Background: The C terminus of prosecretory mitogen lacritin targets the first 50 amino acids of syndecan-1 in a heparanase-dependent manner.
Results: The amphipathic α-helix of lacritin ligates the sequence Gly-Ala-Gly-Ala-Leu and N-terminal chondroitin and heparan sulfate chains of SDC1.
Conclusion: Ligation requires all three binding elements.
Significance: This hybrid binding domain helps explain the remarkable cell selectivity of lacritin and may have relevance in dry eye.
Keywords: Cornea, Epithelium, Eye, Glycobiology, Glycosaminoglycan, Heparan Sulfate, Protein Complexes, Protein Domains, Proteoglycan, Receptors
Abstract
Cell surface heparan sulfate (HS) proteoglycans shape organogenesis and homeostasis by capture and release of morphogens through mechanisms largely thought to exclude the core protein domain. Nevertheless, heparanase deglycanation of the N-terminal HS-rich domain of syndecan-1 (SDC1), but not SDC2 or -4, is a prerequisite for binding of the prosecretory mitogen lacritin (Ma, P., Beck, S. L., Raab, R. W., McKown, R. L., Coffman, G. L., Utani, A., Chirico, W. J., Rapraeger, A. C., and Laurie, G. W. (2006) Heparanase deglycanation of syndecan-1 is required for binding of the epithelial-restricted prosecretory mitogen lacritin. J. Cell Biol. 174, 1097–1106). We now report that the conserved and hydrophobic GAGAL domain in SDC1, adjacent to predicted HS substitution sites, is necessary to ligate and substantially enhance the α-helicity of the amphipathic C terminus of lacritin. Swapping out GAGAL for GADED in SDC2 or for GDLDD in SDC4 (both less hydrophobic) abrogated binding. HS and chondroitin sulfate are also essential. Both are detected in the N terminus, and when incubated with antibodies HS4C3 (anti-HS) or IO3H10 (anti-chondroitin sulfate), binding was absent, as occurred when all three N-terminal glycosaminoglycan substitution sites were mutated to alanine or when cells were treated with 4-methylumbelliferyl-β-d-xylopyranoside or chlorate to suppress glycosaminoglycan substitution or sulfation, respectively. SDC1 interacts with the hydrophobic face of lacritin via Leu-108/Leu-109/Phe-112 as well as with Glu-103/Lys-107 and Lys-111 of the largely cationic face. Carving a hybrid hydrophobic/electrostatic docking site out of SDC1 in a manner dependent on endogenous heparanase is a dynamic process appropriate for subtle or broad epithelial regulation in morphogenesis, health, and disease.
Introduction
Cell surface proteoglycans are carbohydrate-rich regulators of epithelial morphogenesis that change cell behavior by binding or stabilizing soluble growth factors, cytokines, extracellular matrix, and signaling receptors. Consisting of a ∼200–850-amino acid core protein to which are attached long anionic glycosaminoglycan side chains, largely heparan sulfate (HS),3 the cell surface proteoglycans of the syndecan (SDC1) and glypican families are complex multidomain structures that have shaped metazoan evolution (1).
Many of their activities have been historically attributed to HS, a glucuronic acid/N-acetylglucosamine-rich polymer generated by stepwise Golgi biosynthetic and modifying catalysis with enzymes arranged in a quaternary complex (2, 3). However, examples of core protein-driven activities have now emerged, most dramatically in fly morphogenesis (4–7), but also in mouse (8, 9) and cell systems (10–15).
Recently, we described a novel mechanism whereby ligation of the SDC1 core protein by lacritin (16, 17) requires heparanase deglycanation (18). Lacritin is a prosecretory mitogen (16, 17) whose discovery emerged from a functional screen for exocrine differentiation and prosecretory factors (16). Expression is abundant in lacrimal and salivary glands (16), with reports in breast cancer (19, 20). Lacritin is a secreted glycoprotein of ∼25 kDa in tears and saliva with homology to dermcidin, a proposed breast oncogene (20) whose N-terminal half displays affinity for HS of SDC1 (21). The two genes are adjacent on chromosome 12q13 in human. Lacritin promotes constitutive tear secretion by lacrimal acinar cells (16) and basal tearing in normal rabbits (22), although it is itself a tear protein and appears to be selectively down-regulated in tears of patients suffering from dry eye (23), the most common eye disease (24, 25). Lacritin mitogenic (16) signaling is initiated within seconds via Gαi or Gαo/PKCα-PLC/Ca2+/calcineurin/NFATC1 and Gαi or Gαo/PKCα-PLC/PLD/mTOR pathways (17), implying an SDC1 complex probably inclusive of a G-protein-coupled receptor.
The involvement of SDC1 was discovered as a consequence of a screen of cell surface-biotinylated proteins from a lacritin-responsive cell. The 1 m NaCl eluant from lacritin columns was dominated by a discrete higher molecular weight band that was identified as SDC1 by mass spectrometry (18). HS chains cleaved with NaBH4 from lacritin-bound SDC1 were ∼4–5 kDa, versus ∼40 kDa for SDC1 purified on FGF2 (18). Short HS chains were non-existent in cells subjected to heparanase depletion by siRNA, and depleted cells failed to proliferate in the presence of lacritin but could be rescued by exogenous heparanase or heparitinase (18). Similarly, siRNA depletion of SDC1, but not SDC2, abrogated lacritin-dependent proliferation in a dose-dependent manner (18). No lacritin binding was observed to SDC2 or -4, and SDC1 bound to lacritin was resolved in the pellet after digestion with heparitinase I and chondroitin ABC lyase, suggesting that (i) short HS chains were necessary (or long chains obscured the binding site) and (ii) binding probably involved the SDC1 core protein (18). Truncation analysis narrowed mutual binding to the N-terminal 50 amino acids of SDC1 and to an α-helical region within the 15 C-terminal amino acids of lacritin (18).
