Background: Phosphatidic acid (PA) is a class of membrane lipid mediators synthesized in response to various stresses in plants.
Results: PA binds to cytosolic glyceraldehyde-3-phosphate dehydrogenases (GAPCs) and promotes proteolytic cleavage of GAPC2 in Arabidopsis.
Conclusion: PA-GAPC interaction constitutes a mechanism for PA signaling in plants.
Significance: PA-GAPC interaction may provide a molecular link modulating coordinately carbohydrate and lipid metabolism.
Keywords: Cell Signaling, Lipid-binding Protein, Phosphatidic Acid, Plant Biochemistry, Zinc, Arabidopsis, Glyceraldehyde-3-phosphate Dehydrogenase, Lipid Metabolism, Lipid Signaling
Abstract
Phosphatidic acid (PA) is a class of lipid messengers involved in a variety of physiological processes. To understand how PA mediates cell functions in plants, we used a PA affinity membrane assay to isolate PA-binding proteins from Camelina sativa followed by mass spectrometric sequencing. A cytosolic glyceraldehyde-3-phosphate dehydrogenase (GAPC) was identified to bind to PA, and detailed analysis was carried out subsequently using GAPC1 and GAPC1 from Arabidopsis. The PA and GAPC binding was abolished by the cation zinc whereas oxidation of GAPCs promoted the PA binding. PA had little impact on the GAPC catalytic activity in vitro, but the PA treatment of Arabidopsis seedlings induced proteolytic cleavage of GAPC2 and inhibited Arabidopsis seedling growth. The extent of PA inhibition was greater in GAPC-overexpressing than wild-type seedlings, but the greater PA inhibition was abolished by application of zinc to the seedling. The PA treatment also reduced the expression of genes involved in PA synthesis and utilization, and the PA-reduced gene expression was partially recovered by zinc treatment. These data suggest that PA binds to oxidized GAPDH and promotes its cleavage and that the PA and GAPC interaction may provide a signaling link coordinating carbohydrate and lipid metabolism.
Introduction
Biological membranes are composed of different types of lipids with distinct properties and functions. Lipids in the membrane not only structurally compartmentalize the cell but also act as second messengers to transduce cellular signals. Signaling lipids normally exist in a trace amount in the membrane and are synthesized rapidly and transiently in response to external stimuli to activate downstream signaling pathways. The minor membrane lipid phosphatidic acid (PA)2 has been shown as an important lipid messenger that mediates a variety of cellular and physiological processes in plants (1–3). The cellular level of PA is increased under various stress conditions, such as treatments of abscisic acid (ABA) (4), osmotic stress (5–7), oxidative stress (8), pathogen infection (9, 10), and cold and freezing conditions (11–13). In addition, PA is involved in regulating normal plant growth and development, particularly in roots (14, 15) and pollen tubes (16, 17).
One mode of action by which PA mediates cellular effect is by its direct interaction with effector proteins to convey signals (1–3). PA was found to interact with abscisic acid-insensitive 1 (ABI1), a protein phosphatase, constitutive triple response 1 (CTR1), a protein kinase, and phosphoethanolamine N-methyltransferase (PEAMT) and inhibit their phosphatase or kinase activities (18–20). PA stimulates the catalytic activity of a phosphoinositide-dependent protein kinase (PDK1), sphingosine kinases (SPHK1/2), NADPH oxidases (RbohD/F), a mitogen-activated protein kinase (MPK6), and a SNF1-related kinase (SnRK2) by direct interaction (21–24). In addition to regulating the catalytic activity, PA may tether its binding proteins to membranes. PA tethers ABI1 to the plasma membrane, resulting in prevention of ABI1 from translocation into the nucleus where it binds to ATHB6, a transcription factor that negatively regulates ABA responses (18, 25). A similar process was observed in yeast that PA bound and prevented a transcriptional repressor Opi1 from entering the nucleus where it suppresses lipid biosynthesis (26, 27). Thus, PA-mediated sequestration of proteins to membranes seems to be a conserved mechanism for the transcriptional regulation of gene expression in response to PA turnover in different organisms.
To understand how PA functions as a lipid mediator, we undertook an untargeted approach to identify PA-interacting proteins and found cytosolic glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a potential PA-interacting protein. GAPDH catalyzes the conversion of glyceraldehyde 3-phosphate to 1,3-bisphosphoglycerate in glycolysis, which provides substrate for acetyl-CoA for synthesis of various cellular compounds including lipids (28). Arabidopsis has two cytosolic GAPDHs, GAPC1 and GAPC2, and a recent study showed that both GAPCs interact with phospholipase Dδ (PLDδ), which directly produces signaling PA by hydrolyzing common membrane phospholipids (29). These observations raise intriguing questions about the function and significance of PA-GAPC interactions. This report describes the identification and characterization of PA-GAPC interaction and its physiological significance in Arabidopsis seedling growth.
EXPERIMENTAL PROCEDURES
Plant Materials and Growth Conditions
Arabidopsis thaliana wild-type (Col-0) and T-DNA insertion knock-out lines of gapc2 (SALK_016539 and SALK_070902) were obtained from ABRC (Ohio State University). The knock-out mutants were screened for homozygotes as described previously (29). Transgenic lines overexpressing GAPC2 were generated by cloning GAPC2 (At1g13440) cDNA into p35S-FAST/eYFP vector and transforming the DNA constructs into Arabidopsis by floral dipping. Seeds were surface-sterilized with 70% (v/v) ethanol and 20% (v/v) bleach followed by washing four times with water. Seeds were stratified at 4 °C for 2 days and germinated in 1/2 Murashige and Skoog medium with 1.5% (w/v) sucrose under a light cycle of 12-h light/12-h dark at 22 °C. After 5 days, seedlings were transferred to 1/2 Murashige and Skoog medium supplemented with various reagents depending on experiments. For lipid treatment, lipids (Avanti Polar Lipids, Alabaster, AL) were prepared by drying the chloroform under a gentle stream of nitrogen gas and resuspending the dried lipid in water followed by sonication before being added to the medium.
Nitrocellulose Membrane Binding Assay and Protein Identification by Mass Spectrometry
Approximately 1 g of 3-week-old wild-type Camelina (Camelina sativa) seedlings grown in soil was ground with liquid nitrogen. Proteins were extracted by adding 4 ml of protein extraction buffer (50 mm Tris-HCl, pH 7.3, 50 mm NaCl, 5% (v/v) glycerol, 1 mm DTT, 0.5 mm PMSF) and cleared by centrifugation at 1,500 × g for 10 min at 4 °C. Protein concentration in the supernatant was determined by the Bradford assay, and ∼500 μg of total proteins was used for nitrocellulose membrane binding assay. The assay was carried out as described previously (30) with some modifications. Approximately 10 μg of lipid dissolved in chloroform was spotted on a piece of nitrocellulose membrane (0.45-μm pore; Whatman) and air-dried for at least 30 min. The membrane was incubated with TBST buffer (10 mm Tris-HCl, pH 7.4, 140 mm NaCl, 0.1% (v/v) Tween 20) containing 0.5% (w/v) fatty acid-free BSA for 1 h to block the membrane, washed three times with TBST buffer, and incubated with proteins overnight at 4 °C. The membrane was washed three times with TBST buffer to remove unbound proteins, and bound proteins were either eluted or probed by immunoblotting as described below.
Proteins bound to the PA-nitrocellulose membrane were eluted by incubation of the membrane with 9 m urea for 1 h and recovered as described previously (31). The resulting protein pellet was dissolved in 50 μl of SDS-PAGE sample buffer, boiled for 5 min, and subjected to SDS-PAGE separation. The protein bands were carefully excised from the gel, and proteins were in-gel digested with trypsin (Sigma-Aldrich) at 37 °C overnight following the manufacturer's instruction. The digested peptides were run on the LC-tandem MS using an LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific). The database search was done with peptide mass fingerprint data using MASCOT v2.2 database search engine (Matrix Science, Boston, MA) against the NCBI database for A. thaliana. The criteria for a significant protein identification were both at least two unique peptides per protein identified and each peptide showing a probability >80% (MASCOT ion score >30).
Liposome Binding Assay
The assay was performed as described previously (22) with some modifications. Liposomes were prepared according to the Avanti Polar Lipids Technical Support. PC and PA (or PE) were mixed in the molar ratio of 3:1 and with the total amount of 10 μmol and dried with a gentle stream of nitrogen gas. Lipids were rehydrated for 1 h with HBS buffer (20 mm HEPES, pH 7.5, 100 mm NaCl, 0.02% (w/v) sodium azide). Small unilamellar vesicles were produced by a mild sonication using a bath sonicator until the solution became nearly clear, and centrifugation at 50,000 × g for 15 min removed large particles. The liposome pellet was resuspended in binding buffer (25 mm Tris-HCl, pH 7.5, 125 mm KCl, 1 mm DTT, 0.5 mm EDTA) and incubated with 10 μg of purified proteins for 1 h. Liposomes were harvested by centrifuging at 16,000 × g for 30 min, washed three times with the binding buffer, and resuspended in SDS-PAGE sample buffer for immunoblotting.
