Background: Infant gut-associated bifidobacteria possess lacto-N-biosidase, which releases lacto-N-biose I (LNB) from human milk oligosaccharides.
Results: The crystal structures of lacto-N-biosidase complexed with LNB and LNB-thiazoline were determined.
Conclusion: The intermediate analog complex allows the proposal of a conformational reaction coordinate.
Significance: The structures of a key enzyme in the colonization of human commensal bacteria provided its structural basis and insight into the development of inhibitors.
Keywords: Carbohydrate Processing, Glycobiology, Glycoside Hydrolases, Microbiology, X-ray Crystallography, Bifidobacterium bifidum, Glycoside Hydrolase Family 20, Human Milk Oligosaccharides, Lacto-N-biose I, Lacto-N-biosidase
Abstract
Human milk oligosaccharides contain a large variety of oligosaccharides, of which lacto-N-biose I (Gal-β1,3-GlcNAc; LNB) predominates as a major core structure. A unique metabolic pathway specific for LNB has recently been identified in the human commensal bifidobacteria. Several strains of infant gut-associated bifidobacteria possess lacto-N-biosidase, a membrane-anchored extracellular enzyme, that liberates LNB from the nonreducing end of human milk oligosaccharides and plays a key role in the metabolic pathway of these compounds. Lacto-N-biosidase belongs to the glycoside hydrolase family 20, and its reaction proceeds via a substrate-assisted catalytic mechanism. Several crystal structures of GH20 β-N-acetylhexosaminidases, which release monosaccharide GlcNAc from its substrate, have been determined, but to date, a structure of lacto-N-biosidase is unknown. Here, we have determined the first three-dimensional structures of lacto-N-biosidase from Bifidobacterium bifidum JCM1254 in complex with LNB and LNB-thiazoline (Gal-β1,3-GlcNAc-thiazoline) at 1.8-Å resolution. Lacto-N-biosidase consists of three domains, and the C-terminal domain has a unique β-trefoil-like fold. Compared with other β-N-acetylhexosaminidases, lacto-N-biosidase has a wide substrate-binding pocket with a −2 subsite specific for β-1,3-linked Gal, and the residues responsible for Gal recognition were identified. The bound ligands are recognized by extensive hydrogen bonds at all of their hydroxyls consistent with the enzyme's strict substrate specificity for the LNB moiety. The GlcNAc sugar ring of LNB is in a distorted conformation near 4E, whereas that of LNB-thiazoline is in a 4C1 conformation. A possible conformational pathway for the lacto-N-biosidase reaction is discussed.
Introduction
Human milk contains various oligosaccharides collectively termed human milk oligosaccharides (HMOs)7 (1, 2). HMOs function as prebiotics, promoting the growth of bifidobacteria in the gastrointestinal tracts of breastfed infants, which in turn promotes optimal health (3, 4). Most HMOs contain a lactose moiety (Gal-β1,4-Glc) at their reducing end, which is elongated by β1,3-linked lacto-N-biose I (Gal-β1,3-GlcNAc; LNB, to give type I HMOs) and/or β1,3/6-linked N-acetyllactosamine (Gal-β1,4-GlcNAc; LacNAc, to give type II HMOs). A unique feature of the composition of HMOs is the predominance of type I over type II oligosaccharides. Indeed, such a composition has not been observed in milk oligosaccharides from other mammals, including anthropoids (5). Further elongation of these core structures is made by the addition of fucose and sialic acid residues via α1,2/3/4- and α2,3/6-linkages, respectively. HMOs are composed of more than 130 different oligosaccharide structures that account for 2% of the solid components of dried human milk. Of note is that four oligosaccharides constitute 25–33% of total HMOs (6) as follows: 2′-fucosyl-lactose (Fuc-α1,2-Gal-β1,4-Glc); lacto-N-fucopentaose I (Fuc-α1,2-Gal-β1,3-GlcNAc-β1,3-Gal-β1,4-Glc; LNFP I); lacto-N-difucohexaose I (Fuc-α1,2-Gal-β1,3-(Fuc-α1,4-)GlcNAc-β1,3-Gal-β1,4-Glc; LNDFH I), and lacto-N-tetraose (Gal-β1,3-GlcNAc-β1,3-Gal-β1,4-Glc; LNT). Three of these oligosaccharides (LNFP I, LNDFH I, and LNT) contain an LNB unit, highlighting the importance of this component in these systems.
In 2005, a novel metabolic pathway specific to LNB and galacto-N-biose (Gal-β1,3-GalNAc; GNB) was uncovered in bifidobacteria (7) and was termed the GNB/LNB pathway (8). Considering the living environment of bifidobacteria (the gastrointestinal tract of infants), LNB and GNB are hypothesized to originate from HMOs and intestinal mucin glycoproteins, respectively. Proteins related to this pathway have been actively investigated, and crystallographic analyses (9) have uncovered the structures of an extracellular endo-α-N-acetylgalactosaminidase that releases GNB from mucin-type O-glycans (10), a solute-binding protein of an ABC transporter specific to GNB and LNB (11), and an intracellular phosphorylase that cleaves both GNB and LNB (12). In addition, it has been demonstrated that LNB is the “bifidus factor” that selectively promotes growth of infant gut-associated bifidobacteria (13, 14), but interestingly, a free form of LNB has not been found in HMOs (1).
Lacto-N-biosidase (LNBase, EC 3.2.1.140), which liberates LNB from the nonreducing end of oligosaccharides, was first found in the soil actinomycete Streptomyces sp. 142 (15, 16). Since then, LNBase activity has also been found in several strains of Bifidobacterium bifidum and Bifidobacterium longum (17), which are particularly predominant in the intestines of infants (18). Bifidobacterial LNBase is a membrane-anchored extracellular enzyme, which suggests that it may play a key role in the excision of LNB from HMOs to supply LNB to the GNB/LNB pathway. Recently, the gene for LNBase from B. bifidum JCM1254 (BbLNBase) was cloned, and its recombinant protein was characterized in detail. BbLNBase consists of 1,112 amino acids and contains a signal sequence, a glycoside hydrolase (GH) family 20 domain, a carbohydrate-binding module (CBM) family 32 domain, a bacterial Ig-like domain, and a transmembrane region (Fig. 1A). The enzyme's activity favored LNT as a substrate to produce LNB and lactose, and it was found that it could not act on oligosaccharides where the LNB unit is modified with fucose. In addition, it was found that BbLNBase specifically releases LNB at the nonreducing end of type I oligosaccharides but does not hydrolyze type II oligosaccharides. Therefore, a potential use for this enzyme is in identifying type I structures in glycoconjugates. Intriguingly, most of the cancer-associated oligosaccharide antigens (including sialyl Lea, sialyl Lex, and their derivatives) have a core structure containing type I or type II chains (19).
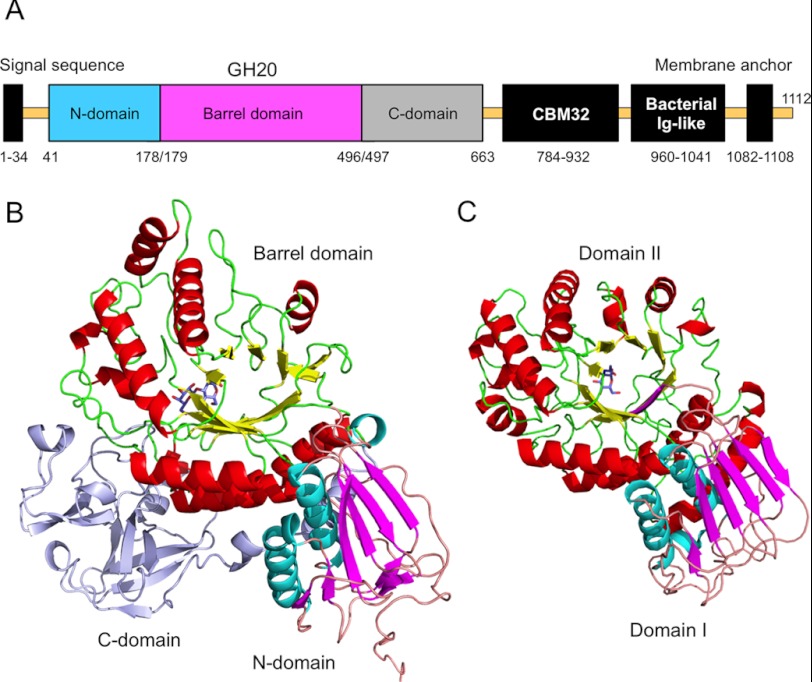
FIGURE 1.
A, domain structure of BbLNBase. B, overall structure of BbLNBase (residues 41–663) complexed with LNB. C, overall structure of the wild-type SpHex complexed with GlcNAc (Protein Data Bank code 1M01). B and C, α-helices and β-strands in the catalytic (α/β)8 barrel domain are shown in red and yellow, respectively, and α-helices and β-strands in the N-terminal domain are shown in cyan and magenta, respectively. Bound ligands are shown as blue sticks.
LNBases are members of the GH20 family of glycoside hydrolases (20) along with β-N-acetylhexosaminidases (β-HexNAcases); however, they exhibit very low sequence homology. For example, BbLNBase exhibits less than 24% amino sequence identity to all the characterized β-HexNAcases. GH20 enzymes cleave the glycosidic linkage at the reducing end of GlcNAc via a retaining substrate-assisted catalytic mechanism in which the 2-acetamido group of the substrate acts as the catalytic nucleophile (21–23). Whereas β-HexNAcases release a monosaccharide (GlcNAc), LNBase releases a disaccharide (LNB), implying that the latter has an extended −2 subsite. The crystal structures of multiple GH20 β-HexNAcases have been reported to date (21, 24–32); however, the three-dimensional structure of LNBase is not yet available.
