Background: Lipid A activation of TLR4 shapes immunity to Gram-negative bacteria.
Results: In Bordetella pertussis lipid A, the genetic basis for longer C3′ acyl chains and glucosamine modification of the phosphate groups was identified; each variation independently increased TLR4 activation.
Conclusion: Minor changes in the penta-acylated B. pertussis lipid A affect TLR4 activation.
Significance: This aids our understanding how lipid A species interact with TLR4.
Keywords: Bacteria, Bacterial Pathogenesis, Endotoxin, Innate Immunity, Lipopolysaccharide (LPS), Toll-like Receptors (TLR), Bordetella pertussis, Lipid A, LpxA, TLR4
Abstract
Lipopolysaccharides (LPS) of Bordetella pertussis are important modulators of the immune system. Interaction of the lipid A region of LPS with the Toll-like receptor 4 (TLR4) complex causes dimerization of TLR4 and activation of downstream nuclear factor κB (NFκB), which can lead to inflammation. We have previously shown that two strains of B. pertussis, BP338 (a Tohama I-derivative) and 18-323, display two differences in lipid A structure. 1) BP338 can modify the 1- and 4′-phosphates by the addition of glucosamine (GlcN), whereas 18-323 cannot, and 2) the C3′ acyl chain in BP338 is 14 carbons long, but only 10 or 12 carbons long in 18-323. In addition, BP338 lipid A can activate TLR4 to a greater extent than 18-323 lipid A. Here we set out to determine the genetic reasons for the differences in these lipid A structures and the contribution of each structural difference to the ability of lipid A to activate TLR4. We show that three genes of the lipid A GlcN modification (Lgm) locus, lgmA, lgmB, and lgmC (previously locus tags BP0399–BP0397), are required for GlcN modification and a single amino acid difference in LpxA is responsible for the difference in C3′ acyl chain length. Furthermore, by introducing lipid A-modifying genes into 18-323 to generate isogenic strains with varying penta-acyl lipid A structures, we determined that both modifications increase TLR4 activation, although the GlcN modification plays a dominant role. These results shed light on how TLR4 may interact with penta-acyl lipid A species.
Introduction
Lipopolysaccharides (LPS) comprise the outer leaflet of the outer membrane in almost all Gram-negative bacteria and consist of two major regions: the hydrophilic polysaccharide and the hydrophobic lipid A region (1). The polysaccharide region of LPS is composed of the polysaccharide core, which is found in most Gram-negative bacteria, and in some cases, a long, repeating O-antigen region. Bordetella pertussis LPS lacks a long O-antigen, and in its place is a short trisaccharide moiety linked to the core polysaccharide (2); it is often referred to as lipooligosaccharide (LOS).4 The lipid A region of LPS plays an important role in modulating the host immune system via interaction with Toll-like receptor 4 (TLR4), which leads to downstream nuclear factor κB (NFκB) activation. Although there are many variations in lipid A structure among different species, and even among different strains of a particular bacterial species, the general structure of lipid A consists of a disaccharide backbone with a number of primary and secondary acyl chains. The canonical lipid A, that of Escherichia coli, has a (1′-6) diglucosamine backbone with the C1 and C4′ carbons modified with a phosphate group and has six acyl chains. However, even within a single bacterium, there is a heterogeneous population of lipid A species present in the outer membrane. For example, many E. coli strains are able to modify lipid A in a number of ways, such as altering the number and length of the acyl chains and adding accessory groups, such as 4-amino-4-deoxy-l-arabinose (l-Ara4N) via ArnT, onto the phosphates. Therefore, within the outer membrane of a single bacterium there are a variety of lipid A structures, with different modifications and acylation patterns (1–4).
Bacteria can use the modification of lipid A as a mechanism of resistance against cationic peptides or to affect the immune response mounted by the host (1). For example, in E. coli and Salmonella, the addition of l-Ara4N to the phosphate groups increases resistance to cationic peptides (5). Other lipid A modifications affect TLR4 activation and complement resistance (1).
B. pertussis lipid A, like E. coli lipid A, consists of (1′-6) diglucosamine with 1- and 4′-phosphate groups, but unlike E. coli lipid A, it has only five acyl chains (2). In addition, B. pertussis lipid A induces lower levels of TLR4 activation when compared with E. coli lipid A, and this difference is generally attributed to the penta-acyl nature of the lipid A, as opposed to the hexa-acyl structure of E. coli lipid A (1, 6). However, even among different B. pertussis strains, there is variation in the ability of LOS to activate TLR4 (6). We have previously shown that LOS from B. pertussis strain BP338 (a derivative of Tohama I) induces greater TLR4 activation than strain 18-323, both of which are commonly used laboratory strains (6). Furthermore, we have shown that 18-323 LOS is antagonistic against E. coli LPS (6). Because lipid A is the region of LPS that interacts with the TLR4-MD2 complex, the variation in lipid A structure between BP338 and 18-323 is likely a contributing factor to the difference in TLR4 activation between these strains.
Analysis of the lipid A structures of BP338 and 18-323 show two differences: 1) BP338 can modify the phosphates of lipid A with glucosamine (GlcN), whereas 18-323 cannot, and 2) the C3′ acyl chain length of BP338 lipid A is 14 carbons, whereas in 18-323, it is only 10 or 12 carbons in length (6). Previously, we have linked the gene BP0398, a homolog of ArnT, with GlcN modification of lipid A in BP338 and shown that a BP338 mutant that lacks this GlcN modification has decreased levels of TLR4 activation (7, 8). Previous work in E. coli suggests that a specific region of the enzyme LpxA, dubbed the “hydrocarbon ruler” region, plays a role in determining the length of the C3 and C3′ acyl chains in lipid A (9, 10). LpxA is the first enzyme in the Raetz lipid A biosynthesis pathway, and it is responsible for the addition of the C3 acyl chain onto the N-acetyl glucosamine (GlcNAc) precursor molecule (11). In B. pertussis LOS, LpxA has been shown to play a role in the flexibility of the C3′ acyl chain length (12).
To determine the contribution of the GlcN modification and C3′ acyl chain length of lipid A on TLR4 activation, we first set out to discover the genetic reasons for these structural differences between B. pertussis strains BP338 and 18-323. In the present study, we show that locus tags BP0399, BP0398, and BP0397 (which we are renaming lipid A GlcN modification A (lgmA), lgmB, and lgmC, respectively) are required for GlcN modification in BP338, and the absence of a complete Lgm locus in 18-323 is responsible for the inability of 18-323 to modify lipid A in this manner. We demonstrate that a single amino acid difference in the hydrocarbon ruler region of LpxA between these two strains is the reason for the difference in the lipid A C3′ acyl chain lengths. Then, to probe the effect these two modifications have on TLR4 activation, we generated strains with varying lipid A structures by adding the lipid A-modifying genes from BP338 into 18-323. Using in vitro assays, we show that both the GlcN modification and the longer C3′ acyl chain independently increase TLR4 activation. We also demonstrate that when both modifications are present, the effect of the GlcN on TLR4 activation supersedes that of the longer acyl chain. These results shed further light on how the TLR4 complex may interact with differently shaped lipid A species, especially penta-acyl lipid A.