SDS-PAGE and Immunoblotting
Protein samples were dissolved in SDS-PAGE sample buffer, boiled for 5 min, and loaded on 10% (v/v) polyacrylamide gel. The gel was run at 100 V for ∼1 h and stained with Coomassie Brilliant Blue for 1 h, followed by washing with methanol:water:acetic acid (3:6:1, v/v/v) to remove background stain. For immunoblotting, proteins were electrophoretically transferred from the gel onto a polyvinylidene fluoride (PVDF) membrane using the Semidry Trans-Blot apparatus (Bio-Rad). The membrane was blocked in TBST buffer containing 5% (w/v) nonfat milk for 1 h, followed by washing three times with TBST buffer. The membrane was incubated with primary antibodies (anti-His6 from Sigma-Aldrich, anti-FLAG and anti-histone H3 from GenScript, and anti-PEPC from Rockland, Gilbertsville, PA) for 1 h. After washing three times with TBST buffer, the membrane was incubated with secondary antibodies from mouse or rabbit conjugated with alkaline phosphatase (Sigma-Aldrich) for 1 h, followed by colorimetric detection of the proteins using alkaline phosphatase conjugate substrate (Bio-Rad) according to the manufacturer's instruction.
Surface Plasmon Resonance (SPR) Analysis
SPR assay was performed as described previously (22). Briefly, a sensor chip preimmobilized with nitrilotriacetic acid was used to capture purified, His-tagged GAPC1 or GAPC2. Response unit (RU) was monitored using a Biacore 2000 system as liposomes composed of 18:1PA+18:1PC (1:3 in molar ratio) or 18:1PC only were injected onto the chip. The sensorgrams were plotted by Microsoft Office Excel (2007), and kinetic constants were calculated by Prism v5 (GraphPad Software).
Gene Cloning, Protein Purification, and Activity Assay
GAPC was cloned into pET-28a-c(+) vector, expressed in BL21(DE3)pLysS, and purified using nickel-nitrilotriacetic acid-agarose (Qiagen) as described previously (29). An NAD-dependent GAPDH activity assay was performed by spectrophotometric quantification (340 nm) of NADH produced in the reaction according to the method described previously (32). SPHK1 was expressed in Escherichia coli and purified as described previously (22).
Nuclear Protein Isolation
Nuclear proteins were extracted as described previously (33) with some modifications. Plant tissues were ground with extraction buffer (50 mm MES, pH 8.5, 25 mm NaCl, 5 mm MgCl2, 30% (v/v) glycerol, 5% (w/v) sucrose, 0.1% (v/v) β-mercaptoethanol, 0.5% (v/v) Triton X-100, 0.5 mm spermidine) and centrifuged at 3,400 × g for 20 min at 4 °C. The pellet was washed with the extraction buffer without Triton X-100 four times at 2,200 × g for 10 min, 1,700 × g for 10 min, 1,700 × g for 8 min, 1,700 × g for 6 min, all at 4 °C. The final pellet was resuspended in homogenization buffer (100 mm Tris-HCl, pH 7.2, 0.1% (v/v) β-mercaptoethanol, 10% (w/v) sucrose, 5 mm EDTA) and used as a nuclear fraction. Supernatant from the centrifugation at 3,400 × g was centrifuged at 150,000 × g for 30 min at 4 °C, and the resulting supernatant was used as a cytosolic fraction.
Reverse Transcription (RT)-PCR and Quantitative Real-time (qRT) PCR
Total RNA was extracted from plant tissues using TRIzol® Reagent (Invitrogen) according to the manufacturer's instructions. RNA was quantified by Nanodrop 2000 spectrophotometer (Thermo Scientific) and checked for integrity by agarose gel electrophoresis. Genomic DNA was digested by RQ1 RNase-free DNase (Promega) at 37 °C for 30 min, and the enzyme was inactivated with RQ1 DNase Stop Solution at 65 °C for 10 min. cDNA was synthesized by a qScript cDNA Synthesis Kit (Quanta BioSciences, Gaithersburg, MD) with a blend of random and oligo(dT) primers. The reaction was at 42 °C for 30 min with preincubation at 22 °C for 5 min and enzyme inactivation at 85 °C for 5 min. The cDNA was amplified with a Taq polymerase using gene-specific primers (supplemental Table S1) through the following thermal cycling conditions: predenaturation at 95 °C for 5 min, 40 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s, and final polymerization at 72 °C for 10 min. PCR products were resolved and visualized on 1% (w/v) agarose gel. RNA extraction and cDNA synthesis were confirmed by amplifying ubiquitin 10. For qRT-PCR, PCR progress was monitored by adding SYBR Green dye using the Step One Plus Real-Time PCR System (Applied Biosystems). The gene expression was normalized with ubiquitin 10 as an internal standard.
RESULTS
Identification of PA-binding Proteins
To discover PA-binding proteins, we used a PA-on-nitrocellulose membrane to incubate with total proteins extracted from an oilseed crop C. sativa in an initial attempt to study the PA signaling in seed lipid metabolism. Proteins bound to the membrane were eluted and subjected to SDS-PAGE (Fig. 1A). Several protein bands were present or increased in the PA affinity membrane fraction, but they were absent or detected in a lower level in the membrane applied with the carrier solvent (chloroform) only. These proteins were sequenced by mass spectrometry. Because the Camelina genome has not been fully sequenced, the protein sequences were blasted against the Arabidopsis data base based on genetic similarity between the two closely related plant species (>90% in genomic identity) (34).
FIGURE 1.
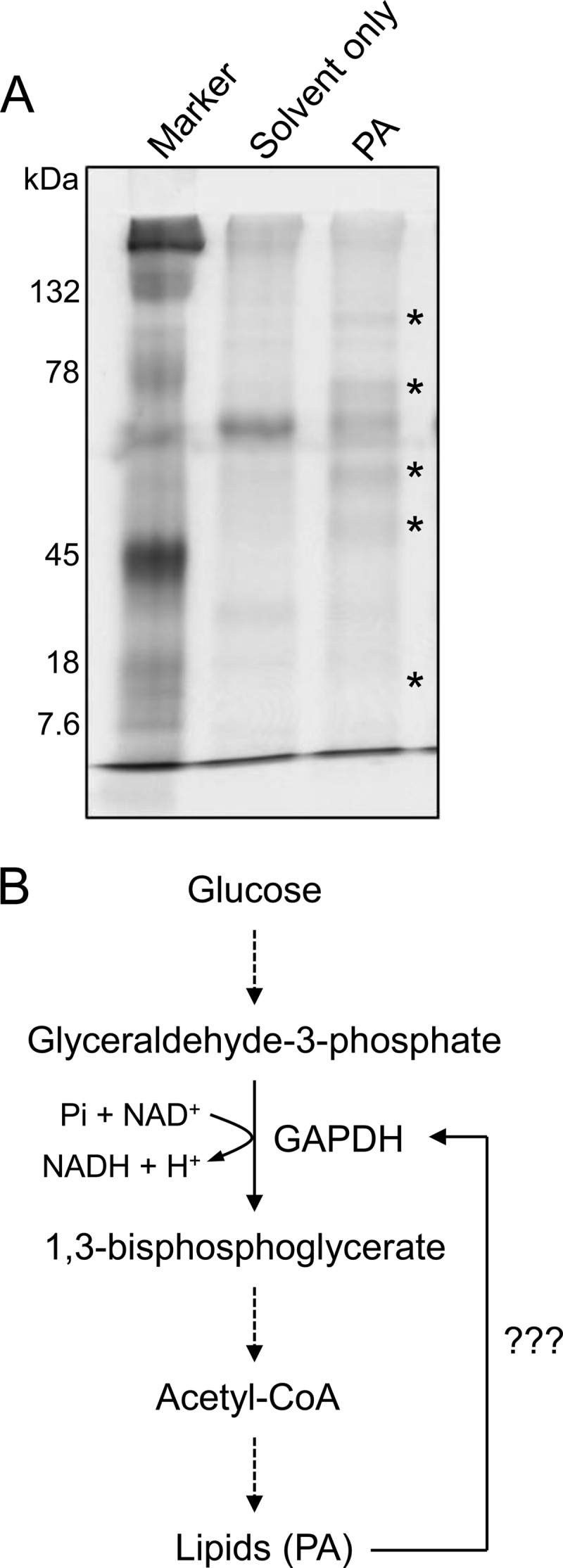
Identification of PA-binding proteins. A, SDS-PAGE image of the candidate PA-binding proteins. Total proteins from C. sativa were incubated with PA-spotted nitrocellulose membrane, and proteins bound to the membrane were resolved by SDS-PAGE. Asterisks indicate the proteins present in the PA-spotted membrane but not in the solvent-only spotted membrane. B, metabolic pathway showing the potential role of GAPDH in lipid biosynthesis. Dashed arrows indicate multistep pathway.
