Background: Serogroup W-135 meningococci express a highly unusual capsular polysaccharide with sialic acid as an internal sugar conjugated by heteroglycans.
Results: The capsule polymerase SiaDW-135 contains two successively active domains comprising galactosyltransferase and sialyltransferase activity in one polypeptide chain.
Conclusion: The capsule polymerase SiaDW-135 is a chimeric enzyme.
Significance: With the characterization of SiaDW-135 a basis has been established for its exploitation in vaccine developmental approaches.
Keywords: Glycoconjugate, Glycosyltransferases, NMR, Polysaccharide, Sialyltransferase, Neisseria meningitidis Serogroup W-135 and Y, Bacterial Capsule polymerases, Capsule Polysaccharides, Glycosyltransferases, Sialyltransferases
Abstract
Neisseria meningitidis (Nm) is a leading cause of bacterial meningitis and sepsis. Crucial virulence determinants of pathogenic Nm strains are the polysaccharide capsules that support invasion by hindering complement attack. In NmW-135 and NmY the capsules are built from the repeating units (→6)-α-d-Gal-(1→4)-α-Neu5Ac-(2→)n and (→6)-α-d-Glc-(1→4)-α-Neu5Ac-(2→)n, respectively. These unusual heteropolymers represent unique examples of a conjugation between sialic acid and hexosyl-sugars in a polymer chain. Moreover, despite the various catalytic strategies needed for sialic acid and hexose transfer, single enzymes (SiaDW-135/Y) have been identified to form these heteropolymers. Here we used SiaDW-135 as a model system to delineate structure-function relationships. In size exclusion chromatography active SiaDW-135 migrated as a monomer. Fold recognition programs suggested two separate glycosyltransferase domains, both containing a GT-B-fold. Based on conserved motifs predicted folds could be classified as a hexosyl- and sialyltransferase. To analyze enzyme properties and interplay of the two identified glycosyltransferase domains, saturation transfer difference NMR and mutational studies were carried out. Simultaneous and independent binding of UDP-Gal and CMP-Sia was seen in the absence of an acceptor as well as when the catalytic cycle was allowed to proceed. Enzyme variants with only one functionality were generated by site-directed mutagenesis and shown to complement each other in trans when combined in an in vitro test system. Together the data strongly suggests that SiaDW-135 has evolved by fusion of two independent ancestral genes encoding sialyl- and galactosyltransferase activity.
Introduction
Bacterial meningitis remains a serious threat to global health, accounting for an estimated 170,000 annual deaths worldwide (WHO website). Despite the availability of potent antimicrobial agents, case fatality rates are high (>10%) and survivors frequently suffer from sequelae such as neurologic disability or limb loss and deafness (1, 2). Although most cases of meningococcal disease are sporadic, outbreaks are frequently observed and large epidemics occur in the African meningitis belt (2, 3). The predominant serogroups are B and C followed by W-135 and Y (1). Serogroup W-135 meningococci caused an outbreak of disease among pilgrims in 2000 (4) and an epidemic in Burkina Faso in 2002 with over 13,000 cases and more than 1,400 fatalities (5, 6). Furthermore, an increase in serogroup Y disease was observed in the 1990s in North America (7).
Crucial virulence determinants of disease causing Nm species are their extracellular polysaccharide capsules (CPSs)4 that are essential for meningococcal survival in human serum (8). Based on the chemical composition of these polysaccharides at least 12 different serogroups of Nm have been identified, but only six account for virtually all cases of disease. Of these serogroups B, C, Y, and W-135 express CPS structures that contain the acidic nonulose sialic acid (Sia or Neu5Ac). Although the capsules expressed by serogroups B and C represent sialic acid homopolymers conjugated by α(2,8)- and α(2,9)-glycosidic linkages, respectively, the capsules of serogroups W-135 and Y represent heteropolymers built from disaccharide repeating units with sialic acid α(2,6)-linked to galactose (W-135) or glucose (Y). For example the repeating unit in NmW-135 is (→6)-α-d-Gal-(1→4)-α-Neu5Ac-(2→)n (9, 10). With sialic acid as an internal sugar conjugated by heteroglycans, the NmW-135 and NmY represent unique structures not found in any other organism so far.
The biosynthesis of these unusual glycoconjugates is achieved by the capsule polymerases, SiaDW-135 and SiaDY (11). The genes encoding these proteins have been cloned (11) and the function of the expressed recombinant proteins was initially characterized in vitro and in vivo (12). Overall the two proteins share 98% identity and differences are limited to the N-terminal domains. Already after cloning, the remarkable size of the genes had stimulated the hypothesis that they have evolved by fusion of ancestral genes encoding sialyl- and hexosyltransferase activities (11). In fact, this hypothesis was supported when the isolated recombinant full-length proteins were shown to have the capacity to synthesize W-135 and Y capsule polymers in vitro (12). Moreover, using site-directed mutagenesis this latter study demonstrated that a single amino acid position (amino acid 310 in both proteins) determines specificity for the UDP-sugar (12).
In the CAZy classification system for glycosyltransferases (cazy.org), SiaDW-135 and SiaDY were assigned to family GT-4, comprising retaining hexosyltransferases, many of bacterial origin (13–15). Allocation to this family was based on primary sequence similarities that the N-terminal domains exhibit with members of the group. A fundamental characteristic in this respect is the motif EX7E, which is highly conserved in retaining hexosyltransferases (16) and present also in the neisserial capsule polymerases SiaDW-135 and SiaDY (12). In contrast, no similarities to CAZy GT-families that contain sialyltransferases or polysialyltransferases were identified. Considering, however, that SiaDW-135 and SiaDY are bifunctional enzymes that not only use donor substrates of significant chemical diversity (this is true with respect to the nucleotide and sugar moieties) but in addition are likely to use different catalytic mechanisms (retaining for the hexose and inverting for the sialic acid transfer) the CAZy assignment must be reassessed.
Accordingly, the present work was focused on bringing new insight to the structure-function relationships of the bifunctional meningococcal capsule polymerases. Using SiaDW-135 as a model system, we have employed fold recognition programs to re-investigate the protein organization. Our refined structural model suggested a series of biochemical and biophysical studies that were carried out and consequently SiaDW-135 was identified as a monomeric protein that comprises two active sites: a galactosyltransferase and a monosialyltransferase. Using natural and artificial acceptors we show that the synthesis of the NmW-135 polymer depends on the alternate activity of these two catalytic domains.
EXPERIMENTAL PROCEDURES
Expression Plasmids and Site-directed Mutagenesis
The construction of the plasmid pET22b-Strep-NmW-135, to express SiaDW-135 as N-terminal Strep-II and C-terminal His6-tagged protein, was described earlier (12). Expression constructs lacking the N-terminal Strep-II tag were constructed by amplifying the siaDW-135 insert from pHC4 (11). PCR products obtained with the oligonucleotides KS422/KS273 were ligated between the NdeI and XhoI sites of the expression vector pET22b (Novagen). Mutagenesis of siaDW-135 was performed using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The plasmid pET22b-Strep-NmW-135 (12) was used as template in all mutagenesis reactions. Primer sequences are given in Table 1 with the mutated base triplets underlined. Mutations and corresponding primer pairs are E307A: KS350/KS351; H934A: KS364/KS365; P935A: KS366/KS367; S972A: KS370/KS371; and T973A: KS372/KS373. The mutated siaDW-135 were subsequently subcloned into the BamHI and NotI sites of pET22b-Strep (17) and the sequence identity of all constructs was confirmed by sequencing.
TABLE 1.
Primers used in this study
| Primer pair | |
|---|---|
| Primer pair used for cloning | |
| KS422/KS273 | 5′-GCATCTCATATGGCTGTTATTATATTTGTTAACG-3′ |
| 5′-CCGCTCGAGTTTTTCTTGGCCAAAAAACTG-3′ | |
| Primer pairs used for the introduction of mutations | |
| KS350/KS351 | 5′-CTGATCATGACATCAGAAAGTGCGGGATTTCCATATATATTTATG-3′ |
| 5′-CATAAATATATATGGAAATCCCGCACTTTCTGATGTCATGATCAG-3′ | |
| KS364/KS365 | 5′-CAACTACAGCTTTTATTTTCTAAGGCTCCAGATGAAAATATAGATTTAAAG-3′ |
| 5′-CTTTAAATCTATATTTTCATCTGGAGCCTTAGAAAATAAAAGCTGTAGTTG-3′ | |
| KS366/KS367 | 5′-CTACAGCTTTTATTTTCTAAGCATGCAGATGAAAATATAGATTTAAAGAAC-3′ |
| 5′-GTTCTTTAAATCTATATTTTCATCTGCATGCTTAGAAAATAAAAGCTGTAG-3′ | |
| KS370/KS371 | 5′-ATCTCGCGTTGCTGTAGGTGTTTATGCAACTAGCTTATTTG-3′ |
| 5′-CAAATAAGCTAGTTGCATAAACACCTACAGCAACGCGAGAT-3′ | |
| KS372/KS373 | 5′-CGTTGCTGTAGGTGTTTATTCAGCTAGCTTATTTGAGGCATTAG-3′ |
| 5′-CTAATGCCTCAAATAAGCTAGCTGAATAAACACCTACAGCAACG-3′ | |
Expression and Purification of Recombinant SiaDW-135
Wild-type and mutant variants of SiaDW-135 carrying a C-terminal His6 tag (the wild-type protein was produced in two forms with C-terminal His6 tag and as double-tagged protein carrying in addition a N-terminal Strep tag), were expressed in Escherichia coli BL21(DE3) in the presence of 100 μg/ml of carbenicillin. Bacteria were cultivated in autoinducing ZYM-5052 medium (18) and grown for 78 h at 15 °C. Cells were harvested (4000 × g; 30 min; 4 °C), re-suspended in binding buffer (50 mm Tris/HCl, pH 8.0, 300 mm NaCl, 40 mg/ml of bestatin, 1 μg/ml of pepstatin, 1 mm PMSF), and disrupted by sonication (BRANSON SONIFIER® W-450D; amplitude 50%; 8 × 30 s). Recombinant proteins were bound to HisTrap affinity columns (GE Healthcare) using binding buffer (50 mm Tris/HCl, pH 8.0, 300 mm NaCl) and eluted in a two-step gradient (50 and 150 mm imidazole in binding buffer). Fractions containing SiaDW-135 were pooled and applied to a HiLoad 16/60 Superdex 200 pg column (GE Healthcare) for further purification by size exclusion chromatography. Proteins were eluted (50 mm Tris/HCl, pH 8.0, 300 mm NaCl, 2 mm DTT) at a flow rate of 0.5 ml/min and subsequently concentrated to 2 mg/ml using Amicon Ultracentrifugal devices (Millipore; 50 kDa MWCO). The purified proteins were flash-frozen in liquid nitrogen and stored at −80 °C.