Here, we report two crystal structures of BbLNBase complexed with LNB and LNB-thiazoline, a potent inhibitor of BbLNBase (Ki = 125 ± 8 nm at pH 4.5) (33). Using the sugar ring conformations of LNB and LNB-thiazoline molecules found in the active site, the reaction mechanism and possible conformational changes of the substrate are discussed.
EXPERIMENTAL PROCEDURES
Protein Production and Purification
The overexpression vector for N-terminally His6-tagged BbLNBase (residues 41–663) was constructed by inserting the PCR-amplified fragment of the lnbB gene (17) into the NdeI and EcoRI sites of the pET28b plasmid (kanr; Novagen, Madison, WI). The primers used were 5′-gggaattccatatggggtacagtgccacggctccc-3′ and 5′-ccggaattctcagtcgctgaccaggtcag-5′ (restriction sites are underlined). The plasmid was introduced into Escherichia coli BL21 CodonPlus (DE3)-RIL (Stratagene, La Jolla, CA) for native protein expression. For selenomethionine-labeled protein expression, an NcoI-XhoI fragment of the pET28b-based expression plasmid was inserted into the pET19b plasmid (ampr), which was subsequently introduced into E. coli BL21 CodonPlus (DE3)-RIL-X (kanr; Stratagene). The transformants were cultured in Luria-Bertani medium (native protein) or LeMaster medium (selenomethionine-labeled protein) containing 100 mg/liter kanamycin or ampicillin and 20 mg/liter chloramphenicol at 25 °C for 20 h. Isopropyl 1-thio-β-d-galactopyranoside was added to a final concentration of 1.0 mm to induce protein expression. Following an additional incubation at 25 °C for 20 h, the cells were harvested by centrifugation and suspended in 50 mm HEPES-NaOH (pH 7.5). Cell extracts were obtained by sonication followed by centrifugation to remove cell debris. The protein was purified to homogeneity by sequential column chromatography involving nickel-nitrilotriacetic acid superflow (Qiagen, Hilden, Germany), Mono Q 10/100 GL, and Superdex 200 pg 16/60 column chromatography (GE Healthcare). The protein concentration was determined using the BCA protein assay kit (Thermo Fisher Scientific) with bovine serum albumin as a standard.
Crystallography
Crystals of both native and selenomethionine-labeled BbLNBase complexed with LNB were obtained at 20 °C using the sitting drop vapor diffusion method, by mixing 0.5 μl of protein solution containing 7 mg/ml BbLNBase and 10 mm LNB with an equal volume of reservoir solution (0.2 m potassium sodium tartrate tetrahydrate, 0.1 m sodium citrate (pH 5.6), and 2.0 m ammonium sulfate). Crystals of BbLNBase complexed with LNB-thiazoline (synthesized as described previously (33)) were obtained in a similar manner, except that the concentration of LNB-thiazoline used was 0.1 mm. Diffraction data were collected at 100 K using a charge-coupled device camera on beamline BL17A at the Photon Factory of the High Energy Accelerator Research Organization (KEK, Tsukuba, Japan) and processed using HKL2000 (34). The selenomethionine sites were determined using SnB (35), and the initial phases were calculated using Solve/Resolve (36). Automated model building was performed using ARP/wARP (37). Manual model rebuilding and refinement was achieved using Coot (38) and REFMAC5 (39). The statistics for data collection and refinement are provided in Table 1. Molecular graphic images were prepared using PyMOL (DeLano Scientific, Palo Alto, CA).
TABLE 1.
Data collection and refinement statistics
| Dataset | Selenomethionine-labeled peak | LNB | LNB-thiazoline |
|---|---|---|---|
| Protein Data Bank entry | 4H04 | 4JAW | |
| Data collection statistics | |||
| Wavelength | 0.97898 Å | 1.00000 Å | 1.00000 Å |
| Space group | P21212 | P21212 | P21212 |
| Unit cell | a = 117.3 Å | a = 116.8 Å | a = 116.5 Å |
| b = 131.6 Å | b = 131.0 Å | b = 131.6 Å | |
| c = 104.6 Å | c = 104.4 Å | c = 104.8 Å | |
| Resolutiona | 50.0 to 2.20 Å (2.28 to 20 Å) | 50.0 to 1.80 Å (1.86 to 1.80 Å) | 50.0 to 1.80 Å (1.83 to 1.80 Å) |
| Total reflections | 607,378 | 1,107,867 | 1,106,951 |
| Unique reflections | 158,438 | 148,717 | 150,089 |
| Completenessa | 100% (99.9%) | 100% (100%) | 100% (100%) |
| Redundancya | 3.8% (3.8%) | 7.4% (7.4%) | 7.4% (7.3%) |
| Mean I/σ(I)a | 14.22% (3.41%) | 27.64% (4.67%) | 23.97% (4.85%) |
| Rmergea | 10.0% (31.2%) | 7.9% (28.1%) | 10.2% (48.5%) |
| Refinement statistics | |||
| Resolution range | 47.51–1.80 Å | 43.61–1.80 Å | |
| No. of reflections | 141,195 | 141,665 | |
| R-factor/Rfree | 15.5/19.1% | 18.2/21.3% | |
| r.m.s.d. from ideal values | |||
| Bond lengths | 0.028 Å | 0.027 Å | |
| Bond angles | 2.503° | 2.365° | |
| Average B-factor | |||
| Protein (chain A/B) | 16.0/15.7 Å2 | 11.6/11.3 Å2 | |
| Ligand (chain A/B) | 11.5/10.1 Å2 | 11.6/9.12 Å2 | |
| Water | 26.6 Å2 | 12.3 Å2 | |
| SO42− | 32.3 Å2 | 23.8 Å2 | |
| Ramachandran plotb | |||
| Favored (chain A/B) | 97.8/97.3% | 98.3/98.4% | |
| Allowed (chain A/B) | 1.7/2.4% | 1.7/1.4% | |
| Outlier (chain A/B) | 0.5/0.3% | 0.0/0.2% | |
a Values in parentheses are for the highest resolution shell.
b Data were calculated by RAMPAGE (71).
Enzymatic Characterization of Wild-type and Mutant LNBases
C-terminally His6-tagged proteins (residues 35 to 1064) were used for kinetic analysis. Purification and expression of wild-type enzyme were carried out as described previously (17). H263F, D320A, D320N, and Y419F mutants were constructed by using the QuikChange site-directed mutagenesis method (Stratagene) with the plasmid pET23b-lnbB as the template. The following primers and their complementary strands were used: 5′-aactccccgggcttcatgaacgtctgg-3′ (H263F), 5′-cacatgggcgccgcggagtacatgatc-3′ (D320A), 5′-cacatgggcgccaacgagtacatgatc-3′ (D320N), and 5′-acccaggccctgttctggtcccgttcg-3′ (Y419F). The entire sequence used for later manipulation was sequenced to check that no base change other than those designed had occurred. The mutants were expressed and purified using a similar procedure as described for the wild-type enzyme.
Activity measurements of the enzymes were conducted using p-nitrophenyl (pNP)-LNB (Sigma) as a substrate. The assay mixture (50 μl) contained substrate dissolved in 50 mm McIlvaine buffer (pH 4.5) and enzymes (13 nm for WT, 1.3 μm for H263F and Y419F, and 6.3 μm for D320A and D320N). The substrate concentrations were varied from 0.3- to 2-fold of the Km values of the respective enzymes. The reaction was carried out at 30 °C and stopped by adding an equal volume of 1 m Na2CO3. The amounts of liberated 4-nitrophenolate were determined by measuring the absorbance at 400 nm.
The hydrolysis of LNT (Dextra Laboratory, Reading, UK) by wild-type enzyme was monitored by high performance anion-exchange chromatography with a CarboPac PA1 column, followed by pulsed amperometric detection (Dionex ICS3000). The elution was performed by a linear gradient of 0–0.5 m sodium acetate in 125 mm NaOH at a flow rate of 1 ml/min for 30 min. The kinetic parameters were calculated by curve-fitting the experimental data using the Michaelis-Menten equation in KaleidaGraph 4.0 (Synergy Software).
RESULTS AND DISCUSSION
Structural Determination of BbLNBase
We first constructed various deletion mutants to determine a minimal region that retained activity toward the substrate pNP-LNB (supplemental Table S1 and supplemental Fig. S1). Constructs 37–663, 41–663, and 46–663 (numbers indicate the residues) exhibited virtually the same activity as the full-length construct (31–1064, which has only the signal and membrane anchor regions deleted). Thus, these three constructs were selected for crystallization screening, but only diffraction-quality crystals were obtained with the 41–663 construct in the presence of LNB. The crystal structure of BbLNBase was determined by the single-wavelength anomalous dispersion method using a selenomethionine derivative. Subsequently, we determined the crystal structure of native (nonlabeled) protein crystals (complexed with LNB and LNB-thiazoline) both at 1.8-Å resolution (Table 1 and Fig. 1B). The crystal contains two molecules in the asymmetric unit, and the final model contains residues from Ser-30 to Ser-662 of the A chain and from Ser-30 to Val-661 of the B chain. The protein has an N-terminal His6 tag containing 21 amino acid residues derived from the pET-28b vector (MGSSHHHHHHSSGLVPRGSHM), in which 11 amino acids (SSGLVPRGSHM, residues 30–40) were visible in the electron density map. A region of the artificial tag sequence (Ser-30 to Arg-36) contributes to the packing of the dimer in the asymmetric unit (supplemental Fig. S2). The molecular masses of the 41–663 construct of BbLNBase as deduced from the amino acid sequence, estimated by SDS-PAGE and calibrated gel filtration chromatography, were 71.8, 72.4, and 64.9 kDa, respectively (data not shown), suggesting that it is monomeric in solution. The root mean square deviations (r.m.s.d.) for the Cα atoms of all pairs of the four molecules (two chains in the two crystal structures) are less than 0.5 Å. The two ligand molecules bound to chains A and B are also virtually identical (r.m.s.d. = 0.047 and 0.069 Å for LNB and LNB-thiazoline, respectively). Hence, our subsequent descriptions will refer to chain A, unless otherwise noted.