The mass spectrometry-based protein sequencing identified several candidate PA-binding proteins. Some of them were enzymes involved in carbohydrate metabolism, such as GAPDH, fructose bisphosphate aldolase, and phosphoglycerate kinase. We also identified known PA-binding proteins, such as annexin, a PA-binding protein found in animals (35), and some isoforms of heat shock protein (Hsp81-3), and tubulins (α2/4) previously identified (36). Among the protein candidates identified, GAPDH was particularly of interest because it was recently found to interact directly with PLDδ (29), an enzyme responsible for PA production. In addition, GAPDH is one of the key enzymes involved in the glycolytic pathway that is critical for energy production and biosynthesis for various compounds, including acetyl-CoA and lipids (Fig. 1B). Thus, the present study is focused on the characterization of the PA and GAPDH interaction.
PA Binds to Two Cytosolic GAPDHs
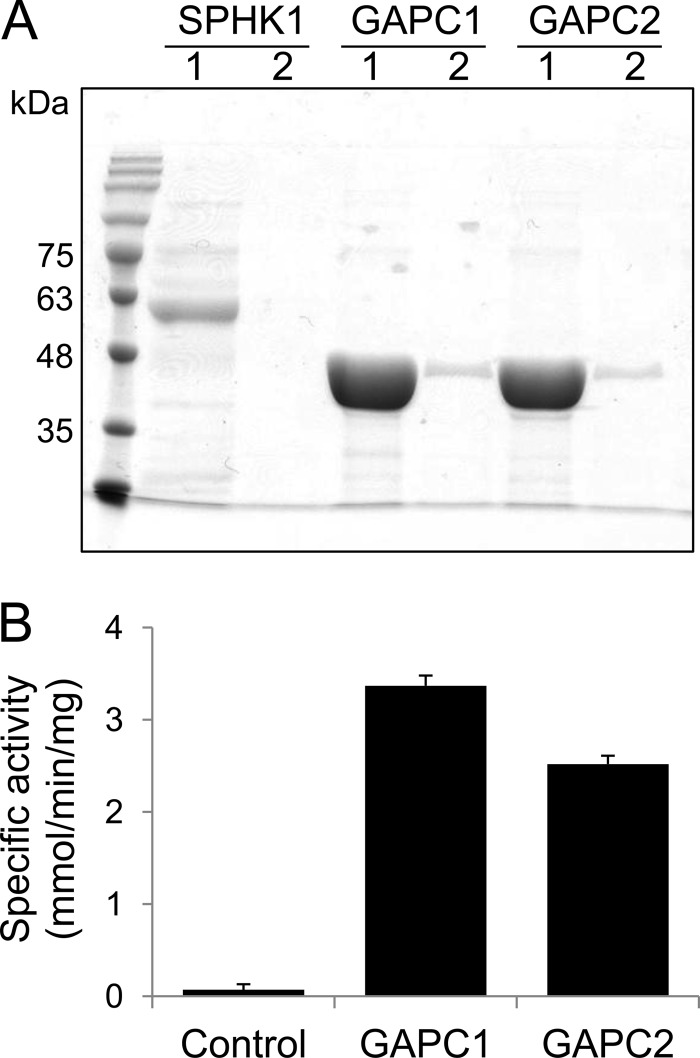
Arabidopsis has two cytosolic GAPDHs, GAPC1 and GAPC2, which share high amino acid sequence identify (>95%; supplemental Fig. S1). GAPC1 and GAPC2 cDNAs were cloned from Arabidopsis, expressed in E. coli as recombinant proteins, and affinity-purified to near homogeneity (Fig. 2A). Both purified GAPC1 and GAPC2 effectively converted their substrate (glyceraldehyde 3-phosphate) into product (1,3-bisphosphoglycerate), and thus they were catalytically active (Fig. 2B).
FIGURE 2.
Purification of GAPC from E. coli. A, SDS-PAGE image of the purified proteins. Proteins were purified by affinity chromatography and subjected to SDS-PAGE. Proteins were eluted from the column twice with the identical volume of elution buffer. 1, first eluent; 2, second eluent; SPHK1, sphingosine kinase 1. B, GAPDH activity assay. The assay was performed by quantifying NADH formed by GAPC in the reaction. Protein expressed from the empty vector was used as the control. Error bars represent means ± S.D. (n = 3).
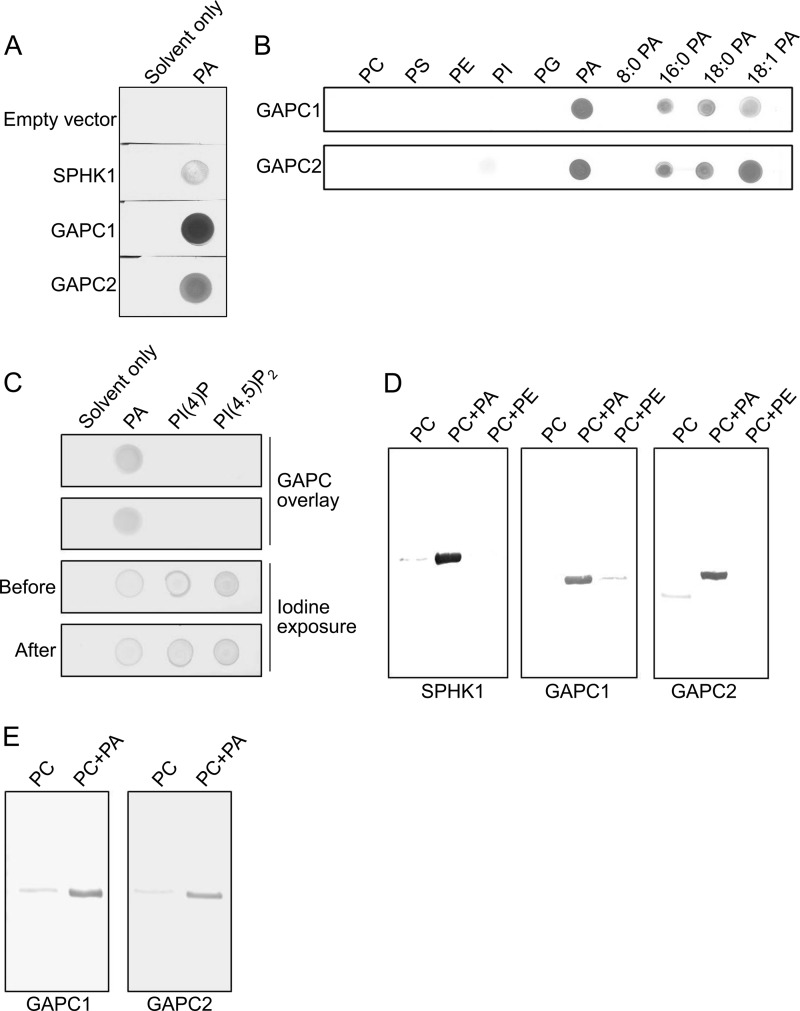
The nitrocellulose membrane binding assay using the purified proteins demonstrated that both GAPCs bound to PA on the membrane, but not to other phospholipids tested, including PC, PE, PS, PI, or PG (Fig. 3, A and B). No binding was detected in the empty vector control. SPHK1, a sphingosine kinase known to bind to PA (22), was included as a positive control for the PA binding assay. In addition, GAPCs did not bind to either of two common polyphosphoinositides PI(4)P or PI(4,5)P2 (Fig. 3C). Exposure of the membrane to iodine indicated the presence of the two phosphoinositide lipids on the membrane, suggesting that the failure of detecting positive binding was not due to absence of the lipids from the membrane (Fig. 3C). We then performed a liposome binding assay to further confirm the interaction between PA and GAPC. Because liposomes are unlikely to be formed by PA alone due to the small size of its head group forming a cone-shaped structure (37), we generated liposomes composed of PA and PC. Both GAPCs bound to PA-containing liposomes as SPHK1 did, but negligibly to those composed of either PC only or PC and PE (Fig. 3D), suggesting that the binding was specific to PA. The membrane binding assay using PA species with different acyl chain compositions indicated that both GAPCs bound virtually to all PA types tested except dioctanoyl (8:0) PA (Fig. 3B). However, GAPCs bound to liposomes containing 8:0 PA and PC (Fig. 3E). One explanation for the difference in the binding assays is that unlike other PA species, the highly soluble nature of 8:0 PA may have caused solubilization of the lipid from the nitrocellulose membrane in the process of assay.
FIGURE 3.
Confirmation of the PA-GAPC interaction. A, nitrocellulose membrane binding assay. The purified proteins were incubated with PA-spotted membranes and probed with anti-His6 antibody. The spots on membrane indicate positive interaction between PA and the proteins. B, membrane binding assay showing PA-specific interaction of GAPC. The purified proteins were incubated with various phospholipids and different PA-spotted membranes and probed with anti-His6 antibody. Numbers indicate acyl chains on both sn-1 and sn-2 positions. C, membrane binding assay showing no interaction with polyphosphoinositides. The assay was performed as in A using PI(4)P and PI(4,5)P2. A mixture of chloroform and methanol (20:1, v/v) was used as solvent to dissolve the lipids. The lipids spotted on the membrane were visualized by exposing the membrane to iodine vapor before and after blotting with proteins to demonstrate the lipids were still present on the membrane after blotting. D, liposome-binding assay. The purified proteins were pulled down with liposomes and detected by immunoblotting. The bands on blot indicate the proteins co-pulled down with PA liposomes. Lipid composition of the liposomes is indicated above the images. E, liposome binding assay showing 8:0 PA-GAPC interaction. The assay was performed as in D using 8:0 PA.