EXPERIMENTAL PROCEDURES
Bacterial Strain and Growth Conditions
The strains and plasmids used in this work are shown in Table 1. B. pertussis strains were grown on Bordet-Gengou agar supplemented with 15% defibrinated sheep's blood (Dalynn) at 37 °C or in Stainer-Scholte broth with 0.06% bovine serum albumin (Sigma-Aldrich) at 180 rpm at 37 °C (13). All BP338 strain derivatives were grown in the presence of 30 μg/ml nalidixic acid. E. coli strains were grown in Luria Bertani (LB) broth at 200 rpm or on LB agar at 37 °C. Strains containing derivatives of pMMB67HE were grown with 100 μg/ml ampicillin, strains containing derivatives of pBBR1MCS were grown with 34 μg/ml chloramphenicol, and strains containing derivatives of pSS4245 or pBBR1MCS2 were grown with 50 μg/ml kanamycin. E. coli strain DH5α (Invitrogen) was used for cloning, and S17-1 (14) was used as the donor strain for conjugation.
TABLE 1.
Strains and constructs
IPTG, isopropyl-1-thio-β-d-galactopyranoside.
| Description | Source or reference | |
|---|---|---|
| Plasmids | ||
| pBBR1MCS | Broad-range vector, ChlorR | 34 |
| pNMLgmAB | pBBR1MCS containing lgmA and lgmB of BP338 | This work |
| pNMLgmCD | pBBR1MCS containing lgmC and lgmD of BP338 | This work |
| pNMLgmABCD | pBBR1MCS containing lgmA, lgmB, lgmC, and lgmD of BP338 | This work |
| pMMB67HE | Ptac promoter (IPTG-inducible), AmpR | 35 |
| pPtacLgmABCD | pMMB67HE containing lgmA, lgmB, lgmC, and lgmD of BP338 | This work |
| pPtacLgmAB | pMMB67HE containg lgmA and lgmB | This work |
| pPtacLpxA338 | pMMB67HE containing lpxA of BP338 | This work |
| pPtacLgmABCDLpxA338 | pMMB67HE containing lgmA, lgmB, lgmC, lgmD, and lpxA of BP338 | This work |
| pBBR1MCS2 | Broad-range vector, KanR | 36 |
| pBBR2Pcpn | pBBR1MCS2 containing the Pcpn heat shock promoter, KanR | This work |
| pBBRPcpnBrkA | pBBR1MCS containing promoter region of cpn10 (Pcpn), CmR | 37 |
| pBBR2LgmA | pBBR2Pcpn containing lgmA of BP338 | This work |
| pBBR2LgmC | pBBR2Pcpn containing lgmC of BP338 | This work |
| pSS4245 | Suicide vector (replicates in E. coli, but not in B. pertussis), KanR | S. Stibitz (38) |
| pSS4245LgmADKO | pSS4245 containing the lgmA upstream region and the lgmD downstream region; for generation of BP338lgmADKO | This work |
| pSS4245LgmAKO | pSS4245 containing the lgmA upstream and downstream regions; for generation of BP338lgmAKO | This work |
| pSS4245LgmBKO | pSS4245 containing the lgmB upstream and downstream regions; for generation of BP338lgmBKO | This work |
| pSS4245LgmCKO | pSS4245 containing the lgmC upstream and downstream regions; for generation of BP338lgmCKO | This work |
| pSS4245LgmDKO | pSS4245 containing the lgmD upstream and downstream regions; for generation of BP338lgmDKO | This work |
| Bordetella pertussis strains | ||
| BP338 | NalR derivative of Tohama I wild-type strain | A. Weiss (39) |
| BP338LgmABCDKO | ΔlgmABCD | This work |
| BP338LgmAKO | ΔlgmA | This work |
| BP338LgmBKO | ΔlgmB | This work |
| BP338LgmCKO | ΔlgmC | This work |
| BP338LgmDKO | ΔlgmD | This work |
| 18–323 | Wild type strain | A. Weiss (6) |
Cloning of Vectors and Generation of Deletion Mutants
Standard molecular cloning techniques were used (15). All restriction enzymes were from New England Biolabs. Vectors were introduced into E. coli strains via transformation (15) and into B. pertussis strains via electroporation (16) or conjugation, as described previously (8), in which E. coli strain S17-1 acted as the donor strain. See the supplemental text for more details on the generation of the vectors and mutant strains, and see supplemental Table S1 for a list of primers. All constructs were verified by PCR and sequencing. The pSS4245 vector was a generous gift from Scott Stibitz (Center for Biologics Evaluation and Research, United States Food and Drug Administration) and pBBR1MCS2 was generously provided by Robert Hancock (University of British Columbia).
PCR Amplification of the Lgm Locus Genes in B. pertussis Strains
Internal fragments of the lgm genes were amplified with PCR using genomic DNA from strain BP338 or 18-323 as templates and the following primer pairs: lgmA (BP0399fw1 and BP0399rev1), lgmB (BP0398fw1 and BP0398rev1), lgmC (BP397-RTfw and BP397-RTrev), and lgmD (BP396-RTfw and BP396-RTrev).
Preparation of Bacterial Cells for Lipid A Isolation and Biological Assays
B. pertussis strains were grown on Bordet-Gengou agar at 37 °C for 3–4 days, and these cells were used to inoculate Stainer-Scholte broth, with the appropriate antibiotics, at an A600 of 0.01. Cultures were grown at 180 rpm at 37 °C until an A600 of 0.6–0.9, and samples from each culture were grown on Bordet-Gengou agar to confirm the hemolytic phenotype. Bacterial cells were harvested into phosphate-buffered saline (PBS) pH = 7.4 to an A600 of 5 and heat-inactivated by incubation at 56 °C for 1 h. To confirm heat inactivation, 50 μl of each cell suspension was spotted onto Bordet-Gengou agar and incubated at 37 °C for 5 days. Heat-killed cells were stored at −20 °C. These cells were used for: stimulation in a TLR4 activation assay, generation of highly purified LOS for use as the stimulus in the TLR4 activation assay, and direct lipid A isolation for use in MALDI-MS analysis.
Highly Purified LOS Preparation
B. pertussis LOS preparations were extracted by an ammonium hydroxide-isobutyric acid method (40). Primary extracts were subjected to a standard enzyme treatment (DNase, RNase, and proteinase K) and finally repurified with the acidified chloroform-methanol-water procedure as described (17). To be sure that no specific LOS molecular species were discriminated during the process, all intermediate and final products were analyzed by MALDI-MS. High purity of the resulting LOS preparations was evidenced by three different methods. The absence of contaminating (non-LOS) peaks was attested by positive-ion MALDI mass spectra analysis; the absence of detectable protein contaminants was demonstrated by Tricine-SDS-PAGE (18) and silver staining (19) loading up to 250 ng of LOS from each preparation; and the absence of lipoprotein content was further demonstrated based on the lack of detectable levels of cysteine by analysis with an amino acid analyzer (Hitachi L-8800, equipped with a 2620MSC-PS column, ScienceTec, Les Ulis, France).
MALDI-MS Analysis
LOS samples were dispersed in water at 1 μg/μl. Lipid A extracts in chloroform-methanol-water were used directly. In both cases, a few microliters of sample solution were desalted with a few grains of ion-exchange resin Dowex 50W-X8 (H+). 0.5–1-μl aliquots of the solution were deposited on the target, covered with matrix solution, and allowed to dry. Dihydroxybenzoic acid (Sigma-Aldrich) was used as matrix. It was dissolved at 10 mg/ml in 0.1 m citric acid solution in the same solvents as those used for the analytes (20). Different analyte/matrix ratios (1:2, 1:1, 2:1, v/v) were tested to obtain the best spectra. Analyses were performed on a PerSeptive Voyager-DE STR time-of-flight mass spectrometer (Applied Biosystems) in linear mode, with delayed extraction. Negative- and positive-ion mass spectra were recorded. The ion-accelerating voltage was set at −20 kV, and the extraction delay time was adjusted to obtain the best resolution and signal-to-noise ratio.
