of which were conserved between both species. To determine whether this region alone could mediate Neurog3 autoregulation, we cloned a 670-bp fragment (−3885 nt to −3217 nt) containing all 8 E boxes in front of the SV40 promoter and co-transfected it along with Neurog3 and/or E47 in mPAC cells. As depicted in Fig. 1D, Neurog3+E47 significantly stimulated the activity of the Neurog3 (−3885 to −3217)-SV40 heterologous promoter by 3.5-fold, thus confirming that this region alone (hereafter referred to as −3.2 kb cluster) confers Neurog3 responsiveness to a heterologous promoter.
We next analyzed which E boxes in the −3.2 kb cluster were required for Neurog3 autoregulation. Given the high number of E boxes present in this region and the fact that bHLH factors exhibit preference for specific E box core sequences, we first determined which E box types are specifically activated by Neurog3. To this aim, we used ten different artificial enhancer luciferase reporter constructs containing three copies in tandem of each possible E box type, CANNTG (23), and tested their activation by Neurog3/E47. These assays revealed activation of the CATATG, CAGATG, and CAGCTG boxes by the Neurog3/E47 heterodimer, albeit the latter was more induced by E47 alone, indicating preference of the E47 homodimer for this site (supplemental Fig. S1). Based on these results, we then searched for potential target E boxes within the −3.2 kb cluster region and identified the closely located E boxes E2 and E3 (5′ to 3′ end) as putative Neurog3 binding sites. Thus, we disrupted E2 and E3, individually or in combination, and tested activation of the 5 kb Neurog3 promoter construct by Neurog3+E47. We found that mutation of E2 significantly impaired transcriptional activation of this construct, whereas mutation of E3 resulted in a reduced but still significant response. The effect of mutating both E2 and E3 was undistinguishable from that of E2 alone. Thus, these data suggest that E box E2 likely mediates Neurog3 autoregulation (Fig. 1E). Remarkably, the sequence of E box E2 (CAGATG) coincides with the Neurog3 responsive E boxes identified in the Pax4, NeuroD, and Atoh8 genes (22, 24–25).
Neurog3 Binds and Induces Chromatin Modifications at the Distal −3.2 kb Cluster Region of the Endogenous Neurog3 Gene
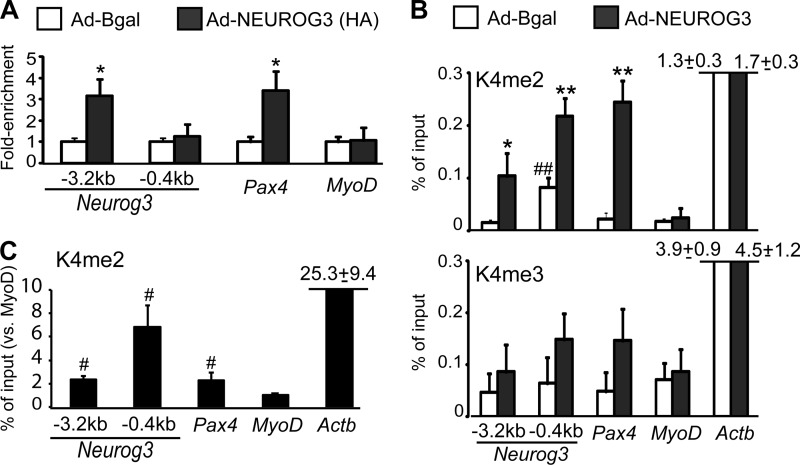
We next assessed whether Neurog3 was directly involved in Neurog3 gene autoregulation using ChIP assays. We determined Neurog3 binding to genomic sequences of the endogenous Neurog3 gene in mPAC cells transduced with a HA-tagged Neurog3 version. Our studies confirmed Neurog3 binding to the −3.2 kb cluster region but not to the proximal promoter (Fig. 2A). In addition, we confirmed that Neurog3 could bind to the −2 kb region of the Pax4 gene, as previously demonstrated by EMSA (24), whereas it did not bind the Myod promoter, chosen as a negative control, as it is not induced by Neurog3 in mPAC cells (Fig. 2A). Altogether, these data demonstrate that Neurog3 binds and activates its own promoter in vitro.
FIGURE 2.
A and B, chromatin was prepared from mPAC cells treated with the indicated recombinant adenoviruses and immunoprecipitation was carried out using specific antibodies for HA (A), H3K4me2, and H3K4me3 (B). Associated DNA was analyzed by real time PCR using primers that amplified the −3.2 kb cluster region and the proximal promoter (−0.4 kb) of the Neurog3 gene, the Neurog3-binding enhancer element of the Pax4 gene and the proximal promoters of the MyoD and/or Actb genes, which were included as controls for non-target/non-expressed and non-target/active gene, respectively. Percentage of input was calculated as described under “Experimental Procedures” and expressed relative to cells treated with the control adenovirus (set at 1) in A. In B and C, unnormalized values are shown to note differences in basal enrichment levels for these marks among the studied genes. Data represents mean ± S.E. from four independent overexpression experiments. C, chromatin was prepared from 8–10 pooled E15.5 embryonic pancreases and immunoprecipitated with a specific antibody for H3K4me2 or IgG as control. Associated DNA was analyzed as indicated previously. Percentage of input was expressed relative to the MyoD gene (set at 1). Data are mean ± S.E. for three independent chromatin preparations. *, p < 0.05, **, p < 0.01 versus Bgal; #, p < 0.05, ##, p < 0.01 versus MyoD.
Functional activation of developmentally regulated enhancers has been associated with the deposition of active histone H3 lysine 4 methylation marks (H3K4me2/3) within these enhancer regions (26, 27). Consistent with this notion, we found that exogenous NEUROG3 expression resulted in increased H3K4me2 levels at the upstream −3.2 kb cluster region as well as at the Pax4 enhancer in mPAC cells (Fig. 2B). Also correlating with transcriptional induction of the endogenous Neurog3 gene, NEUROG3 increased deposition of H3K4me2 marks at the Neurog3 proximal promoter while it had no effect on the MyoD or Actb proximal promoters, neither of which is activated by Neurog3 (Fig. 2B). On the other hand, NEUROG3 had no statistically significant effects on H3K4me3 levels at any of the genes studied, even though it tended to increase this mark at the Neurog3 proximal promoter and Pax4 upstream enhancer (Fig. 2B). We next assessed whether the −3.2 kb cluster region was enriched in active histone marks in vivo by performing ChIP assays using chromatin prepared from whole pancreases from embryonic day (E) 15.5 mouse embryos. As shown in Fig. 2C, the −3.2 kb cluster region of the Neurog3 gene as well as the Pax4 enhancer exhibited similar and significantly higher H3K4me2 levels than the MyoD promoter (Fig. 2C). Despite that Neurog3 expression peaks at E15.5 (6), it should be noted that endocrine progenitor cells represent a minor fraction of total pancreatic cells, thus explaining the modest enrichment levels observed in endocrine-specific genes relative to Actb. In summary, these data demonstrate deposition of active chromatin marks at the −3.2 kb cluster region in response to exogenous Neurog3 in vitro and confirm the presence of these marks in this region in vivo, supporting the notion that the −3.2 kb cluster becomes activated by this transcription factor and could thus function as a developmentally regulated enhancer.