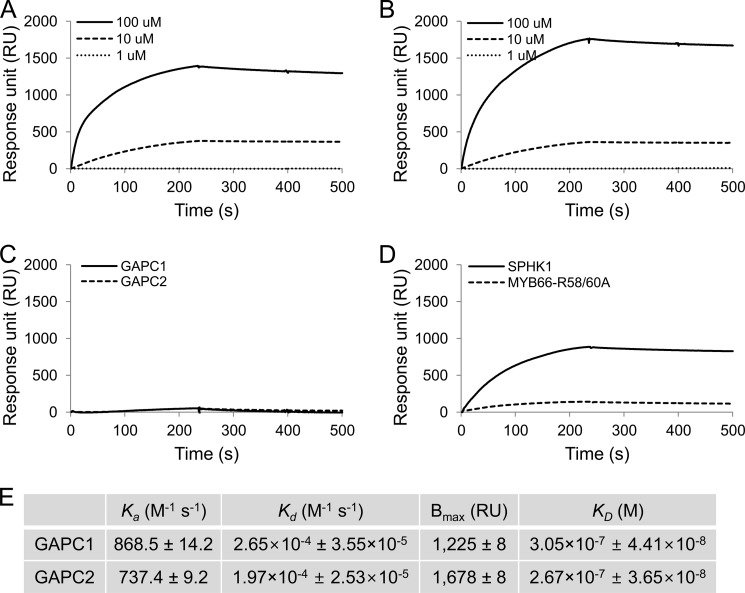
The PA-GAPC interaction was quantified by SPR analysis. Both GAPCs demonstrated an association/dissociation response with an increased RU when PA+PC liposomes were injected and a decreased RU when liposome injection was stopped (Fig. 4, A and B). The response was highly dependent on liposome concentration. The carrier lipid, PC only liposomes, barely increased RU, indicating again that GAPCs interact with PA specifically (Fig. 4C). We tested the SPR response using a mutated Myb protein as a negative control and SPHK1 as a positive control (22). When the same amounts of proteins were used as that of GAPCs, SPHK1 displayed the binding response to PA+PC liposomes as reported previously (22), whereas a negligible response was obtained from the negative control protein (Fig. 4D). GAPC1 had slightly higher association and dissociation rate constants (Ka and Kd) than GAPC2 did (Fig. 4E). The maximum binding (Bmax) was estimated to be 1,225 RU for GAPC1 and 1,678 RU for GAPC2. The equilibrium binding constant (KD) indicates that GAPC2 has a slightly higher affinity for PA than GAPC1 (Fig. 4E).
FIGURE 4.
SPR quantitative analysis of PA-GAPC interaction. His-tagged proteins (0.2 μm) were first immobilized on the nitrilotriacetic acid chip followed by injection of liposomes containing PC only or PC plus PA. The representative sensorgrams show RU values at different liposome concentrations over time starting with the time point that liposomes were injected (0 s). Liposome injection was stopped at 235 s. A–D, PA-GAPC1 (A), PA-GAPC2 (B), PC (100 μm)-GAPC1/GAPC2 (C), PA (100 μm)-SPHK1/Myb (D). E, summary of kinetic constants for the PA-GAPC interaction. Values were averaged from three independent assays and are shown ± S.E.
The PA-GAPC Interaction Is Inhibited by Zn2+ but Enhanced by Oxidation
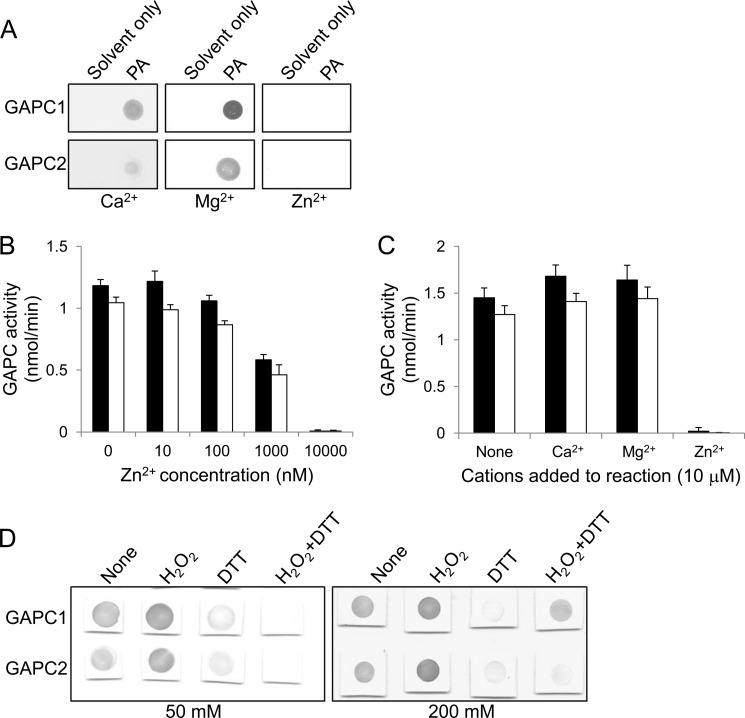
Many lipid-binding domains found in lipid-binding proteins have a cationic surface responsible for the electrostatic attraction to anionic lipids (38). The physical interaction between proteins and lipids are often dependent on the existence of divalent cations, such as Ca2+ and Zn2+, on the cationic surface (38). Thus, we tested the effect of different divalent cations on the PA-GAPC interaction. The interaction was not significantly affected by either Ca2+ or Mg2+, but completely abolished by Zn2+ (Fig. 5A). It has been reported that the catalytic activity of mammalian GAPDH is inhibited by a low nanomolar concentration of Zn2+ (39). Consistent with this property, both Arabidopsis GAPCs were inhibited specifically by Zn2+ in a dose-dependent manner, whereas Ca2+ or Mg2+ displayed no inhibitory effect on their catalytic activity (Fig. 5, B and C).
FIGURE 5.
Effect of cations and redox on the PA-GAPC interaction. A, nitrocellulose membrane binding assay showing effect of cations on the interaction. The proteins were incubated with the PA-spotted membrane along with a 1 mm concentration of the different cations indicated and probed with anti-His6 antibody. B, dose-dependent inhibition of GAPC catalytic activity by Zn2+. The enzymes were preincubated with Zn2+ at the different concentrations indicated prior to the reaction. Black and white bars denote GAPC1 and GAPC2, respectively. Bars represent means ± S.D. (error bars; n = 3). C, effect of different cations on GAPC catalytic activity. The enzymes were preincubated with a 10 μm concentration of the different cations indicated prior to the reaction. Black and white bars denote GAPC1 and GAPC2, respectively. Bars represent means ± S.D. (error bars; n = 3). D, nitrocellulose membrane binding assay showing effect of GAPC redox state on PA binding. The proteins were incubated with the PA-spotted membrane along with the indicated concentrations of H2O2 or DTT and probed with anti-His6 antibody.
GAPDH is known to be reversibly oxidized at a cysteine residue to lose its catalytic activity depending on the redox state of the cell (40). In addition, our recent result indicates that oxidized GAPCs bind stronger to PLDδ (29). To test in what state GAPC binds to PA, we performed a PA-GAPC binding assay in the presence of hydrogen peroxide (H2O2) and DTT as an oxidant and reductant, respectively. The PA interaction of both GAPCs was slightly enhanced by H2O2 treatment but was substantially compromised in the presence of DTT (Fig. 5D). Adding an excessive amount of H2O2 restored the PA-GAPC1 interaction disrupted by DTT (Fig. 5D), suggesting that PA prefers binding to the oxidized form of GAPCs.
PA Does Not Affect GAPC Catalytic Activity but Promotes Proteolytic Cleavage of GAPC
To investigate whether PA binding affects the catalysis of GAPCs, we measured the enzymatic activity of GAPC in vitro with increasing concentrations of PA. The activity of either GAPC1 or GAPC2 was not significantly affected by PA even at the highest concentration tested (100 μm; Fig. 6A). We then investigated whether PA affected the subcellular association of GAPC. In mammalian cells under certain growth conditions, a portion of GAPDH is localized in the nucleus, where it binds to transcription factors to regulate gene expression (41). Thus, we examined whether GAPC was localized in the nucleus of plant cells and whether PA would have any effect on the nuclear localization. Proteins from Arabidopsis seedlings were fractionated into cytosolic and nuclear fractions. Phosphoenolpyruvate carboxylase 1 (PEPC1) and histone H3 were used as the marker for the cytosolic and nuclear fractions, respectively (Fig. 6B). The absence of PEPC1 in the nuclear fraction and the lack of histone H3 in the cytosolic fraction indicate that the cytosolic and nuclear fractions were well separated (Fig. 6B). Using 35S::YFP-GAPC2 transgenic Arabidopsis, GAPC2 was detected in both the cytosolic and nuclear fractions, with a majority being in the cytosol (Fig. 6B). The nuclear localization of GAPC is consistent with previous reports (42, 43). Semiquantification by densitometric analysis suggests that ∼98% of total GAPC2 is in the cytosol and 2% in the nucleus.