Overall Structure
The BbLNBase monomer consists of three domains as follows: an N-terminal domain (N-domain, residues 41–178); a catalytic (β/α)8 barrel domain (barrel domain, residues 179–496), and a C-terminal domain (C-domain, residues 497–662) (Fig. 1B). The N-domain has an α/β topology with a seven-stranded β-sheet exposed to the surface, and two α-helices buried in the interface with the subsequent barrel domain. The N-domain and the barrel domain correspond to the two conserved domains of typical GH20 β-HexNAcases (21, 24, 25, 27–32). The N-domain and the barrel domain of BbLNBase are structurally similar to the domains I and II, respectively, of β-HexNAcase from Streptomyces plicatus (SpHex) (Fig. 1C). The N-domain is conserved in most GH20 enzymes, although its function remains unknown.
The C-domain, however, is not common to GH20 enzymes. Two examples of GH20 β-HexNAcases that have a distinct domain in the C-terminal region of the catalytic barrel domain are the β-HexNAcases from Serratia marcescens (chitobiase) and Streptococcus gordonii (GcnA), which have a small (67 amino acids) immunoglobulin-like β-sandwich domain (domain IV) and a large (227 amino acids) α-helical domain (domain III), respectively (24, 28). In the case of the C-domain of BbLNBase, there is no resemblance to these GH20 C-domain structures. The C-domain of BbLNBase is located on the side of the barrel domain that is farthest from the active site. A deletion of only five residues at the C terminus of this domain (construct 37–658) significantly reduced the catalytic activity, and further deletions caused complete inactivation (supplemental Table S1). These results indicate that the C-domain of BbLNBase is essential for protein stability and catalytic activity. Of interest is that the C-domain possesses a broken β-trefoil fold. According to a structural similarity search using the DALI server (40), the C-domain is slightly similar to an R-type lectin (MOA) from Marasmius oreades (CBM13) (Z-score = 11.0, r.m.s.d. = 2.0 Å for 102 Cα atoms), which also has a typical β-trefoil fold (41). The β-trefoil fold of MOA is composed of three subdomains that are similar in structure and are assembled at a 3-fold axis (supplemental Fig. S3C), and each subdomain contains four-stranded β-hairpin turns (β1–β4) (supplemental Fig. S3E). The C-domain of BbLNBase can be divided into an α-subdomain (residues 522–550), β-subdomain (residues 551–615), and γ-subdomain (residues 616–662) (supplemental Fig. S3, A and B), with each subdomain having at least one additional α-helix. The β-subdomain retains the typical topology of the β-trefoil fold with a complete set of four β-strands (supplemental Fig. S3D). However, the α- and γ-subdomains lack one or two β-strand(s), and the C-domain is largely broken at the interface between these subdomains. In the β-subdomain, a disulfide bond is formed between Cys-564 (within β2) and Cys-589 (loop after β3). Interestingly, these structural features (a disulfide bond in the β-subdomain and an additional α-helix after β3) are also present in a CBM13-related arabinose-binding domain (CBM42) of the fungal GH54 α-l-arabinofuranosidase (42). The CBM42 domain also partially lacks the 3-fold symmetry, and its α-subdomain lacks the arabinose-binding site. However, the C-domain of BbLNBase appears to be more broken than CBM42.
Active Site of BbLNBase
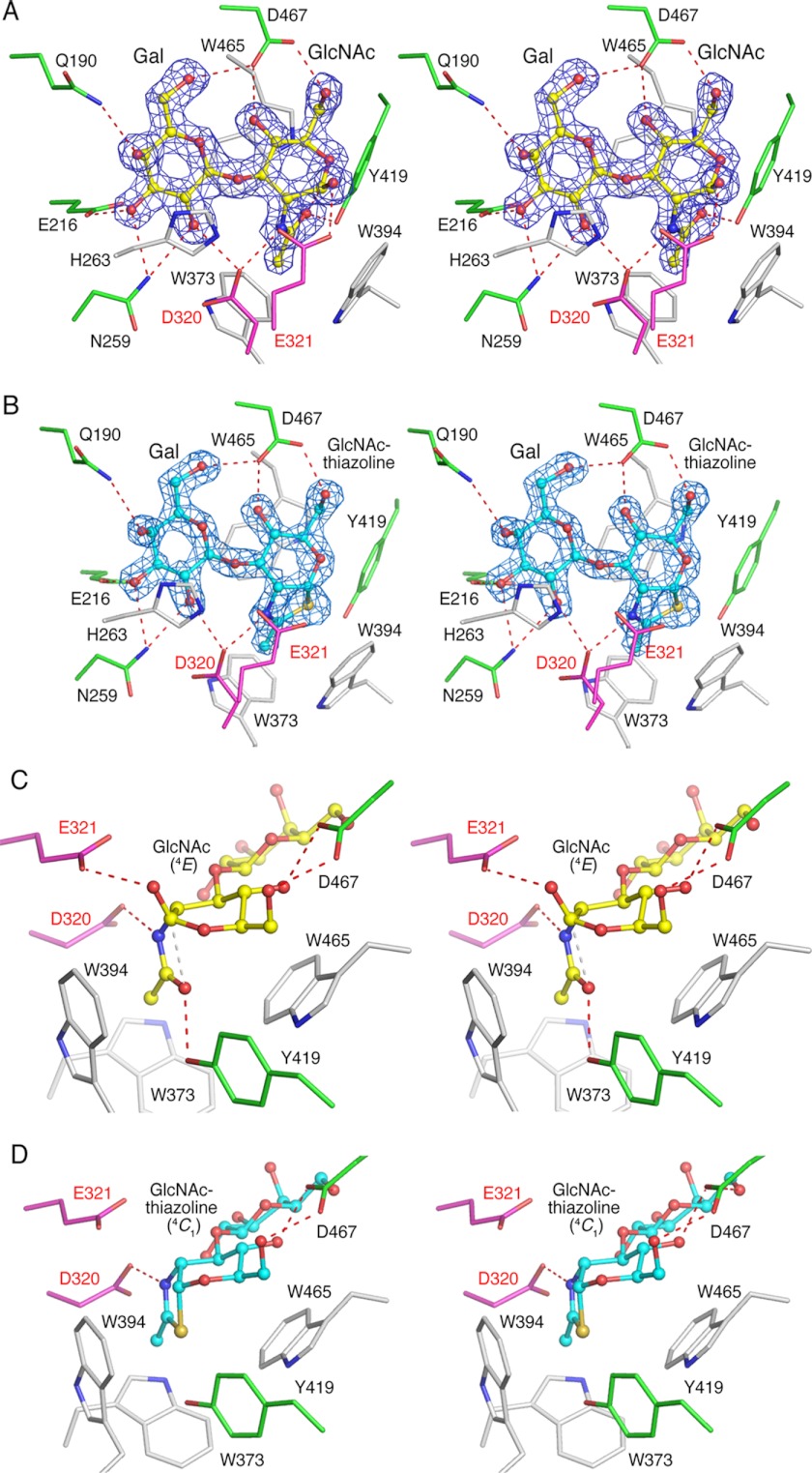
We observed a clear electron density for ligands at the center of the barrel domain (Fig. 2, A and B). The pyranose ring of the GlcNAc residue, in LNB, is in the 4E conformation and that of GlcNAc-thiazoline, in LNB-thiazoline, is in the 4C1 conformation (Fig. 2, C and D, discussed below). For LNB, GlcNAc and Gal are bound to the −1 and −2 subsites, respectively, with all the hydroxyl groups of LNB forming hydrogen bonds with the surrounding amino acids. Such extensive recognitions give evidence for the strict substrate specificity observed for this enzyme (17) as demonstrated for pNP-β-GNB, the 4-epimer of pNP-β-LNB, that shows highly reduced activity. In the BbLNBase structure, the C4-hydroxyl group of GlcNAc of LNB forms a hydrogen bond with the side chain of Asp-467 (Fig. 2), and this amino acid would block GalNAc-containing substrates from binding such as GNB. In accordance with other biochemical studies, BbLNBase does not hydrolyze fucosylated substrates such as pyridylamino (PA)-LNFP I and PA-LNFP II (17), which can be rationalized as the active site of BbLNBase, and it has no space for the fucosyl group at the C2-hydroxyl of Gal (blocked by Asn-259 and Asp-320).
FIGURE 2.
Stereoviews of LNB (A and C, yellow) and LNB-thiazoline (B and D, cyan) bound to the active site of BbLNBase. Catalytic residues, residues forming hydrogen bonds, and residues forming hydrophobic interactions are shown in magenta, green, and white, respectively. Hydrogen bonds are shown as red dashed lines. A and B, |Fo| − |Fc| omit electron density map (contoured at 4σ) and interactions with protein atoms. C and D, GlcNAc and GlcNAc-thiazoline sugar ring and surrounding residues.