Here we provide evidence for a hybrid binding site involving three essential elements: (i) the hydrophobic and conserved GAGAL sequence in the SDC1 N terminus that promotes α-helicity of the lacritin amphipathic C terminus, probably by interacting with lacritin residues Leu-108/Leu-109/Phe-112, without which no binding occurs, and (ii) HS proximal to GAGAL, probably as heparanase-modified stubs (18), that together with (iii) co-substituted chondroitin sulfate (CS) in the N terminus of SDC1 may bind required Glu-103/Lys-107 and Lys-111 on the largely cationic face of lacritin. This heparanase-dependent and hybrid hydrophobic/electrostatic docking site thus appropriates a widely expressed HS proteoglycan and transforms it into a lacritin-selective binding protein.
EXPERIMENTAL PROCEDURES
Cell Culture, Plasmid Constructs, and Transfection
HEK293-EBNA1 (293E) cells (26) were kindly provided by Yves Durocher (National Research Council, Montreal, Canada) and both cultured and transiently transfected as described (26) for suspension culture expression. Suspension culture expression avoids cellular adhesion problems associated with manipulation of SDC1. For this purpose, hS1-pcDNA3 (18) was subcloned into pTT5 (26) (hS1-pTT5) using HindIII and BamHI sites generated via DNA forward primer 5′-CTGAAAGCTTATGAGGCGCGCGGCGCTCTGG-3′ and reverse primer 5′-CAGGATCCTCAGGCATAGAATTCCTCCTGTTTGGTGGG-3′. hS1-pTT5 was transiently transfected into poorly adhesive 293E cells using linear polyethyleneimine (25-kDa Linear, powder; Polysciences Inc., Warrington, PA). Transfected and normal 293E cells were propagated in suspension by continuous rotation (125 rpm) in glycol-modified polyethylene terephthalate (PETG) flasks (Nalgene, Rochester, NY) containing F17 medium (05-0092DK, Invitrogen) supplemented with 4 mm l-glutamine and 0.1% of Pluronic F-68.
Numbering of SDC1 and lacritin constructs excludes the signal peptide, whose location was defined by SignalP version 4.1. Human SDC1 deletion constructs lacking 20 or 30 amino acids from the N terminus of mature SDC1 (del 1–20 or 1–30), respectively, were generated from hS1-pTT5 by long range reverse PCR (see primers in supplemental Table 1). Human SDC1 double point mutants S15A/23A, S15A/25A, S184A/S194A, and triple point mutant S15A/S23A/S25A were generated from hS1-pTT5 using the QuikChange site-directed mutagenesis kit (Stratagene/Agilent Technologies, Santa Clara CA) (supplemental Table 1).
Two human SDC1-swapping constructs were developed from hS1-pTT5 by replacing the GAGAL sequence (amino acids 26–30) with the corresponding regions GADED and GDLDD from human SDC2 (amino acids 40–44) and SDC4 (amino acids 46–50), respectively. A two-step process was used for each (supplemental Table 1). Plasmids were then sequenced and transfected into suspension 293E cells, and transient transfectants were developed.
Development of human lacritin-intein (pLAC) and lacritin C-25-intein constructs was described previously (17). N-terminal lacritin deletions of 24, 35, 45, 55, 65, 71, and 75 amino acids were developed from human pLAC (see primers in supplemental Table 2).
Human lacritin point mutants K66S, I68S, V69S, E70S, I73S, L74S, L75S, V91S, I98S, G101S, F104S, L108S, L109S, K110S, K111S, F112S, I68S/I73S, V91S/L98S, V91S/L109S, E103S/K107S, and L108S/L109S/F112S were generated from pLAC using the QuikChange site-directed mutagenesis kit (Stratagene/Agilent Technologies) (supplemental Table 2). All constructs were confirmed by DNA sequencing. Bacterially expressed lacritin-intein and mutants were enriched on chitin columns, as detailed by Wang et al. (17), and further purified on DEAE. An FGF2-GST construct kindly provided by William J. Chirico (State University of New York, Brooklyn, NY) was subcloned into pTYB1 (New England Biolabs, Inc., Ipswich, MA) with an intein tag and bacterially expressed (17).
Synthetic Peptides
Peptides corresponding to human SDC1 amino acids 1–20 (Pep1–20, QIVATNLPPEDQDGSGDDSD), 10–30 (Pep10–30, DQDGSGDDSDNFSGSGAGAL), 20–40 (Pep20–40, NFSGSGAGALQDITLSQQTP), 20–30 (Pep20–30, NFSGSGAGAL), 19–30 (Pep19–30, DNFSGSGAGAL), and 30–50 (Pep30–50, QDITLSQQTPSTWKDTQLLT) as well as scrambled 10–30 (PepScram10–30, FDGADSSSLGQGGDSGDAND) and 19–30 (PepScram19–30, AGFGLSNSADG) were synthesized by GeneScript USA Inc. (Piscataway, NJ), 95% purified, lyophilized, and aliquoted. Also synthesized were SDC2 amino acids 34–43 (SDC2 Pep34–43, SASGSGADED), SDC4 amino acids 41–50 (SDC4 Pep41–50, ELSGSGDLDD), and lacritin C-terminal amino acids 95–119 (LacPep95–119, KQFIENGSEFAQKLLKKFSLLKPWA). Each was N-terminally acetylated and C-terminally amidated, with the exception of C-terminal LacPep95–119. Residue 19 (Asp) in PepScram19–30 was included to increase solubility.
Modification and Expression of SDC1s and Affinity Precipitation
Transiently expressed human SDC1 from suspension-cultured 293E cells and human SDC1 deletion and point mutants from 293E cells were harvested on ice (18), solubilized in 1 ml of lysis buffer (50 mm Tris-HCl, pH 7.4, 100 mm NaCl, 5 mm MnCl2, 2 mm PMSF, 200 mm n-octyl-β-d-glucopyranoside, and protease inhibitors (Roche Applied Science)) for 30 min, and centrifuged at 18,407 × g for 20 min at 4 ºC. Cleared supernatants (referred to in the figures as the “input lysate”) were then aliquoted and stored at −80 ºC or applied immediately to affinity precipitation. Some hS1-pTT5-transfected 293E cells were cultured for several days in medium containing 30, 100, or 300 mm 4-methylumbelliferyl-β-d-xylopyranoside (xyloside; 30, 100, 300 mm) to suppress glycosaminoglycan assembly (both heparan and chondroitin sulfate) (27) and then lysed. Lysates were also collected from hS1-pTT5-transfected 293E cells grown in sodium chlorate (25, 50, or 100 mm)-supplemented F17 medium lacking sulfates and sulfur (with the exception of l-cysteine; Invitrogen) to suppress sulfation (28). Lacritin-intein, lacritin-intein truncation or point mutant, or FGF2-intein fusion proteins were bound to chitin beads. Beads were incubated overnight (4 ºC) with SDC1-enriched lysates (18) in a final binding buffer volume of 500 μl and then washed three times (each wash with 30 times the bead volume) with the same buffer.