Activity Assays
Radioactive incorporation assays were performed as described (12, 19). Briefly, purified recombinant proteins (7.5 μg) were assayed in reaction buffer (20 mm Tris/HCl, pH 8.0, 10 mm MgCl2, 1 mm DTT) in the presence of 1 mm radiocarbon labeled CMP-[14C]Neu5Ac (0.13 mCi/mmol, GE Healthcare) and 2 mm UDP-Gal (Sigma). Additionally, 2 mm of an artificial oligosaccharide acceptor (i.e. sialyllactose, Sigma; Neu5Ac, and the dimer and trimer of α(2,8)-Neu5Ac, Nacalai Tesque) or 0.4 mg/ml of hydrolyzed W-135 CPS were included in a total volume of 25 μl. Samples were incubated at room temperature and enzymatic activity was determined at appropriate time intervals by mixing 5-μl aliquots of the reaction solution with 5 μl of chilled ethanol (96%). Samples were spotted on Whatman 3MM CHR paper and the chromatographically immobile 14C-labeled reaction products were quantified by scintillation counting following descending paper chromatography in 96% ethanol, 1 m ammonium acetate, pH 7.5 (7:3, v/v).
To visualize the size distribution of the synthesized W-135 polysaccharides, purified SiaDW-135 (5–15 μg) was assayed in reaction buffer (20 mm Tris/HCl, pH 8.0, 10 mm MgCl2, 1 mm DTT) in the presence of 1 mm CMP-Neu5Ac (GERBU), 2 mm UDP-Gal, and hydrolyzed W-135 CPS (0.16 μg/μl) as the oligosaccharide acceptor structure in a total volume of 37.5 μl. Samples were incubated at room temperature and reactions were stopped at appropriate time intervals by addition of 1 m sucrose. Synthesized products were separated by PAGE (25%) and subsequently detected using a combined Alcian blue/silver staining procedure as described (20).
For the in vivo testing of mutants strain Nm-WUE171 was used and the plasmids were generated on the basis of pJP2 as previously described (12). Briefly, the HpaI/SphI fragment of wild-type siaDW-135 comprising position 19–2943 was replaced by the respective fragments of the pET22b-Strep-NmW-135 mutants. Correct integration of the mutated DNA into the meningococcal chromosome was confirmed by PCR and subsequent DNA sequencing. A siaD knock-out mutant designated WUE2661, used as a negative control in this study, resulted from the transformation of strain WUE171 with pHC4siaD::TnMax5 (21). Phenotypic analysis of capsule expression was performed as described recently with mAb1509 specific to serogroup W-135 and PorA antibody P1.10 (12).
STD NMR Experiments
All NMR experiments were performed on a Bruker 600 UltrashieldTM at 280 K, equipped with a standard triple resonance CryoProbe, in deuterated 20 mm Tris buffer, supplemented with 20 mm MgCl2 (D2O), pH 8.4. 200 μl of sample in Shigemi tubes (Shigemi Co.) were used for all acquisitions. STD spectra were acquired using 150 μg of SiaDW-135 and saturation time of 2 s at an on-resonance frequency of −1 ppm and off-resonance of 33 ppm. A WATERGATE sequence was used to suppress the residual HDO signal. The on- and off-resonance spectra free induction decays were stored and processed separately and the subtraction of the on-resonance and off-resonance spectrum resulted in the STD NMR spectrum. All STD NMR effects were calculated according to the formula ASTD = (I0 × Isat)/I0 = ISTD/I0 and the strongest STD NMR signal was set to 100%.
RESULTS
Fold Recognition Tools Predict Two GT-B-like Glycosyltransferase Domains in SiaDW-135
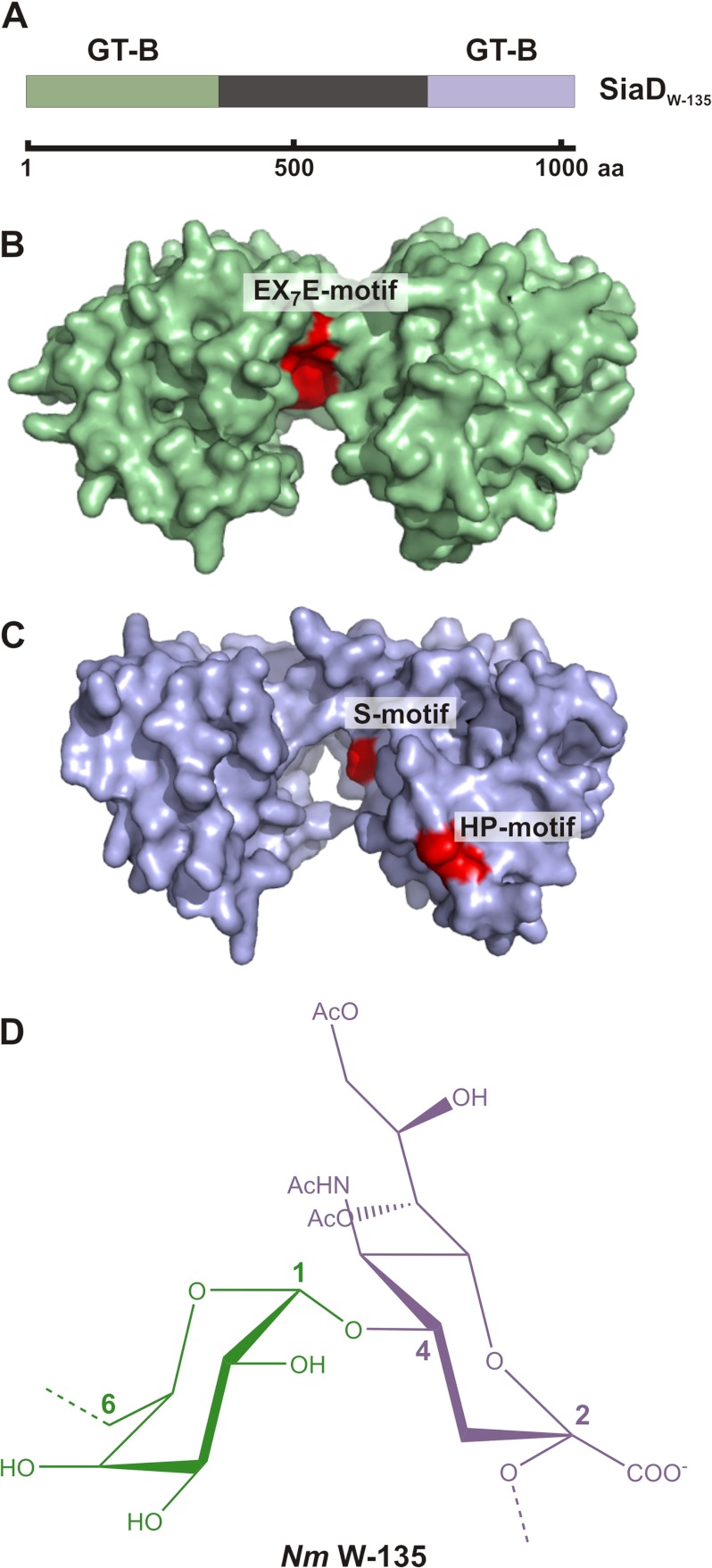
The homology of the N-terminal domain with hexosyltransferases of CAZy family GT-4, led to the grouping of SiaDW-135 into this GT class (11, 12). Considering, however, that SiaDW-135 catalyzes the formation of a heteropolymer (Fig. 1D) by use of two significantly different donor substrates (UDP-Gal and CMP-Sia) and two different catalytic mechanisms (galactosyltransferases are retaining and sialyltransferases inverting enzymes), the organization of SiaDW-135 was reinvestigated with the help of the structure prediction tool Phyre (22). Two GT-B folds were identified, the first spanning N-terminal residues 1–399 and the second residues 763–1037 in the C-terminal domain (Fig. 1, A–C). Although primary sequence identities to the respective template structures were low (≤13 and ≤10%, respectively), high confidence matches (100% each) were reported and indicated an accurate prediction of the overall fold (22). Closer inspection of the predicted models revealed in addition that highly conserved amino acid clusters known to be part of the active sites in either hexosyl- (EX7E motif (16)) or sialyltransferases (HP- and S-motif (23–25) localize along the deep active site crevices that separate the two β/α/β Rossmann-like domains in GT-B glycosyltransferases (Fig. 1, B and C). Subsequent experiments therefore addressed the question if SiaDW-135 represents a chimera with two enzymatic functions.
FIGURE 1.
GT-B folds predicted in SiaDW-135 and structure of the biosynthetic product. A, schematic illustration of full-length SiaDW-135. The protein comprises 1037 amino acids. The N- and C-terminal domains (shaded in green and blue, respectively) have been predicted to individually attain the GT-B fold. No structural information is available for the linker region (black) that connects the N- and C-terminal domains. B and C, GT-B folds as identified by Phyre. Based on the presence of the conserved motifs EX7E as well as S and HP, hexosyl- and sialyltransferase activity could be attributed to the GT-B folds. D, chemical structure of the repeating unit forming the W-135 capsular polymer.