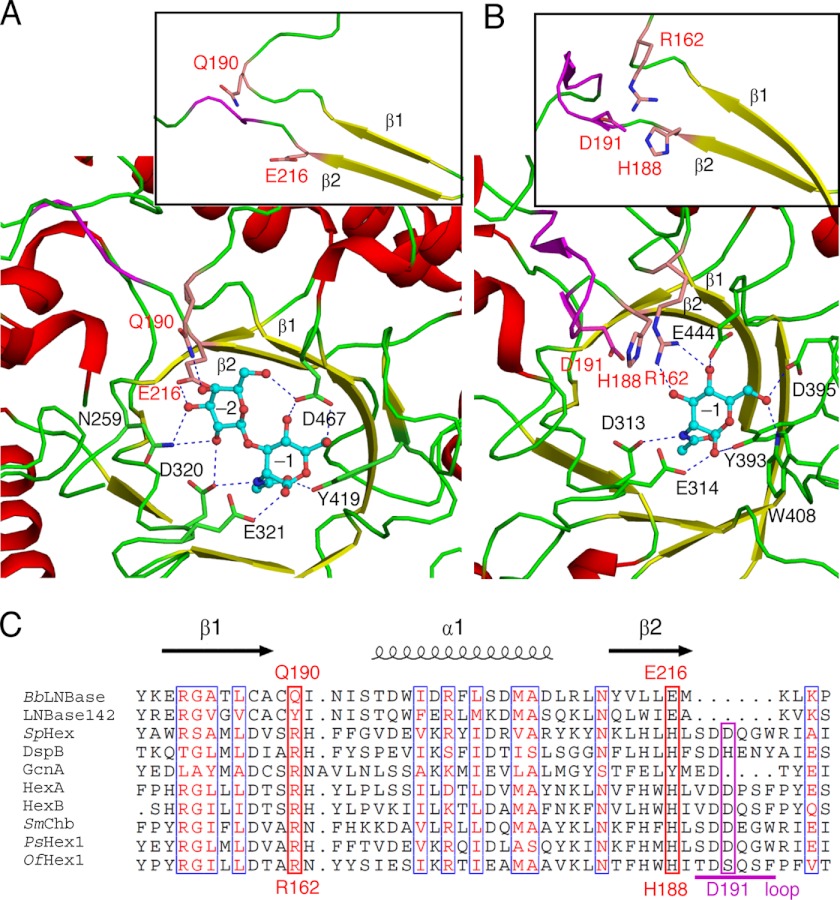
The substrate-binding site of BbLNBase was compared with that of SpHex, which is one of the most well studied GH20 β-HexNAcases (Fig. 3) (21–23). Amino acids surrounding the GlcNAc at −1 subsite (Asp-320, Glu-321, Tyr-419, and Asp-467 in BbLNBase) are highly conserved with SpHex and among GH20 enzymes (supplemental Fig. S4). The two catalytic residues of GH20, Asp-320 (polarizing residue) and Glu-321 (acid/base catalytic residue), form hydrogen bonds with the amide nitrogen of the 2-acetamido group and the O1-hydroxyl, respectively. Tyr-419 is a highly conserved residue in GH20 enzymes, and its side-chain hydroxyl group forms a hydrogen bond with the carbonyl oxygen atom of the 2-acetamido group. Asp-467 forms bifurcated hydrogen bonds with the O4- and O6-hydroxyl groups of the GlcNAc residue. The corresponding residue is often substituted with Glu in β-HexNAcases (Glu-444 in SpHex), which forms a hydrogen bond with the O4-hydroxyl group alone. The hydrogen bond with Asp-467 fixes the O6-hydroxyl group in a gauche-gauche orientation in BbLNBase (Fig. 3A), whereas the O6-hydroxyl group of GlcNAc in SpHex is in the gauche-trans orientation due to hydrogen bonds with Asp-395 and Trp-408 (Fig. 3B). In addition, His-263, Trp-373, Trp-394, and Trp-465 form hydrophobic interactions with the substrate in the active site (Fig. 2) and are highly conserved in GH20 (supplemental Fig. S4).
FIGURE 3.
Comparison of the active sites of GH20 enzymes. A and B, active sites of BbLNBase (A) and SpHex (B), respectively. Inset, loop regions following β1 and β2 that determine the presence or absence of −2 subsite. C, amino acid sequence alignment of the loop regions of GH20 enzymes (two LNBases and eight β-HexNAcases). Residues important for the presence or absence of −2 subsites are labeled with red letters. Six-amino acid loop insertion of β-HexNAcases is indicated by a magenta bar. LNBase142, Streptomyces sp. 142 LNBase; DspB, Actinobacillus actinomycetemcomitans dispersin B; GcnA, S. gordonii GcnA; HexA, human lysosomal β-HexNAcase A; HexB, human lysosomal β-HexNAcase B; SmChb, S. marcescens chitobiase; PsHex1, Paenibacillus sp. TS12 Hex1; OfHex1, O. furnacalis β-HexNAcase.
To investigate the importance of these residues in the −1 subsite, we constructed site-directed mutants D320A, D320N, Y419F, and H263F (Table 2). Mutations at the catalytically important Asp-320 residue (D320A and D320N) did not affect the Km values, but they exhibited significantly reduced kcat values, respectively, which is consistent with those of the corresponding mutants of SpHex (D313A and D313N) (22), human HexB (D354N) (43), and Paenibacillus sp. TS12 Hex1 (D321N) (29). In the case of Y419F, the Km and kcat values were both significantly reduced compared with the wild-type enzyme. The mutation at His-263 also showed a reduced kcat value, which is consistent with this residue being critical in the hydrophobic interaction with GlcNAc.
TABLE 2.
Kinetic parameters of wild-type and mutants of BbLNBase
Activities of pNP-LNB hydrolysis were measured in 50 mm McIlvaine buffer (pH 4.5) at 30 °C.
| Enzyme | Km | kcat | kcat/Km |
|---|---|---|---|
| ×1−6 m | s−1 | ×103 s−1/m | |
| Wild type | 99 ± 11 | 15 ± 1 | 160 ± 10 |
| H263F | 120 ± 20 | 0.15 ± 0.01 | 1.3 ± 0.1 |
| D320A | 130 ± 20 | 0.0068 ± 0.0005 | 0.052 ± 0.006 |
| D320N | 380 ± 50 | 0.011 ± 0.001 | 0.028 ± 0.002 |
| Y419F | –a | 0.086b | – |
a The Km value was too low to be determined.
b The v/[E]0 value at 100 μm substrate.
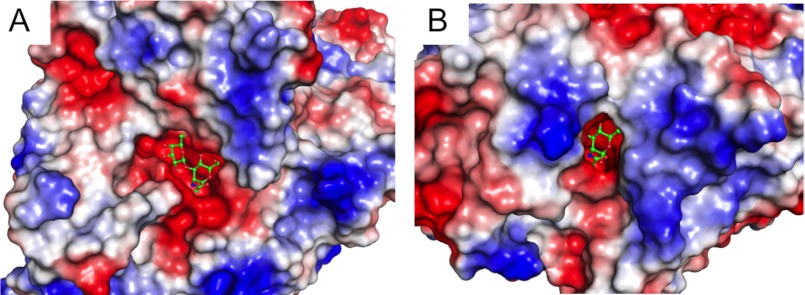
As expected from the disaccharide-releasing characteristics of BbLNBase, a −2 subsite specific for β-1,3-linked Gal is clearly defined in the crystal structure (Fig. 3A). The side chains of Gln-190 and Glu-216 form hydrogen bonds with the O4- and O3-hydroxyl groups of Gal, respectively. Gln-190 and Glu-216 in BbLNBase are replaced by Arg-162 and His-188, respectively, in SpHex (Fig. 3C) and jointly block the space for a potential −2 subsite in this enzyme (Fig. 3B). Furthermore, in most β-HexNAcases, a 6-amino acid loop insertion is present just after the His-188 residue (Fig. 3C), and a relatively conserved Asp residue (Asp-191 in SpHex) located in the loop occupies a position corresponding to the −2 subsite (Gal binding site) of BbLNBase (Fig. 3B). Comparison of the molecular surfaces of BbLNBase and SpHex (Fig. 4) illustrates pockets that are suitable in size for binding disaccharide and monosaccharide units, respectively. However, we were unable to determine the positive subsites in the BbLNBase structure. Attempts at co-crystallization and soaking experiments with lactose, Gal, or Glc were undertaken, but no electron densities for these sugars were found (data not shown). BbLNBase has been previously shown to efficiently hydrolyze PA-LNT as well as pNP-β-LNB (17). However, the enzymatic activity against LNT, the natural form of a major HMO component, has not been fully studied. Thus, we measured the kinetic parameters for hydrolysis of LNT by BbLNBase. The Km, kcat, and kcat/Km values were determined to be 626 ± 23 μm, 42.1 ± 0.8 s−1, and 67.2 ± 1.9 s−1 mm−1, respectively. The Km and kcat values were higher compared with those for pNP-LNB (Table 2), demonstrating that this enzyme is an exo-acting enzyme and does not have positive subsites specific for sugar moieties. In addition, the entrance of the substrate-binding pocket of BbLNBase appears to be wider than that of SpHex (Fig. 4). This is in agreement with the finding that LNBase from Streptomyces sp. 142 releases LNB from the nonreducing end of various oligosaccharides, including a large triantennary sugar chain (15, 16).
FIGURE 4.
Molecular surfaces of BbLNBase (A) and SpHex (B) showing the substrate binding pockets. According to the electrostatic potential, the surface is colored blue for positive and red for negative charges. LNB and GlcNAc bound to the pocket are shown as green sticks.