mPAC Cells Express a Set of Transcription Factors Present in Embryonic MPCs
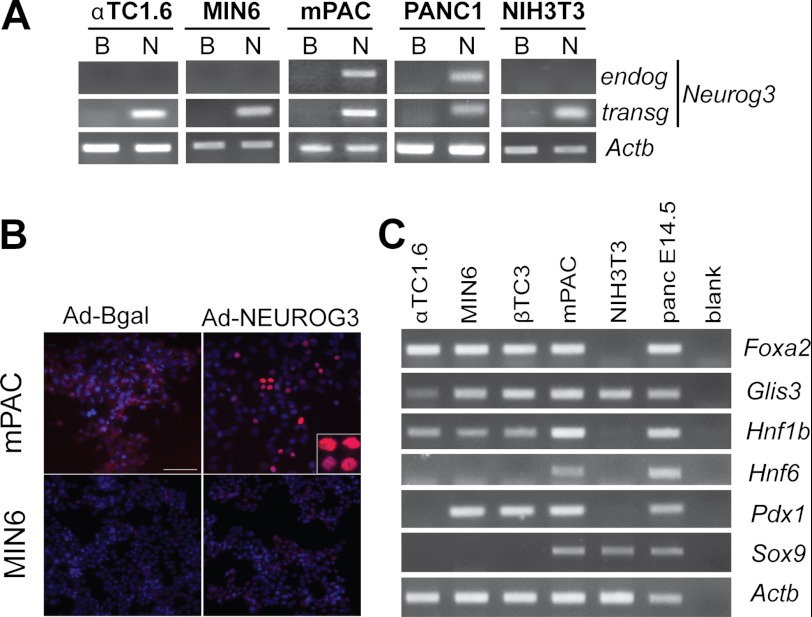
To further understand the molecular cues underlying Neurog3 autoactivation, we assessed whether the autoregulatory loop was dependent upon cell context. We found that, among the tested cell lines, only mouse ductal mPAC and human ductal PANC-1 cells exhibited positive Neurog3 autoregulation (Fig. 3A). Indeed, despite correct NEUROG3 transgene expression, neither the endocrine cell lines αTC1.6 (glucagon-expressing) and MIN6 (insulin-expressing) nor NIH3T3 fibroblasts induced the endogenous mouse Neurog3 gene (Fig. 3A). Furthermore, Neurog3 positive nuclei were detected by immunocytochemistry using an antibody against mouse Neurog3 in mPAC cells treated with the adenovirus encoding human NEUROG3 but not in control cells (Fig. 3B), confirming endogenous mouse Neurog3 protein accumulation in addition to mRNA induction upon NEUROG3 transgene introduction.
FIGURE 3.
A, indicated cell lines were infected with recombinant adenoviruses expressing β-galactosidase (B) or NEUROG3 (N). Total RNA was extracted 48 h after infection and expression of the endogenous Neurog3 mRNA and the NEUROG3 transgene was determined by RT-PCR. Note that the endogenous Neurog3 gene was only activated in ductal mPAC (mouse) and PANC-1 (human) cells despite the fact that the NEUROG3 transgene was present in all studied cell lines. The housekeeping Actb gene was used as loading control. Images are representative of a minimum of two independent experiments. B, immunocytochemistry using an antibody specific for mouse Neurog3 in mPAC and MIN6 cells 48 h after treatment with the indicated adenoviruses. C, RT-PCR showing expression of endogenous mRNAs encoding pancreatic progenitor transcription factors in the indicated mouse cell lines. Total RNA from E14.5 embryonic pancreas was included as a positive control for all assayed genes.
Pancreatic ductal cells are known to express several of the transcription factors found in MPCs in the embryonic pancreas, namely HNF6, HNF1b, Sox9, Pdx1, and Foxa2. Hence, we postulated that the responsiveness of mPAC cells to Neurog3 autoregulation might be related to the presence of these pancreatic progenitor transcription factors. We compared expression of these and other known Neurog3 upstream regulators among different cell lines, and found that mPAC cells expressed the complete set of mRNAs encoding these proteins (Foxa2, Glis3, HNF6, HNF1b, Pdx1, and Sox9), whereas the endocrine and non-pancreatic tested cell lines expressed some but not all of these regulators (Fig. 3C). Therefore, it is plausible that because of this particular expression profile, mPAC cells are uniquely equipped to allow Neurog3-dependent autoactivation. Moreover, it may be argued that the similarities in pancreatic progenitor factor expression between mPAC cells and MPCs may also underlie the reported ability of these cells and of primary duct cells to recapitulate the endocrine differentiation program upon Neurog3 expression (7, 8).
Neurog3 Synergizes with Foxa2 to Autoactivate Its Own Gene
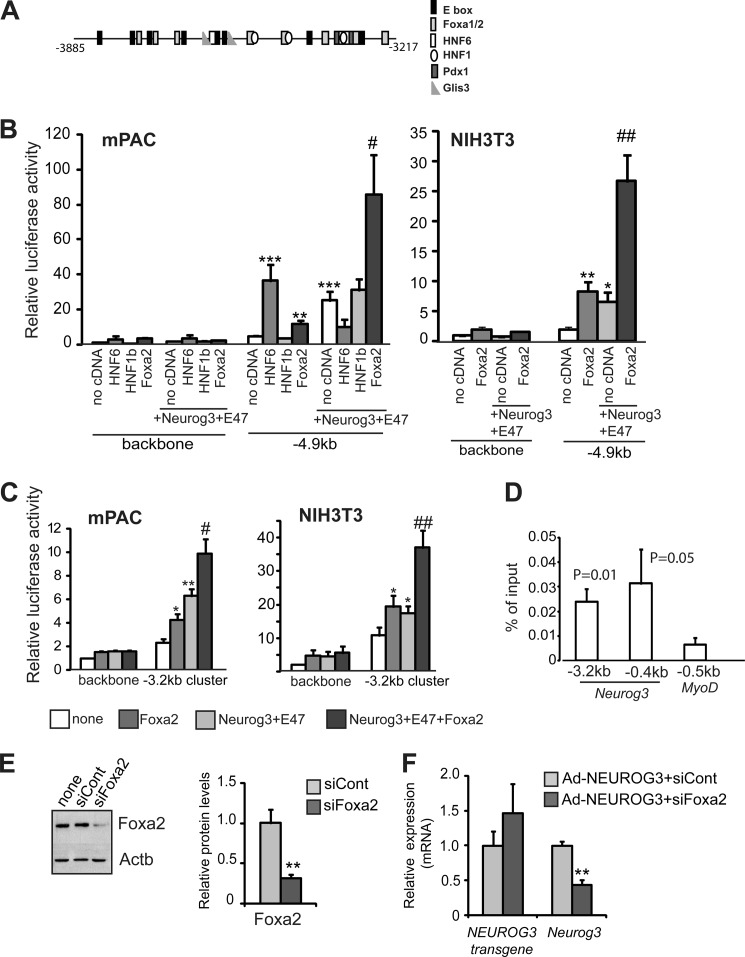
In addition to E boxes, the −3.2 kb cluster region contains potential binding sites for several of the pancreatic transcription factors previously identified as Neurog3 upstream regulators, including the HNF factors HNF6, HNF1b, and Foxa2 (Fig. 4A). HNF6 is required for Neurog3 expression in the embryonic pancreas (11), while HNF1b and Foxa2 have been connected to Neurog3 expression through co-immunolocalization (28) and in vitro DNA binding assays (12), respectively. Furthermore, as shown in Fig. 3C, these three factors are endogenously expressed in mPAC cells. To investigate their possible involvement in Neurog3 autoregulation, we co-transfected expression vectors for HNF6, HNF1b, and Foxa2 in the presence or absence of Neurog3+E47 along with the 5 kb Neurog3 promoter reporter construct into mPAC cells. We found that both HNF6 and Foxa2 individually transactivated the 5kb Neurog3 promoter, albeit with different potencies (8-fold by HNF6 and 2-fold by Foxa2), whereas HNF1b did not (Fig. 4B). Conversely, co-transfection of Neurog3 and its partner E47 with each of these factors revealed that only Foxa2 functioned in a synergistic manner with the bHLH heterodimer to transactivate the Neurog3 promoter (Fig. 4B). The synergism between Foxa2 and the bHLH heterodimer was corroborated in NIH3T3 fibroblasts (Fig. 4B). As expected, the synergistic action of Neurog3 and Foxa2 was specific for the 5 kb construct and was not detected on the 3 kb promoter luciferase construct in either mPAC or NIH3T3 cells (data not shown). To determine if the distal Neurog3 autoregulatory region was sufficient for synergistic activation by Foxa2, we used the Neurog3 (−3885 to −3217)-SV40 heterologous promoter in transient transfection assays as in Fig. 1D. We found that Foxa2 exerted a more moderate effect on Neurog3 promoter autoactivation from this reporter vector than from the 5 kb Neurog3 promoter construct in mPAC and NIH3T3 cells (Fig. 4C), which may indicate that additional sequences are needed for full response of the Neurog3 promoter to Foxa2+Neurog3+E47. In fact, the mouse Neurog3 gene harbors a Foxa2 site at the proximal promoter, conserved in the human gene, which has been shown to compete equally for Foxa2 binding as the distal site by shift assays (12). Further corroborating these evidences, we found that Foxa2 bound both the upstream and proximal Neurog3 promoter regions in mPAC cells using ChIP assays (Fig. 4D). However, mutation of the Foxa2 proximal site had no significant effect on the induction by Foxa2+Neurog3+E47 of the 5kb Neurog3 promoter construct in luciferase assays (supplemental Fig. S2), precluding a major involvement of this site in the Foxa2 synergistic effect.