Direct Lipid A Isolation from Bacterial Cells
For MALDI-MS analysis, lipid A was isolated directly by hydrolysis of bacterial cells as described previously (21, 22). Briefly, lyophilized bacterial cells (10 mg) were suspended in 200 μl of a mixture of isobutyric acid, 1 m ammonium hydroxide (5:3, v/v) and were kept for 2 h at 100 °C in a screw-cap test tube under magnetic stirring. The suspension was cooled in ice water and centrifuged (2000 × g, 10 min). The recovered supernatant was diluted with 2 volumes of water and lyophilized. The sample was then washed once with 200 μl of methanol (by centrifugation at 2000 × g for 10 min). Finally, lipid A was extracted from the pellet in 100 μl of a mixture of chloroform, methanol, and water (3:1.5:0.25, v/v/v). In some of the spectra, peaks corresponding to small contaminants were identified and marked with an “X” (peaks at m/z 1349 and 1377).
Cell Culture
HEK-Blue (InvivoGen) cell lines hTLR4 and Null2 cells were maintained in complete Dulbecco's modified Eagle's medium (DMEM) containing 10% heat-inactivated fetal calf serum, 100 units/ml penicillin, 100 μg/ml streptomycin (Gibco, Life Technologies), 2 mm GlutaMAX, 1 mm pyruvate (Life Technologies), and 100 μg/ml Normocin (InvivoGen). Both the Null2 and the hTLR4 cell lines express secreted alkaline phosphatase under the control of an NFκB promoter. In addition, the hTLR4 cell line is stably transfected with human TLR4, MD-2, and CD14 co-receptor genes, such that TLR4 activation leads to transcription from the NFκB promoter and expression of secreted alkaline phosphatase. Null2 cells were grown in the presence of 100 μg/ml Zeocin (InvivoGen), and hTLR4 cells were grown in the presence of 100 μg/ml Zeocin, 200 μg/ml Hygrogold, and 30 μg/ml Blasticidin (InvivoGen). Cells were incubated at 37 °C in humid air with 5% CO2.
HEK-Blue TLR4 Activation Assay
The HEK-Blue (InvivoGen) manufacturer's guidelines were followed to assay TLR4 activity of purified LPS or heat-killed bacterial samples. Briefly, HEK-Blue hTLR4 and Null2 were seeded in 96-well plates at 25,000 or 50,000 cells/well, 100 μl/well. All media used in this assay were pyrogen-free and consisted of complete DMEM, with the absence of the following antibiotics: Normocin, Zeocin, Hygrogold, and Blasticidin. HEK-Blue cells were incubated at 37 °C, as described, and at 24 h, 100 μl of fresh medium was added to each well, and the cells were incubated for another 24 h. HEK-Blue cells were then washed with 100 μl of medium and stimulated with 100, 10, or 1 ng/ml highly purified LOS or heat-killed B. pertussis cells (prepared as described), at a 1/10 dilution. After 24 h of incubation, the supernatants were assayed for secreted alkaline phosphatase activity by mixing 20 μl of supernatant with 180 μl of QUANTI-Blue (InvivoGen) reagent in a 96-well plate and incubating at 37 °C in the dark until the color of the mixture started turning blue. Absorbance at 650 nm was used as a readout of alkaline phosphatase activity, which indicates NFκB activation via TLR4. In each assay, the Null2 cell line and unstimulated hTLR4 cells were used as negative controls.
Statistical Analysis
All TLR4 activation assay data were analyzed using one-way analysis of variance with a Bonferroni's post test to compare groups. GraphPad Prism 5 software was used for the analyses.
RESULTS
LgmA, LgmB, and LgmC Are Required for GlcN Modification of Lipid A in B. pertussis
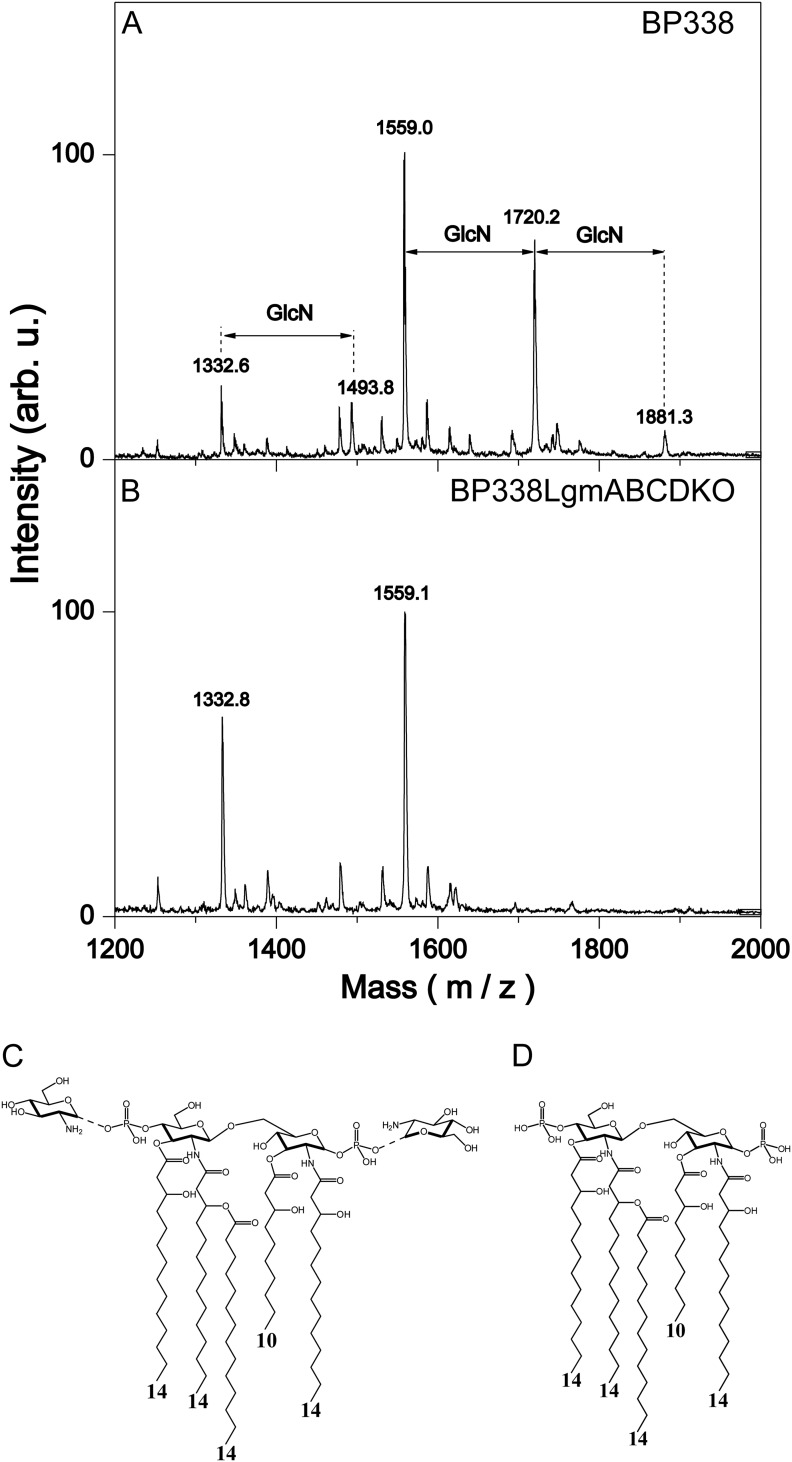
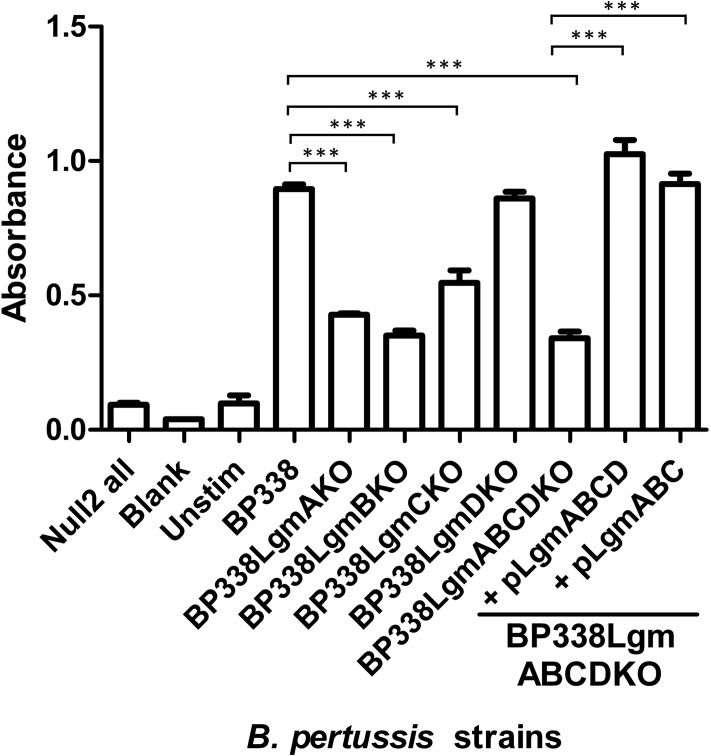
Previously, we have shown that B. pertussis strain BP338 can modify the phosphates of its lipid A with GlcN, and when locus tag BP0398 is disrupted with an insertion mutation, this GlcN modification is abrogated (7). Locus tag BP0398 is part of a four-gene cluster (locus tags BP0399–BP0396), which we propose renaming the Lgm (lipid A glucosamine modification) locus, consisting of lgmA, lgmB, lgmC, and lgmD (locus tags BP0399–BP0396, respectively). Bioinformatic analysis shows that LgmA and LgmB are homologs of ArnC and ArnT, respectively. ArnC and ArnT are glycosyl transferases in the pathway that modifies the phosphate of lipid A with aminoarabinose (l-Ara4N) in species such as Salmonella (5, 7, 23). LgmC has predicted structural similarity to the YdjC-like superfamily of proteins, members