In competition assays, SDC1 lysates were incubated with increasing amounts of Pep1–20, Pep10–30, Pep20–40, Pep20–30, Pep30–50, PepScram10–30, or PepScram19–30 (each reconstituted in binding buffer). In other competition assays, SDC1 lysates were incubated with increasing concentrations of anti-HS single chain variable fragment antibodies HS4C3, MPB49, AO4B08, and IO3H10 that had been purified on Protein A from the periplasmic fraction (29). SDC1 lysates with competitors were then added to lacritin immobilized on beads and further analyzed. Syndecan-1 pulled down by lacritin(s) or FGF2 was digested for 18–24 h with heparitinase I (Seikagaku America) and chondroitin ABC lyase (Sigma) at 0.0001 and 0.005 units, respectively (18), and centrifuged. Lacritin-bound SDC1 resides in the pellet, whereas FGF2-bound SDC1 is in the supernatant (18). Pellets of lacritin affinity precipitates or, in some cases, both pellets of lacritin and supernatants of FGF2 affinity precipitates were separated by SDS-PAGE and immunoblotted using anti-human SDC1 mAb B-B4 (Serotec), as previously described (18) with minor changes. B-B4 detects core protein epitope LPEV (amino acids 85–88 of mature SDC1) (30). LPEV is not affected by any of the introduced mutations. Blotting was also performed with mAb 3G10 (kindly provided by G. David, KULeuven, Leuven Belgium) with specificity for desaturated uronates in heparitinase-cleaved HS (31) and with mAb 2030, which targets chondroitin-4 sulfate in chondroitinase ABC-cleaved CS (Millipore, Billerica, MA). In some cases, affinity precipitations were performed in radioimmune precipitation assay buffer (Cell Signaling Technology, Danvers, MA) with pellets and supernatants separated and blotted as above. All experiments were performed at least three times.
Circular Dichroism of Lacritin C-terminal Peptide without and with SDC1, SDC2, or SDC4 Peptides
Lacritin LacPep95–119 (81 μm) in 10 mm dodecylphosphocholine without or with 57 μm SDC1 Pep20–30, SDC1 Pep19–30, SDC1 ScrambPep19–30, SDC2 Pep34–43, or SDC4 Pep41–50 was subjected to circular dichroism using a Jasco 710 spectropolarimeter (1-mm path length quartz cuvette) at room temperature. The background contribution of solvent and SDC1, SDC2, or SDC4 peptide was subtracted to obtain the spectra plots displayed that are presented as the mean ± S.E. Data were analyzed by the paired two-tailed t test.
RESULTS
Lacritin Targets SDC1 Amino Acids 20–30
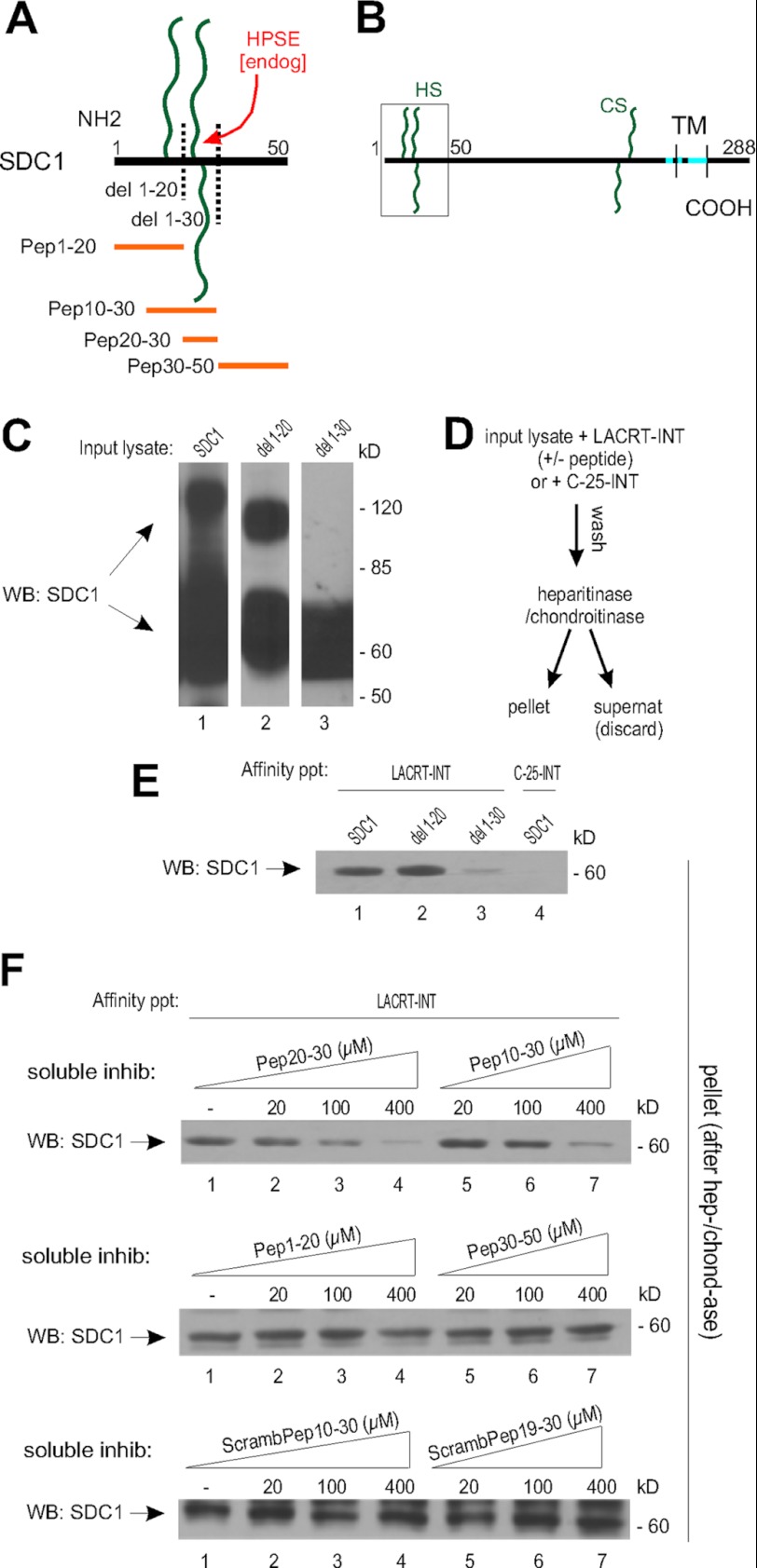
Lacritin mitogenic activity is dependent on binding a subpopulation of cell surface SDC1 (but not SDC2 or -4) molecules with heparanase-shortened heparan sulfate chains (18). Mutual binding elements appear to be contained within an amphipathic α-helix in the C terminus of lacritin and within the N-terminal 50 amino acids of SDC1 (18) (Fig. 1, A and B). To narrow the site, we generated 293E suspension cultures transiently expressing native SDC1 or SDC1 