FIGURE 4.
A, schematic representation of the −3.2 kb cluster region of the mouse Neurog3 distal promoter showing potential binding sites for several pancreatic progenitor transcription factors. B, luciferase reporter vector containing the −4.9 kb Neurog3 promoter fragment was transiently cotransfected with a CMV promoter-driven Renilla luciferase reporter plasmid and expression vectors encoding the indicated transcription factors or an empty vector (no cDNA) into mPAC and NIH3T3 cells. Reporter gene activity is expressed relative to the activity of the promoter-less parental vector pFOXLuc (backbone) and the empty expression vector, set at 1. All data are presented as the mean ± S.E. from 4–10 independent transfection experiments. *, p < 0.05, **, p < 0.01, ***, p < 0.001 versus no cDNA (empty bar) and #, p < 0.05, ##, p < 0.01 versus Neurog3+E47. C, luciferase reporter vector carrying the −3.2 kb cluster region of the Neurog3 promoter ligated in front of the SV40 promoter (same as used in Fig. 1D) was transiently cotransfected with a CMV promoter-driven Renilla luciferase reporter plasmid and expression vectors encoding the indicated transcription factors or an empty vector into mPAC and NIH3T3 cells. Data shown represent the fold-activation relative to the activity of the parental reporter vector and the empty expression vector (set at 1) and are expressed as mean ± S.E. from 6–8 independent transfection experiments. *, p < 0.05, **, p < 0.01 versus no cDNA (empty bar), and #, p < 0.05, ##, p < 0.01 versus Neurog3+E47. D, chromatin was prepared from mPAC cells and immunoprecipitation was carried out using an anti-Foxa2 antibody. Associated DNA was analyzed by real time PCR using primers that amplified the distal conserved region and proximal promoter of the Neurog3 gene and the MyoD proximal promoter, which was used as negative control. Percentage enrichment of input was calculated as described under “Experimental Procedures.” Data represent mean ± S.E. from five independent experiments. p values relative to MyoD are shown on top of each bar. E, mPAC cells were transfected with a siRNA directed against Foxa2 (siFoxa2) or with a control siRNA (siCont). Foxa2 protein levels were analyzed by immunoblot analysis 48 h after transfection. Representative immunoblot and densitometric quantification (normalized by Actb). Data are mean ± S.E. of three independent experiments. F, mPAC cells were transfected with siFoxa2 or siCont and infected with an adenovirus encoding NEUROG3. 48 h later, total RNA was extracted and expression of the NEUROG3 transgene and the endogenous Neurog3 gene were assayed by real time PCR. Results were normalized to the Tbp gene and expressed relative to levels in mPAC cells transfected with siCont (set at 1). Data are mean ± S.E. for five independent experiments. ***, p < 0.001 versus siCont.
We next investigated whether Foxa2 is necessary for autoregulation of the endogenous Neurog3 gene by using siRNA-based transient knockdown of Foxa2 expression in mPAC cells. Transfection of the Foxa2-specific siRNA resulted in a significant reduction of Foxa2 protein levels in mPAC cells (Fig. 4E) and a concomitant marked reduction (∼60%) in the activation of the Neurog3 gene upon exogenous NEUROG3 expression in these cells (Fig. 4F). Therefore, these data show that Foxa2 is required to induce the expression of the endogenous Neurog3 gene. All together, these findings point to a possible role of Foxa2 in endocrine fate specification through the modulation of Neurog3 expression levels in pancreatic progenitors.
Foxa2 Protein Localization during the Secondary Transition
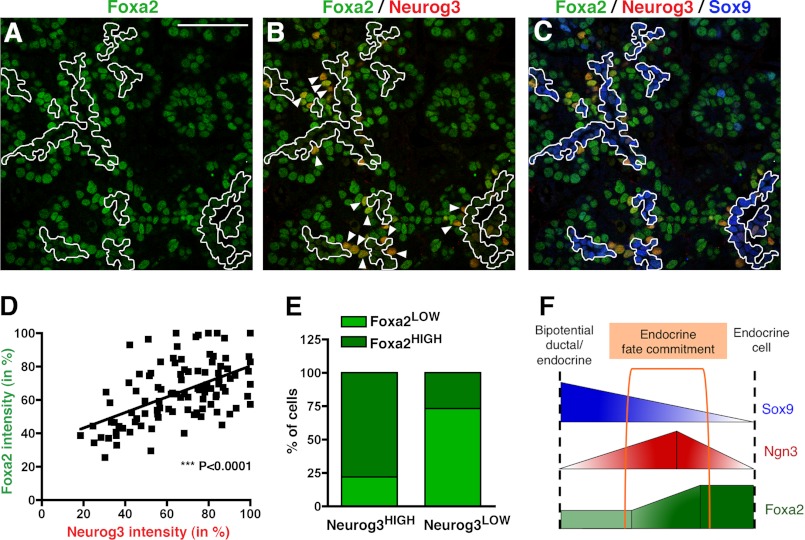
While Foxa2 protein expression has been well characterized in early stages of pancreas development and in adult islets (29), its expression pattern during the secondary transition, the major endocrine differentiation wave that begins at approximately E14 in the mouse, has not been investigated in detail. To further define the expression pattern of Foxa2 during this period and determine whether Neurog3 and Foxa2 co-exist in endocrine progenitors, we performed a series of immunofluorescence staining using antibodies specific to Foxa2 in E13.5, E14.5, and E15.5 pancreata. We found that Foxa2 is expressed and readily detectable throughout the pancreatic epithelium at the three stages analyzed, but with distinct expression patterns between them (Fig. 5A and supplemental Fig. S3). Thus, at E13.5, Foxa2 was homogeneously expressed throughout the branching pancreatic epithelium, with the exception of a few cells displaying higher levels of this transcription factor (Foxa2HIGH) (supplemental Fig. S3). These cells appeared both grouped in clusters (corresponding to glucagon-expressing cells, data not shown) or as single cells within or in close proximity to the pancreatic epithelial chords (supplemental Fig. S3). In contrast, at E14.5-E15.5, Foxa2 was expressed at lower levels in the cells lining the lumen of the epithelial chords, while Foxa2HIGH cells were detected within or adjacent to the epithelial chords (Fig. 5A and supplemental Fig. S3). Simultaneous staining for Foxa2 and Neurog3 revealed that Neurog3 positive cells were also positive for Foxa2 from E13.5 to E15.5 (Fig. 5B and supplemental Fig. S3). Remarkably, cells expressing high levels of Neurog3 (Neurog3HIGH, arrowheads in Fig. 5B) largely localized with Foxa2HIGH cells. To confirm a possible correlation between Neurog3 and Foxa2 expression levels, we quantified their respective fluorescence intensities in a series of confocal images from E15.5 pancreata. We found that indeed, there is a statistically significant correlation between Neurog3 and Foxa2 expression levels (Fig. 5D). In addition, cell quantification revealed that the ratio of Foxa2HIGH/Foxa2LOW cells is dramatically different in Neurog3LOW and Neurog3HIGH endocrine progenitor populations (Fig. 5E), indicating that Neurog3HIGH endocrine progenitors express Foxa2 at higher levels. Altogether, these studies show that Foxa2 and Neurog3 co-exist in vivo in endocrine progenitor cells and support a role for Foxa2 in enhancing the Ngn3 auto-regulatory loop during endocrine differentiation in vivo.