of which are deacetylases (24–26), and LgmD is predicted to be a small integral membrane protein with four transmembrane-spanning segments whose function is not known. Further analysis shows that the stop codon of lgmA overlaps with the start codon of lgmB, as do the stop and start codons of lgmC and lgmD, suggesting that all four genes function together. To determine whether the four Lgm genes are involved in GlcN modification of lipid A in B. pertussis, we generated clean deletion mutants of each of the individual Lgm locus genes and the full Lgm locus, resulting in the following mutant strains: BP338LgmAKO, BP338LgmBKO, BP338LgmCKO, BP338LgmDKO, and BP338LgmABCDKO (full Lgm locus deletion mutant). The lipid A from these strains was analyzed via negative-ion MALDI mass spectrometry (Fig. 1 and supplemental Fig. S1, A–D). The presence of the GlcN modification is represented by peaks of [M-H]− molecular ions observed at m/z 1720 (one GlcN at either 1′-phosphate or 4′-phosphate) and 1881 (GlcN at both 1′-phosphate and 4′-phosphate), i.e. 161 and 322 mass units higher than the major nonmodified penta-acyl lipid A species observed at m/z 1559 (Fig. 1). Wild type BP338 and the lgmD mutant have GlcN-modified lipid A, whereas the lgmA, lgmB, lgmC, and the full lgmABCD locus mutants lack this modification (Fig. 1, supplemental Fig. S1, A–D). This suggests that only lgmA, lgmB, and lgmC are required for GlcN modification of lipid A in B. pertussis. We confirmed that these results were not due to downstream polar effects by complementing each mutant (BP338LgmAKO, BP338LgmBKO, and BP338LgmCKO) with a vector containing the deleted gene (pBBR2LgmA, pPtacLgmAB, and pBBR2LgmC, respectively), and analyzed the lipids A of these strains via negative-ion MALDI mass spectrometry. We found, in each case, that complementation of the missing gene resulted in restoration of the lipid A GlcN modification phenotype (supplemental Fig. S1, E–G). To further confirm the dispensability of lgmD, we complemented the full Lgm locus mutant either with lgmABCD or with only lgmABC (vectors pPtacLgmABCD and pPtacLgmABC, respectively) and tested these strains for lipid A GlcN modification in a similar manner. We found that GlcN modification was restored in both strains, therefore confirming that lgmD is not required for modification of lipid A with GlcN (supplemental Fig. S1, H and I). Furthermore, we used the HEK-Blue TLR4 activation assay to determine the TLR4 activity of these strains by stimulating the HEK cells with heat-killed bacteria (Fig. 2). These results confirm our previously published data (8), showing that strains with the GlcN-modified lipid A (BP338, BP338lgmDKO, and BP338lgmABCDKO complemented with either pPtacLgmABCD or pPtacLgmABC) have higher TLR4 activity when compared with strains without the GlcN modification (BP338lgmAKO, BP338lgmBKO, BP338lgmCKO, and BP338lgmABCDKO).
FIGURE 1.
Structural analysis of lipid A with negative-ion MALDI mass spectrum analysis. Mass spectra of BP338 (A) and BP338LgmABCDKO (B), the full lgm locus deletion mutant. Peaks at m/z 1559 represent penta-acyl lipid A that lack GlcN modification, peaks at m/z 1720 represent penta-acyl lipid A with one GlcN modification at either phosphate group, and peaks at m/z 1881 represent penta-acyl lipid A with a GlcN modification at both phosphate groups. The peaks at m/z 1333 and 1494 represent tetra-acyl species. arb. u., arbitrary units. C and D, the lipid A structures present in BP338 (C) and BP338LgmABCDKO (D) as derived by mass spectral analysis. The numbers at the bottom of the structures indicate the length of the acyl chains.
FIGURE 2.
TLR4 activity of heat-killed bacterial cells as measured with the HEK-Blue NFκB TLR4 activity assay. Null2 all, stimulation of HEK-Blue Null2 cell line that lacks TLR4 expression with all LPS variants; Blank, medium only with no HEK-Blue cells; Unstim, HEK-Blue hTLR4 cells stimulated with medium only; BP338LgmABCDKO, the full Lgm locus deletion mutant; BP338LgmABCDKO + pLgmABCD, full Lgm locus deletion mutant complemented with lgmABCD; BP338LgmABCDKO + pLgmABC, full Lgm locus deletion mutant complemented with lgmABC. Graph shows the results of one representative experiment of three, n = 6 replicates per experiment. p values: < 0.001 (***). Error bars indicate S.D.