lacking 20 (del 1–20) or 30 (del 1–30) N-terminal amino acids (Fig. 1C). As previously observed, SDC1 blotting can include a high molecular weight multimer whose variable formation was independent of lacritin or FGF2 targeting (18). We incubated the precipitates with heparitinase and chondroitinase (Fig. 1D) to remove the variably long heparan and chondroitin sulfate chains, with many of the heparan sulfate chains shortened by endogenous heparanase (18). The pellet of the digest was then blotted for SDC1 core protein using mAb B-B4. Lacritin bound SDC1 and del 1–20 but not del 1–30 (Fig. 1E), implicating a contribution by amino acids 20–30. No affinity was displayed by lacritin “C-25” lacking the C-terminal amphipathic α-helix (Fig. 1E) (18).
FIGURE 1.
Lacritin binding requires SDC1 N-terminal amino acids 20–30. A, linear diagram of the human SDC1 N terminus with glycosaminoglycan chains; truncation mutants del 1–20 and del 1–30; and synthetic peptides Pep1–20, Pep10–30, Pep20–30, and Pep30–50. B, schematic diagram of human SDC1 with boxed N terminus. TM, transmembrane domain. C, lysates from HEK293E cells transiently expressing SDC1, SDC1 del 1–20, and SDC1 del 1–30 were blotted (WB) with mAb B-B4 for SDC1 core protein. D, affinity precipitation scheme. E, lacritin-intein beads were incubated with lysates from HEK293E cells transiently expressing SDC1, SDC1 del 1–20, or SDC1 del 1–30. Lacritin C-25-intein (lacking SDC1 binding domain) beads were incubated with lysates from HEK293E cells transiently expressing SDC1. After incubation, beads were washed extensively and treated with heparitinase I/chondroitinase ABC (hep-/chond-ase). Digests were centrifuged, and the pellets were blotted with mAb B-B4 for SDC1 core protein. F, lacritin-intein beads were incubated with lysates from HEK293E cells transiently expressing SDC1 in the presence of increasing amounts of SDC1 Pep20–30, Pep10–30, Pep1–20, Pep30–50, ScrambPep10–30, or ScrambPep19–30. After incubation, beads were thoroughly washed and treated with heparitinase I/chondroitinase ABC. Digests were centrifuged, and the pellets were blotted with mAb B-B4 for SDC1 core protein.
We then challenged lacritin affinity precipitations with synthetic peptides together spanning this region (Fig. 1A). Pep10–30 (amino acids 10–30) and Pep20–30 (amino acids 20–30), but not Pep1–20 (amino acids 1–20) or Pep30–50 (amino acids 30–50) inhibited binding. Most inhibitory was Pep20–30, whereas randomly scrambled Pep10–30 and Pep19–30 had no effect (Fig. 1F). Collectively, these data suggest that correctly ordered amino acids 20–30 (NFSGSGAGAL) in the N terminus of SDC1 are necessary for lacritin binding. Interestingly, NFSGSGAGAL serves as an important region for glycosaminoglycan attachment at serines 23 and 25 (32). Glycosaminoglycan steric hindrance might therefore in part explain the requirement of heparanase or heparitinase digestion (18) for lacritin binding.