FIGURE 6.
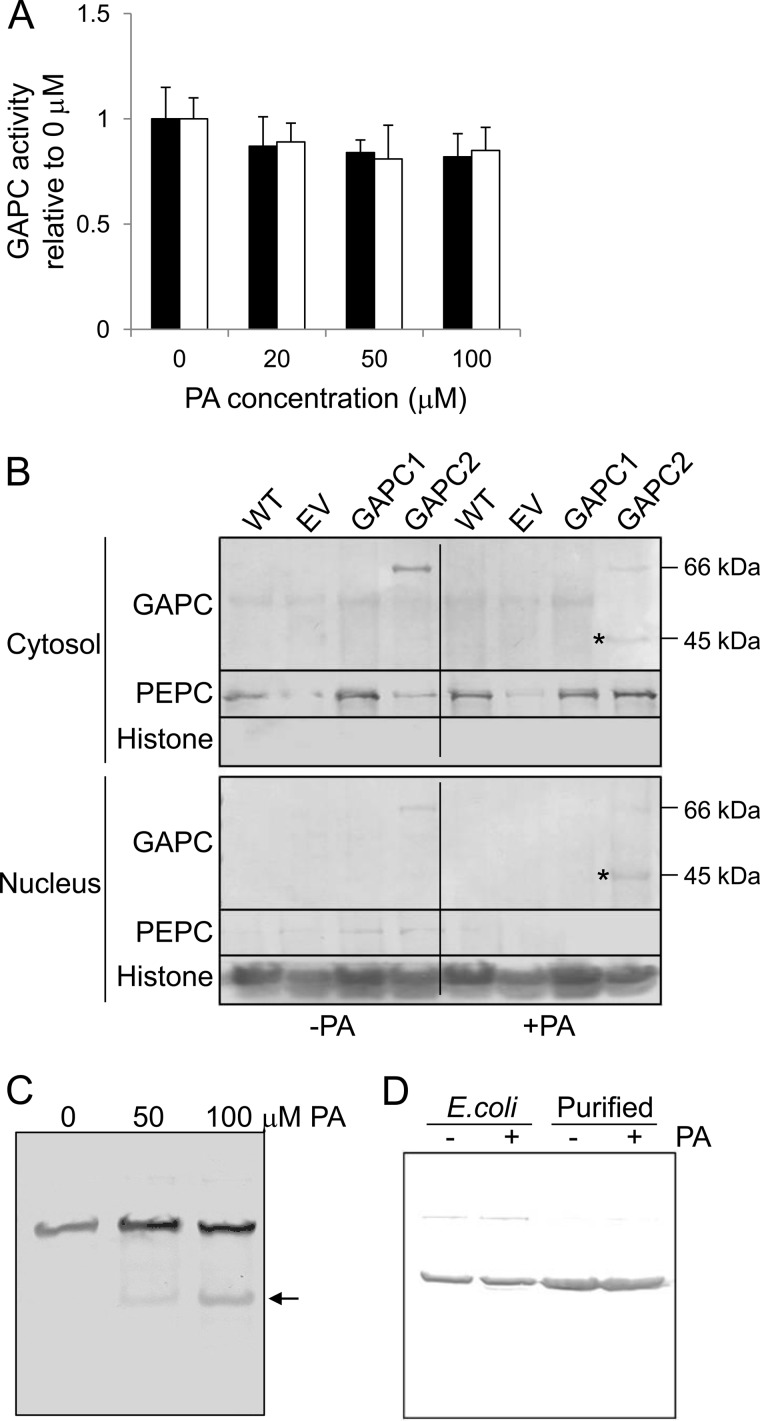
Effect of PA on GAPC activity and subcellular association. A, effect of PA on GAPC catalytic activity. The enzymes were preincubated with PA at different concentrations indicated prior to the reaction. Black and white bars denote GAPC1 and GAPC2, respectively. Bars represent means ± S.D. (error bars; n = 3). B, effect of PA on cytosolic/nuclear localization of GAPC. Transgenic Arabidopsis lines overexpressing GAPC2 were grown with (+) and without (−) 100 μm PA for 2 days and fractionated into cytosol and nucleus. The proteins were probed by immunoblotting using anti-FLAG antibody. PEPC1 and histone H3 were used as cytosolic and nuclear protein markers, respectively. Asterisks denote the proteolytic fragment of GAPC2. Molecular masses of the corresponding proteins are indicated on the right. EV, empty vector control; GAPC2, GAPC2 overexpressor. C, dose-dependent proteolytic fragmentation of GAPC2 by PA. Transgenic Arabidopsis line overexpressing GAPC2 was grown with PA for 2 days at different concentrations indicated, and total proteins were probed by immunoblotting using anti-FLAG antibody. The position of proteolytic fragments is indicated by an arrow. D, in vitro assay of GAPC2 proteolysis. E. coli lysate expressing GAPC2 and affinity-purified GAPC2 were incubated with (+) or without (−) 100 μm PA overnight and probed using anti-His6 antibody on the immunoblot.
Applications of PA to Arabidopsis seedlings decreased the amount of intact GAPC2 (66 kDa including tags) in the cytosol, but had no apparent effect on the 66-kDa banding intensity in the nucleus (Fig. 6B). In the PA-treated samples, however, a GAPC2 fragment with a molecular mass of ∼45 kDa including tags was observed in both cytosol and nucleus, and this fragment was detected only when the plants were treated with PA (Fig. 6B). Semiquantification by densitometric analysis revealed that ∼50% of cytosolic GAPC2 and approximately 80% of nuclear GAPC2 were cleaved following PA treatment. The proteolytic cleavage of GAPC2 increased with the increase of PA concentrations applied to Arabidopsis seedlings (Fig. 6C). However, no GAPC cleavage fragment was observed when E. coli protein extracts containing GAPC2 or purified GAPC2 were incubated with PA in vitro (Fig. 6D). These results indicate that the PA-induced cleavage occurs in vivo and requires other effectors for the cleavage.
PA Inhibits Seedling Growth
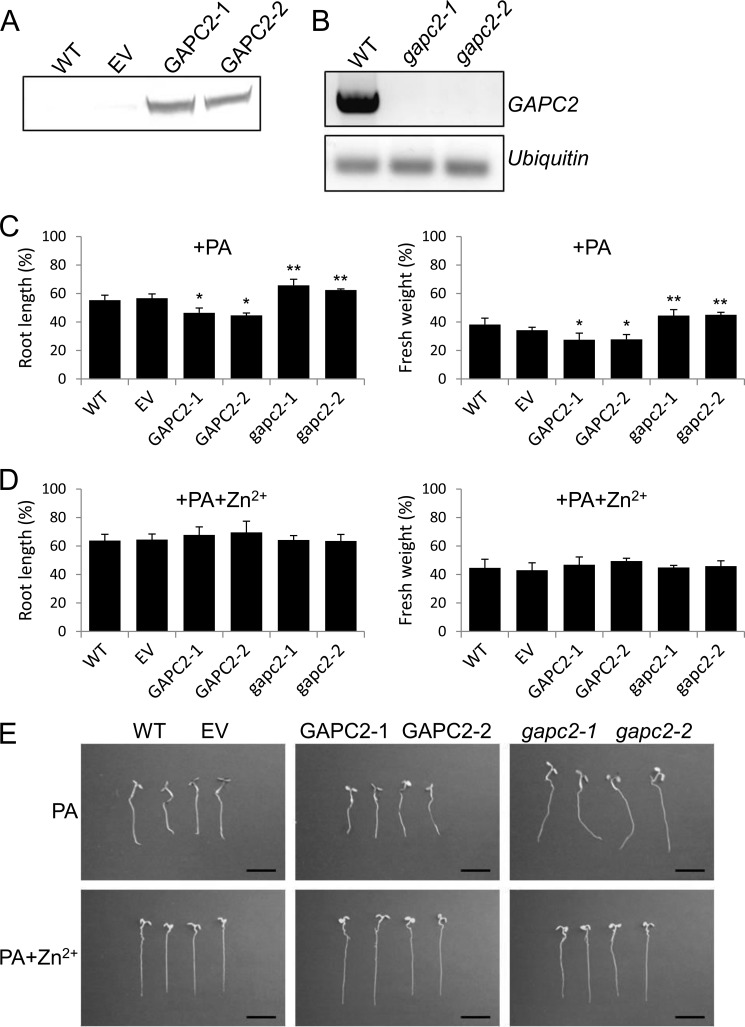
To gain insights into the physiological effect of the PA-GAPC interaction and PA-induced cleavage of GAPC, we determined the effect of PA on seedling growth in GAPC-altered plants, using two GAPC2-overexpressing lines (GAPC2-1 and GAPC2-2) and two homozygous T-DNA insertional mutants with GAPC2 disruption at different positions (gapc2-1 and gapc2-2). Both GAPC2-1 and GAPC2-2 produced GAPC2 protein at the expected size, and gapc2-1 and gapc2-2 both lost GAPC2 transcript, as shown by immunoblotting and RT-PCR analyses, respectively (Fig. 7, A and B). Without added PA, seedlings of GAPC2-altered lines were indistinguishable from either wild-type (WT) or a transgenic line with empty vector (EV; supplemental Fig. S2).