Introducing the Lgm Locus from BP338 into 18-323 Allows for the GlcN Modification of Lipid A
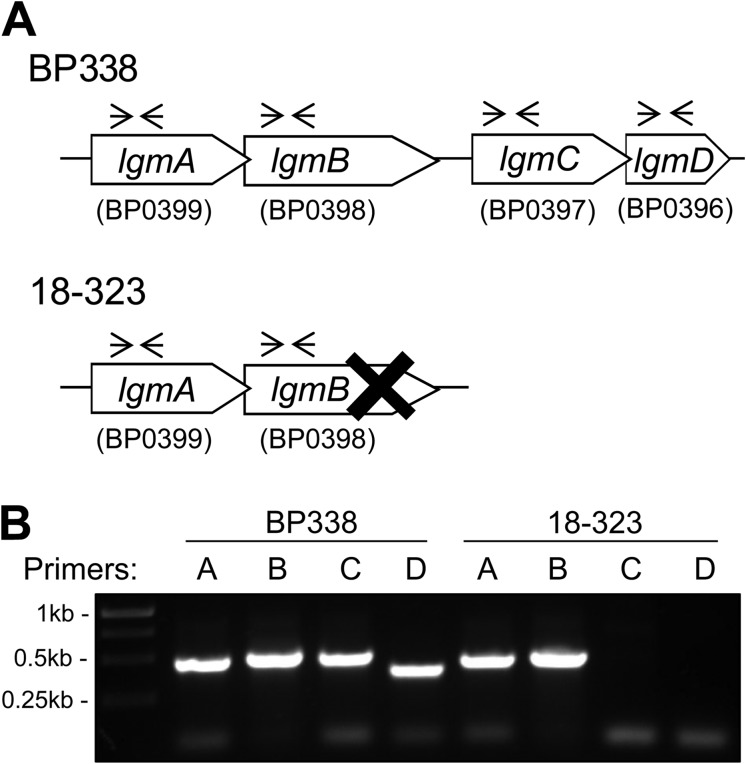
We have previously shown that BP338 can modify lipid A with GlcN, whereas the GlcN modification is absent in 18-323 lipid A (6). We analyzed and compared specific genes and loci in BP338 and 18-323 to determine the genetic reasons for this structural difference. Sequence analysis of the Tohama I (27) and 18-323 (28) genomes shows that although BP338 has a complete Lgm locus, consisting of lgmA, lgmB, lgmC, and lgmD, 18-323 only possesses a full copy of the lgmA gene. Its lgmB gene has a mutation at bp 981 where bases TT are deleted, resulting in a frameshift, and an early stop codon (Fig. 3A). No matches to lgmC or lgmD were found in the 18-323 genome sequence. The absence of lgmC and lgmD in the 18-323 genome was confirmed via PCR using primers to amplify internal fragments of each of the Lgm locus genes (Fig. 3B). In addition, the existence of the dinucleotide deletion in lgmB was verified by sequencing using primers spanning the mutation site. The absence of a complete Lgm locus in 18-323 thus explains the lack of GlcN-modified lipid A in this strain.
FIGURE 3.
Genetic analysis of the Lgm locus BP338 and 18-323 strains of B. pertussis. A, the Lgm loci of BP338 and 18-323, as determined by sequence analysis. These data were provided by the pathogen genomics group at the Wellcome Trust Sanger Institute and can be obtained from the Sanger Institute website. X represents a TT deletion mutation at bp 981 of lgmB in 18-323; the forward arrows and the reverse arrows illustrate the annealing sites of the primers used for the PCR shown in B. B, PCR of the Lgm locus genes in BP338 and 18-323 using gene-specific primers. Expected positive bands: 0.48 kb (lgmA primers), 0.51 kb (lgmB primers), 0.50 kb (lgmC primers), and 0.40 kb (lgmD primers).
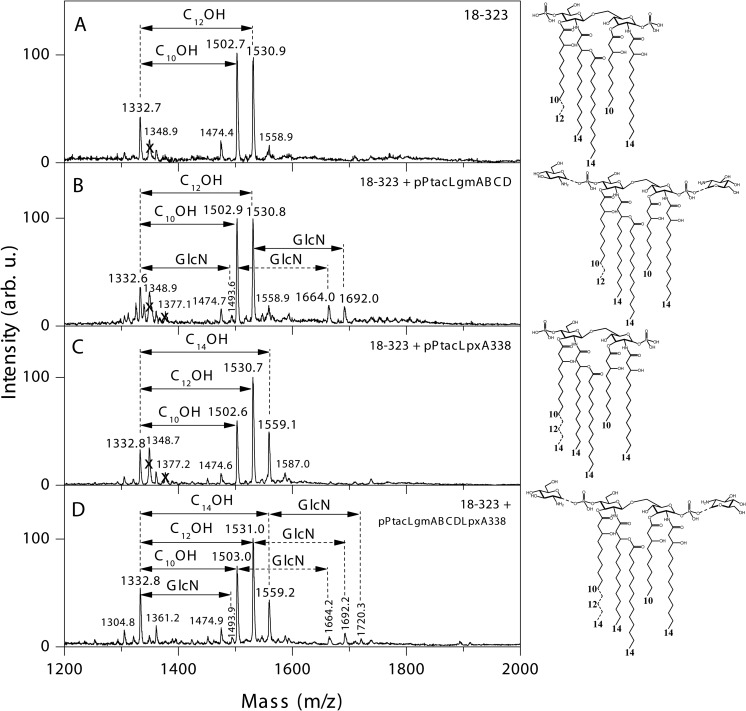
To corroborate these results, we complemented 18-323 with pPtacLgmABCD, thereby introducing the Lgm locus of BP338 into 18-323. Negative-ion MALDI-MS analysis of the lipid A from this strain shows an extra set of peaks at m/z 1664 and 1692, i.e. 161 mass units (the mass of GlcN) higher relatively to the corresponding peaks of major nonmodified penta-acylated lipid A molecular species observed in the wild type strain spectrum at m/z 1503 and 1531 (Fig. 4, A and B). These additional peaks represent the presence of 18-323 lipid A with C10OH or C12OH C3′ acyl chains with the addition of a GlcN modification on the lipid A.
FIGURE 4.
Negative-ion MALDI mass spectra and structure of lipid A from B. pertussis 18-323 strains containing BP338 lipid A-modifying genes. A–D, 18-323 (A), 18-323 + pPtacLgmABCD (complemented with the Lgm locus of BP338) (B), 18-323 + pPtacLpxA338 (complemented with lpxA of BP338) (C), and 18-323 + pPtacLgmABCDLpxA338 (complemented with the Lgm locus of BP338 and lpxA of BP338) (D). Arrows labeled with C10OH, C12OH, or C14OH indicate the 10-, 12-, or 14-carbon acyl chains absent in the tetra-acyl and present in the respective penta-acyl lipid A species. Arrows labeled with GlcN indicate the addition of GlcN at a phosphate group. The lipid A structures are summarized to the right of the mass spectra. Numbers at the bottom of the structures indicate the length of the acyl chains. Structures with GlcN modifications (B and D) have one GlcN added to either of the phosphate groups (peaks at m/z 1664, 1692, or 1720). arb. u., arbitrary units.
Introducing lpxA from BP338 into 18-323 Increases the Length of the C3′ Acyl Chain of Lipid A
BP338 has C14OH C3′ acyl chains, and 18-323 has C10OH and C12OH C3′ acyl chains. This difference in C3′ acyl chain length between BP338 and 18-323 can be explained by the single amino acid difference in LpxA (Fig. 5), which is the first enzyme of the Raetz lipid A biosynthesis pathway (11). LpxA is an essential enzyme and catalyzes the reaction that adds the C3 acyl chain onto UDP-GlcNAc, and this C3 acyl chain can then become the C3′ acyl chain, when two GlcN backbone subunits are joined by LpxB, to form the di-GlcN moiety of lipid A (11). Amino acid 173 is a serine in BP338 LpxA and a leucine in 18-323 LpxA. This amino acid is equivalent to E. coli LpxA Gly-176, which is positioned at the tip of the active site in the vicinity of the C14 carbon of the 14-carbon-long acyl chain that is added onto GlcNAc by E. coli LpxA (9)(Fig. 5). We hypothesize that the larger Leu-173 in 18-323 LpxA occludes the tip of the active site, therefore allowing only C10OH and C12OH acyl chains into the active site, whereas the smaller Ser-173 in BP338 LpxA allows C14OH acyl chains.