Mutual Hydrophobic Binding Specified by GAGAL
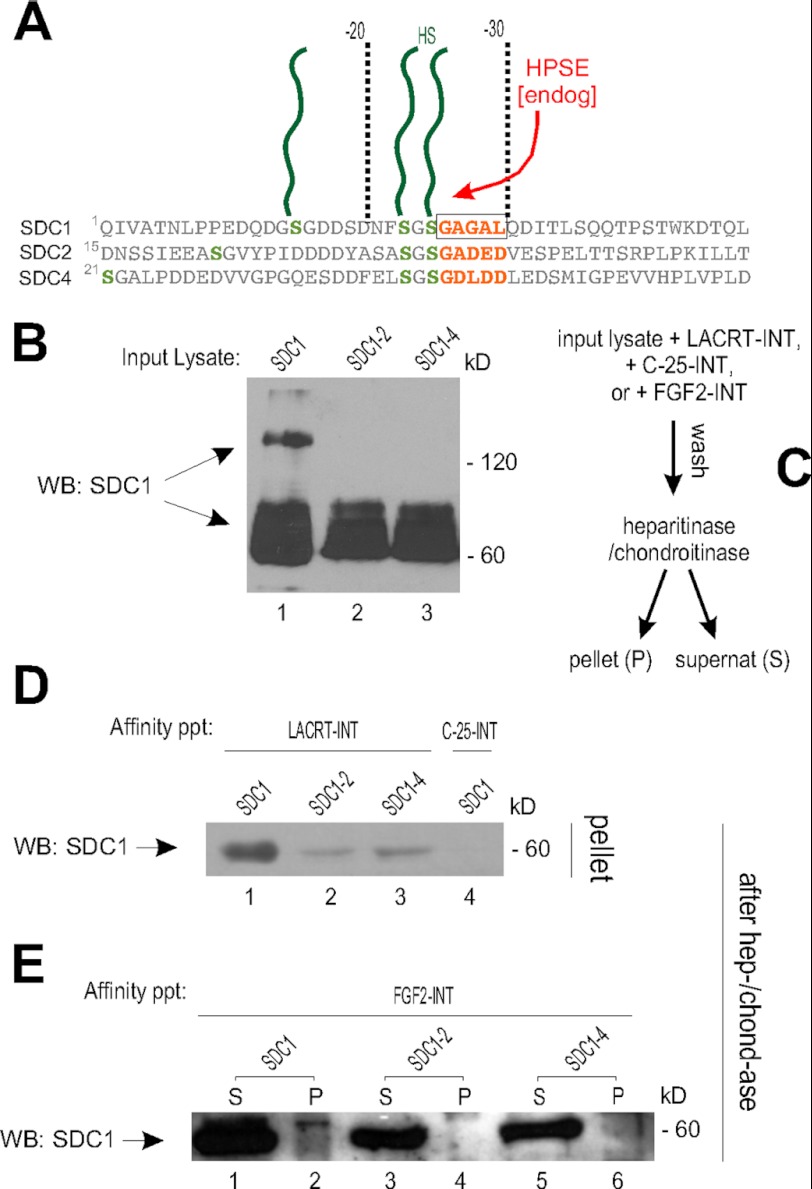
Because lacritin targets SDC1, but not SDC2 or -4 (18), we wondered whether the corresponding sequence in SDC2 or -4 differed, in keeping with low ectodomain identity among syndecans (Fig. 2). SDC1 serines 23 and 25 are followed by GAGAL (Fig. 2A). The homologous serines in SDC2 and -4 are followed by the less hydrophobic GADED and GDLDD, respectively (Fig. 2A). GAGAL was swapped out for GADED or GDLDD. Replacing GAGAL with GADED (SDC1-2) or with GDLDD (SDC1-4) in otherwise unaltered SDC1 (Fig. 2B) respectively reduced binding substantially (Fig. 2, C and D). Disruption of HS substitution might be an unintended consequence of swapping. FGF2 is entirely dependent on HS for ligation (28), with some interaction with CS (33) that can substitute in place of HS in this region (34). Accordingly, FGF2 affinity precipitates were treated with heparitinase and chondroitinase. Both the supernatant and pellet of the digest were then blotted for SDC1 core protein. FGF2 bound each, although somewhat less SDC1-2 and SDC1-4 (Fig. 2E), and was detected in the digest supernatant in keeping the specificity of FGF2 for heparan sulfate independent of the core protein (28).
FIGURE 2.
SDC1 GAGAL within amino acids 20–30 is necessary for lacritin binding. A, GAGAL in N-terminal SDC1 and GADED in SDC2 (amino acids 1–48) with HS(/CS) substitution sites, respectively, at serines 15, 23, 25, and 22, 36, 38. Also shown are SDC4 amino acids 20–67 with GDLDD and HS(/CS) substitution sites at 21, 13, and 45. All numbering excludes the signal peptide, as defined by SignalP version 4.1. B, lysates from HEK293E cells transiently expressing SDC1, SDC1-2 (GAGAL replaced with GADED), or SDC1-4 (GAGAL replaced with GDLDD) were blotted with mAb B-B4 for SDC1 core protein. WB, Western blot. C, affinity precipitation scheme. D, lacritin-intein beads were incubated with lysates from SDC1-, SDC1-2-, or SDC1-4-expressing HEK293E cells. C-25-intein beads were similarly incubated with lysates from SDC1-expressing HEK293E cells. After incubation, beads were extensively washed and treated with heparitinase I/chondroitinase ABC. Digests were centrifuged, and the pellets were blotted with mAb B-B4 for SDC1 core protein. E, FGF2-intein beads were incubated with lysates from SDC1-, SDC1-2-, or SDC1-4-expressing HEK293E cells, thoroughly washed, and treated with heparitinase I/chondroitinase ABC. Digests were centrifuged, and the supernatants were blotted with mAb B-B4 for SDC1 core protein.
SDC1, SDC1-2, and SDC1-4 display substantially different hydropathic profiles (Fig. 3A), as per the domination of GAGAL by amino acids with nonpolar side chains. Only three other ectodomain regions are as hydrophobic (not shown; ExPASy ProtScale analysis (Kyte and Doolittle)).