FIGURE 5.
Co-expression of Foxa2 and Neurog3 in the mouse embryonic pancreas by immunofluorescence. Staining for Foxa2 is shown alone (green, A), with Neurog3 (red, B), or with Neurog3 (red) and Sox9 (blue, C) at E15.5. D, linear regression analysis and (E) bar graph displaying the positive correlation between Neurog3 and Foxa2 expression levels. The signal intensities were normalized for each channel to the maximum intensity (100%) for each of the images. F, diagram depicting the conclusions from the expression analysis in vivo. Cells from the bipotential ductal/endocrine cells that adopt and endocrine cell fate are Neurog3HIGH/Foxa2HIGH/Sox9LOW/−. Arrowheads in B indicate Neurog3HIGH cells lying outside of the Foxa2LOW/Sox9HIGH cell domain lining the ducts (outlined). Scale bar: 50 μm.
The fact that Foxa2 is present throughout the pancreatic epithelium at E14.5 and E15.5, prompted us to investigate its expression in the different pancreatic cell lineages. Thus, we performed a series of simultaneous immunofluorescence staining to evaluate the presence of low and/or high levels of Foxa2 together with well-known pancreatic markers. Co-staining of Foxa2 and Nkx6.1, which at these stages is excluded from tip cells, revealed that Foxa2 is expressed in this domain, which mainly contributes to acinar fates by E14.5 (30) (supplemental Fig. S4). Notably, some of the scattered double positive cells for Nkx6.1 and Foxa2HIGH were detected adjacent to the epithelial chords, suggestive of cells in an intermediate state of β-cell differentiation (supplemental Fig. S4). Given that Foxa2 levels appeared to be more variable in the cells located in the trunk of the developing pancreas, we performed a more extensive analysis on endocrine (Pdx-1, insulin, ChromograninA) and ductal (Sox9, mucin-1) markers, the cell lineages distinguishable in the trunk domain at E14.5. Staining against Foxa2 and Pdx1 in E14.5 pancreata revealed an almost complete overlap between these two factors. At this stage, Pdx1 is found in all pancreatic epithelial cells, albeit at much higher levels in β-cells. Most of the Pdx1HIGH cells did not display the highest levels of Foxa2, indicating that Foxa2HIGH cells did not correspond to β-cells (supplemental Fig. S4). Accordingly, immunodetection of insulin showed that most of the Foxa2HIGH cells are not β-cells (supplemental Fig. S4). However, detection of the pan-endocrine marker Chromogranin A (ChgA), which is temporarily expressed after Neurog3, revealed an almost complete overlap with Foxa2HIGH cells (supplemental Fig. S4), showing their likely specification toward an endocrine fate.
During the secondary transition, Sox9 and mucin-1 serve as markers of a bipotential endocrine/ductal progenitor population, and are absent in both young and mature endocrine cells. Simultaneous antibody staining against Sox9 and Foxa2 revealed that Sox9+ bipotential ductal/endocrine progenitors contained Foxa2 at low levels. Conversely, Foxa2HIGH cells did not express Sox9 (Fig. 5C and supplemental Fig. S4). Triple immunofluorescence staining against Foxa2, Sox9, and Neurog3 further confirmed that most endocrine progenitors do not localize in the Foxa2LOW/Sox9+ ductal domain (Fig. 5C). Similarly, Foxa2LOW cells lining the epithelial chords contained mucin-1 on its apical side, thus confirming the low Foxa2 expression levels in ductal/endocrine progenitors (supplemental Fig. S4). In summary, we found that Foxa2 is expressed in low abundance in the ductal/endocrine progenitors lining the epithelial chords, but its expression level seems to peak in the Neurog3HIGH endocrine precursors supporting a role for Foxa2, together with Neurog3, in the specification of endocrine cells.
Foxa2 and Foxa1 Display Similar Abilities for Neurog3 Autoactivation
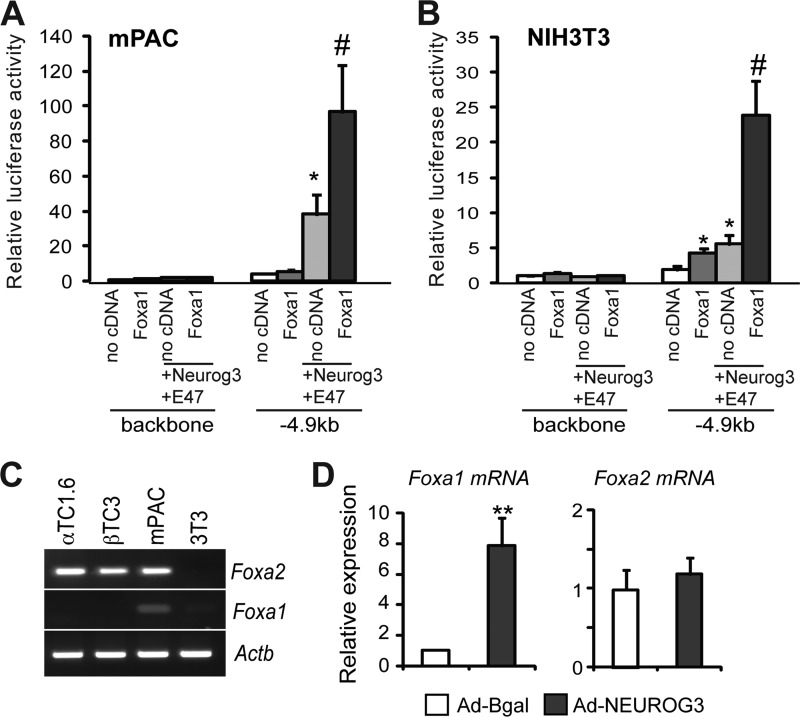
The closely related transcription factor Foxa1 shares gene targets with Foxa2 in multiple tissues including the pancreas. Both factors are thought to exert both common and distinct functions during pancreatic development as well as in the maintenance of adult islet functionality (31, 32). Hence, we postulated that Foxa1 and Foxa2 might also share the ability to enhance Neurog3 gene autoactivation. Consistent with this notion, we found that Foxa1 synergistically cooperated with Neurog3+E47 in activating the Neurog3 promoter in transient transfection assays in mPAC and NIH3T3 cells (Fig. 6, A and B), thus demonstrating that Foxa1, like Foxa2, can regulate Neurog3 promoter activity in vitro.