FIGURE 5.
Alignment of LpxA sequences from various Gram-negative species. Species (top to bottom) are: E. coli K-12 (EC), P. aeruginosa (PA), B. bronchiseptica (Bbronch), Bordetella parapertussis (Bpara), B. pertussis Tohama I BP338 (Tohama), B. pertussis 18-323 (18323), R. sphaeroides (RS), and P. gingivalis (Pging). The alignment was constructed with ClustalW (32). The down arrow indicates the single amino acid difference between LpxA of BP338 and 18-323 (amino acid 173), which corresponds to Gly-176 in E. coli LpxA. The bottom right inset shows the active site region between chains A (gray) and B (brown) of the E. coli LpxA homo-trimer and the amino acids surrounding the 14-carbon acyl chain (pink). E. coli LpxA amino acids: His-160 (green), Gly-173 (blue), Gly-176 (red), and His-191 (orange). E. coli LpxA structure PDB ID 2QIA (9) was used to generate this figure in Swiss-Pdb Viewer (ExPASy) (33).
To demonstrate that the difference at amino acid 173 of B. pertussis LpxA is the reason for the difference in C3′ acyl chain lengths between BP338 and 18-323, we introduced BP338 lpxA into 18-323 via the vector pPtacLpxA338. Analysis of the lipid A structural modifications of this strain via negative-ion MALDI-MS shows that introduction of BP338 lpxA into 18-323 generates a new major peak at m/z 1559, which corresponds to the addition of C3′ acyl chains with 14 carbons, as opposed to wild type 18-323, which only has peaks at m/z 1503 and 1531 (C10OH and C12OH C3′ acyl chains, respectively) (Fig. 4, A and C). This shows that the LpxA from BP338 (with the single amino acid difference) alone is sufficient to introduce C14OH C3′ acyl chains onto the lipid A of 18-323.
Gradual Modification of 18-323 Lipid A Structure Affects TLR4 Activity
We set out to explore the effect of these minute structural differences in lipid A (the presence of C14OH acyl chains at the C3′ position and the GlcN modification) on TLR4 activation. As such, we first generated a strain of 18-323 that was complemented with both lpxA from BP338 and the full Lgm locus (18-323 + pPtacLgmABCDLpxA338), and our mass spectral analysis shows that lipid A from this strain displays a mixture of lipid A species, including species that have longer C14OH C3′ acyl chains (peaks m/z 1559 and 1720) and species that contain the GlcN modification (peaks m/z 1664, 1692, and 1720) (Fig. 4D). We tested the TLR4 activation of these 18-323-derived strains that contain a variety of lipid A structures with the HEK-Blue TLR4 activation assay by stimulating the HEK cells with purified LOS from these strains (Fig. 6, supplemental Fig. S2). Our results show that 18-323 with BP338lpxA, which now has longer acyl chains at the C3′ position, has greater TLR4 activation activity than wild type 18-323. When 18-323 lipid A is modified with GlcN at the phosphate groups, there is also a significant increase in TLR4 activation over the wild type levels. If both these modifications are added to 18-323 to produce a longer C3′ acyl chain and the GlcN modification (18-323 + pPtacLpxA338LgmABCD), there is a significant increase in TLR4 activation over both wild type (18-323) levels and 18-323 with the longer acyl chain (18-323 + pPtacLpxA338) levels, but there is no difference when compared with TLR4 activation by 18-323 LOS that only has the GlcN modification (18-323 + pPtacLgmABCD). This shows that each modification alone is sufficient to increase TLR4 activation of B. pertussis LOS. These results also suggest that when the GlcN modification is present, increasing the C3′ acyl chain length does not increase TLR4 activation. However, because there are varying levels of these modifications in each of the 18-323 strains, the relative contribution of each modification is difficult to determine.
FIGURE 6.
TLR4 activity of purified LPS from 18-323 strains as measured with the HEK-Blue NFκB TLR4 activity assay. Null2 all, stimulation of HEK-Blue Null2 cell line that lacks TLR4 expression with all LPS variants; Blank, medium only with no HEK-Blue cells; Unstim, HEK-Blue hTLR4 cells stimulated with medium only; 18-323 + pPtacLpxA338, 18-323 complemented with lpxA from BP338; 18-323 + pPtacLgmABCD, 18-323 complemented with Lgm locus from BP338; 18-323 + pPtacLgmABCDLpxA338, 18-323 complemented with both lpxA and Lgm locus from BP338. Graph shows the results of one representative experiment of three, n = 6. p values: <0.01 (**), < 0.001 (***). Error bars indicate S.D.
DISCUSSION
Minute changes in the structure of lipid A can have great effects on how these lipid A species are able to interact with and cause dimerization of the TLR4 complex and downstream NFκB signaling (1). Coats et al. (29) suggest that in penta-acyl lipid A species, longer fatty acid chain lengths correlate with greater TLR4 activity. This hypothesis is based on the observation that penta-acyl E. coli LPS with acyl chain lengths of C12 and C14 is a weaker TLR4 agonist in comparison with Porphyromonas gingivalis penta-acyl LPS, which contains acyl chain lengths of C15, C16, and C17 (29). In the present study, we have elucidated the genetic reasons for the structural differences between B. pertussis strain BP338 and 18-323 lipid A. lgmA, lgmB, and lgmC are responsible for the GlcN modification present on phosphate groups in BP338, but absent in 18-323, and the single amino acid difference at position 173 of B. pertussis LpxA is responsible for the presence of 14 carbon acyl chains at the C3′ position in BP338 and only 10 and 12 carbon acyl chains at this position in 18-323. Furthermore, we have shown that each difference in lipid A structure between BP338 and 18-323 effects TLR4 activation in penta-acyl B. pertussis lipid A.
Previously, we had suggested that LgmA and LgmB (encoded by locus tags BP0399 and BP0398, respectively), which are homologs of glycosyl transferases ArnC and ArnT, respectively, were involved in the modification of B. pertussis lipid A with GlcN (7). Here, we show that lgmA and lgmB, along with the downstream gene lgmC (locus tag BP0397), are required for GlcN modification of lipid A, but not lgmD (locus tag BP0396), which has a start codon overlapping with the stop codon of lgmC. LgmC has a structural fold similarity to the YdjC-like superfamily of proteins (25, 26). NaxD, a member of this protein super family, was recently shown to function as a deacetylase as part of a pathway that modifies the lipid A 1-phosphate of Francisella novicida with galactosamine (24). Interestingly, the authors also showed that Bordetella bronchiseptica locus tag BB4247 (i.e. lgmC) is required for deacetylation of undecaprenyl phosphate (C55P)-GlcNAc and for the presence of GlcN-modified lipid A in total lipid extracts of B. bronchiseptica (24). Taken together, we hypothesize that LgmA functions to transfer GlcNAc to C55P, followed by deacetylation of this product by LgmC, and finally transfer of the GlcN from C55P-GlcN onto the phosphate of lipid A by LgmB. Because lgmD is closely linked with lgmC, LgmD may play a role in this pathway. If this is the case, in the absence of lgmD, as seen in the lgmD mutant, another B. pertussis protein would carry out the function of LgmD, thereby allowing the GlcN modification of lipid A.