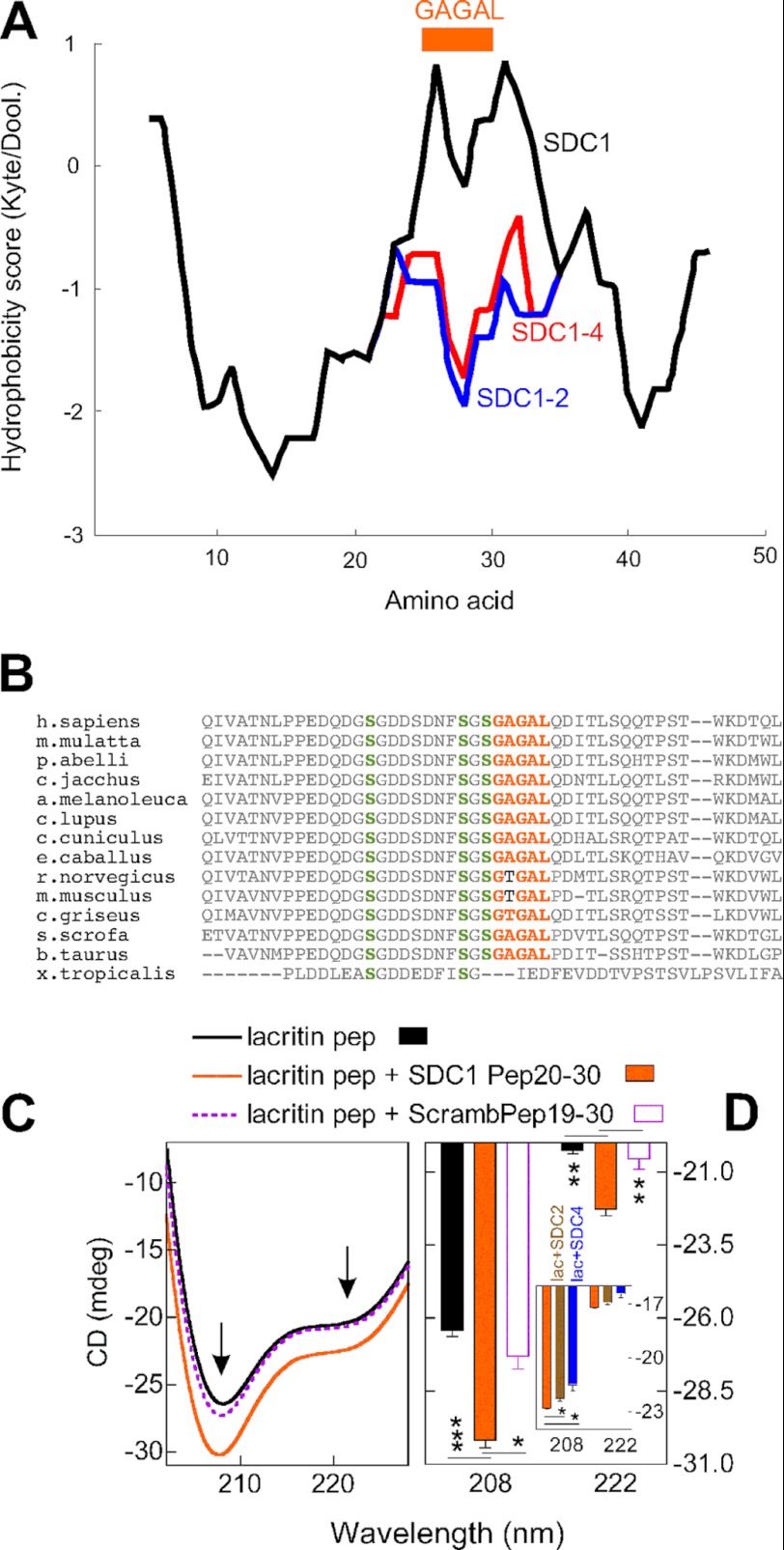
FIGURE 3.
GAGAL of SDC1 enhances lacritin C-terminal α-helicity. A, N-terminal 50 amino acids of SDC1, SDC1-2, and SDC1-4 were analyzed using ExPASy ProtScale (hydropathicity scale of Kyte and Doolittle with default Window size of 9). B, orthologs from BLAST of the N-terminal 50 amino acids of SDC1 were aligned using ClustalW (version 2.1). C, lacritin peptide LacPep95–119 comprising C-terminal amino acids 95–119 was subjected to circular dichroism in 10 mm dodecylphosphocholine without or with syndecan peptide Pep20–30 (amino acids 20–30) or scrambled peptide ScrambPep19–30 (scrambled amino acids 19–30). Shown is a mean profile of three independent experiments after subtraction of SDC1 Pep20–30 and ScrambPep 19–30 (both random coil) profiles. D, mean 208 and 222 signals from C with S.E. (n = 3). **, p < 0.005; ***, p < 0.0005. Inset, LacPep95–119 was subjected to circular dichroism in 10 mm dodecylphosphocholine without or with syndecan SDC1 Pep20–30, SDC2 Pep34–43, or SDC4 Pep41–50. Shown is a mean profile with S.E. (error bars) of three independent experiments after subtraction of SDC1 Pep20–30, SDC2 Pep34–43, or SDC4 Pep41–50 profiles.
GAGAL is well conserved in all SDC1 orthologs, as detected by an SDC1 protein BLAST search of the non-redundant database, with the exception of Xenopus tropicalis, where it is absent. In mice and rats, threonine is substituted for the first alanine (Fig. 3B). Prior analysis with lacritin truncation mutants C-5, -10, -15, and -25 implicated a potential hydrophobic binding face approximately centered on lacritin amino acids 100–109 as part of an amphipathic α-helix (17, 18).
Could mutual hydrophobicity drive SDC1-lacritin ligation, as respectively occurs in receptor-ligand pairs, such as PTH1R (parathyroid hormone 1 receptor)-PTHLH (parathyroid hormone-like hormone), PTH1R−PTH (35, 36), CRFR1 (corticotropin-releasing hormone receptor 1)-CRF (corticotropin-releasing hormone) (37), GLP1R (glucagon-like peptide 1 receptor)-GCG (glucagon) (38), and GLP1R-GIP (gastric inhibitory polypeptide) (39)? Indeed, an initially low affinity complex between CRFR1 and CRF drives assembly of a CRF amphipathic α-helix that solidifies the interaction (37). We compared the α-helicity of lacritin C-terminal peptide LacPep95–119 (amino acids 95–119) in 10 mm dodecylphosphocholine (17) without or with SDC1, -2, or -4 peptides (Fig. 3, C and D). Background contribution of each syndecan peptide in dodecylphosphocholine (random coil) was subtracted. α-Helicity was substantially enhanced by GAGAL containing Pep20–30 (Fig. 3, C and D) or Pep19–30 (not shown), whereas scrambled ScrambPep19–30 had no effect (Fig. 3, C and D). Significantly less effective were GADED or GDLDD containing SDC2 and SDC4 peptides (Fig. 3D, inset).