The Purified Recombinant SiaDW-135 Is Monomeric and an Active Bifunctional Polymerase
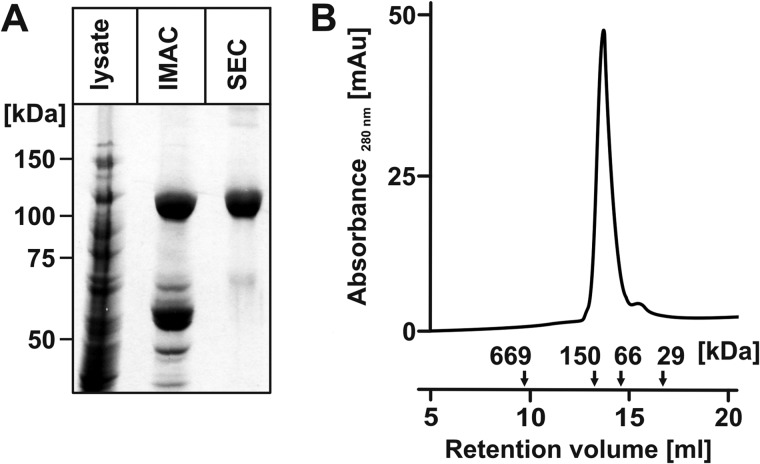
To obtain a suitable enzyme source for the planned biochemical and biophysical studies, SiaDW-135 carrying a C-terminal His6 tag was purified in two steps (IMAC and SEC) (Fig. 2A). The protein eluted as a monomer with an apparent molecular mass of 120 kDa (calculated molecular mass 121.137 kDa) from the calibrated SEC column (Fig. 2B). Activity of the purified enzyme was confirmed in a radiocarbon incorporation assay using CPS isolated from NmW-135 (for reference see CPS in Fig. 3, A and B) as acceptor. As already described for the partially purified SiaDW-135 (12) significant incorporation was only detected when CMP-[14C]Neu5Ac was combined with the second donor sugar UDP-Gal (data not shown).
FIGURE 2.
Recombinant epitope-tagged SiaDW-135 is a monomer. A, SiaDW-135 was expressed with C-terminal His6 tag. The Coomassie-stained SDS-PAGE shows that a highly purified protein fraction was obtained in two steps including Ni2+ chelating (IMAC) and size exclusion chromatography (SEC). B, the oligomerization status of recombinant SiaDW-135 was determined after calibration of the Superdex 200 column used in SEC. In the presence of 300 mm NaCl, SiaDW-135 eluted with an apparent molecular mass of 120 kDa, corresponding to the monomeric protein. Standards were: thyroglobulin (669 kDa), alcohol dehydrogenase (150 kDa), albumin (66 kDa), and carbonic anhydrase (29 kDa).
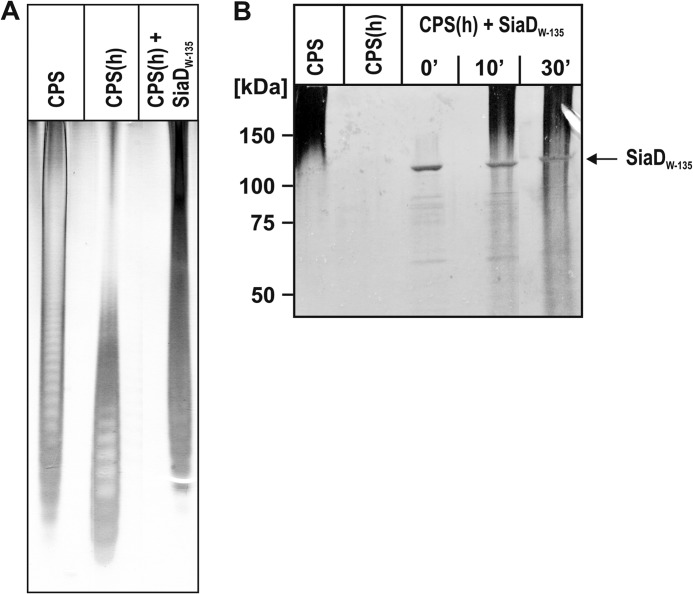
FIGURE 3.
Recombinant SiaDW-135 synthesizes high molecular weight W-135 polymer chains from short oligosaccharide primers. A, to test if recombinant SiaDW-135 can elongate short oligomer primers, CPS isolated from NmW-135 was hydrolyzed (CPS(h)) and used to prime the polymerase reaction. The production of long polymer chains demonstrated an active enzyme. High percentage PAGE and Alcian blue/silver staining was used to display starting, as well as synthesis products. B, CPS(h) produced from CPS was used in a time course experiment. Here products were analyzed on a 10% SDS-PAGE to additionally visualize the enzyme.
To unequivocally demonstrate that the recombinant SiaDW-135 catalyzes the synthesis of W-135 polymers under the in vitro conditions, the isolated CPS (CPS in Fig. 3, A and B) was partially hydrolyzed to yield oligomers (CPS(h) in Fig. 3A) and the oligomers were used to prime the polymerase reaction in the presence of “cold” substrates. Long chain products obtained in this reaction were displayed in high percentage PAGE (Fig. 3A) and, following a time course experiment, in 10% SDS-PAGE (Fig. 3B). To visualize polymerization products and enzyme the 10% SDS-PAGE was simultaneously stained with Alcian blue and silver. Oligomeric acceptors were not retained in the low percentage gel, but long chain polymers were detectable after 10 min. Moreover, this analysis demonstrated that the protein band corresponding to purified SiaDW-135 remained unaltered during the reaction.
Mutagenesis Studies Indicate Two Distinct Active Sites in SiaDW-135
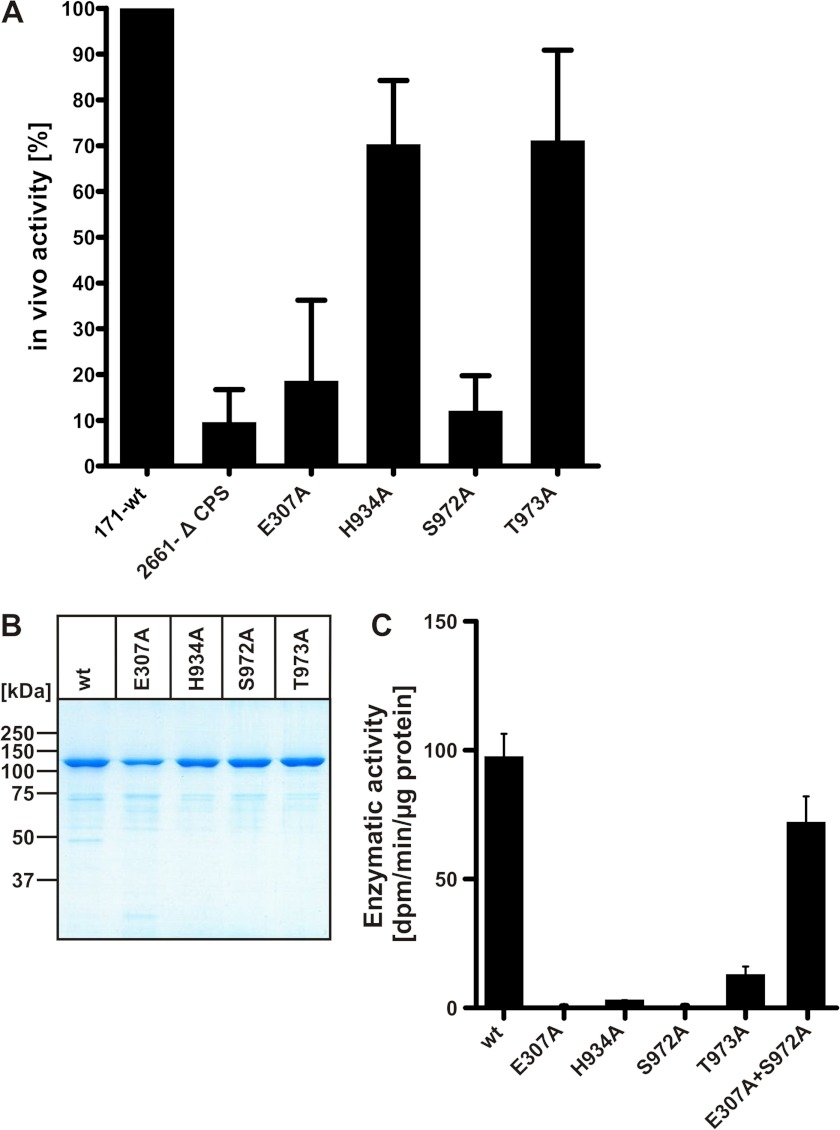
Site-specific mutations were generated to interrogate the functional nature of the identified GT-B like domains in SiaDW-135. Targets in this study were the highly conserved EX7E motif that represents a characteristic of the nucleotide recognition domain in retaining GTs (16) and the recently identified bacterial “sialyl motifs” HP and SS/T (23–25) presumed to be involved in substrate binding (26) (see Fig. 1). To inactivate these motifs Glu-307 (the first residue in the EX7E motif), His-934 (HP motif) as well as Ser-972 and Thr-973 (SS/T motif) were individually exchanged to alanine. Obtained mutants were tested in vivo and, with CPS(h) as acceptor, also in vitro (Fig. 4).
FIGURE 4.
Site-directed mutagenesis identified amino acid residues crucial for enzymatic activity. A, in vivo activity of the mutant capsule polymerases (as indicated) was tested with Nm strain WUE171 as recipient. Mutant genes were introduced by homologous recombination and capsule production was quantified in a whole cell ELISA. Values are given as means of three independent experiments in relationship to the wild-type recipient (100%). The capsule-deficient strain WUE 2661-Δ CPS served as a negative control. B, display of recombinant purified wild-type and mutant forms of SiaDW-135 in a Coomassie Blue-stained 10% SDS-PAGE. C, activity of recombinant purified wild-type and mutant capsule polymerases (proteins as shown in B) was determined in a radioactive incorporation assay with CPS(h) as acceptor. Of note, combination of the inactive mutants E307A and S972A in one reaction mixture restored activity to 70% of wild-type.