Differences between enzymes in the Raetz lipid A biosynthesis pathway (11) can also be responsible for the variations observed in lipid A structures between different strains and species. LpxA, the first enzyme in this pathway, catalyzes the addition of an acyl chain onto the C3 carbon of GlcNAc, and when the di-GlcN backbone of lipid A is formed, this acyl chain can be in the C3 or the C3′ position (11). Williams and Raetz (9) suggest that the length of the acyl chain at the C3 and C3′ positions of lipid A is controlled by the hydrocarbon ruler region of LpxA, that is, the amino acids located near the active site in proximity to the acyl chain of the substrate, such as Gly-173 and Gly-176 in E. coli LpxA. Previous work, where mutating Gly-173 of E. coli LpxA changed the acyl chain length specificity of the enzyme, supports this theory (10). Our data also support this theory because amino acid 173 in B. pertussis LpxA is equivalent to Gly-176 in E. coli (Fig. 5), and if B. pertussis LpxA has a serine in this position (as seen in BP338), a C14OH acyl chain can be added at the C3′ position, whereas if a leucine is found in position 173 (as seen in 18-323), only C10OH and C12OH acyl chains are found. Therefore, we hypothesize that the larger leucine is obscuring the tip of the active site of LpxA, thus only allowing 10 or 12 carbon acyl chains to be added. An alignment of LpxA sequences from several Gram-negative species suggests a similar correlation between strains with shorter acyl chains at the C3 and/or C3′ positions (e.g. Pseudomonas aeruginosa and Rhodobacter sphaeroides) and the presence of a larger amino acid at positions equivalent to E. coli LpxA Gly-173, Gly-176, or both (Fig. 5). However, for P. gingivalis LpxA, this correlation, which is mostly based on sequence alignment, does not hold true and may be a reflection of the decreased sequence similarity of the P. gingivalis LpxA. Thus, although this analysis generally supports the role of both Gly-173 and Gly-176 of E. coli LpxA as a hydrocarbon ruler for the length of the acyl chain added to lipid A at the C3 and C3′ positions, there are likely more complexities involved in determining acyl chain length, especially in more distantly related LpxA species.
In this study, we have shown that slight variations in the structure of B. pertussis penta-acyl lipid A result in differences in TLR4 activation, suggesting that these structural modifications of lipid A affect how it interacts with the TLR4-MD2 complex. The solved crystal structure of dimerized TLR4-MD2 in complex with E. coli lipid A or lipid IVa (a tetra-acyl precursor of lipid A that acts as a TLR4 antagonist) can give us clues as to which amino acids may be important in this interaction (30, 31). In the case of E. coli lipid A, Park et al. (30) show that lipid IVa, which harbors only four acyl chains, sits deeper in the MD2 pocket when compared with hexa-acyl lipid A, and therefore, suggest that the phosphates of lipid IVa cannot interact with key positive amino acids that the phosphates of lipid A interact with to cause TLR4 dimerization. In the case of B. pertussis lipid A, which has only five acyl chains, we propose, in the absence of the GlcN modification on the phosphate groups and with shorter C3′ acyl chains, as seen in 18-323, that the phosphate groups of lipid A are not able to interact with the key positively charged residues in TLR4 to cause dimerization and activation. However, when the C3′ acyl chain length is increased, this may position the lipid A molecule slightly more out of the MD2 pocket and slightly closer to the positively charged residues in TLR4, thus significantly increasing TLR4 dimerization and activation. Alternatively, when the phosphates are modified with positively charged GlcN, we hypothesize the lipid A must now interact with negatively charged residues in TLR4 to cause dimerization. Because B. pertussis lipid A modified with only GlcN has greater TLR4 activation when compared with nonmodified lipid A, we propose that the penta-acyl lipid A is able to interact with negatively charged residues in TLR4 to promote dimerization. Adding both GlcN and longer acyl chain modifications to 18-323 lipid A also increases TLR4 activation when compared with both wild type 18-323 and 18-323 with only the longer acyl chain modification, but this level is statistically equal to that of 18-323 with only the GlcN modification. This suggests that the slight increase in the C3′ acyl chain length in the GlcN-modified lipid A does not change the position within the TLR4 complex enough to significantly affect the interaction between the negatively charged residues in TLR4 and the GlcNs.
The structure of the TLR4-MD-2 complex bound to E. coli hexa-acyl lipid A has shed light on the interactions between LPS and TLR4 that lead to TLR4 dimerization and activation, especially the key interactions between the negatively charged 1- and 4′-phosphate groups of LPS and positively charged amino acid residues within TLR4 (30). However, many Gram-negative species have penta-acyl lipid A, and the structure of TLR4-MD2 bound to penta-acyl lipid A has yet to be determined. A greater understanding of how different lipid A structures interact with the TLR4-MD2 complex and the effect of these structural differences on TLR4 activation will reveal new insights on how the immune system recognizes different bacteria. This may lead to the generation of tools to specifically modify the TLR4 activation potential of bacterial strains, and therefore, the ability of these strains to modulate the immune response.
Acknowledgment
We thank J. Felberg for contributions to the project.
This work was funded by an operating grant from the Canadian Institutes of Health Research (Grant MOP-102706) and a Programme International de Collaboration Scientifique (Franco-Canadien) grant from the CNRS to fund exchanges between the two countries.
We are indebted to Christian Raetz for his pioneering work in the field and dedicate this paper to his memory.

This article contains supplemental text, Table S1, and Figs. S1 and S2.
- LOS
- lipooligosaccharide
- TLR4
- Toll-like receptor 4
- hTLR4
- human TLR4
- Lgm
- lipid A glucosamine modification
- l-Ara4N
- 4-amino-4-deoxy-l-arabinose
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- C55P
- undecaprenyl phosphate.
REFERENCES
- 1. Trent M. S., Stead C. M., Tran A. X., Hankins J. V. (2006) Diversity of endotoxin and its impact on pathogenesis. J. Endotoxin Res. 12, 205–223 [DOI] [PubMed] [Google Scholar]
- 2. Caroff M., Brisson J., Martin A., Karibian D. (2000) Structure of the Bordetella pertussis 1414 endotoxin. FEBS Lett. 477, 8–14 [DOI] [PubMed] [Google Scholar]
- 3. Caroff M., Karibian D. (2003) Structure of bacterial lipopolysaccharides. Carbohydr. Res. 338, 2431–2447 [DOI] [PubMed] [Google Scholar]
- 4. Raetz C. R., Reynolds C. M., Trent M. S., Bishop R. E. (2007) Lipid A modification systems in Gram-negative bacteria. Annu. Rev. Biochem. 76, 295–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Trent M. S., Ribeiro A. A., Lin S., Cotter R. J., Raetz C. R. (2001) An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-l-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J. Biol. Chem. 276, 43122–43131 [DOI] [PubMed] [Google Scholar]
- 6. Marr N., Novikov A., Hajjar A. M., Caroff M., Fernandez R. C. (2010) Variability in the lipooligosaccharide structure and endotoxicity among Bordetella pertussis strains. J. Infect. Dis. 202, 1897–1906 [DOI] [PubMed] [Google Scholar]