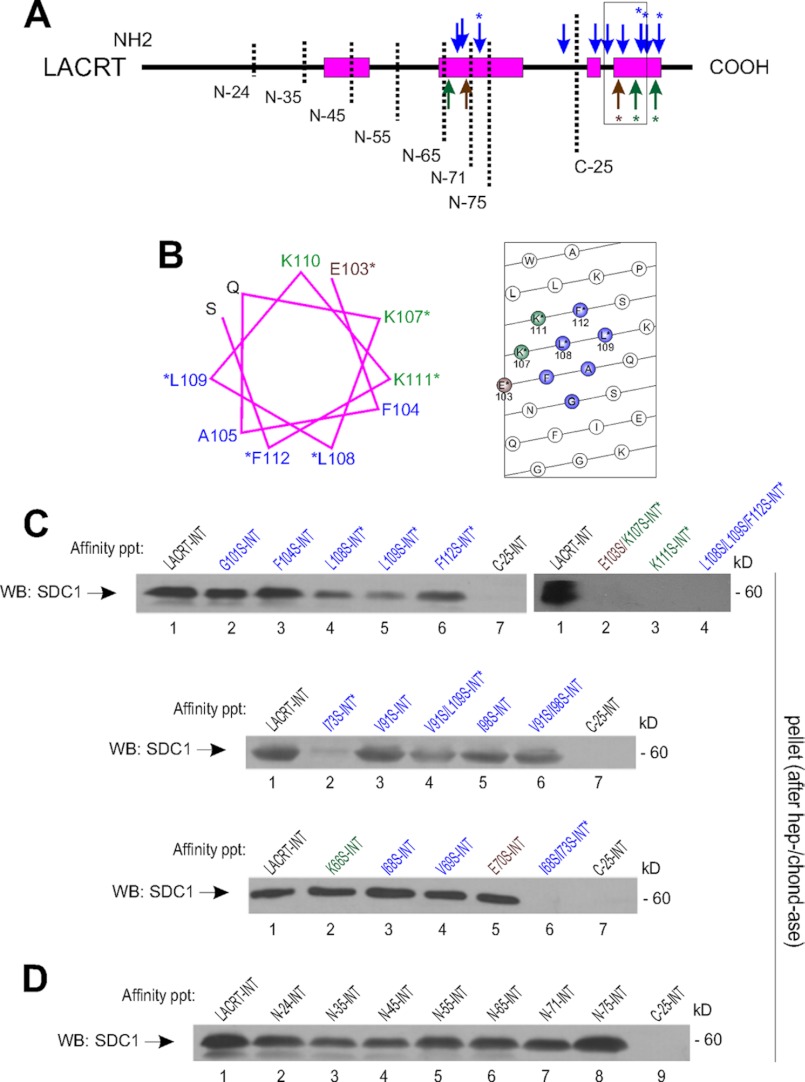
To further address the role of mutual hydrophobicity, lacritin hydrophobic residues within or flanking this region (Fig. 4, A and B) were selectively mutated. Others in an adjacent α-helix were also mutated. SDC1 binding was unaffected by lacritin point mutations I68S, V69S, V91S, I98S, G101S, or F104S (Fig. 4C). In contrast, less SDC1 binding was observed to L108S, L109S, V91S/L109S, and F112S lacritin (Fig. 4C), and triple mutant L108S/L109S/F112S completely lacked affinity (Fig. 4C), although bacterial expression of each exceeded that of unaltered lacritin (40). Further mutation at Ala-105 rendered lacritin insoluble (not shown). Ala-105, Leu-108, Leu-109, and Phe-112 are all predicted to contribute to the C-terminal amphipathic α-helix of lacritin.
FIGURE 4.
Lacritin C-terminal Leu-108, Leu-109, and Phe-112 and Glu-103, Lys-107, and Lys-111 are necessary for SDC1 binding. A, linear diagram of lacritin truncations (vertical dashed lines) and point mutants (arrows; blue, nonpolar; green, positively charged; brown, negatively charged side chains) with filled boxes representing α-helices predicted by PSIPRED (version 3.0) and/or demonstrated by CD. Numbering excludes the signal peptide. B, helical wheel (left) and helical net (right) projections of the open box region in A, with flanking amino acids. C, lacritin- or lacritin G101S-, F104S-, L108S-, L109S, F112S-, C-25-, E103S/K107S-, K111S-, L108S/L109S/F112S-, I73S-, V91S-, V91S/L109S-, I98S-, V91S/I98S-, K66S-, I68S-, V69S-, E70S-, or I68S/I73S-intein beads were incubated with lysates from HEK293E cells transiently expressing SDC1. After incubation, beads were washed extensively and treated with heparitinase I/chondroitinase ABC (hep-/chond-ase). Digests were centrifuged, and the pellets were blotted (WB) with mAb B-B4 for SDC1 core protein. D, lacritin- or lacritin N-24-, N-35-, N-45-, N-55-, N-65-, N-71-, N-75-, or C-25-intein beads were incubated with lysates from HEK293E cells transiently expressing SDC1. After incubation, beads were thoroughly washed and treated with heparitinase I/chondroitinase ABC. Digests were centrifuged, and the pellets were blotted with mAb B-B4 for SDC1 core protein.
Other C-terminal sites were considered. Ile-73 and Ile-68 reside in an adjacent predicted α-helix. Binding was unaffected by I68S, whereas neither I68S/I73S nor I73S displayed any affinity for SDC1 (Fig. 4C), although bacterial expression of I73S exceeded that of unaltered lacritin (40). Ile-73 is available in negative control C-25, and lacritin lacking 75 amino acids from the N terminus binds SDC1 as well as unaltered lacritin (Fig. 4D), implying that Ile-73 is probably not an SDC1 binding component. Some similarities may be drawn to parathyroid hormone-like hormone that captures parathyroid hormone 1 receptor via its C-terminal amphipathic α-helix. Truncation of six N-terminal amino acids had little effect, yet binding was largely abrogated by mutation of the third serine to glutamic acid by steric hindrance of proximal N and C termini (35). Similarly, an NMR model of the C-terminal 48 amino acids of lacritin homolog DCD (dermcidin) (Protein Data Bank entry 2KSG) suggests that the same region in lacritin could assume a folded conformation with Ile-73 in proximity to the hydrophobic binding face. Taken together, these data point to an interaction between SDC1 and lacritin that derives specificity from GAGAL and lacritin amino acids 108, 109, and 112, the latter of which may be adjacent to Ile-73 by folding.