FIGURE 6.
A and B, −4.9 kb mouse Neurog3 promoter luciferase reporter vector or the backbone luciferase vector were cotransfected with a CMV promoter-driven Renilla luciferase reporter plasmid and CMV-driven expression vectors encoding Foxa1, Neurog3, E47, or an empty vector into mPAC (A) and NIH3T3 (B) cells as indicated. Reporter gene activity is expressed relative to the activity of the promoter-less parental vector pFOXLuc and the empty expression vector, set at 1. All data are presented as the mean ± S.E. from 3–4 independent experiments. *, p < 0.05 versus no cDNA (empty bar), #, p < 0.05 versus Neurog3+E47. C, endogenous Foxa1 and Foxa2 mRNA expression in the indicated mouse cell lines was assessed by RT-PCR. Actb is included as loading control. Image is representative of two independent experiments. D, total RNA from mPAC cells was collected 44–46 h after infection with recombinant adenoviruses expressing B-gal or NEUROG3. Foxa1 and Foxa2 mRNAs were quantified relative to the Tbp gene by real time RT-PCR and expressed as fold-increase versus expression in cells treated with B-gal set at 1. Results are mean ± S.E. from four independent experiments. p < 0.01 versus B-gal.
The Foxa1-expressing cells in the fetal pancreas have not been precisely defined and hence, we performed immunofluorescence staining against Foxa1 in E14.5 pancreata. In agreement with previous reports (29, 33), we failed to detect Foxa1 protein in endocrine progenitors at this stage (data not shown), thus ruling out a relevant role of this protein in the regulation of Neurog3 expression levels in vivo. Interestingly, Foxa1 mRNA is faintly expressed in mPAC cells (Fig. 6C), but it is significantly induced upon NEUROG3 introduction in these cells (Fig. 6D), uncovering Foxa1 as a potential Neurog3 downstream target during endocrine cell differentiation. In this regard, it should be noted that Foxa1 expression is enriched in islets but negligible in acinar cells in the adult pancreas (29). Thus, our results indicate that although Foxa2 and Foxa1 are both equally capable of potentiating the autoactivation of Neurog3 in in vitro luciferase assays, in vivo, Foxa1 probably functions at later stages of endocrine differentiation.
Foxa2 Knockdown Impairs Neurog3-induced Activation of the Endocrine Differentiation Program
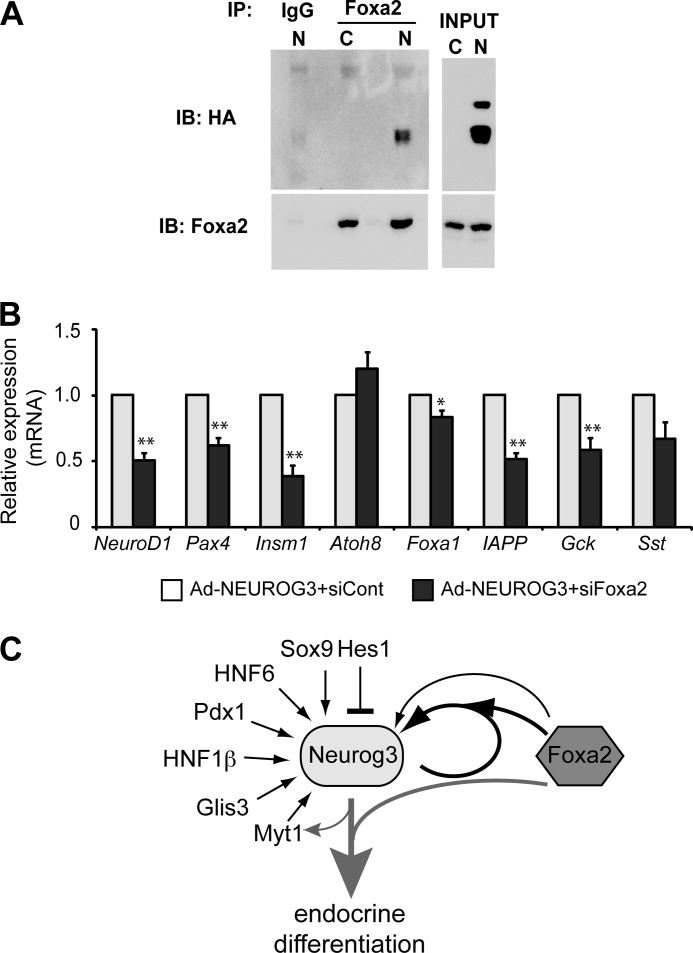
To determine the molecular basis of the synergy between Neurog3 and Foxa2, we investigated whether Foxa2 physically interacts with Neurog3. We used an antibody against Foxa2 to immunoprecipitate Foxa2-interacting partners from cell extracts of mPAC cells transduced with an adenovirus expressing a HA-tagged version of Neurog3 or cells left untreated. The anti-HA antibody detected Neurog3 in the Foxa2 immunoprecipitate, whereas it failed to detect it in the IgG immunoprecipitate (Fig. 7A). These results indicate that Foxa2 interacts with Neurog3 both physically and functionally to regulate Neurog3 gene transcription.
FIGURE 7.
A, mPAC cells were infected with a recombinant adenovirus expressing a HA-tagged NEUROG3 (N) or were left untreated (Control, C). At 48 h after infection, cell lysates were prepared and immunoprecipitated with an anti-Foxa2 antibody or IgG control. Immunoprecipitates were analyzed by immunoblot using goat anti-Foxa2 and mouse anti-HA antibodies. Image is representative of three independent experiments. B, mPAC cells were transfected with a siRNA against Foxa2 (siFoxa2) or control siRNA (siCont). 24 h after transfection, cells were treated with AdCMV.NEUROG3 or left untreated. Total RNA from mPAC cells was collected 44–46 h after infection. Levels of endogenous mRNAs encoding the indicated genes relative to Tbp gene were quantified by real time RT-PCR and expressed as fold-increase versus expression in NEUROG3+siCont-expressing cells, set at 1. Note that all genes tested except for Atoh8 are not expressed in untreated mPAC cells (not shown). All data are presented as the mean ± S.E. from five independent experiments. *, p < 0.05, **, p < 0.01 versus siCont. C, based on our present data we propose a model whereby Neurog3 synergizes with Foxa2 to autoactivate its own expression and to induce the endocrine differentiation program. Foxa2 may also directly regulate the Neurog3 gene. Other known transcriptional regulators of the Neurog3 gene are also depicted.