Reaction Mechanism and Conformational Changes of the Substrate
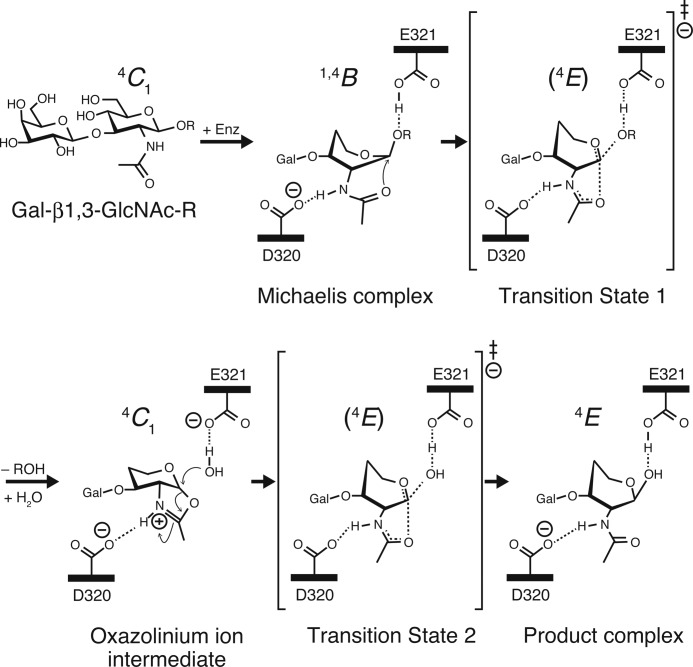
In the widely accepted mechanism of GH20 (Fig. 5), which is one of substrate-assisted catalysis, the 2-acetamido group of a substrate is polarized and oriented by a deprotonated Asp residue that is located at the neighboring position of the acid/base Glu residue (44). The oxygen atom of the carbonyl group acts as the nucleophile, which attacks the anomeric carbon, resulting in the formation of an oxazoline or oxazolinium ion intermediate. The protonated acid/base Glu residue facilitates bond cleavage by providing a proton to the glycosidic oxygen. In the second step of the reaction, the acid/base Glu activates a water molecule, thereby facilitating its nucleophilic attack at the anomeric carbon. During the course of this reaction, two oxocarbenium ion-like transition states are thought to exist one on each side of the oxazoline or oxazolinium ion intermediate.
FIGURE 5.
Proposed reaction pathway of BbLNBase and possible conformations of the GlcNAc sugar ring. R = a leaving group.
When investigating the catalytic mechanism of carbohydrate-active enzymes, the dynamic behavior of a substrate sugar molecule during the catalytic route in the active site is of particular interest. In this work, we observed distorted GlcNAc sugars of LNB in the −1 subsite of the BbLNBase structure (Fig. 2C), whereas the GlcNAc-thiazoline group of the LNB-thiazoline complex is in a 4C1 conformation (Fig. 2D). Therefore, to gain insight into the conformations of the sugar at binding, we analyzed the sugar ring conformations in detail using the Cremer-Pople system, which is widely used to define the puckering conformations of a pyranose (45). The Cremer-Pople parameters of GlcNAc of LNB in chains A and B were φ = 245.9°, θ = 60.1°, and Q = 0.595, and φ = 252.6°, θ = 56.7°, and Q = 0.616, respectively, indicating that they are in a conformation close to 4E (ideally, φ = 240° and θ = 54.7°). On the other hand, the parameters of GlcNAc-thiazoline of LNB-thiazoline in chains A and B were φ = 237.6°, θ = 13.2°, and Q = 0.547, and φ = 265.4°, θ = 17.9°, and Q = 0.597, respectively, indicating that they are in a conformation close to 4C1.
The 2-acetamido group of GlcNAc in LNB is nestled in among three aromatic residues (Trp-373, Trp-394, and Trp-465) and fixed by two hydrogen bonds with Asp-320 and Tyr-419 (Fig. 2C). The 2-acetamido group at its fixed position elevates the C1 atom of GlcNAc, and the hydrogen bond between Asp-467 and O4-hydroxyl fixes the C4 atom of GlcNAc in an elevated position. These interactions potentially distort the conformation of GlcNAc to 4E, which is not intrinsically stable as evidenced by crystallographic analysis and molecular simulation (46, 47).
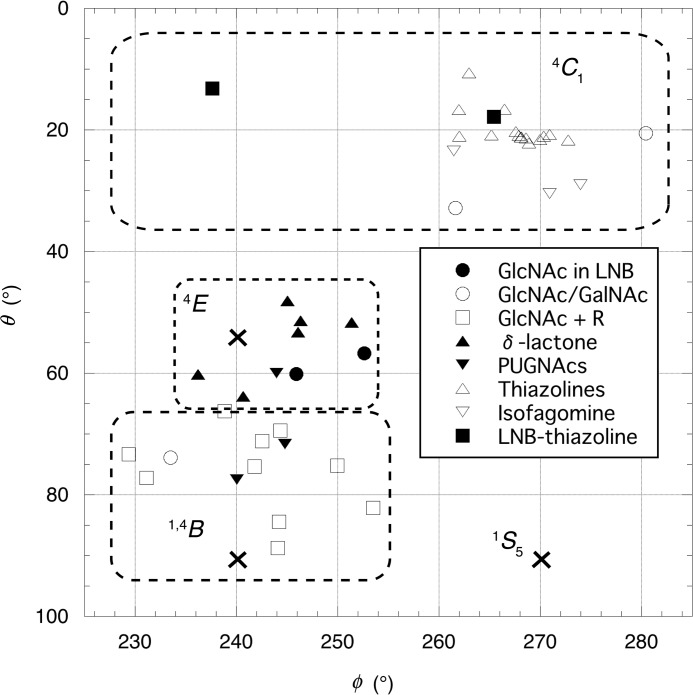
Currently, various substrates (e.g. chitobiose), inhibitors (e.g. GlcNAc-thiazoline), and reaction products (e.g. GlcNAc) of β-HexNAcases have been observed in the crystal structures of other GH20 enzymes (supplemental Table S2) (21, 22, 24, 25, 27–31, 48–52). In an effort to gain insight into the global binding of these ligands against GH20 enzymes, their Cremer-Pople parameters (φ and θ) were evaluated and plotted (Fig. 6) together with those of the GlcNAc sugars of LNB and LNB-thiazoline in BbLNBase.
FIGURE 6.
Distribution map of sugar ring conformations observed in the −1 subsite of GH20 enzymes as analyzed by Cremer-Pople angle parameters. Cross symbols indicate ideal positions of 1,4B (φ = 240°, θ = 90°), 4E (φ = 240°, θ = 54.7°), and 1S5 (φ = 270°, θ = 90°) conformations. Open squares (GlcNAc + R) include chitobiose, GlcNAc-β1,2-Man, GMMG, NGAB2, and TMG-chitotriomycin. Closed downward triangles (PUGNAcs) include PUGNAc and Gal-PUGNAc. Open upward triangles (thiazolines) include GlcNAc-thiazoline and GalNAc-thiazoline. Open downward triangles (isofagomine) indicate GalNAc-isofagomine. Closed squares indicate LNB-thiazoline. Details of the sugar compound names and their parameters are listed in supplemental Table S2.
GlcNAc-thiazoline and GalNAc-thiazoline are specific inhibitors of GHs that utilize the substrate-assisted mechanism (53, 54) because they structurally resemble the oxazoline or oxazolinium ion intermediate. A recent study using ab initio molecular dynamic simulations predicts that the oxazoline or oxazolinium ion intermediate is distorted to a conformational region of 4H3/4E/4H5 (44). However, in GH20 β-HexNAcases, the thiazoline inhibitors always adopt conformations close to 4C1 (θ < 23°) (21, 25, 27, 52). Our crystallographic result also indicates that the LNB-thiazoline in LNBase also adopt a 4C1 conformation (Fig. 6). These ample crystallographic data indicate that the oxazolinium ion intermediate adopts the 4C1 conformation (21).
Various substrates that carry sugar moieties at positive subsites have also been observed in crystal structures of GH20 β-HexNAcases. Complexes of a chitobiase (β-HexNAcase) from S. marcescens with chitobiose (di-N-acetyl-d-glucosamine) were trapped using the mutants D537A and E540A (catalytic residues) (48) or by a simple soaking method (24). The complex structures of the N- and C-terminal modules (GH20A and GH20B) of the large multimodular β-HexNAcase StrH from Streptococcus pneumoniae with disaccharide (in GH20A), tetrasaccharide (in GH20B), and bisected glycan heptasaccharide (in GH20B) substrates were reported using their acid/base residue mutants (E361Q of GH20A and E805Q of GH20B) (31). The GlcNAc moieties of these substrates bound to the −1 subsite were all in conformations close to a 1,4B conformation (229° < φ < 254° and θ > 66°; ideally φ = 240° and θ = 90°). Moreover, the complex structure of TMG-chitotriomycin, a linear tetrasaccharide inhibitor (55), bound to the β-HexNAcase from Ostrinia furnacalis was reported (30) with the N,N,N-trimethyl-d-glucosamine group at the nonreducing end of TMG-chitotriomycin also adopting a 1,4B-like conformation (φ = 244.2° and θ = 84.5°). Therefore, the substrate of the Michaelis complex for GH20 enzymes is thought to adopt a 1,4B conformation.