For the in vivo testing capsule expression of a NmW-135 wild-type strain and the respective SiaDW-135 mutants generated via homologous recombination were quantified in a well established whole cell ELISA (12, 27). In Fig. 4A results are shown as relative values with the starting strain (NmW-135 171-wt) set to 100%. The capsule deficient mutant 2661-Δ CPS was used as a negative control. The point mutants E307A and S972A reduced capsule expression to the level of the negative control. In contrast, mutants H934A and T973A, both affecting the putative sialyltransferase domain, had only minor effects if tested in vivo (capsule expression reduced by ∼30%), but decreased activity to <3 and <10%, respectively, if the recombinant soluble enzymes were tested in vitro (Fig. 4C). As this difference could not be explained by reduced protein expression, stability, or purity (see Fig. 4B), it is likely that integration of mutant proteins into the capsule synthesis complex in vivo creates conditions that favor functionality (12). Importantly, however, if the purified mutants were brought together in one reaction mixture activity was restored (>70% of the wild-type). Because each mutant still contains one functional domain (sialyltransferase in the case of mutant E307A and galactosyltransferase in the case of S972A), this complementation in trans strongly supported the hypothesis that SiaDW-135 represents a chimeric enzyme.
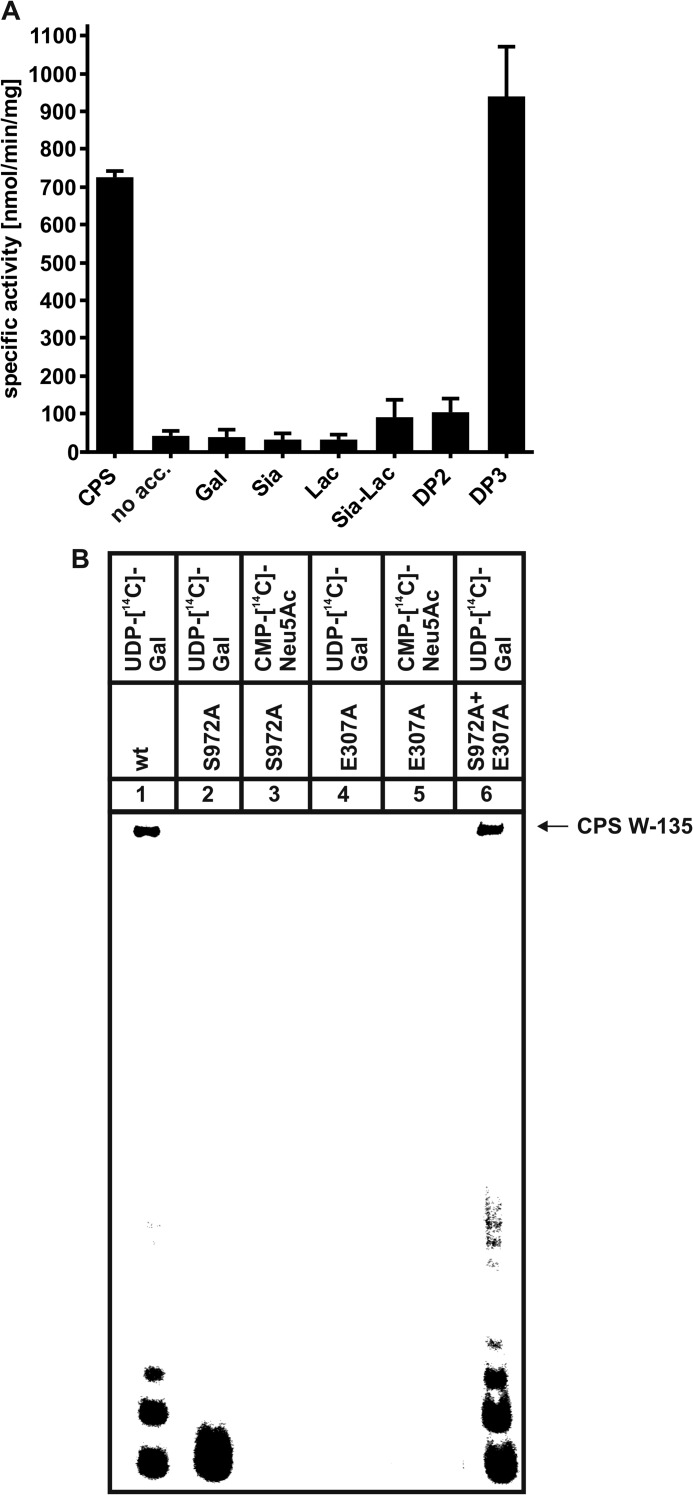
SiaDW-135 Elongates α(2,8)-Linked Sialic Acid Trimers
Because defined oligosaccharides consisting of (→6)-α-d-Gal-(1→4)-α-Neu5Ac-(2→)n were not readily available, further analysis of the catalytic functions of SiaDW-135 was preceded by the search for a low molecular weight acceptor molecule. The compounds tested are indicated in Fig. 5A. Neither the free monosaccharides (Sia and Gal) nor lactose were extended by the enzyme, whereas a slight but reproducible activity was seen with DP2 (Neu5Ac-α(2→8)-Neu5Ac) and 2,3-sialyllactose (Neu5Ac-α(2→3)-d-Gal-β(1→4)-d-Glc). However, if DP3 (trimer of α(2,8)-linked Neu5Ac) was used as acceptor, the activity escalated. To better understand this unexpected finding, the binding modes of DP1, DP2, and DP3 were investigated by STD NMR. This method records saturation transfer by spin diffusion from the target protein to binding ligands. Ligand protons making close contacts to the protein surface receive higher saturation resulting in high STD NMR signal intensities. Substrate binding profiles can thus be determined based on the relative saturation of ligand epitopes (28).
FIGURE 5.
A, the trimer of α(2,8)-linked sialic acid residues (DP3) is efficiently elongated by SiaDW-135. As defined W-135 CPS oligomers were not readily available, the compounds as listed were analyzed for their capacity to prime the SiaDW-135 reaction in the radioactive incorporation assay. In this experiment DP3 was found to be efficiently elongated by SiaDW-135, whereas DP2 and 2,3-sialyllactose (Sia-Lac) were only weak acceptors. The monosaccharides Gal and Sia as well as the disaccharide lactose (Lac) were not used by the enzyme. Specific activities were calculated from three independent experiments. B, asking if DP3 is an acceptor for both glycosyltransferase domains present in SiaDW-135, the transfer reactions catalyzed by the single mutants E307A and S972A were analyzed in comparison to the wild-type enzyme. Both nucleotide sugars were present in these reactions, with only one radioactively labeled as indicated. Long polymer chains were synthesized in reaction mixtures with wild-type SiaDW-135 (lane 1) and both single mutants (lane 6). In contrast, a single radioactive spot, representing the transfer of one [14C]Gal onto DP3 (lane 2) by S972A (comprises an active galactosyltransferase domain) was observed, whereas all other reactions (lanes 3-5) remained negative. These data clearly demonstrate that DP3 is selectively recognized by the galactosyltransferase domain and that formation of long CPS requires the iterative activity between galactosyl- and sialyltransferase domain.
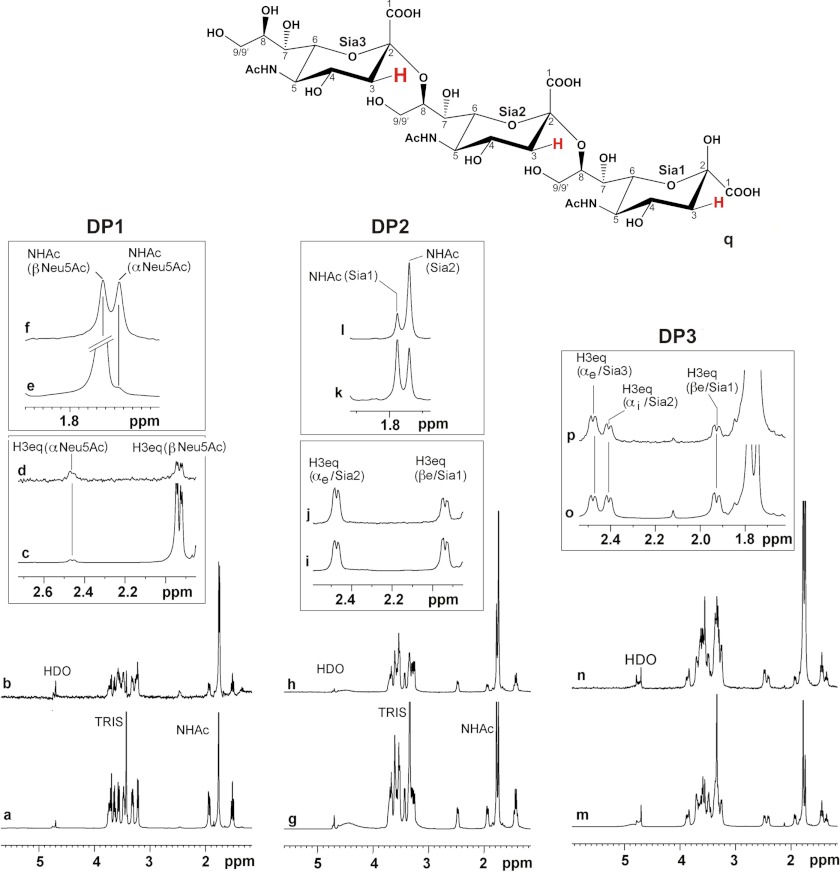
In Fig. 6 the NMR spectra of the compounds are shown. The bottom panels (Fig. 6, a, g, and m) display the 1H NMR spectra of the soluble compounds. The respective STD NMR spectra obtained for ligands in complex with the recombinant SiaDW-135 are shown in Fig. 6, b, h, and n. From the combined spectra it is immediately clear that all three ligands (DP1, DP2, and DP3) bind to SiaDW-135. A closer inspection of the data, however, revealed that the α-anomer of the free sialic acid was bound with much higher affinity than the β-anomer. This effect is easiest seen for the H3eq protons. Although in solution ∼5% of DP1 attains α- (at 2.45 ppm) and ∼95% β-configuration (at 1.95 ppm), signals of comparable intensities were observed if the protein was present (Fig. 6d). A similar effect could be seen for the singlet of the N-acetamido group at 1.76 (β-anomer) and 1.73 ppm (α-anomer) (Fig. 6, e and f). The STD NMR data thus strongly suggest that free Neu5Ac (DP1) is bound to the nucleotide sugar binding pocket.