Disruption of Foxa2 prevents full differentiation of fetal α- and β-cells (34, 35). These evidence together with our observation that Neurog3 and Foxa2 co-immunoprecipate in the same complexes prompted us to investigate whether Foxa2 is broadly involved in Neurog3-dependent activation of the endocrine differentiation program. To this aim, we compared activation of several Neurog3 gene targets in mPAC cells transduced with AdCMV-NEUROG3 in the presence of the siRNA against Foxa2 or a siRNA control as established previously (see Fig. 4). We found that Foxa2 knockdown resulted in significantly reduced induction of genes encoding markers of differentiating (NeuroD1, Pax4, Insm1) as well as fully differentiated (IAPP, Gck) endocrine cells (Fig. 7B). However, not all Neurog3 targets were blocked by siFoxa2 (i.e. Atoh8, Sst), disclosing gene-specific effects of Foxa2 knockdown. Interestingly, siFoxa2 did reduce Foxa1 activation but to a lesser extent (16%) than the other studied genes, suggesting that Foxa1 inhibition is unlikely to be primarily responsible for the impairment in endocrine gene activation in response to siFoxa2. Taken together, these results uncover the possible direct contribution of Foxa2 to the establishment of the Neurog3-dependent endocrine transcriptional program (Fig. 7C).
DISCUSSION
Neurog3 is both essential and sufficient for endocrine cell differentiation in the pancreas, and understanding how Neurog3 is regulated will be valuable not only for attaining mechanistic insights into pancreatic endocrine cell genesis during development, but also for devising better in vitro differentiation protocols for β-cell replacement therapies in diabetes. In this study, we define a positive direct autoregulatory mechanism for Neurog3, which involves binding of Neurog3 to a highly conserved 0.7 kb region located at −3.2 to −3.9 kb of the Neurog3 promoter. In addition, we show that the forkhead-containing transcription factor Foxa2, which is expressed in MPCs, synergizes with Neurog3 to enhance this autoactivation loop, revealing its potential contribution to regulation of Neurog3 expression in endocrine progenitors. Furthermore, our study shows that the functional interaction between Neurog3 and Foxa2 extends to other Neurog3 target genes in vitro, thus uncovering a possible cooperation of these transcription factors in the implementation of the endocrine differentiation program in the pancreas.
Positive autoregulation is a transcriptional control mechanism used by many cell fate-determining bHLH factors in development (36–39), which is thought to sustain expression of these proteins throughout specific time windows so that they can reliably perform their developmental functions. In addition, because autoactivation can release these factors from control by external stimuli, it may also ensure cell fate commitment (40). Another suggested function of positive autoregulatory loops is to raise the expression levels of cell fate-determining factors over a threshold necessary for initiation of their respective developmental programs (41). The observation that low Neurog3 expression in MPCs results in their allocation to exocrine fates (18) underlines the potential importance of this autoamplification function so that Neurog3 can reach the threshold needed to activate endocrine differentiation. Importantly, autoregulatory loops are not irreversible and can be modified or extinguished by additional regulatory inputs. In this regard, using Escherichia coli as a model, autoactivation has been shown to make systems more sensitive to inhibition and thus advantageous when sharp repression is needed (42), an enticing concept that might be applicable to transcription factors such as Neurog3 that are only expressed during a narrow time window (20, 19). Hence, based on these notions, positive autoregulation can provide several functional advantages to the control of cell fate determination by Neurog3. Importantly, while this manuscript was under review, in vivo findings by Shih et al. were published that support the concept that Neurog3 enforces its own expression in the embryonic pancreas (43). In their study, the authors found decreased Neurog3 promoter activity in Neurog3-deficient embryos, consistent with the existence of Neurog3 autoactivation in vivo. However, whether this occurs through direct positive autofeedback, is mediated by additional Neurog3-dependent factors, or both, it is difficult to discriminate using in vivo models. Our present findings demonstrate that Neurog3 can activate its own promoter in vitro, thus validating at a molecular level the existence of a direct autoregulatory mechanism. Even so, we cannot exclude the possibility that additional positive feedback loops control Neurog3 expression in the embryonic pancreas (21). In any case, promoter occupancy data using chromatin from purified endocrine progenitors will be needed to provide definite confirmation of the existence of direct autoregulation.
In many instances, autoregulatory loops involving cell-fate determining factors rely on extrinsic cues that promote cell fate decisions. For example, in Drosophila's sensory organ development, proneural bHLH autoregulation is contingent on the activation of the EGFR pathway (39). Conversely, Notch inhibits sensory organ precursor recruitment by directly antagonizing proneural bHLH autoactivation (44). Based on present and previously available data, we propose a model where inhibition of the Notch pathway would activate Neurog3 expression in progenitor cells (17, 12, 43, 45), while Neurog3 autoactivation would act to boost Neurog3 levels after transcription has been initiated. This last process would be independent of Notch activity as recently suggested (43). Nonetheless, we cannot rule out that, in addition to Neurog3 itself, other signals are required to trigger Neurog3 autoactivation in vivo. In other words, Neurog3 autoregulation may be restricted to some cells by additional signals, leading to differences in the amount of Neurog3 and, in turn, in different cell fate outcomes. In this regard, it is noteworthy that Neurog3 mRNA is more broadly expressed than Neurog3 protein in the pancreatic epithelium (46), implying that not all cells that activate the Neurog3 gene end up producing detectable amounts of this factor. Both the Notch and the GDF11/TGFβ signaling pathways, which are known to regulate the number of Neurog3+ cells in the pancreas (17, 47), could potentially interact with Neurog3 self-regulation.
In addition to extrinsic signals, intrinsic signals can also influence autoregulatory loops. Accordingly, our present data indicate that Neurog3 self-regulation depends on the cellular context. We show that Neurog3 autoactivation occurs in pancreatic ductal cell lines. Likewise, Neurog3 autoactivation has recently been reported in primary cultures of adult human pancreatic duct cells (48). We postulate that the presence of key transcription factors normally found in MPCs (and implicated as Neurog3 gene activators) makes duct cells competent to respond to Neurog3 autoregulation. It is possible that binding of these factors to cis-acting elements of the Neurog3 gene directs recruitment of Neurog3 to its own gene (note the clustering of progenitor factor binding sites within the −3.2 kb region), and/or warrants a chromatin structure permissive of transcription. Particularly, we show that Foxa2 affects Neurog3 autoactivation both in reporter luciferase assays and on activation of the endogenous Neurog3 gene in mPAC cells. Remarkably, one of the key functions of the Foxa family of forkhead-containing transcriptional regulators is to serve as pioneer factors that initiate regulatory control of transcription through direct opening of chromatin (49). In this regard, we have found that the Neurog3 proximal promoter, which harbors a conserved Foxa binding site (12), exhibits moderate enrichment in the active histone mark H3K4me2 in control mPAC cells, indicating that despite being silent, the Neurog3 gene is transcriptionally competent in these cells. However, the sole presence of Foxa2 is not sufficient to confer competency for Neurog3 autoactivation as Foxa2 is expressed in all pancreatic cell lines studied, but not all cells exhibit Neurog3 self-regulation. Therefore, Neurog3 autoregulation likely requires several components, both transcriptional and epigenetic, to operate in some cells and not in others.
Cooperative interactions between transcription factors are instrumental to the regulation of gene expression. Here we show that Foxa2 synergizes with the Neurog3/E47 bHLH heterodimer to regulate not only Neurog3 autoactivation but also the induction of other Neurog3 target genes, uncovering a specific role of Foxa2 in the induction of endocrine cell differentiation. Foxa2 deletion in the early pancreatic epithelium results in normal endocrine differentiation, but α-cells fail to completely mature (34). However, single knockouts may not evidence the full complement of functions exerted by individual Foxa factors due to functional compensation by remaining Foxa proteins (50). Thus, compound Foxa2 and Foxa1 mutants exhibit severe pancreatic hypoplasia and impaired acinar and endocrine cell development (31). Nevertheless, whether the endocrine differentiation defects in this mouse model are due to the specific absence of Foxa2 in the prospective endocrine precursor or secondary to impaired expansion of the MPC population is unclear at present. Conditional deletion of Foxa genes in endocrine progenitors will be necessary to elucidate the role of these factors in the activation of the endocrine developmental program.