Transition state analogs that have an sp2-hybridized planar anomeric carbon were also used to study β-HexNAcases and related enzymes as these compounds are found to be potent inhibitors of these enzymes. In the complex structure of human lysosomal β-HexNAcase B, 2-acetamido-2-deoxy-d-glucono-1,5-lactone (δ-lactone) was bound in conformations close to 4E (236° < φ < 253° and 48° < θ < 64°) (50). O-(2-Acetamido-2-deoxy-d-glucopyranosylidene)-amino-N-phenylcarbamate (PUGNAc) is a potent inhibitor of a wide range of β-HexNAcases that utilize the classical double displacement mechanism (e.g. GH3) as well as the substrate-assisted mechanism used by GH20 enzymes. In β-HexNAcases from O. furnacalis and Paenibacillus sp. TS12 (Hex1), PUGNAc adopts conformations ranging from 4E to 1,4B (240° < φ < 245° and 60° < θ < 78°) (29, 51). Our results strongly support the hypothesis that the conformation at the transition states of GH20 enzymes is nearly 4E, which is in compliance with the requirement of coplanarity at the C2, C1, O5, and C5 pyranose ring atoms to attain the oxocarbenium ion-like transition state. 4E-Like transition states are also predicted for other GH enzymes that utilize the substrate-assisted mechanism, namely a GH18 chitinase (by a QM/MM modeling study) (56), and a GH84 O-GlcNAcase (57).
According to previous knowledge regarding GH20 β-HexNAcases and the present crystal structures, we propose a conformational itinerary pathway for the BbLNBase-catalyzed reaction that obeys the “principle of least nuclear motion” (21, 58): 1,4B (Michaelis complex)-4E (transition state 1)-4C1 (oxazolinium ion intermediate)-4E (transition state 2)-4E (product complex) (Fig. 5). Of interest is the 4E product complex observed in this study. In GH20 β-HexNAcases, several complexes observed with reaction products (GlcNAc or GalNAc) have been reported. However, the ring conformations and binding modes of these molecules differ depending on the conditions of complex formation. In the structures of β-HexNAcase (Hex1) from Paenibacillus sp. TS12, GlcNAc is in a 4C1 conformation, whereas GalNAc is in a 1,4B conformation (29). In SpHex, GlcNAc is bound in a 4C1 conformation with the wild-type enzyme, but it adopts the alternative conformations of 1,4B and 4C1 in the D313A mutant (22). In the D313N mutant, the bound GlcNAc is in a 4C1 conformation but is tilted and dropped from its normal position in the wild-type complex. In BbLNBase, we speculate that there are two major factors that fix LNB with its GlcNAc moiety in a 4E conformation. First, the Gal moiety bound to −2 subsite acts as an anchor to lock in place the LNB disaccharide in the active site. This anchor is lacking in other GH20 β-HexNAcases as they only have confined space in the active site to accommodate a GlcNAc/GalNAc residue. Second, the bifurcated hydrogen bond between Asp-467 and the O4/O6-hydroxyls of the GlcNAc moiety strongly fix it in the 4E and gauche-gauche conformations. These factors are unique to LNBases and have to date not been shown in β-HexNAcases.
C-terminal Domains
In the crystal structure of BbLNBase, we observed a novel β-trefoil-like domain (C-domain) that is required for protein stability. One of our most striking findings was that the C-domain has several features similar to those of the arabinose-binding domain CBM42. However, there is currently no evidence indicating that the C-domain in BbLNBase has carbohydrate-binding ability. We attempted co-crystallization and soaking experiments with various sugars such as lactose, Gal, or Glc at high concentrations (up to 1 m), but we could not observe any extra electron density in this domain (data not shown). In a study examining β-HexNAcases from Paenibacillus sp. TS12, the C-terminal region of one of the two β-HexNAcases (residues 503–978 of Hex1) was suggested to help the enzyme in its interaction with glycosphingolipid substrates in the absence of detergent (29). However, the three-dimensional structure of this domain remains unknown because only a truncated structure of Hex1 is available (deletion mutant 1–502), and there is no amino acid sequence homology between the C-terminal region of Hex1 and the C-domain of BbLNBase. The full-length BbLNBase has a CBM32 domain (residues 784–932) in a region adjacent to the C-domain (Fig. 1A). CBM32 domains of GHs from enteric bacteria have been shown to recognize a terminal Gal or GalNAc, or disaccharide motifs such as LacNAc and GlcNAc-α1,4-Gal (59). Therefore, the CBM32 domain of BbLNBase may also function in binding β-1,3-linked Gal, LNB, or other moieties of HMOs.
Biological Implications
In the type I HMO-degradation system of B. bifidum JCM1254, BbLNBase plays a central role in releasing LNB from HMOs (9, 60). However, the assistance of other enzymes that remove modifying sugars such as fucose or sialic acid is required. Many extracellular glycosidases involved in HMO degradation have been found in B. bifidum JCM1254. GH95 1,2-α-l-fucosidase (AfcA) (61, 62), GH29 1,3–1,4-α-l-fucosidase (AfcB) (63, 64), and GH33 exo-α-sialidases (SiaBb1 and SiaBb2) (65) all share roles in the complete degradation of α1,2- and α1,3/4-fucosylated and sialylated HMOs. In this study, we provide a structural basis for the substrate specificity of BbLNBase. This enzyme cannot accommodate any modified LNB moieties because of a substrate binding pocket that is constrained to allow only LNB binding. This feature is suitable for the specificity of the solute-binding protein of the GNB/LNB transporter (GL-BP) in the GNB/LNB pathway, which specifically binds unmodified LNB disaccharide (11). GL-BP binds LNB and GNB with low Kd values (< 0.09 μm), whereas LNT, a major HMO tetrasaccharide containing LNB, exhibits a significantly higher Kd value (11 μm). Following uptake, LNB is subsequently metabolized by GNB/LNB phosphorylase and other enzymes, including N-acetylhexosamine kinase, UDP-glucose hexose-1-phosphate uridylyltransferase, and UDP-glucose 4-epimerase (8). The positive subsites of BbLNBase are wider and appear to be capable of accommodating various groups, suggesting that this enzyme acts on various type I HMOs after the action of fucosidases and sialidases. Moreover, B. bifidum JCM1254 has three extracellular enzymes, including one GH2 β-galactosidase (BbgIII) and two GH20 β-HexNAcases (BbhI and BbhII) (66). Two of these enzymes (BbgIII and BbhI) are suggested to play essential roles in degrading type II HMOs because they catalyze the complete hydrolysis of lacto-N-neotetraose (Gal-β1,4-GlcNAc-β1,3-Gal-β1,4-Glc) to monosaccharides. BbgIII hydrolyzes β-1,4- and β-1,6-linked Gal but not β-1,3-linked Gal in LNB and LNT, suggesting that it is also involved in the complete degradation of type I HMOs after the action of LNBase (e.g. cleavage of lactose released from LNT).
In contrast to B. bifidum JCM1254, B. longum subsp. infantis ATCC15697 lacks the LNBase gene. A genomic analysis of B. longum subsp. infantis identified a unique gene cluster, the HMO cluster, containing various intracellular GHs that lack signal sequences (67, 68). A GH29 fucosidase, a GH95 fucosidase, a GH33 sialidase, a GH20 β-HexNAcase, a GH2 β-galactosidase, and at least four putative sugar transporters were found in the HMO cluster. The GH2 β-galactosidase is specific for lactose and type II HMOs (69). Furthermore, the intracellular GH42 β-galactosidase Bga42A, which is distant from the HMO cluster, is highly specific for LNT and functions as the sole β-galactosidase acting on type I HMOs. In the HMO degradation system of this strain, it was suggested that low molecular weight HMOs (degree of polymerization ranging from 3 to 8) are directly imported into the cells by transporters and subsequently cleaved by intracellular GHs (70). This analysis as well as recent research on commensal bacteria is unveiling the different HMO consumption modes of bifidobacterial strains. Extracellular bifidobacterial LNBases may also aid in the HMO consumption of other bifidobacterial strains that lack endogenous LNBases. Importantly, in this study, we identified residues that are important for β-1,3-linked Gal recognition at the −2 subsite (viz. Gln-190 and Glu-216). Conservation of these residues is a critical marker that helps to predict putative LNBase genes. A BLAST search against protein databases indicates that putative LNBase genes are present in many bacteria, mainly in the Actinobacteridae subclass. The presence (or absence) of LNBase in the genome of microbes in an infant's gastrointestinal tract will provide important information that will advance our understanding of how these microbes metabolize HMOs.
Acknowledgments
We thank Drs. M. Nishimoto and M. Kitaoka (National Food and Research Institute, National Agriculture and Food Research Organization, Japan) for providing LNB and for helpful discussion. We thank the staff of the Photon Factory and SPring-8 for the x-ray data collection. M. H. and K. A. S. thank the Centre for Microscopy, Characterization, and Analysis at the University of Western Australia, which is supported by University, State, and Federal Government funding.
This work was supported in part by the Program for the Promotion of Basic Research Activities for Innovative Bioscience, Japan (to S. F., K. Y., and T. K.), by Japan Society for the Promotion of Science KAKENHI Grant 24380053 (to S. F. and H. A.), and by the Australian Research Council. Part of this work was performed under the Priority Program for Disaster-affected Quantum Beam Facilities Proposals PF 2011G080 and SPring-8 2011A1908.

This article contains supplemental Tables S1 and S2, Figs. S1–S4, and additional references.
The atomic coordinates and structure factors (codes 4H04 and 4JAW) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- HMO
- human milk oligosaccharide
- LNB
- lacto-N-biose I, (Gal-β1,3-GlcNAc)
- LacNAc
- N-acetyllactosamine, (Gal-β1,4-GlcNAc)
- LNFP I
- lacto-N-fucopentaose I (Fuc-α1,2-Gal-β1,3-GlcNAc-β1,3-Gal-β1,4-Glc)
- LNDFH I
- lacto-N-difucohexaose I (Fuc-α1,2-Gal-β1,3-(Fuc-α1,4-)GlcNAc-β1,3-Gal-β1,4-Glc)
- LNT
- lacto-N-tetraose (Gal-β1,3-GlcNAc-β1,3-Gal-β1,4-Glc)
- GNB
- galacto-N-biose (Gal-β1,3-GalNAc)
- LNBase
- lacto-N-biosidase
- BbLNBase
- LNBase from B. bifidum JCM1254
- GH
- glycoside hydrolase
- CBM
- carbohydrate-binding module
- β-HexNAcase
- β-N-acetylhexosaminidase
- pNP
- p-nitrophenyl
- PA
- pyridylamino
- r.m.s.d.