FIGURE 6.
DP1–DP3 are entirely bound by SiaDW-135. 1H and STD NMR were used to study the binding of Sia and oligoSia structures to SiaDW-135. All three compounds (DP1, DP2, and DP3) were entirely bond to the enzyme. However, in the case of DP1 ∼50% of the sugar attained β-configuration (at 1.95 ppm), indicating that the monomer binds to the donor sugar (CMP-Sia) binding site (a–f). In DP2, the dimer of α(2,8)-linked sialic acid residues (g–l), and DP3, the trimer of α(2,8)-linked sialic acid residues (m–p), all sugar units are in contact with the protein. The signals are highlighted for H3eq and N-acetamido protons (NHAc). Although in both compounds the nonreducing end sugar receives the highest energy transfer, only DP3 was found to be an efficient primer to start the enzyme reaction. q, schematic representation of DP3. Differences in saturation transfer are illustrated by the letter size of 3Heq protons. Spectra were recorded at 280 K, 600 MHz using deuterated Tris buffer (20 mm, pH 8.3, and 20 mm MgCl2). Protein resonances were saturated using 40 Gaussian pulses of 5 ms duration at −1.00 ppm. The off-resonance frequency was set to 33 ppm.
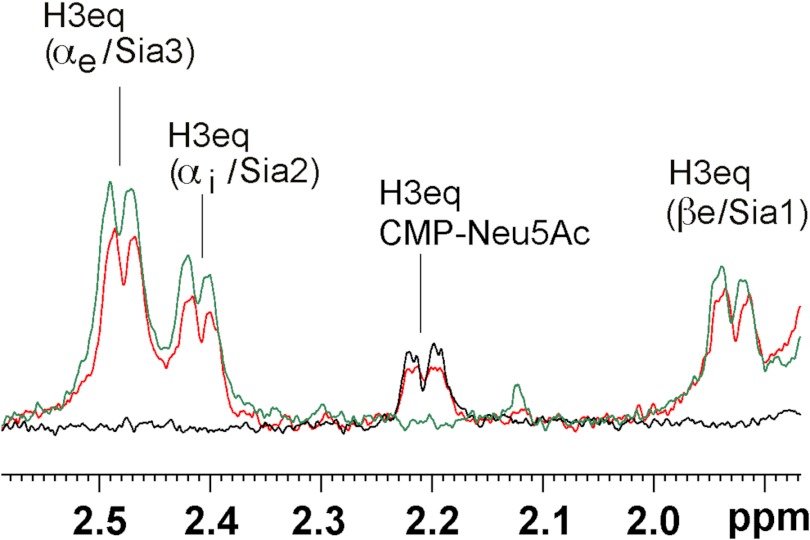
The spectra obtained with DP2 and DP3 revealed that the nonreducing sugar received more saturation than the middle (Sia2 in DP3) and reducing sugar (Sia1). This effect again was reflected by both the H3eq and the N-acetamido group protons (see Fig. 6, i–l for DP2 and o and p for DP3).
Although the data collected so far do not suffice to draw a detailed picture of the acceptor binding site in SiaDW-135, the observation that DP3 is an efficient acceptor and fully accommodated in the binding pocket supports the notion that only DP3 has the capacity to make contacts to all sites involved in binding and presenting the acceptor to the catalytic center.
Moreover, with this defined acceptor at hand we re-evaluated the hypothesis of consecutive polymer building by the two GT-B folds. Therefore, the wild-type SiaDW-135 as well as mutants E307A and S972A were analyzed in vitro with DP3 as acceptor. With UDP-[14C]Gal and cold CMP-Neu5Ac long radioactively labeled polymers (CPS) were synthesized in the reactions with wild-type enzyme (lane 1) and if the two single mutants E307A and S972A were present in one reaction mixture (lane 6). A single transfer of [14C]Gal onto the acceptor DP3 (lane 2) was catalyzed by mutant S972A, harboring an intact galactosyltransferase domain, whereas mutant E307A, harboring a defective galactosyltransferase, did not produce a radioactive product (lane 4). Importantly, with CMP-[14C]Neu5Ac and cold UDP-Gal none of the mutants produced a radioactive signal (lanes 3 and 5), thus clearly evidencing that Gal transfer to DP3 is prerequisite to generate the acceptor recognized by the sialyltransferase domain. The data provided in this paragraph prove the iterative activity of the two GT-B folds in SiaDW-135.
Independent Acceptor and Nucleotide-Sugar Binding to SiaDW-135
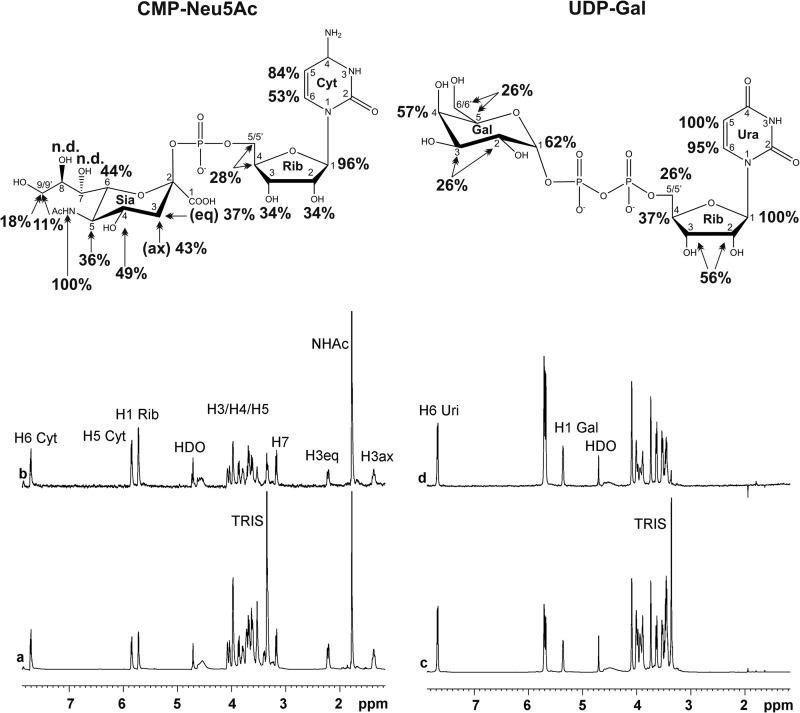
To further analyze the concept of two glycosyltransferase domains in one polypeptide chain the binding of the nucleotide sugars (UDP-Gal and CMP-Neu5Ac) to the protein was investigated. 1H NMR spectra of UDP-Gal and CMP-Neu5Ac (Fig. 7, a and c) and STD NMR spectra of UDP-Gal and CMP-Neu5Ac in the presence of SiaDW-135 (Fig. 7, b and d) were acquired and demonstrated in line with earlier data (29) the relevance of H1 Rib in the interactions of sugar nucleotides with proteins. H1 Rib as well as H5 in the nucleotide base received high saturation in both UDP-Gal and CMP-Neu5Ac. Of note, H5 Ura and H1 Rib STD NMR signals were overlapping in UDP-Gal and could not be separated. Confirming the specificity of SiaDW-135 for galactose H4 Gal received 57% saturation and, remarkably, also the anomeric proton in Gal received considerable saturation (62%). In the case of CMP-Neu5Ac, the N-acetamido group (1.82 ppm) was found to be strongly involved in the recognition event by the enzyme. Presence of equimolar concentrations of UDP-Gal did not change the STD NMR signals obtained for CMP-Neu5Ac (Fig. 8), thus excluding co-location of the nucleotide-sugar binding sites. This fact is particularly highlighted by the methyl protons of the N-acetamido group of CMP-Neu5Ac (Fig. 8c).
FIGURE 7.
STD NMR spectra of SiaDW-135 in complex with the nucleotide sugar donor substrates. To determine epitopes in the donor sugars CMP-Neu5Ac (a and b) and UDP-Gal (c and d) that make contact to the protein, an STD NMR study was performed. In line with previous data this experiment demonstrated the importance of H1 Rib for nucleotide sugar binding. This position together with H5 in the nucleotide base received the highest saturation in both UDP-Gal and CMP-Neu5Ac. In accordance with the specificity of SiaDW-135 for galactose H4 Gal received 57% saturation and, remarkably, also the opposite site of the pyranose ring, the anomeric proton H1 Gal, received considerable saturation (62%). For experimental details, see Fig. 6.
FIGURE 8.
CMP-Neu5Ac and UDP-Gal bind independently to SiaDW-135. To find out if binding of one nucleotide sugar impacts binding of the second, the STD NMR spectrum of CMP-Neu5Ac in complex with SiaDW-135 was recorded in the absence (a) and presence (b) of an equimolar concentration of UDP-Gal. No change in the position or intensity of any signal was seen, thus indicating independent binding of the nucleotide sugars. To highlight this fact, an overlay is shown for the intense signal produced by the tightly bound N-acetamido group (c). For experimental details, see Fig. 6.