It has been argued that autoregulation positively correlates with the developmental and/or physiological importance of a given transcription factor (41). Because of the crucial role of Neurog3 in endocrine cell differentiation, it is expected that multiple direct and indirect positive and negative feedback loops will cooperate to control Neurog3 gene expression. In this study, we have characterized one of these loops that involve direct transcriptional autoactivation by Neurog3 of its own promoter. Further work aimed at extending our knowledge on the autoregulatory and crossregulatory circuits that regulate Neurog3 levels is needed to better comprehend how endocrine cell fate is established during pancreatic development.
Acknowledgments
We thank Dr. M. Párrizas for critical reading of this manuscript. The mouse monoclonal antibodies against Neurog3 and Nkx6.1 were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242.
This work was supported by the Spanish Ministerio de Ciencia e Innovación (BFU2008–02299/BMC) (to R. G.). Predoctoral fellowships were provided by IDIBAPS (to M. E.) and by the Spanish Ministerio de Ciencia e Innovación (to G. P.). The research leading to these results has received funding from the European Community's Seventh Framework Programme (FP7/2009-2013) under Grant Agreement No 229673 (to S. C.).

This article contains supplemental Figs. S1—S4 and Tables.
- bHLH
- basic helix-loop-helix
- Neurog
- Neurogenin
- MPC
- multipotent pancreatic progenitor cell
- ChgA
- Chromogranin A.
REFERENCES
- 1. Gradwohl G., Dierich A., LeMeur M., Guillemot F. (2000) Neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc. Natl. Acad. Sci. U.S.A. 97, 1607–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gu G., Dubauskaite J., Melton D. A. (2002) Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development 129, 2447–2457 [DOI] [PubMed] [Google Scholar]
- 3. Wang J., Cortina G., Wu S. V., Tran R., Cho J. H., Tsai M. J., Bailey T. J., Jamrich M., Ament M. E., Treem W. R., Hill I. D., Vargas J. H., Gershman G., Farmer D. G., Reyen L., Martín M. G. (2006) Mutant neurogenin-3 in congenital malabsorptive diarrhea. N. Engl. J. Med. 355, 270–280 [DOI] [PubMed] [Google Scholar]
- 4. Pinney S. E., Oliver-Krasinski J., Ernst L., Hughes N., Patel P., Stoffers D. A., Russo P., De León D. D. (2011) Neonatal diabetes and congenital malabsorptive diarrhea attributable to a novel mutation in the human neurogenin-3 gene coding sequence. J. Clin. Endocrinol. Metab. 96, 1960–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rubio-Cabezas O., Jensen J. N., Hodgson M. I., Codner E., Ellard S., Serup P., Hattersley A. T. (2011) Permanent Neonatal Diabetes and Enteric Anendocrinosis Associated With Biallelic Mutations in NEUROG3. Diabetes 60, 1349–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schwitzgebel V. M., Scheel D. W., Conners J. R., Kalamaras J., Lee J. E., Anderson D. J., Sussel L., Johnson J. D., German M. S. (2000) Expression of neurogenin3 reveals an islet cell precursor population in the pancreas. Development 127, 3533–3542 [DOI] [PubMed] [Google Scholar]
- 7. Heremans Y., Van De Casteele M., in't Veld P., Gradwohl G., Serup P., Madsen O., Pipeleers D., Heimberg H. (2002) Recapitulation of embryonic neuroendocrine differentiation in adult human pancreatic duct cells expressing neurogenin 3. J. Cell Biol. 159, 303–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gasa R., Mrejen C., Leachman N., Otten M., Barnes M., Wang J., Chakrabarti S., Mirmira R., German M. (2004) Proendocrine genes coordinate the pancreatic islet differentiation program in vitro. Proc. Natl. Acad. Sci. U.S.A. 101, 13245–13250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yechoor V., Liu V., Paul A., Lee J., Buras E., Ozer K., Samson S., Chan L. (2009) Gene therapy with neurogenin 3 and betacellulin reverses major metabolic problems in insulin-deficient diabetic mice. Endocrinology 150, 4863–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang S., Jensen J. N., Seymour P. A., Hsu W., Dor Y., Sander M., Magnuson M. A., Serup P., Gu G. (2009) Sustained Neurog3 expression in hormone-expressing islet cells is required for endocrine maturation and function. Proc. Natl. Acad. Sci. U.S.A. 106, 9715–9720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jacquemin P., Durviaux S. M., Jensen J., Godfraind C., Gradwohl G., Guillemot F., Madsen O. D., Carmeliet P., Dewerchin M., Collen D., Rousseau G. G., Lemaigre F. P. (2000) Transcription factor hepatocyte nuclear factor 6 regulates pancreatic endocrine cell differentiation and controls expression of the proendocrine gene ngn3. Mol. Cell Biol. 20, 4445–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee J. C., Smith S. B., Watada H., Lin J., Scheel D., Wang J., Mirmira R. G., German M. S. (2001) Regulation of the pancreatic pro-endocrine gene neurogenin3. Diabetes 50, 928–936 [DOI] [PubMed] [Google Scholar]
- 13. Lynn F. C., Smith S. B., Wilson M. E., Yang K. Y., Nekrep N., German M. S. (2007) Sox9 coordinates a transcriptional network in pancreatic progenitor cells. Proc. Natl. Acad. Sci. U.S.A. 104, 10500–10505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oliver-Krasinski J. M., Kasner M. T., Yang J., Crutchlow M. F., Rustgi A. K., Kaestner K. H., Stoffers D. A. (2009) The diabetes gene Pdx1 regulates the transcriptional network of pancreatic endocrine progenitor cells in mice. J. Clin. Invest. 119, 1888–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang Y., Chang B. H., Yechoor V., Chen W., Li L., Tsai M. J., Chan L. (2011) The Kruppel-like zinc finger protein GLIS3 transactivates neurogenin 3 for proper fetal pancreatic islet differentiation in mice. Diabetologia 54, 2595–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smith S. B., Watada H., German M. S. (2004) Neurogenin3 activates the islet differentiation program while repressing its own expression. Mol. Endocrinol. 18, 142–149 [DOI] [PubMed] [Google Scholar]
- 17. Apelqvist A., Li H., Sommer L., Beatus P., Anderson D. J., Honjo T., Hrabe de Angelis M., Lendahl U., Edlund H. (1999) Notch signalling controls pancreatic cell differentiation. Nature 400, 877–881 [DOI] [PubMed] [Google Scholar]
- 18. Wang S., Yan J., Anderson D. A., Xu Y., Kanal M. C., Cao Z., Wright C. V., Gu G. (2010) Neurog3 gene dosage regulates allocation of endocrine and exocrine cell fates in the developing mouse pancreas. Dev. Biol. 339, 26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Beucher A., Martín M., Spenle C., Poulet M., Collin C., Gradwohl G. (2012) Competence of failed endocrine progenitors to give rise to acinar but not ductal cells is restricted to early pancreas development. Dev. Biol. 361, 277–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miyatsuka T., Li Z., German M. S. (2009) Chronology of islet differentiation revealed by temporal cell labeling. Diabetes 58, 1863–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang S., Hecksher-Sorensen J., Xu Y., Zhao A., Dor Y., Rosenberg L., Serup P., Gu G. (2008) Myt1 and Ngn3 form a feed-forward expression loop to promote endocrine islet cell differentiation. Dev. Biol. 317, 531–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pujadas G., Felipe F., Ejarque M., Sanchez L., Cervantes S., Lynn F. C., Gomis R., Gasa R. (2011) Sequence and epigenetic determinants in the regulation of the Math6 gene by Neurogenin3. Differentiation 82, 66–76 [DOI] [PubMed] [Google Scholar]