- root mean square deviation
- SpHex
- β-HexNAcase from Streptomyces plicatus
- PUGNAc
- O-(2-acetamido-2-deoxy-d-glucopyranosylidene)-amino-N-phenylcarbamate.
REFERENCES
- 1. Urashima T., Fukuda K., Kitaoka M., Terabayashi T., Kobata A. (eds) (2011) Milk Oligosaccharides, Nova Science Publishers, New York [Google Scholar]
- 2. Urashima T., Asakuma S., Leo F., Fukuda K., Messer M., Oftedal O. T. (2012) The predominance of type I oligosaccharides is a feature specific to human breast milk. Adv. Nutr. 3, 473S-482S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zivkovic A. M., German J. B., Lebrilla C. B., Mills D. A. (2011) Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc. Natl. Acad. Sci. U.S.A. 108, 4653–4658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Venema K. (2012) Intestinal fermentation of lactose and prebiotic lactose derivatives, including human milk oligosaccharides. Int. J. Dairy 22, 123–140 [Google Scholar]
- 5. Urashima T., Odaka G., Asakuma S., Uemura Y., Goto K., Senda A., Saito T., Fukuda K., Messer M., Oftedal O. T. (2009) Chemical characterization of oligosaccharides in chimpanzee, bonobo, gorilla, orangutan, and siamang milk or colostrum. Glycobiology 19, 499–508 [DOI] [PubMed] [Google Scholar]
- 6. Asakuma S., Urashima T., Akahori M., Obayashi H., Nakamura T., Kimura K., Watanabe Y., Arai I., Sanai Y. (2008) Variation of major neutral oligosaccharides levels in human colostrum. Eur. J. Clin. Nutr. 62, 488–494 [DOI] [PubMed] [Google Scholar]
- 7. Kitaoka M., Tian J., Nishimoto M. (2005) Novel putative galactose operon involving lacto-N-biose phosphorylase in Bifidobacterium longum. Appl. Environ. Microbiol. 71, 3158–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nishimoto M., Kitaoka M. (2007) Identification of N-acetylhexosamine 1-kinase in the complete lacto-N-biose I/galacto-N-biose metabolic pathway in Bifidobacterium longum. Appl. Environ. Microbiol. 73, 6444–6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fushinobu S. (2010) Unique sugar metabolic pathways of bifidobacteria. Biosci. Biotechnol. Biochem. 74, 2374–2384 [DOI] [PubMed] [Google Scholar]
- 10. Suzuki R., Katayama T., Kitaoka M., Kumagai H., Wakagi T., Shoun H., Ashida H., Yamamoto K., Fushinobu S. (2009) Crystallographic and mutational analyses of substrate recognition of endo-α-N-acetylgalactosaminidase from Bifidobacterium longum. J. Biochem. 146, 389–398 [DOI] [PubMed] [Google Scholar]
- 11. Suzuki R., Wada J., Katayama T., Fushinobu S., Wakagi T., Shoun H., Sugimoto H., Tanaka A., Kumagai H., Ashida H., Kitaoka M., Yamamoto K. (2008) Structural and thermodynamic analyses of solute-binding protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I. J. Biol. Chem. 283, 13165–13173 [DOI] [PubMed] [Google Scholar]
- 12. Hidaka M., Nishimoto M., Kitaoka M., Wakagi T., Shoun H., Fushinobu S. (2009) The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: a large deformation of a TIM barrel scaffold. J. Biol. Chem. 284, 7273–7283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kiyohara M., Tachizawa A., Nishimoto M., Kitaoka M., Ashida H., Yamamoto K. (2009) Prebiotic effect of lacto-N-biose I on bifidobacterial growth. Biosci. Biotechnol. Biochem. 73, 1175–1179 [DOI] [PubMed] [Google Scholar]
- 14. Xiao J. Z., Takahashi S., Nishimoto M., Odamaki T., Yaeshima T., Iwatsuki K., Kitaoka M. (2010) Distribution of in vitro fermentation ability of lacto-N-biose I, a major building block of human milk oligosaccharides, in bifidobacterial strains. Appl. Environ. Microbiol. 76, 54–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sano M., Hayakawa K., Kato I. (1992) An enzyme releasing lacto-N-biose from oligosaccharides. Proc. Natl. Acad. Sci. U.S.A. 89, 8512–8516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sano M., Hayakawa K., Kato I. (1993) Purification and characterization of an enzyme releasing lacto-N-biose from oligosaccharides with type 1 chain. J. Biol. Chem. 268, 18560–18566 [PubMed] [Google Scholar]
- 17. Wada J., Ando T., Kiyohara M., Ashida H., Kitaoka M., Yamaguchi M., Kumagai H., Katayama T., Yamamoto K. (2008) Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure Appl. Environ. Microbiol. 74, 3996–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Turroni F., Peano C., Pass D. A., Foroni E., Severgnini M., Claesson M. J., Kerr C., Hourihane J., Murray D., Fuligni F., Gueimonde M., Margolles A., De Bellis G., O'Toole P. W., van Sinderen D., Marchesi J. R., Ventura M. (2012) Diversity of bifidobacteria within the infant gut microbiota. PLoS One 7, e36957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Magnani J. L., Steplewski Z., Koprowski H., Ginsburg V. (1983) Identification of the gastrointestinal and pancreatic cancer-associated antigen detected by monoclonal antibody 19–9 in the sera of patients as a mucin. Cancer Res. 43, 5489–5492 [PubMed] [Google Scholar]
- 20. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mark B. L., Vocadlo D. J., Knapp S., Triggs-Raine B. L., Withers S. G., James M. N. (2001) Crystallographic evidence for substrate-assisted catalysis in a bacterial β-hexosaminidase. J. Biol. Chem. 276, 10330–10337 [DOI] [PubMed] [Google Scholar]
- 22. Williams S. J., Mark B. L., Vocadlo D. J., James M. N., Withers S. G. (2002) Aspartate 313 in the Streptomyces plicatus hexosaminidase plays a critical role in substrate-assisted catalysis by orienting the 2-acetamido group and stabilizing the transition state. J. Biol. Chem. 277, 40055–40065 [DOI] [PubMed] [Google Scholar]
- 23. Vocadlo D. J., Withers S. G. (2005) Detailed comparative analysis of the catalytic mechanisms of β-N-acetylglucosaminidases from families 3 and 20 of glycoside hydrolases. Biochemistry 44, 12809–12818 [DOI] [PubMed] [Google Scholar]
- 24. Tews I., Perrakis A., Oppenheim A., Dauter Z., Wilson K. S., Vorgias C. E. (1996) Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease. Nat. Struct. Biol. 3, 638–648 [DOI] [PubMed] [Google Scholar]
- 25. Mark B. L., Mahuran D. J., Cherney M. M., Zhao D., Knapp S., James M. N. (2003) Crystal structure of human β-hexosaminidase B: understanding the molecular basis of Sandhoff and Tay-Sachs disease. J. Mol. Biol. 327, 1093–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ramasubbu N., Thomas L. M., Ragunath C., Kaplan J. B. (2005) Structural analysis of dispersin B, a biofilm-releasing glycoside hydrolase from the periodontopathogen Actinobacillus actinomycetemcomitans. J. Mol. Biol. 349, 475–486 [DOI] [PubMed] [Google Scholar]
- 27. Lemieux M. J., Mark B. L., Cherney M. M., Withers S. G., Mahuran D. J., James M. N. (2006) Crystallographic structure of human β-hexosaminidase A: interpretation of Tay-Sachs mutations and loss of GM2 ganglioside hydrolysis. J. Mol. Biol. 359, 913–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Langley D. B., Harty D. W., Jacques N. A., Hunter N., Guss J. M., Collyer C. A. (2008) Structure of N-acetyl-β-d-glucosaminidase (GcnA) from the endocarditis pathogen Streptococcus gordonii and its complex with the mechanism-based inhibitor NAG-thiazoline. J. Mol. Biol. 377, 104–116 [DOI] [PubMed] [Google Scholar]
- 29. Sumida T., Ishii R., Yanagisawa T., Yokoyama S., Ito M. (2009) Molecular cloning and crystal structural analysis of a novel β-N-acetylhexosaminidase from Paenibacillus sp. TS12 capable of degrading glycosphingolipids. J. Mol. Biol. 392, 87–99 [DOI] [PubMed] [Google Scholar]
- 30. Liu T., Zhang H., Liu F., Wu Q., Shen X., Yang Q. (2011) Structural determinants of an insect β-N-acetyl-d-hexosaminidase specialized as a chitinolytic enzyme. J. Biol. Chem. 286, 4049–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pluvinage B., Higgins M. A., Abbott D. W., Robb C., Dalia A. B., Deng L., Weiser J. N., Parsons T. B., Fairbanks A. J., Vocadlo D. J., Boraston A. B. (2011) Inhibition of the pneumococcal virulence factor StrH and molecular insights into N-glycan recognition and hydrolysis. Structure 19, 1603–1614 [DOI] [PubMed] [Google Scholar]
- 32. Jiang Y. L., Yu W. L., Zhang J. W., Frolet C., Di Guilmi A. M., Zhou C. Z., Vernet T., Chen Y. (2011) Structural basis for the substrate specificity of a novel β-N-acetylhexosaminidase StrH protein from Streptococcus pneumoniae R6. J. Biol. Chem. 286, 43004–43012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hattie M., Debowski A. W., Stubbs K. A. (2012) Development of tools to study lacto-N-biosidase: an important enzyme involved in the breakdown of human milk oligosaccharides. ChemBioChem 13, 1128–1131 [DOI] [PubMed] [Google Scholar]