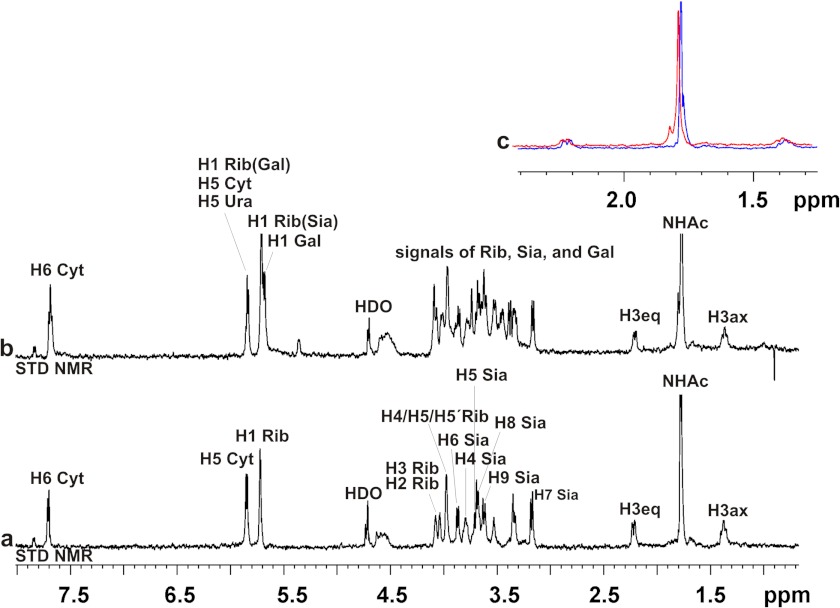
Next we analyzed if addition of the acceptor DP3 alters nucleotide sugar binding profiles. As previous experiments (see Fig. 5B) had shown that DP3 is not elongated by the sialyltransferase domain, data were recorded in the absence of UDP-Gal to prevent the start of the catalytic reaction. In Fig. 9 three STD NMR spectra are superimposed: 1) SiaDW-135 in complex with CMP-Neu5Ac (black), 2) SiaDW-135 in complex with DP3 (green), and 3) SiaDW-135 in complex with equimolar amounts of DP3 and CMP-Neu5Ac (red). Although the simultaneous presence of acceptor (DP3) and nucleotide-sugar (CMP-Neu5Ac) induced a slight reduction in the absolute STD NMR intensities (∼15% for the H3eq proton of CMP-Neu5Ac and ∼20% for all H3eq protons in DP3) the STD NMR signal intensities remained predominately unchanged and suggested that DP3 binds to a distinct domain.
FIGURE 9.
STD NMR spectra of SiaDW-135 in complex with CMP-Neu5Ac and the acceptor DP3. To evaluate if binding of the acceptor DP3 influences the binding of CMP-Neu5Ac, three STD NMR spectra were overlaid. In black and green the spectra obtained for SiaDW-135 in complex with CMP-Neu5Ac and DP3, respectively, are shown. The spectrum obtained when both substrates were present in equimolar concentrations is shown in red. The absolute STD NMR intensities revealed only a minor (∼15%) reduction in the saturation transfer to the H3eq proton of CMP-Neu5Ac when DP3 was present. Similarly, a reduction of ∼20% was seen for the H3eq protons of DP3 under these conditions. The faintness of the differences observed in the presence of the second substrate argued for independent binding sites. For experimental details, see Fig. 6.
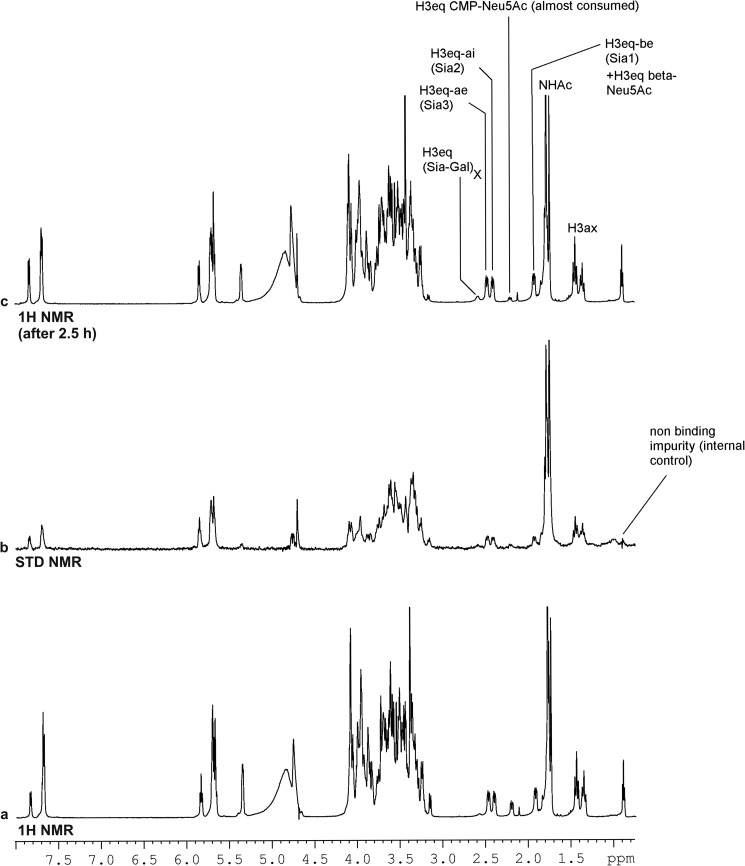
The above result was confirmed when the catalytic reaction of SiaDW-135 was allowed to proceed in the presence of equimolar concentrations of UDP-Gal, CMP-Neu5Ac, and DP3 (Fig. 10a). After 2.5 h acquisition time, CMP-Neu5Ac was almost completely consumed and a product peak representing the W-135 polymer became visible by 1H NMR (Fig. 10c). The STD NMR spectrum recorded for the active enzyme in the presence of all substrates clearly revealed independent binding sites for all three substrates (Fig. 10b).
FIGURE 10.
1H and STD NMR analysis of SiaDW-135 in complex with acceptor and donor substrates. The STD NMR analysis was carried out with SiaDW-135 in complex with CMP-Neu5Ac, UDP-Gal, and DP3 (a and b). The absolute STD NMR intensities revealed specific and independent binding of all three substrates (b). Consumption of CMP-Neu5Ac and parallel appearance of a product peak after the 2.5-h acquisition time (c) clearly showed that the enzyme reaction proceeded. For experimental details, see Fig. 6.
DISCUSSION
In this study we show that bifunctionality of SiaDW-135 is the result of the presence of two catalytic domains in one polypeptide chain. Based on the conserved structural motifs catalytic functions could be predicted for the two GT-B folds identified by bioinformatics tools. Rational mutations confirmed the predictions and proved galactosyl- and sialyltransferase activity for the N and C terminally located folds, respectively. Moreover, by combining point mutated proteins, in which one of the putative catalytic functions was disturbed, 70% of wild-type activity was restored. In size exclusion chromatography the active recombinant enzyme migrated with an apparent molecular mass of 120 kDa, indicating that the monomer is the functional unit. In STD NMR studies, the two GT-B folds were shown to bind their sugar-nucleotide substrates (CMP-Neu5Ac and UDP-Gal) independently and simultaneously. With DP3, a trimer of α(2,8)-linked sialic acids, a suitable minimal primer for the polymerase reaction has been identified and used in in vitro activity assays as well as in STD NMR studies to demonstrate that chain elongation needs the successive activity of the hexosyl- and sialyltransferase domains.
Glycosyltransfeases (GTs) catalyze the synthesis of complex products (e.g. polymers) in the absence of templates. Product control is an enzyme-inherent feature installed by the specificity of the GTs for their substrates (nucleotide-sugar and acceptor) and the type of catalyzed glycosidic linkage. In 1968 this observation led to the formulation of the paradigm of “one enzyme-one linkage” (30). Although the identification of GTs that in the absence of a second catalytic center conjugate one sugar with two types of glycosidic bonds (31–33) challenged the paradigm, SiaDW-135 and SiaDY remain unique because these enzymes conjugate sugars of very different chemical nature (the disparity is even higher in the GT substrates CMP-Neu5Ac and UDP-Gal/Glc) and use different catalytic mechanisms to form the glycosidic bonds (inversion of the anomeric center in the case of sialic acid and retention in the case of the hexoses). Not surprising thus that these large enzymes were suggested to represent fusion products of ancestral genes (11, 12).
The N-terminal GT-B fold in SiaDW-135 and SiaDY exhibits sequence similarity with retaining hexosyltransferases of CAZy-family GT-4. Common to members of this group is the EX7E motif, a part of the so called nucleotide recognition domain 1α (16, 34). Mutation of the first conserved glutamic acid residue in the EX7E motif has been demonstrated to inactivate retaining GTs (35–38). In line with these earlier data we show here that the position (Glu-307) is essential also in SiaDW-135. The replacement E307A abolished activity in vitro and in vivo. Of importance, position four within the EX7E motif (aa 310; proline in NmW-135 and glycine in NmY) was previously shown to determine UDP-sugar specificity in these bifunctional enzymes (12). The STD NMR data obtained in the current study show that H4 Gal receives high saturation (Fig. 7). The specificity for galactose in SiaDW-135 can thus be explained by a tight contact formed between the C-4 hydroxyl group of Gal and proline 310 in the sugar nucleotide binding pocket.
For the second GT-B fold, shown in this study to comprise an α(2,6)-monosialyltransferase, no similarity to members in any of the CAZy families could be identified. Nevertheless, the used fold recognition tool properly located the previously identified conserved motifs HP and SS/T (23–25) along the active site cleft that separates the two Rossmann subdomains (see Fig. 1C). The HP motif is broadly conserved in bacterial sialyltransferases and was shown to be involved in CMP binding and/or enzyme catalysis (39). The more C terminally located S-motif contains a highly conserved serine and a second less conserved serine or threonine residue. In CAZy family GT-80 this motif was shown to coordinate the CMP-phosphate group in the intermediate state carbenium ion (24, 26, 40). In accordance with this essential function, point mutation of the crucial Ser-972 in SiaDW-135 fully inactivated the enzyme.