FIGURE 7.
Effect of GAPC2 expression on PA-reduced seedling growth. A, confirmation of GAPC2 overexpression by immunoblotting. The proteins from 10-day-old transgenic seedlings were probed using anti-FLAG antibody. EV, empty vector control. B, confirmation of gapc2 disruption by RT-PCR. Total RNA was extracted and reverse-transcribed. The resulting cDNA was amplified by GAPC2 primers and visualized on an agarose gel. Ubiquitin was used as a loading control. C and D, growth phenotype of the GAPC2-altered lines grown with PA (C) or PA plus Zn2+ (D). Arabidopsis seedlings were grown with 100 μm PA and 1 mm ZnCl2 for 5 days, harvested, and measured in terms of root length (left) and fresh weight (right). Values are percentage of untreated seedlings, indicating the PA suppression of seedling growth. Bars represent means ± S.D. (error bars; n = 10). Single and double asterisks denote a statistical significance from Student's t test paired with EV and WT, respectively (p < 0.01). E, representative images of controls (WT and EV), GAPC2 overexpressors (GAPC2-1 and GAPC2-2), and GAPC2-knock-outs (gapc2-1 and gapc2-2) grown as in C and D. Scale bars, 1 cm.
When seedlings were transferred to a medium containing 100 μm PA for 5 days, however, all lines including WT and EV exhibited shorter roots and reduced seedling biomass (Fig. 7C). GAPC2-1 and GAPC2-2 were significantly more sensitive than EV control to the growth inhibitory effect of PA whereas gapc2-1 and gapc2-2 were less responsive to PA than WT control. This result raised the possibility that the applied PA interacted with overxpressed GAPC2 that contributes to the growth inhibition. To test that, we treated the GAPC2-altered plants with PA and Zn2+ and compared their growth with WT and EV treated identically. We reasoned that because Zn2+ disrupted the PA-GAPC interaction (Fig. 5A), the Zn2+ treatment should alleviate the PA inhibition of seedling growth. Indeed, the application of Zn2+ restored fully the PA-suppressed growth of GAPC2-1 and GAPC2-2 seedlings to that of EV (Fig. 7, D and E). Zn2+-treated WT and EV were slightly less sensitive to PA inhibition than non-zinc-treated seedlings, nearly to the level of the knock-out mutants (Fig. 7, C–E), presumably due to disruption of the interaction between PA and GAPC2 expressed in basal level. More importantly, PA-treated gapc2-1 or gapc2-2 did not noticeably respond to the ion. Zn2+ by itself severely reduced growth of all plants (supplemental Fig. S3A). These results suggest that PA-GAPC2 interaction is involved in seedling growth.
We also tested whether suppression of endogenous PA produced by PLD could have an effect opposite that of the PA treatment on seedling growth. The seedlings of GAPC2-knock-outs and overexpression lines, as well as EV and WT, were treated with n-butyl alcohol that diverts part of PLD-produced PA to phosphatidylbutanol (44). n-Butyl alcohol at 0.4% substantially reduced growth of all lines as reported previously (45), but there was no significant difference among WT and GAPC2-altered seedlings (supplemental Fig. S3B). This result could mean that PLD-produced PA may operate in a manner different from exogenous PA and/or could be explained alternatively as under “Discussion.”
Effect of the PA-GAPC Interaction on Gene Expression
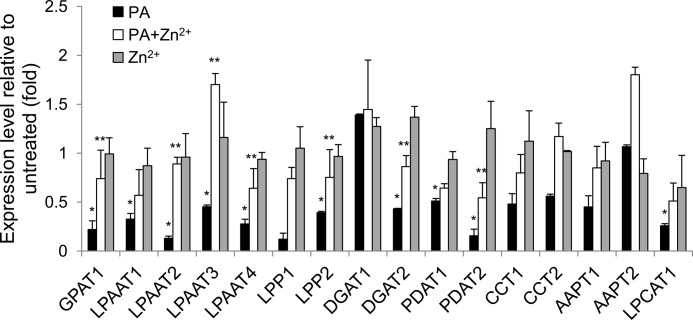
To gain an insight into whether the PA-GAPC interaction is involved in the regulation of gene expression, this study tested several genes involved in PA biosynthesis and degradation. The application of PA substantially reduced the expression of most genes tested, with a lysophosphatidic acid acyltransferase (LPAAT2), a lipid phosphate phosphatase (LPP1), and a phospholipid diacylglycerol acyltransferase (PDAT2) being most down-regulated (<15% of untreated; Fig. 8). PG, another acidic phospholipid, at the same concentration of PA, did not exert as profound effect as PA did on the gene expression (supplemental Fig. S4), indicating that the PA-suppressed gene expression was not due to the pH effect of an acidic lipid and/or nonspecific effect of an excessive exogenous compound. The only exception was LPP1 being more significantly down-regulated by PG than by PA.
FIGURE 8.
Expression of the genes involved in PA metabolism. Total RNA was extracted from seedlings treated with 100 μm PA and/or 1 mm Zn2+ for 2 days and reverse-transcribed. The resulting cDNA was amplified by gene-specific primers (supplemental Table S1) and monitored by qRT-PCR. The gene expression was normalized with ubiquitin 10 as an internal standard. Bars represent means ± S.D. (error bars; n = 3). Single and double asterisks denote a statistical significance from Student's t test paired with PG-treated (see supplemental Fig. S4) and PA-treated samples, respectively (p < 0.01). GPAT, glycerolphosphate acyltransferase; LPP, lipid phosphate phosphatase; DGAT, diacylglycerol acyltransferase; PDAT, phospholipid diacylglycerol acyltransferase; CCT, cholinephosphate cytidyltransferase; AAPT, aminoalcohol phosphotransferase; LPCAT, lysophosphatidylcholine acyltransferase.
Furthermore, we reasoned that if the PA-reduced gene expression is through direct interaction with GAPC, PA should not be able to affect the gene expression when the interaction is disrupted. Thus, the Zn2+ effect on the PA-suppressed gene expression was tested. The gene expression was not significantly affected by Zn2+ only; however, application of Zn2+ together with PA to Arabidopsis seedlings partially recovered the expression of many genes suppressed by the PA treatment (Fig. 8).
DISCUSSION
Results of this study show that PA binds to GAPCs, and the binding is enhanced by oxidation of GAPCs. GAPDH is susceptible to reversible oxidation at a reactive cysteine residue in the active site to lose its catalytic activity and proposed to act as a molecular sensor to perceive the redox sate of the cells (40, 46). A recent study shows that oxidized inactive form of GAPCs interacts directly with and stimulates PLDδ, whose immediate product is PA (29). The GAPC-PLDδ interaction has been implicated in plant response to oxidative stress and water deficits (29). These observations could mean that reduced, active GAPC is involved in the glycolysis for cellular energy production whereas its oxidized, inactive form may be utilized for cell signaling through its interaction with other cellular compounds. PLDδ, together with PLDα1, is known to be involved in ABA signaling (18, 23, 29). The PA produced by oxidized GAPC-enhanced PLDδ activity may associate with GAPC in a feedback loop to modulate the PLDδ activity, possibly by degrading GAPC as described below, to desensitize the ABA signaling machinery (29).
The treatment of PA induces a partial proteolytic cleavage of GAPC2. The PA-induced GAPC2 cleavage did not occur in vitro when tested using either purified protein or E. coli lysate expressing GAPC2, suggesting that the GAPC2 cleavage requires other factors(s) in plants in addition to PA. GAPC was reported to bind to a 14-3-3 protein under sugar-sufficient conditions but lose its binding and undergo a selective partial cleavage in Arabidopsis cells starved of sugars (47). The proteolytic fragment of GAPC was ∼30 kDa which was close to the size of PA-induced proteolytic fragment of GAPC observed here, raising the possibility of whether a similar proteolytic activity is used for both the sugar starvation-induced and PA-mediated proteolytic cleavages of GAPC. In another study, two members of the 14-3-3 protein family were identified as putative PA-binding proteins in a PA affinity pulldown assay (36). These observations may offer a possible mechanism for the PA-induced GAPC cleavage; 14-3-3 proteins may serve as a molecular scaffold converging downstream effectors from sugar- and PA-signaling pathways, where either PA binding or sugar deprivation induces the dissociation of GAPC from 14-3-3 and exposure of the proteolytic cleavage site on GAPC protein.
We found that exogenously added PA suppressed Arabidopsis seedling growth and that overexpression and knock out of GAPC2 had an opposite effect on the PA-reduced seedling growth. GAPC could mediate the PA-triggered growth reduction by direct interaction with PA and transducing growth-slowing signal from PA. This proposition is supported by the finding that Zn2+, which disrupts the PA-GAPC binding, restored the growth of all lines nearly to the levels of knock-out mutants. In the GAPC2-knock-out mutants, this signaling pathway is attenuated by the loss of GAPC-PA interaction, thus rendering the mutant plants less sensitive to the PA-reduced growth than WT. Conversely, the GAPC overexpression may allow to meet the stoichiometry between GAPC and PA molecules so that the GAPC molecules overexpressed can more actively transduce the growth regulatory signal from the excessive PA (100 μm). The GAPC expression-dependent PA inhibition of growth and its restoration by Zn2+-mediated disruption of PA-GAPC interaction clearly support a connection between biochemical events of the PA-GAPC interaction and physiological consequence of the altered growth phenotype.