- 34. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 35. Rappleye J., Innus M., Weeks C. M., Miller R. (2002) SnB version 2.2: an example of crystallographic multiprocessing. J. Appl. Crystallogr. 35, 374–376 [Google Scholar]
- 36. Terwilliger T. C., Berendzen J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Perrakis A., Morris R., Lamzin V. S. (1999) Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 38. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 40. Holm L., Sander C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233, 123–138 [DOI] [PubMed] [Google Scholar]
- 41. Grahn E., Askarieh G., Holmner A., Tateno H., Winter H. C., Goldstein I. J., Krengel U. (2007) Crystal structure of the Marasmius oreades mushroom lectin in complex with a xenotransplantation epitope. J. Mol. Biol. 369, 710–721 [DOI] [PubMed] [Google Scholar]
- 42. Miyanaga A., Koseki T., Matsuzawa H., Wakagi T., Shoun H., Fushinobu S. (2004) Crystal structure of a family 54 α-l-arabinofuranosidase reveals a novel carbohydrate-binding module that can bind arabinose. J. Biol. Chem. 279, 44907–44914 [DOI] [PubMed] [Google Scholar]
- 43. Hou Y., Vocadlo D. J., Leung A., Withers S. G., Mahuran D. (2001) Characterization of the Glu and Asp residues in the active site of human β-hexosaminidase B. Biochemistry 40, 2201–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Greig I. R., Zahariev F., Withers S. G. (2008) Elucidating the nature of the Streptomyces plicatus β-hexosaminidase-bound intermediate using ab initio molecular dynamics simulations. J. Am. Chem. Soc. 130, 17620–17628 [DOI] [PubMed] [Google Scholar]
- 45. Cremer D., Pople J. A. (1975) A general definition of ring puckering coordinates. J. Am. Chem. Soc. 97, 1354–1358 [Google Scholar]
- 46. Wada M., Kobayashi K., Nishimoto M., Kitaoka M., Noguchi K. (2009) 2-Acetamido-2-deoxy-3-O-β-d-galactopyranosyl-d-glucose dihydrate. Acta Crystallogr. E 65, 1781–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sattelle B. M., Almond A. (2011) Is N-acetyl-d-glucosamine a rigid 4C1 chair? Glycobiology 21, 1651–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Prag G., Papanikolau Y., Tavlas G., Vorgias C. E., Petratos K., Oppenheim A. B. (2000) Structures of chitobiase mutants complexed with the substrate Di-N-acetyl-d-glucosamine: the catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540. J. Mol. Biol. 300, 611–617 [DOI] [PubMed] [Google Scholar]
- 49. Mark B. L., Vocadlo D. J., Zhao D., Knapp S., Withers S. G., James M. N. (2001) Biochemical and structural assessment of the 1-N-azasugar GalNAc-isofagomine as a potent family 20 β-N-acetylhexosaminidase inhibitor. J. Biol. Chem. 276, 42131–42137 [DOI] [PubMed] [Google Scholar]
- 50. Maier T., Strater N., Schuette C. G., Klingenstein R., Sandhoff K., Saenger W. (2003) The x-ray crystal structure of human β-hexosaminidase B provides new insights into Sandhoff disease. J. Mol. Biol. 328, 669–681 [DOI] [PubMed] [Google Scholar]
- 51. Liu T., Zhang H., Liu F., Chen L., Shen X., Yang Q. (2011) Active-pocket size differentiating insectile from bacterial chitinolytic β-N-acetyl-d-hexosaminidases. Biochem. J. 438, 467–474 [DOI] [PubMed] [Google Scholar]
- 52. Sumida T., Stubbs K. A., Ito M., Yokoyama S. (2012) Gaining insight into the inhibition of glycoside hydrolase family 20 exo-β-N-acetylhexosaminidases using a structural approach. Org. Biomol. Chem. 10, 2607–2612 [DOI] [PubMed] [Google Scholar]
- 53. Knapp S., Vocadlo D. J., Gao Z., Kirk B., Lou J., Withers S. G. (1996) NAG-thiazoline, an N-acetyl-β-hexosaminidase inhibitor that implicates acetamido participation. J. Am. Chem. Soc. 118, 6804–6805 [Google Scholar]
- 54. Amorelli B., Yang C., Rempel B., Withers S. G., Knapp S. (2008) N-Acetylhexosaminidase inhibitory properties of C-1 homologated GlcNAc- and GalNAc-thiazolines. Bioorg. Med. Chem. Lett. 18, 2944–2947 [DOI] [PubMed] [Google Scholar]
- 55. Usuki H., Nitoda T., Ichikawa M., Yamaji N., Iwashita T., Komura H., Kanzaki H. (2008) TMG-chitotriomycin, an enzyme inhibitor specific for insect and fungal β-N-acetylglucosaminidases, produced by actinomycete Streptomyces anulatus NBRC 13369. J. Am. Chem. Soc. 130, 4146–4152 [DOI] [PubMed] [Google Scholar]
- 56. Jitonnom J., Lee V. S., Nimmanpipug P., Rowlands H. A., Mulholland A. J. (2011) Quantum mechanics/molecular mechanics modeling of substrate-assisted catalysis in family 18 chitinases: conformational changes and the role of Asp-142 in catalysis in ChiB. Biochemistry 50, 4697–4711 [DOI] [PubMed] [Google Scholar]
- 57. Rao F. V., Dorfmueller H. C., Villa F., Allwood M., Eggleston I. M., van Aalten D. M. (2006) Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis. EMBO J. 25, 1569–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sinnott M. L. (1984) On the antiperiplanar lone pair hypothesis and its application to catalysis by glycosidases. Biochem. J. 224, 817–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ficko-Blean E., Boraston A. B. (2012) Insights into the recognition of the human glycome by microbial carbohydrate-binding modules. Curr. Opin. Struct. Biol. 22, 570–577 [DOI] [PubMed] [Google Scholar]
- 60. Asakuma S., Hatakeyama E., Urashima T., Yoshida E., Katayama T., Yamamoto K., Kumagai H., Ashida H., Hirose J., Kitaoka M. (2011) Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J. Biol. Chem. 286, 34583–34592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Katayama T., Sakuma A., Kimura T., Makimura Y., Hiratake J., Sakata K., Yamanoi T., Kumagai H., Yamamoto K. (2004) Molecular cloning and characterization of Bifidobacterium bifidum 1,2-α-l-fucosidase (AfcA), a novel inverting glycosidase (glycoside hydrolase family 95). J. Bacteriol. 186, 4885–4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nagae M., Tsuchiya A., Katayama T., Yamamoto K., Wakatsuki S., Kato R. (2007) Structural basis of the catalytic reaction mechanism of novel 1,2-α-l-fucosidase from Bifidobacterium bifidum. J. Biol. Chem. 282, 18497–18509 [DOI] [PubMed] [Google Scholar]
- 63. Ashida H., Miyake A., Kiyohara M., Wada J., Yoshida E., Kumagai H., Katayama T., Yamamoto K. (2009) Two distinct α-l-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology 19, 1010–1017 [DOI] [PubMed] [Google Scholar]
- 64. Sakurama H., Fushinobu S., Hidaka M., Yoshida E., Honda Y., Ashida H., Kitaoka M., Kumagai H., Yamamoto K., Katayama T. (2012) 1,3–1,4-α-l-Fucosynthase that specifically introduces Lewis a/x antigens into type-1/2 chains. J. Biol. Chem. 287, 16709–16719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kiyohara M., Tanigawa K., Chaiwangsri T., Katayama T., Ashida H., Yamamoto K. (2011) An exo-α-sialidase from bifidobacteria involved in the degradation of sialyloligosaccharides in human milk and intestinal glycoconjugates. Glycobiology 21, 437–447 [DOI] [PubMed] [Google Scholar]
- 66. Miwa M., Horimoto T., Kiyohara M., Katayama T., Kitaoka M., Ashida H., Yamamoto K. (2010) Cooperation of β-galactosidase and β-N-acetylhexosaminidase from bifidobacteria in assimilation of human milk oligosaccharides with type 2 structure. Glycobiology 20, 1402–1409 [DOI] [PubMed] [Google Scholar]
- 67. Sela D. A., Chapman J., Adeuya A., Kim J. H., Chen F., Whitehead T. R., Lapidus A., Rokhsar D. S., Lebrilla C. B., German J. B., Price N. P., Richardson P. M., Mills D. A. (2008) The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. U.S.A. 105, 18964–18969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sela D. A., Mills D. A. (2010) Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 18, 298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yoshida E., Sakurama H., Kiyohara M., Nakajima M., Kitaoka M., Ashida H., Hirose J., Katayama T., Yamamoto K., Kumagai H. (2012) Bifidobacterium longum subsp. infantis uses two different β-galactosidases for selectively degrading type-1 and type-2 human milk oligosaccharides. Glycobiology 22, 361–368 [DOI] [PubMed] [Google Scholar]
- 70. Barboza M., Sela D. A., Pirim C., Locascio R. G., Freeman S. L., German J. B., Mills D. A., Lebrilla C. B. (2009) Glycoprofiling bifidobacterial consumption of galacto-oligosaccharides by mass spectrometry reveals strain-specific, preferential consumption of glycans. Appl. Environ. Microbiol. 75, 7319–7325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Structure validation by Cα geometry: φ, ψ, and Cβ deviation. Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]