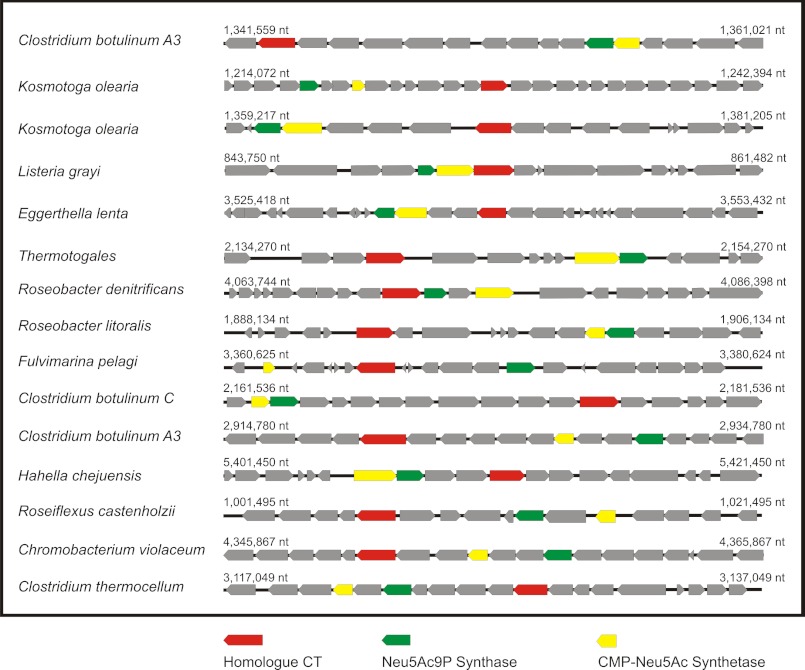
Still searching for homologues of the SiaDW-135/Y sialyltransferase domain we performed an extended BLAST analysis and identified weak homologies to several bacterial open reading frames encoding proteins of unknown function. All identified open reading frames contain the HP and S motifs and, as schematically shown in Fig. 11, occur in the company of two genes encoding enzymes that locate immediately upstream of the sialyltransferase activity in the sialylation pathway. The accompanying enzymes are the sialic acid synthase (41) and the CMP-sialic acid synthetase (42). Because functionally linked genes are often organized in operons in bacteria, the identified ORFs are likely to encode sialyltransferases. Surprisingly, the multifunctional sialyltransferase PmST1 from Pasteurella multocida was not identified in this initial screen although the enzyme has been demonstrated to attain a GT-B fold (43).
FIGURE 11.
Bacterial genome regions comprising putative sialyltransferases with homology to SiaDW-135. With the sialyltransferase domain identified in SiaDW-135 as template, a BLAST search was carried out and returned a number of bacterial proteins with weak homology. Although no function has yet been attributed to these proteins, all are encoded by genes (designated homologue CT and marked in red) that in the genome occur in close company with two other genes encoding enzymes of the sialylation pathway: the Neu5Ac9P-synthase (gene marked in green) and the CMP-Neu5Ac synthetase (gene marked in yellow). Bacteria harboring these homologues are named and the respective gene regions are schematically shown.
Besides identification of bifunctionality in single domain GTs the presence of two independent binding sites has also been shown. Examples are the hyaluronan synthase from P. multocida (44) and the E. coli K4 chondroitin polymerase (K4CP) (45). However, in marked difference to SiaDW-135/Y these polymerases are hexosyltransferases only and belong to the GT-A structural superfamily.
Binding analyses carried out by STD NMR provided conclusive evidence that the nucleotide sugar binding pockets in SiaDW-135 are independent from each other (Fig. 8). In both domains the nucleotide moieties (CMP and UDP) receive highest saturation, whereas the sugar moieties bind weaker. This binding mode has been previously described and was suggested to facilitate the transfer process (29, 46, 47). A particular feature in CMP-Neu5Ac binding is the tight contact that the N-acetamido group of sialic acid makes to the protein. This tight contact may be necessary to prevent the negatively charged acetamido group from interfering with the transfer process.
Unexpectedly we found DP3, but neither DP2 nor 2,3-sialyllactose, an efficient acceptor to start the polymerization reaction in SiaDW-135. Although additional experimental work is needed to understand the primer quality of DP3, the current STD NMR data demonstrate that DP3 like DP2 is entirely bound to the acceptor site of the enzyme. Because DP2 is not an efficient acceptor, we speculate that only DP3 has the necessary geometry (e.g. length and topology of functional groups) to make contacts to all sites involved in the binding and presentation of the acceptor to the catalytic center.
The nonreducing end sialic acid of DP3 (the sugar where transfer occurs) received the highest saturation when bound to SiaDW-135. In contrast, if DP3 was complexed with the recombinant NmB capsule polymerase (a sialyltransferase produces a homopolymer of α(2,8)-linked sialic acids), the saturation was found to be lowest for the nonreducing end sugar (47). Understanding the molecular reasons for these differences needs additional work, but it should be mentioned at this point that DP3 in the case of SiaDW-135 is the acceptor of the galactosyltransferase domain, which may have different stereochemical requirements compared with sialyltransferases.
Not clear at this moment is if the two GT-domains in SiaDW-135 comprise individual acceptor binding sites or share a common one. Although the STD NMR experiments carried out in the presence of two (CMP-Neu5Ac and DP3) or even three (CMP-Neu5Ac, UDP-Gal and DP3) substrates demonstrate an independent binding of acceptor and donor substrates, the way the growing polymer is bound to the enzyme remains an open question. Initial trials made to separately express the hexosyl- and sialyltransferase domains failed to produce active proteins. Combined with the knowledge that full-length proteins with only one functional domain were able to complement each other in trans (Fig. 4C) this latter finding suggests that the large linker domain that connects the GT-B fold in SiaDW-135 (see Fig. 1A) may have a function in forming a proper acceptor binding pocket. Our current work focuses at answering this question. With the current study we identified the bifunctional SiaDW-135 as a chimeric enzyme comprising two independent catalytic domains that work in succession to produce the highly unusual W-135 capsular polymer.
The work was supported by LOM (impact oriented funding) funds to the Institute for Cellular Chemistry and the Australian Research Council through a Federation Fellowship (to M. v. I.).
- CPS
- capsule polysaccharide
- Nm
- Neisseria meningitidis
- DP3
- trimer of α(2,8)-linked sialic acid
- CMP-Neu5Ac and/or CMP-Sia
- CMP-N-acetylneuraminic acid
- GT
- glycosyltransferase
- STD NMR
- saturation transfer difference nuclear magnetic resonance.
REFERENCES
- 1. Rosenstein N. E., Perkins B. A., Stephens D. S., Popovic T., Hughes J. M. (2001) Meningococcal disease. N. Engl. J. Med. 344, 1378–1388 [DOI] [PubMed] [Google Scholar]
- 2. Stephens D. S., Greenwood B., Brandtzaeg P. (2007) Epidemic meningitis, meningococcaemia, and Neisseria meningitidis. Lancet 369, 2196–2210 [DOI] [PubMed] [Google Scholar]
- 3. Raymond N. J., Reeves M., Ajello G., Baughman W., Gheesling L. L., Carlone G. M., Wenger J. D., Stephens D. S. (1997) Molecular epidemiology of sporadic (endemic) serogroup C meningococcal disease. J. Infect. Dis. 176, 1277–1284 [DOI] [PubMed] [Google Scholar]
- 4. Taha M. K., Achtman M., Alonso J. M., Greenwood B., Ramsay M., Fox A., Gray S., Kaczmarski E. (2000) Serogroup W135 meningococcal disease in Hajj pilgrims. Lancet 356, 2159. [DOI] [PubMed] [Google Scholar]
- 5. Decosas J., Koama J. B. (2002) Chronicle of an outbreak foretold. Meningococcal meningitis W135 in Burkina Faso. Lancet Infect. Dis. 2, 763–765 [DOI] [PubMed] [Google Scholar]
- 6. Raghunathan P. L., Jones J. D., Tiendrebéogo S. R., Sanou I., Sangaré L., Kouanda S., Dabal M., Lingani C., Elie C. M., Johnson S., Ari M., Martinez J., Chatt J., Sidibe K., Schmink S., Mayer L. W., Kondé M. K., Djingarey M. H., Popovic T., Plikaytis B. D., Carlone G. M., Rosenstein N., Soriano-Gabarró M. (2006) Predictors of immunity after a major serogroup W-135 meningococcal disease epidemic, Burkina Faso, 2002. J. Infect. Dis. 193, 607–616 [DOI] [PubMed] [Google Scholar]
- 7. Pollard A. J., Scheifele D. (2001) Meningococcal disease and vaccination in North America. J. Paediatr. Child Health 37, S20-S27 [DOI] [PubMed] [Google Scholar]
- 8. Tzeng Y. L., Stephens D. S. (2000) Epidemiology and pathogenesis of Neisseria meningitidis. Microbes. Infect. 2, 687–700 [DOI] [PubMed] [Google Scholar]
- 9. Bhattacharjee A. K., Jennings H. J., Kenny C. P., Martin A., Smith I. C. (1975) Structural determination of the sialic acid polysaccharide antigens of Neisseria meningitidis serogroups B and C with carbon 13 nuclear magnetic resonance. J. Biol. Chem. 250, 1926–1932 [PubMed] [Google Scholar]
- 10. Bhattacharjee A. K., Jennings H. J., Kenny C. P., Martin A., Smith I. C. (1976) Structural determination of the polysaccharide antigens of Neisseria meningitidis serogroups Y, W-135, and BO1. Can. J. Biochem. 54, 1–8 [DOI] [PubMed] [Google Scholar]
- 11. Claus H., Vogel U., Mühlenhoff M., Gerardy-Schahn R., Frosch M. (1997) Molecular divergence of the sia locus in different serogroups of Neisseria meningitidis expressing polysialic acid capsules. Mol. Gen. Genet. 257, 28–34 [DOI] [PubMed] [Google Scholar]
- 12. Claus H., Stummeyer K., Batzilla J., Mühlenhoff M., Vogel U. (2009) Amino acid 310 determines the donor substrate specificity of serogroup W-135 and Y capsule polymerases of Neisseria meningitidis. Mol. Microbiol. 71, 960–971 [DOI] [PubMed] [Google Scholar]