We used an alcohol treatment in an attempt to corroborate whether PLD-produced PA in plants was involved in the GAPC-mediated effect. Arabidopsis has 12 PLDs, and there are no mutants deficient in all PLDs or inhibitors that are specific to plant PLDs. Primary alcohols, such as n-butyl alcohol, are sometimes used to suppress PLD-produced PA. The alcohol treatment makes use of the PLD transphosphatidylation activity that forms phosphatidylalcohol at the expense of PA. It should be noted that the treatment of primary alcohols to plants often activates PLD, even though it diverts part of the phosphatidyl moiety from PA to phosphatidylalcohol (1). The alcohol-induced increase in phospholipid hydrolysis may have other consequences. Thus, the data with alcohol treatment need to be interpreted with caution. The lack of the opposite effect of the n-butyl alcohol treatment on seedling growth from that of applied PA on GAPC2-overexpressing seedling could result from the nonspecific effect of the alcohol. In addition to PLD, PA can be produced by other routes, such as diacylglycerol kinases and de novo biosynthesis (1). Therefore, the cellular source of PA that interacts with GAPCs requires further investigation.
GAPCs have been regarded as two phosphorylating cytosolic forms of GAPDH, but our results show that GAPC2 and its cleavage product are present both in the cytosol and the nucleus. This raises questions on the function of the nuclear GAPCs. In mammalian cells, GAPDH, in addition to the role as a glycolytic enzyme, has multiple functions, such as the regulation of gene expression, cell signaling, and membrane trafficking (41). GAPDH functions in both transcriptional and posttranscriptional regulations of gene expression through diverse molecular mechanisms. GAPDH is able to regulate gene expression by maintaining DNA integrity in the nucleus and modulating mRNA stability in the cytosol, via protein-nucleic acid interaction, as well as by associating with transcription factors (48–50). Glycolytic activity is not required for many of these novel functions of GAPDH. The present study tested the effect of PA and potential PA-GAPC interaction on the expression of several genes involved in PA biosynthesis and degradation. Treatment with PA inhibited the expression of many genes whereas PA/Zn2+ co-treatment prevented PA inhibition. Among them, LPAAT2 was most responsive to both PA inhibition and Zn2+ restoration. LPAAT is an endomembrane-associated enzyme that catalyzes the synthesis of PA by incorporating an acyl chain to lysoPA. Thus, LPAAT expression may be suppressed by PA as a feedback inhibition, possibly being mediated by GAPC. GAPC is likely to behave in a manner similar to the mammalian system to regulate the gene expression in Arabidopsis cells, and this process may be modulated by the PA binding through the PA-induced proteolysis of GAPC. PA may compromise GAPC protein integrity by inducing proteolysis, possibly through being mediated by 14-3-3 as described, to impede its regulatory activity. Otherwise, the cleavage products of GAPC may have an activity in the regulation of gene expression. The small size of fragments may permit the polypeptides to enter the nucleus where they have influence on the gene expression, which is supported by a slightly more abundance of the fragment in the nucleus than in the cytosol demonstrated in this study. Alternatively, the cytosolic fragment of GAPC may regulate the gene expression posttranscriptionally, possibly by modulating mRNA stability in the cytosol as mammalian GAPDH does. It will be of interest in future studies to elucidate the detailed mechanism for the alteration of GAPC regulatory function by PA binding.
We propose that PA physically interacts with oxidized, inactive GAPC to induce proteolysis of the protein. GAPC catalyzes a reaction in the glycolytic pathway important for energy production and for providing metabolic intermediates for other biosynthetic processes, including lipids. PA is a key intermediate in glycerolipid biosynthesis. The identification of GAPC as a PA-binding protein in Camelina and Arabidopsis raises an intriguing question of whether the PA-GAPC interaction provides a metabolic link coordinating carbohydrate and lipid metabolism. It will be of great interest in further studies to determine whether and how the PA-GAPC interaction plays a role in plant metabolism, biomass accumulation, and stress response.
Addendum
While the work described in the paper was submitted for publication, another study screening for PA-binding proteins under NaCl stress in Arabidopsis also identified GAPC interacting with PA (51).
This work was supported primarily by the Center for Advanced Biofuels Systems, an Energy Frontier Research Center funded by the United States Department of Energy, Office of Science, Basic Energy Sciences, under Award DE-SC0001295 (lipid-protein interactions) and by National Science Foundation Award 0818740 (mutant isolation).

This article contains supplemental Figs. S1–S4 and Table S1.
- PA
- phosphatidic acid
- ABA
- abscisic acid
- EV
- empty vector
- GAPC
- cytosolic glyceraldehyde-3-phosphate dehydrogenase
- LPAAT
- lysophosphatidic acid acyltransferase
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PEPC
- phosphoenolpyruvate carboxylase
- PG
- phosphatidylglycerol
- PLD
- phospholipase D
- PS
- phosphatidylserine
- qRT-PCR
- quantitative real-time PCR
- RU
- response unit
- SPHK
- sphingosine kinase
- SPR
- surface plasmon resonance.
REFERENCES
- 1. Wang X., Devaiah S. P., Zhang W., Welti R. (2006) Signaling functions of phosphatidic acid. Prog. Lipid Res. 45, 250–278 [DOI] [PubMed] [Google Scholar]
- 2. Li M., Hong Y., Wang X. (2009) Phospholipase D- and phosphatidic acid-mediated signaling in plants. Biochim. Biophys. Acta 1791, 927–935 [DOI] [PubMed] [Google Scholar]
- 3. Testerink C., Munnik T. (2011) Molecular, cellular, and physiological responses to phosphatidic acid formation in plants. J. Exp. Bot. 62, 2349–2361 [DOI] [PubMed] [Google Scholar]
- 4. Jacob T., Ritchie S., Assmann S. M., Gilroy S. (1999) Abscisic acid signal transduction in guard cells is mediated by phospholipase D activity. Proc. Natl. Acad. Sci. U.S.A. 96, 12192–12197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Munnik T., Meijer H. J., Ter Riet B., Hirt H., Frank W., Bartels D., Musgrave A. (2000) Hyperosmotic stress stimulates phospholipase D activity and elevates the levels of phosphatidic acid and diacylglycerol phosphate. Plant J. 22, 147–154 [DOI] [PubMed] [Google Scholar]
- 6. Katagiri T., Takahashi S., Shinozaki K. (2001) Involvement of a novel Arabidopsis phospholipase D, AtPLDδ, in dehydration-inducible accumulation of phosphatidic acid in stress signaling. Plant J. 26, 595–605 [DOI] [PubMed] [Google Scholar]
- 7. Hong Y., Zhang W., Wang X. (2010) Phospholipase D and phosphatidic acid signaling in plant response to drought and salinity. Plant Cell Environ. 33, 627–635 [DOI] [PubMed] [Google Scholar]
- 8. Zhang W., Wang C., Qin C., Wood T., Olafsdottir G., Welti R., Wang X. (2003) The oleate-stimulated phospholipase D, PLDδ, and phosphatidic acid decrease H2O2-induced cell death in Arabidopsis. Plant Cell 15, 2285–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van der Luit A. H., Piatti T., van Doorn A., Musgrave A., Felix G., Boller T., Munnik T. (2000) Elicitation of suspension-cultured tomato cells triggers the formation of phosphatidic acid and diacylglycerol pyrophosphase. Plant Physiol. 123, 1507–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Raho N., Ramirez L., Lanteri M. L., Gonorazky G., Lamattina L., ten Have A., Laxalt A. M. (2011) Phosphatidic acid production in chitosan-elicited tomato cells, via both phospholipase D and phospholipase C/diacylglycerol kinase, requires nitric oxide. J. Plant Physiol. 168, 534–539 [DOI] [PubMed] [Google Scholar]
- 11. Ruelland E., Cantrel C., Gawer M., Kader J. C., Zachowski A. (2002) Activation of phospholipase C and D is an early response to a cold exposure in Arabidopsis suspension cells. Plant Physiol. 130, 999–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Welti R., Li W., Li M., Sang Y., Biesiada H., Zhou H. E, Rajashekar C. B., Williams T. D., Wang X. (2002) Profiling membrane lipids in plant stress responses: role of phospholipase Dα in freezing-induced lipid changes in Arabidopsis. J. Biol. Chem. 277, 31994–32002 [DOI] [PubMed] [Google Scholar]
- 13. Gómez-Merino F. C., Brearley C. A., Ornatowska M., Abdel-Haliem M. E., Zanor M. I., Mueller-Roeber B. (2004) AtDGK2, a novel diacylglycerol kinase from Arabidopsis thaliana, phosphorylates 1-stearoyl-2-arachidonoyl-sn-glycerol and 1,2-dioleoyl-sn-glycerol and exhibits cold-inducible gene expression. J. Biol. Chem. 279, 8230–8241 [DOI] [PubMed] [Google Scholar]