- 7. Marr N., Tirsoaga A., Blanot D., Fernandez R., Caroff M. (2008) Glucosamine found as a substituent of both phosphate groups in Bordetella lipid A backbones: role of a BvgAS-activated ArnT ortholog. J. Bacteriol. 190, 4281–4290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marr N., Hajjar A. M., Shah N. R., Novikov A., Yam C. S., Caroff M., Fernandez R. C. (2010) Substitution of the Bordetella pertussis lipid A phosphate groups with glucosamine is required for robust NF-κB activation and release of proinflammatory cytokines in cells expressing human but not murine Toll-like receptor 4-MD-2-CD14. Infect. Immun. 78, 2060–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Williams A. H., Raetz C. R. (2007) Structural basis for the acyl chain selectivity and mechanism of UDP-N-acetylglucosamine acyltransferase. Proc. Natl. Acad. Sci. U.S.A. 104, 13543–13550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wyckoff T. J., Lin S., Cotter R. J., Dotson G. D., Raetz C. R. (1998) Hydrocarbon rulers in UDP-N-acetylglucosamine acyltransferases. J. Biol. Chem. 273, 32369–32372 [DOI] [PubMed] [Google Scholar]
- 11. Raetz C. R., Guan Z., Ingram B. O., Six D. A., Song F., Wang X., Zhao J. (2009) Discovery of new biosynthetic pathways: the lipid A story. J. Lipid Res. 50, (suppl.) S103–S108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sweet C. R., Preston A., Toland E., Ramirez S. M., Cotter R. J., Maskell D. J., Raetz C. R. (2002) Relaxed acyl chain specificity of Bordetella UDP-N-acetylglucosamine acyltransferases. J. Biol. Chem. 277, 18281–18290 [DOI] [PubMed] [Google Scholar]
- 13. Stainer D. W., Scholte M. J. (1970) A simple chemically defined medium for the production of phase I Bordetella pertussis. J. Gen. Microbiol. 63, 211–220 [DOI] [PubMed] [Google Scholar]
- 14. Simon R., Priefer U., Puhler A. (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat. Biotechnol. 1, 784–791 [Google Scholar]
- 15. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Second Ed., pp. 1.53–1.84 and 5.10–5.11, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 16. Weingart C. L., Broitman-Maduro G., Dean G., Newman S., Peppler M., Weiss A. A. (1999) Fluorescent labels influence phagocytosis of Bordetella pertussis by human neutrophils. Infect. Immun. 67, 4264–4267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tirsoaga A., Novikov A., Adib-Conquy M., Werts C., Fitting C., Cavaillon J. M., Caroff M. (2007) Simple method for repurification of endotoxins for biological use. Appl. Environ. Microbiol. 73, 1803–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schägger H., von Jagow G. (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379 [DOI] [PubMed] [Google Scholar]
- 19. Tsai C. M., Frasch C. E. (1982) A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119, 115–119 [DOI] [PubMed] [Google Scholar]
- 20. Therisod H., Labas V., Caroff M. (2001) Direct microextraction and analysis of rough-type lipopolysaccharides by combined thin-layer chromatography and MALDI mass spectrometry. Anal. Chem. 73, 3804–3807 [DOI] [PubMed] [Google Scholar]
- 21. El Hamidi A., Tirsoaga A., Novikov A., Hussein A., Caroff M. (2005) Microextraction of bacterial lipid A: easy and rapid method for mass spectrometric characterization. J. Lipid Res. 46, 1773–1778 [DOI] [PubMed] [Google Scholar]
- 22. Tirsoaga A., El Hamidi A., Perry M. B., Caroff M., Novikov A. (2007) A rapid, small-scale procedure for the structural characterization of lipid A applied to Citrobacter and Bordetella strains: discovery of a new structural element. J. Lipid Res. 48, 2419–2427 [DOI] [PubMed] [Google Scholar]
- 23. Breazeale S. D., Ribeiro A. A., McClerren A. L., Raetz C. R. (2005) A formyltransferase required for polymyxin resistance in Escherichia coli and the modification of lipid A with 4-amino-4-deoxy-l-arabinose: identification and function oF UDP-4-deoxy-4-formamido-l-arabinose. J. Biol. Chem. 280, 14154–14167 [DOI] [PubMed] [Google Scholar]
- 24. Llewellyn A. C., Zhao J., Song F., Parvathareddy J., Xu Q., Napier B. A., Laroui H., Merlin D., Bina J. E., Cotter P. A., Miller M. A., Raetz C. R., Weiss D. S. (2012) NaxD is a deacetylase required for lipid A modification and Francisella pathogenesis. Mol. Microbiol. 86, 611–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 26. Imagawa T., Iino H., Kanagawa M., Ebihara A., Kuramitsu S., Tsuge H. (2008) Crystal structure of the YdjC-family protein TTHB029 from Thermus thermophilus HB8: structural relationship with peptidoglycan N-acetylglucosamine deacetylase. Biochem. Biophys. Res. Commun. 367, 535–541 [DOI] [PubMed] [Google Scholar]
- 27. Parkhill J., Sebaihia M., Preston A., Murphy L. D., Thomson N., Harris D. E., Holden M. T., Churcher C. M., Bentley S. D., Mungall K. L., Cerdeño-Tárraga A. M., Temple L., James K., Harris B., Quail M. A., Achtman M., Atkin R., Baker S., Basham D., Bason N., Cherevach I., Chillingworth T., Collins M., Cronin A., Davis P., Doggett J., Feltwell T., Goble A., Hamlin N., Hauser H., Holroyd S., Jagels K., Leather S., Moule S., Norberczak H., O'Neil S., Ormond D., Price C., Rabbinowitsch E., Rutter S., Sanders M., Saunders D., Seeger K., Sharp S., Simmonds M., Skelton J., Squares R., Squares S., Stevens K., Unwin L., Whitehead S., Barrell B. G., Maskell D. J. (2003) Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis, and Bordetella bronchiseptica. Nat. Genet. 35, 32–40 [DOI] [PubMed] [Google Scholar]
- 28. Park J., Zhang Y., Buboltz A. M., Zhang X., Schuster S. C., Ahuja U., Liu M., Miller J. F., Sebaihia M., Bentley S. D., Parkhill J., Harvill E. T. (2012) Comparative genomics of the classical Bordetella subspecies: the evolution and exchange of virulence-associated diversity amongst closely related pathogens. BMC Genomics 13, 545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Coats S. R., Berezow A. B., To T. T., Jain S., Bainbridge B. W., Banani K. P., Darveau R. P. (2011) The lipid A phosphate position determines differential host Toll-like receptor 4 responses to phylogenetically related symbiotic and pathogenic bacteria. Infect. Immun. 79, 203–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Park B. S., Song D. H., Kim H. M., Choi B. S., Lee H., Lee J. O. (2009) The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 458, 1191–1195 [DOI] [PubMed] [Google Scholar]
- 31. Ohto U., Fukase K., Miyake K., Satow Y. (2007) Crystal structures of human MD-2 and its complex with antiendotoxic lipid IVa. Science 316, 1632–1634 [DOI] [PubMed] [Google Scholar]
- 32. Thompson J. D., Higgins D. G., Gibson T. J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guex N., Peitsch M. C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 34. Kovach M. E., Phillips R. W., Elzer P. H., Roop R. M., 2nd, Peterson K. M. (1994) pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16, 800–802 [PubMed] [Google Scholar]
- 35. Fürste J. P., Pansegrau W., Frank R., Blöcker H., Scholz P., Bagdasarian M., Lanka E. (1986) Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48, 119–131 [DOI] [PubMed] [Google Scholar]
- 36. Kovach M. E., Elzer P. H., Hill D. S., Robertson G. T., Farris M. A., Roop R. M., 2nd, Peterson K. M. (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176 [DOI] [PubMed] [Google Scholar]
- 37. Marr N., Shah N. R., Lee R., Kim E. J., Fernandez R. C. (2011) Bordetella pertussis autotransporter Vag8 binds human C1 esterase inhibitor and confers serum resistance. PLoS One 6, e20585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Inatsuka C. S., Xu Q., Vujkovic-Cvijin I., Wong S., Stibitz S., Miller J. F., Cotter P. A. (2010) Pertactin is required for Bordetella species to resist neutrophil-mediated clearance. Infect. Immun. 78, 2901–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Weiss A. A., Hewlett E. L., Myers G. A., Falkow S. (1983) Tn5-induced mutations affecting virulence factors of Bordetella pertussis. Infect. Immun. 42, 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Caroff M. (March 20, 2012) Patent WO 2004/062690 A1