- 23. Seo S., Lim J. W., Yellajoshyula D., Chang L. W., Kroll K. L. (2007) Neurogenin and NeuroD direct transcriptional targets and their regulatory enhancers. EMBO J. 26, 5093–5108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smith S. B., Gasa R., Watada H., Wang J., Griffen S. C., German M. S. (2003) Neurogenin3 and hepatic nuclear factor 1 cooperate in activating pancreatic expression of Pax4. J. Biol. Chem. 278, 38254–38259 [DOI] [PubMed] [Google Scholar]
- 25. Huang H. P., Liu M., El-Hodiri H. M., Chu K., Jamrich M., Tsai M. J. (2000) Regulation of the pancreatic islet-specific gene BETA2 (neuroD) by neurogenin 3. Mol. Cell Biol. 20, 3292–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sérandour A. A., Avner S., Percevault F., Demay F., Bizot M., Lucchetti-Miganeh C., Barloy-Hubler F., Brown M., Lupien M., Métivier R., Salbert G., Eeckhoute J. (2011) Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res. 21, 555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pekowska A., Benoukraf T., Zacarias-Cabeza J., Belhocine M., Koch F., Holota H., Imbert J., Andrau J. C., Ferrier P., Spicuglia S. (2011) H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 30, 4198–4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maestro M. A., Boj S. F., Luco R. F., Pierreux C. E., Cabedo J., Servitja J. M., German M. S., Rousseau G. G., Lemaigre F. P., Ferrer J. (2003) Hnf6 and Tcf2 (MODY5) are linked in a gene network operating in a precursor cell domain of the embryonic pancreas. Hum. Mol. Genet. 12, 3307–3314 [DOI] [PubMed] [Google Scholar]
- 29. Besnard V., Wert S. E., Hull W. M., Whitsett J. A. (2004) Immunohistochemical localization of Foxa1 and Foxa2 in mouse embryos and adult tissues. Gene Expr. Patterns 5, 193–208 [DOI] [PubMed] [Google Scholar]
- 30. Zhou Q., Law A. C., Rajagopal J., Anderson W. J., Gray P. A., Melton D. A. (2007) A multipotent progenitor domain guides pancreatic organogenesis. Dev. Cell 13, 103–114 [DOI] [PubMed] [Google Scholar]
- 31. Gao N., LeLay J., Vatamaniuk M. Z., Rieck S., Friedman J. R., Kaestner K. H. (2008) Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes Dev. 22, 3435–3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gao N., Le Lay J., Qin W., Doliba N., Schug J., Fox A. J., Smirnova O., Matschinsky F. M., Kaestner K. H. (2010) Foxa1 and Foxa2 maintain the metabolic and secretory features of the mature beta-cell. Mol. Endocrinol. 24, 1594–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thompson N., Gésina E., Scheinert P., Bucher P., Grapin-Botton A. (2012) RNA profiling and chromatin immunoprecipitation-sequencing reveal that PTF1a stabilizes pancreas progenitor identity via the control of MNX1/HLXB9 and a network of other transcription factors. Mol. Cell Biol. 32, 1189–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee C. S., Sund N. J., Behr R., Herrera P. L., Kaestner K. H. (2005) Foxa2 is required for the differentiation of pancreatic alpha-cells. Dev. Biol. 278, 484–495 [DOI] [PubMed] [Google Scholar]
- 35. Sund N. J., Vatamaniuk M. Z., Casey M., Ang S. L., Magnuson M. A., Stoffers D. A., Matschinsky F. M., Kaestner K. H. (2001) Tissue-specific deletion of Foxa2 in pancreatic beta cells results in hyperinsulinemic hypoglycemia. Genes Dev. 15, 1706–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zingg J. M., Pedraza-Alva G., Jost J. P. (1994) MyoD1 promoter autoregulation is mediated by two proximal E-boxes. Nucleic Acids Res. 22, 2234–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Raft S., Koundakjian E. J., Quinones H., Jayasena C. S., Goodrich L. V., Johnson J. E., Segil N., Groves A. K. (2007) Cross-regulation of Ngn1 and Math1 coordinates the production of neurons and sensory hair cells during inner ear development. Development 134, 4405–4415 [DOI] [PubMed] [Google Scholar]
- 38. Masui T., Swift G. H., Hale M. A., Meredith D. M., Johnson J. E., Macdonald R. J. (2008) Transcriptional autoregulation controls pancreatic Ptf1a expression during development and adulthood. Mol. Cell Biol. 28, 5458–5468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. zur Lage P. I., Powell L. M., Prentice D. R., McLaughlin P., Jarman A. P. (2004) EGF receptor signaling triggers recruitment of Drosophila sense organ precursors by stimulating proneural gene autoregulation. Dev. Cell 7, 687–696 [DOI] [PubMed] [Google Scholar]
- 40. Siciliano V., Menolascina F., Marucci L., Fracassi C., Garzilli I., Moretti M. N., di Bernardo D. (2011) Construction and modelling of an inducible positive feedback loop stably integrated in a mammalian cell-line. PLoS Comput. Biol. 7, e1002074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Crews S. T., Pearson J. C. (2009) Transcriptional autoregulation in development. Curr. Biol. 19, R241–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hermsen R., Ursem B., ten Wolde P. R. (2010) Combinatorial gene regulation using auto-regulation. PLoS Comput. Biol. 6, e1000813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shih H. P., Kopp J. L., Sandhu M., Dubois C. L., Seymour P. A., Grapin-Botton A., Sander M. (2012) A Notch-dependent molecular circuitry initiates pancreatic endocrine and ductal cell proliferation. Development 139, 2488–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Culi J., Modolell J. (1998) Proneural gene self-stimulation in neural precursors: an essential mechanism for sense organ development that is regulated by Notch signaling. Genes Dev. 12, 2036–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Murtaugh L. C., Stanger B. Z., Kwan K. M., Melton D. A. (2003) Notch signaling controls multiple steps of pancreatic differentiation. Proc. Natl. Acad. Sci. U.S.A. 100, 14920–14925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Villasenor A., Chong D. C., Cleaver O. (2008) Biphasic Ngn3 expression in the developing pancreas. Dev. Dyn. 237, 3270–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Harmon E. B., Apelqvist A. A., Smart N. G., Gu X., Osborne D. H., Kim S. K. (2004) GDF11 modulates NGN3+ islet progenitor cell number and promotes beta-cell differentiation in pancreas development. Development 131, 6163–6174 [DOI] [PubMed] [Google Scholar]
- 48. Swales N, Martens G. A., Bonné S., Heremans Y., Borup R., Van de Casteele M., Ling Z., Pipeleers D., Ravassard P., Nielsen F., Ferrer J., Heimberg H. (2012) Plasticity of adult human pancreatic duct cells by neurogenin3-mediated reprogramming. PlosOne 7, e37055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zaret K. S., Carroll J. S. (2011) Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25, 2227–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lantz K. A., Kaestner K. H. (2005) Winged-helix transcription factors and pancreatic development. Clin. Sci. 108, 195–204 [DOI] [PubMed] [Google Scholar]