- 14. Ohashi Y., Oka A., Rodrigues-Pousada R., Possenti M., Ruberti I., Morelli G., Aoyama T. (2003) Modulation of phospholipid signaling by GLABRA2 in root-hair pattern formation. Science 300, 1427–1430 [DOI] [PubMed] [Google Scholar]
- 15. Li G., Xue H. W. (2007) Arabidopsis PLDζ2 regulates vesicle trafficking and is required for auxin response. Plant Cell 19, 281–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Potocký M., Eliás M., Profotová B., Novotná Z., Valentová O., Zárský V. (2003) Phosphatidic acid produced by phospholipase D is required tobacco pollen tube growth. Planta 217, 122–130 [DOI] [PubMed] [Google Scholar]
- 17. Monteiro D., Liu Q., Lisboa S., Scherer G. E., Quader H., Malhó R. (2005) Phosphoinositides and phosphatidic acid regulate pollen tube growth and reorientation through modulation of [Ca2+]c and membrane secretion. J. Exp. Bot. 56, 1665–1674 [DOI] [PubMed] [Google Scholar]
- 18. Zhang W., Qin C., Zhao J., Wang X. (2004) Phospholipase Dα1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc. Natl. Acad. Sci. U.S.A. 101, 9508–9513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Testerink C., Larsen P. B., van der Does D., van Himbergen J. A., Munnik T. (2007) Phosphatidic acid binds to and inhibits the activity of Arabidopsis CTR1. J. Exp. Bot. 58, 3905–3914 [DOI] [PubMed] [Google Scholar]
- 20. Jost R., Berkowitz O., Shaw J., Masle J. (2009) Biochemical characterization of two wheat phosphoethanolamine N-methyltransferase isoforms with different sensitivities to inhibition by phosphatidic acid. J. Biol. Chem. 284, 31962–31971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Anthony R. G., Henriques R., Helfer A., Mészáros T., Rios G., Testerink C., Munnik T., Deák M., Koncz C., Bögre L. (2004) A protein kinase target of a PDK1 signaling pathway is involved in root hair growth in Arabidopsis. EMBO J. 23, 572–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guo L., Mishra G., Taylor K., Wang X. (2011) Phosphatidic acid binds and stimulates Arabidopsis sphingosine kinases. J. Biol. Chem. 286, 13336–13345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Y., Zhu H., Zhang Q., Li M., Yan M., Wang R., Wang L., Welti R., Zhang W., Wang X. (2009) Phospholipase Dα1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell 21, 2357–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu L., Nie J., Cao C., Jin Y., Yan M., Wang F., Liu J., Xiao Y., Liang Y., Zhang W. (2010) Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 188, 762–773 [DOI] [PubMed] [Google Scholar]
- 25. Himmelbach A., Hoffmann T., Leube M., Höhener B., Grill E. (2002) Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J. 21, 3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Loewen C. J., Gaspar M. L., Jesch S. A., Delon C., Ktistakis N. T., Henry S. A., Levine T. P. (2004) Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science 304, 1644–1647 [DOI] [PubMed] [Google Scholar]
- 27. Carman G. M., Henry S. A. (2007) Phosphatidic acid plays a central role in the transcriptional regulation of glycerophospholipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 282, 37293–37297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ohlrogge J., Browse J. (1995) Lipid biosynthesis. Plant Cell 7, 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guo L., Devaiah S. P., Narasimhan R., Pan X., Zhang Y., Zhang W., Wang X. (2012) Cytosolic glyceraldehyde-3-phosphate dehydrogenases interact with phospholipase Dδ to transducer hydrogen peroxide signals in the Arabidopsis response to stress. Plant Cell 24, 2200–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stevenson J. M., Perera I. Y., Boss W. F. (1998) A phosphatidylinositol 4-kinase pleckstrin homology domain that binds phosphatidylinositol 4-monophosphate. J. Biol. Chem. 273, 22761–22767 [DOI] [PubMed] [Google Scholar]
- 31. Wessel D., Flügge U. I. (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 [DOI] [PubMed] [Google Scholar]
- 32. Rius S. P., Casati P., Iglesias A. A., Gomez-Casati D. F. (2008) Characterization of Arabidopsis lines deficient in GAPC-1, a cytosolic NAD-dependent glyceraldehyde-3-phosphate dehydrogenase. Plant Physiol. 148, 1655–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu X. L., Covington M. F., Fankhauser C., Chory J., Wagner D. R. (2001) ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13, 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hutcheon C., Ditt R. F., Beilstein M., Comai L., Schroeder J., Goldstein E., Shewmaker C. K., Nguyen T., De Rocher J., Kiser J. (2010) Polyploid genome of Camelina sativa revealed by isolation of fatty acid synthesis genes. BMC Plant Biol. 10, 233–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stace C. L., Ktistakis N. T. (2006) Phosphatidic acid- and phosphatidylserine-binding proteins. Biochim. Biophys. Acta 1761, 913–926 [DOI] [PubMed] [Google Scholar]
- 36. Testerink C., Dekker H. L., Lim Z. Y., Johns M. K., Holmes A. B., Koster C. G., Ktistakis N. T., Munnik T. (2004) Isolation and identification of phosphatidic acid targets from plants. Plant J. 39, 527–536 [DOI] [PubMed] [Google Scholar]
- 37. Barr F. A., Shorter J. (2000) Membrane traffic: do cones mark sites for fission? Curr. Biol. 10, R141–144 [DOI] [PubMed] [Google Scholar]
- 38. Lemmon M. A., (2008) Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9, 99–111 [DOI] [PubMed] [Google Scholar]
- 39. Dineley K. E., Votyakova T. V., Reynolds I. J. (2003) Zinc inhibition of cellular energy production: implications for mitochondria and neurodegeneration. J. Neurochem. 85, 563–570 [DOI] [PubMed] [Google Scholar]
- 40. Sirover M. A. (1999) New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim. Biophys. Acta 1432, 159–184 [DOI] [PubMed] [Google Scholar]
- 41. Sirover M. A. (2011) On the functional diversity of glyceraldehyde-3-phosphate dehydrogenase: biochemical mechanisms and regulatory control. Biochim. Biophys. Acta 1810, 741–751 [DOI] [PubMed] [Google Scholar]
- 42. Wang Q., Guo L., Sjolund R., Shih M. C. (1997) Immunolocalization of glyceraldehyde-3-phosphate dehydrogenase in Arabidopsis thaliana. Protoplasma 198, 155–162 [Google Scholar]
- 43. Holtgrefe S., Gohlke J., Starmann J., Druce S., Klocke S., Altmann B., Wojtera J., Lindermayr C., Scheibe R. (2008) Regulation of plant cytosolic glyceraldehyde-3-phosphate dehydrogenase isoforms by thiol modifications. Physiol. Plant. 133, 211–228 [DOI] [PubMed] [Google Scholar]
- 44. Munnik T., Arisz S. A., De Vrije T., Musgrave A. (1995) G protein activation stimulates phospholipase D signaling in plants. Plant Cell 7, 2197–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gardiner J., Collings D. A., Harper J. D., Marc J. (2003) The effects of the phospholipase D-antagonist 1-butanol on seedling development and microtubule organization in Arabidopsis. Plant Cell Physiol. 44, 687–696 [DOI] [PubMed] [Google Scholar]
- 46. Hara M. R., Cascio M. B., Sawa A. (2006) GAPDH as a sensor of NO stress. Biochim. Biophys. Acta 1762, 502–509 [DOI] [PubMed] [Google Scholar]
- 47. Cotelle V., Meek S. E., Provan F., Milne F. C., Morrice N., MacKintosh C. (2000) 14-3-3s regulate global cleavage of their diverse binding partners in sugar-starved Arabidopsis cells. EMBO J. 19, 2869–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Demarse N. A., Ponnusamy S., Spicer E. K., Apohan E., Baatz J. E., Ogretmen B., Davies C. (2009) Direct binding of glyceraldehyde-3-phosphate dehydrogenase to telomeric DNA protects telomeres against chemotherapy-induced rapid degradation. J. Mol. Biol. 394, 789–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhou Y., Yi X., Stoffer J. B., Bonafe N., Gilmore-Hebert M., McAlpine J., Chambers S. K. (2008) The multifunctional protein glyceraldehyde-3-phosphate dehydrogenase is both regulated and controls colony-stimulating factor-1 messenger RNA stability in ovarian cancer. Mol. Cancer Res. 6, 1375–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Harada N., Yasunaga R., Higashimura Y., Yamaji R., Fujimoto K., Moss J., Inui H., Nakano Y. (2007) Glyceraldehyde-3-phosphate dehydrogenase enhances transcriptional activity of androgen receptor in prostate cancer cells. J. Biol. Chem. 282, 22651–22661 [DOI] [PubMed] [Google Scholar]
- 51. McLoughlin F., Arisz S. A., Dekker H. L., Kramer G., de Koster C. G., Haring M. A., Munnik T., Testerink C. (2013) Identification of novel candidate phosphatidic acid-binding proteins involved in the salt-stress response of Arabidopsis thaliana roots. Biochem. J. 450, 573–581 [DOI] [PubMed] [Google Scholar]