- 13. Campbell J. A., Davies G. J., Bulone V., Henrissat B. (1997) A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J. 326, 929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coutinho P. M., Deleury E., Davies G. J., Henrissat B. (2003) An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328, 307–317 [DOI] [PubMed] [Google Scholar]
- 15. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The Carbohydrate-Active EnZymes database (CAZy). An expert resource for Glycogenomics. Nucleic Acids Res. 37, D233-D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kapitonov D., Yu R. K. (1999) Conserved domains of glycosyltransferases. Glycobiology 9, 961–978 [DOI] [PubMed] [Google Scholar]
- 17. Schwarzer D., Stummeyer K., Gerardy-Schahn R., Mühlenhoff M. (2007) Characterization of a novel intramolecular chaperone domain conserved in endosialidases and other bacteriophage tail spike and fiber proteins. J. Biol. Chem. 282, 2821–2831 [DOI] [PubMed] [Google Scholar]
- 18. Studier F. W. (2005) Protein production by autoinduction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 19. Weisgerber C., Troy F. A. (1990) Biosynthesis of the polysialic acid capsule in Escherichia coli K1. The endogenous acceptor of polysialic acid is a membrane protein of 20 kDa. J. Biol. Chem. 265, 1578–1587 [PubMed] [Google Scholar]
- 20. Min H., Cowman M. K. (1986) Combined alcian blue and silver staining of glycosaminoglycans in polyacrylamide gels. Application to electrophoretic analysis of molecular weight distribution. Anal. Biochem. 155, 275–285 [DOI] [PubMed] [Google Scholar]
- 21. Ram S., Cox A. D., Wright J. C., Vogel U., Getzlaff S., Boden R., Li J., Plested J. S., Meri S., Gulati S., Stein D. C., Richards J. C., Moxon E. R., Rice P. A. (2003) Neisserial lipooligosaccharide is a target for complement component C4b. Inner core phosphoethanolamine residues define C4b linkage specificity. J. Biol. Chem. 278, 50853–50862 [DOI] [PubMed] [Google Scholar]
- 22. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web. A case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 23. Freiberger F., Claus H., Günzel A., Oltmann-Norden I., Vionnet J., Mühlenhoff M., Vogel U., Vann W. F., Gerardy-Schahn R., Stummeyer K. (2007) Biochemical characterization of a Neisseria meningitidis polysialyltransferase reveals novel functional motifs in bacterial sialyltransferases. Mol. Microbiol. 65, 1258–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ni L., Chokhawala H. A., Cao H., Henning R., Ng L., Huang S., Yu H., Chen X., Fisher A. J. (2007) Crystal structures of Pasteurella multocida sialyltransferase complexes with acceptor and donor analogues reveal substrate binding sites and catalytic mechanism. Biochemistry 46, 6288–6298 [DOI] [PubMed] [Google Scholar]
- 25. Yamamoto T., Ichikawa M., Takakura Y. (2008) Conserved amino acid sequences in the bacterial sialyltransferases belonging to glycosyltransferase family 80. Biochem. Biophys. Res. Commun. 365, 340–343 [DOI] [PubMed] [Google Scholar]
- 26. Kakuta Y., Okino N., Kajiwara H., Ichikawa M., Takakura Y., Ito M., Yamamoto T. (2008) Crystal structure of Vibrionaceae Photobacterium sp. JT-ISH-224 α2,6-sialyltransferase in a ternary complex with donor product CMP and acceptor substrate lactose. Catalytic mechanism and substrate recognition. Glycobiology 18, 66–73 [DOI] [PubMed] [Google Scholar]
- 27. Vogel U., Morelli G., Zurth K., Claus H., Kriener E., Achtman M., Frosch M. (1998) Necessity of molecular techniques to distinguish between Neisseria meningitidis strains isolated from patients with meningococcal disease and from their healthy contacts. J. Clin. Microbiol. 36, 2465–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mayer M., Meyer B. (2001) Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J. Am. Chem. Soc. 123, 6108–6117 [DOI] [PubMed] [Google Scholar]
- 29. Biet T., Peters T. (2001) Angew. Chem. Int. Ed. 40, 4189–4192 [DOI] [PubMed] [Google Scholar]
- 30. Hagopian A., Bosmann H. B., Eylar E. H. (1968) Glycoprotein biosynthesis. The localization of polypeptidyl:N-acetylgalactosaminyl, collagen, glucosyl, and glycoprotein:galactosyltransferases in HeLa cell membrane fractions. Arch. Biochem. Biophys. 128, 387–396 [DOI] [PubMed] [Google Scholar]
- 31. McGowen M. M., Vionnet J., Vann W. F. (2001) Elongation of alternating α2,8/2,9 polysialic acid by the Escherichia coli K92 polysialyltransferase. Glycobiology 11, 613–620 [DOI] [PubMed] [Google Scholar]
- 32. Chiu C. P., Watts A. G., Lairson L. L., Gilbert M., Lim D., Wakarchuk W. W., Withers S. G., Strynadka N. C. (2004) Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog. Nat. Struct. Mol. Biol. 11, 163–170 [DOI] [PubMed] [Google Scholar]
- 33. May J. F., Levengood M. R., Splain R. A., Brown C. D., Kiessling L. L. (2012) A processive carbohydrate polymerase that mediates bifunctional catalysis using a single active site. Biochemistry 51, 1148–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Geremia R. A., Petroni E. A., Ielpi L., Henrissat B. (1996) Towards a classification of glycosyltransferases based on amino acid sequence similarities. Prokaryotic α-mannosyltransferases. Biochem. J. 318, 133–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cid E., Gomis R. R., Geremia R. A., Guinovart J. J., Ferrer J. C. (2000) Identification of two essential glutamic acid residues in glycogen synthase. J. Biol. Chem. 275, 33614–33621 [DOI] [PubMed] [Google Scholar]
- 36. Abdian P. L., Lellouch A. C., Gautier C., Ielpi L., Geremia R. A. (2000) Identification of essential amino acids in the bacterial α-mannosyltransferase aceA. J. Biol. Chem. 275, 40568–40575 [DOI] [PubMed] [Google Scholar]
- 37. Saksouk N., Pelosi L., Colin-Morel P., Boumedienne M., Abdian P. L., Geremia R. A. (2005) The capsular polysaccharide biosynthesis of Streptococcus pneumoniae serotype 8. Functional identification of the glycosyltransferase WciS (Cap8H). Biochem. J. 389, 63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guerin M. E., Kordulakova J., Schaeffer F., Svetlikova Z., Buschiazzo A., Giganti D., Gicquel B., Mikusova K., Jackson M., Alzari P. M. (2007) Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria. J. Biol. Chem. 282, 20705–20714 [DOI] [PubMed] [Google Scholar]
- 39. Audry M., Jeanneau C., Imberty A., Harduin-Lepers A., Delannoy P., Breton C. (2011) Current trends in the structure-activity relationships of sialyltransferases. Glycobiology 21, 716–726 [DOI] [PubMed] [Google Scholar]
- 40. Kim D. U., Yoo J. H., Lee Y. J., Kim K. S., Cho H. S. (2008) Structural analysis of sialyltransferase PM0188 from Pasteurella multocida complexed with donor analogue and acceptor sugar. BMB Rep. 41, 48–54 [DOI] [PubMed] [Google Scholar]
- 41. Hao J., Balagurumoorthy P., Sarilla S., Sundaramoorthy M. (2005) Cloning, expression, and characterization of sialic acid synthases. Biochem. Biophys. Res. Commun. 338, 1507–1514 [DOI] [PubMed] [Google Scholar]
- 42. Kean E. L., Münster-Kühnel A. K., Gerardy-Schahn R. (2004) CMP-sialic acid synthetase of the nucleus. Biochim. Biophys. Acta 1673, 56–65 [DOI] [PubMed] [Google Scholar]
- 43. Ni L., Sun M., Yu H., Chokhawala H., Chen X., Fisher A. J. (2006) Cytidine 5′-monophosphate (CMP)-induced structural changes in a multifunctional sialyltransferase from Pasteurella multocida. Biochemistry 45, 2139–2148 [DOI] [PubMed] [Google Scholar]
- 44. Williams K. J., Halkes K. M., Kamerling J. P., DeAngelis P. L. (2006) Critical elements of oligosaccharide acceptor substrates for the Pasteurella multocida hyaluronan synthase. J. Biol. Chem. 281, 5391–5397 [DOI] [PubMed] [Google Scholar]
- 45. Sobhany M., Kakuta Y., Sugiura N., Kimata K., Negishi M. (2008) The chondroitin polymerase K4CP and the molecular mechanism of selective bindings of donor substrates to two active sites. J. Biol. Chem. 283, 32328–32333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Damerow S., Lamerz A. C., Haselhorst T., Führing J., Zarnovican P., von Itzstein M., Routier F. H. (2010) Leishmania UDP-sugar pyrophosphorylase. The missing link in galactose salvage? J. Biol. Chem. 285, 878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Böhm R., Freiberger F., Stummeyer K., Gerardy-Schahn R., von Itzstein M., Haselhorst T. (2010) Neisseria meningitidis serogroup B polysialyltransferase. Insights into substrate binding. Chembiochem. 11, 170–174 [DOI] [PubMed] [Google Scholar]