Binding Requires HS and CS
Six other GAGAL-containing plasma membrane or extracellular proteins (ADCY9 (adenylate cyclase 9), CD9 (CD9 molecule), CHRNB2 (cholinergic receptor, nicotinic, β2), FLNA (filamin A, α), LRRC26 (leucine rich repeat-containing 26), and ROS1 (c-ros oncogene 1, receptor tyrosine kinase)) are revealed by FASTA and BLAST searches of the human/Refseq database. Twenty-nine others are intracellular (supplemental Table 3). A further 26 plasma membrane or extracellular proteins contain a single amino acid substitution with a Smith-Waterson score of ≥27 (versus a score of 31 for GAGAL; supplemental Table 4). Although there is no evidence that GAGAL is active or available in these proteins, it is nonetheless interesting that lacritin cell targeting (17) and protein binding (18) are so apparently restricted and specific.
GAGAL is adjacent to serine 23 and 25 attachment sites in NFSGS, with binding dependent on digestion by heparanase (18), an endo-β-d-glucuronidase not widely expressed. Could the ∼4–5-kDa cleavage stubs (18) residual from heparanase exposure create a second binding platform for lacritin Lys-107, Lys-110, or Lys-111 on the opposite face of the lacritin amphipathic α-helix? We cultured cells overnight in 30, 100, or 300 mm xyloside to competitively inhibit heparan and chondroitin sulfate assembly (Fig. 5A, right) (37). This progressively diminished the heterogeneity of SDC1. Because xyloside can be toxic and thus might reduce synthesis, we monitored the relative quantity of SDC1 core protein in each lysate by digesting with heparitinase and chondroitinase prior to blotting for SDC1 core protein. No differences were apparent (Fig. 5B). However, affinity for lacritin was completely erased (Fig. 5C).
FIGURE 5.
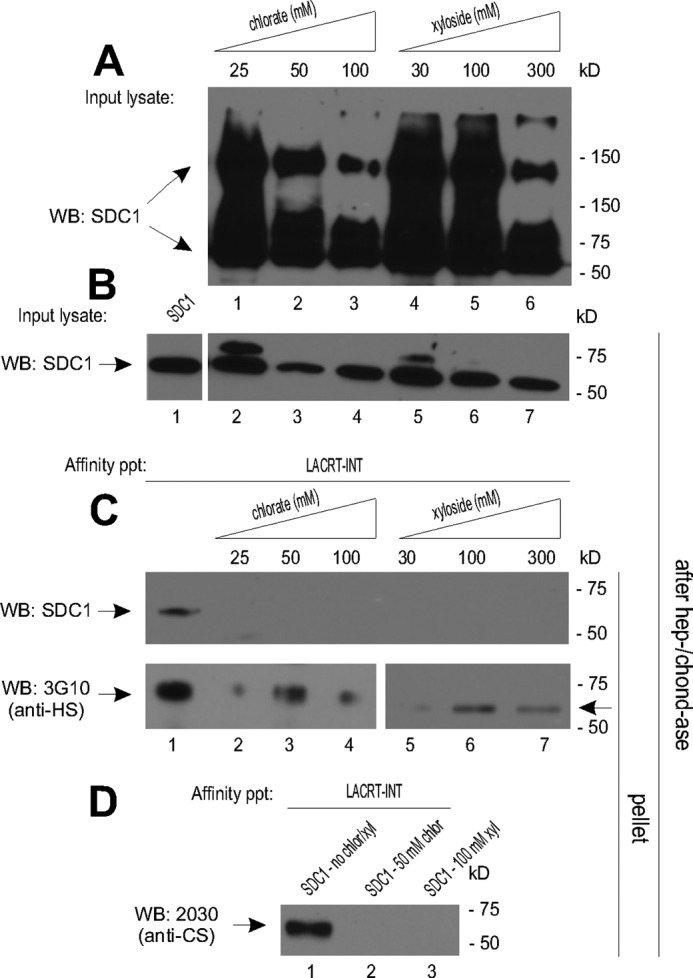
Lacritin binding requires sulfation of SDC1 chains. A, lysates from HEK293E cells transiently expressing SDC1 in the presence of increasing amounts of sodium chlorate or xyloside. Lysates were blotted (WB) with mAb B-B4 for SDC1 core protein. B, lysates from HEK293E cells in A or from HEK293E cells transiently expressing SDC1 without treatment were digested with heparitinase I/chondroitinase ABC (hep-/chond-ase) to control for equal loading by core protein. Lysates were blotted with mAb B-B4 for SDC1 core protein. C, lacritin-intein beads were incubated with lysates from A or with lysate from HEK293E cells transiently expressing SDC1 without treatment. After incubation, beads were washed extensively and treated with heparitinase I/chondroitinase ABC. Digests were centrifuged, and the pellets were blotted with mAb B-B4 for SDC1 core protein or with mAb 3G10 for desaturated uronates in heparitinase-cleaved HS. D, lacritin-intein beads were incubated with lysates from HEK293E cells transiently expressing SDC1 in the presence of 50 mm sodium chlorate or 100 mm xyloside or with lysate from HEK293E cells transiently expressing SDC1 without treatment. After incubation, beads were thoroughly washed and treated with heparitinase I/chondroitinase ABC. Digests were centrifuged, and the pellets were blotted with mAb 2030 for chondroitin 4-sulfate in chondroitinase ABC-cleaved CS.
For validation, we also utilized mAb 3G10 with specificity for desaturated uronates in heparitinase-cleaved HS (31). Some ∼60-kDa 3G10 epitope was detected, although substantially less (Fig. 5C). 4-Methylumbelliferyl-β-d-xylopyranoside is a more efficient inhibitor of CS substitution (40) than HS (41). This is in keeping with residual SDC1 size heterogeneity that is attributable largely to HS and less so to CS (34). We therefore blotted for CS with mAb 2030 for chondroitin-4 sulfate in chondroitinase ABC-cleaved CS (42). A ∼60-kDa chondroitin-4 sulfate band was detected in lacritin affinity precipitates using untreated SDC1 cell lysates but not from SDC1 lysates of 4-methylumbelliferyl-β-d-xylopyranoside-treated cells (Fig. 5D). The implication was that sulfated glycosaminoglycans may support GAGAL binding, possibly both CS- and heparanase-shortened HS stubs.
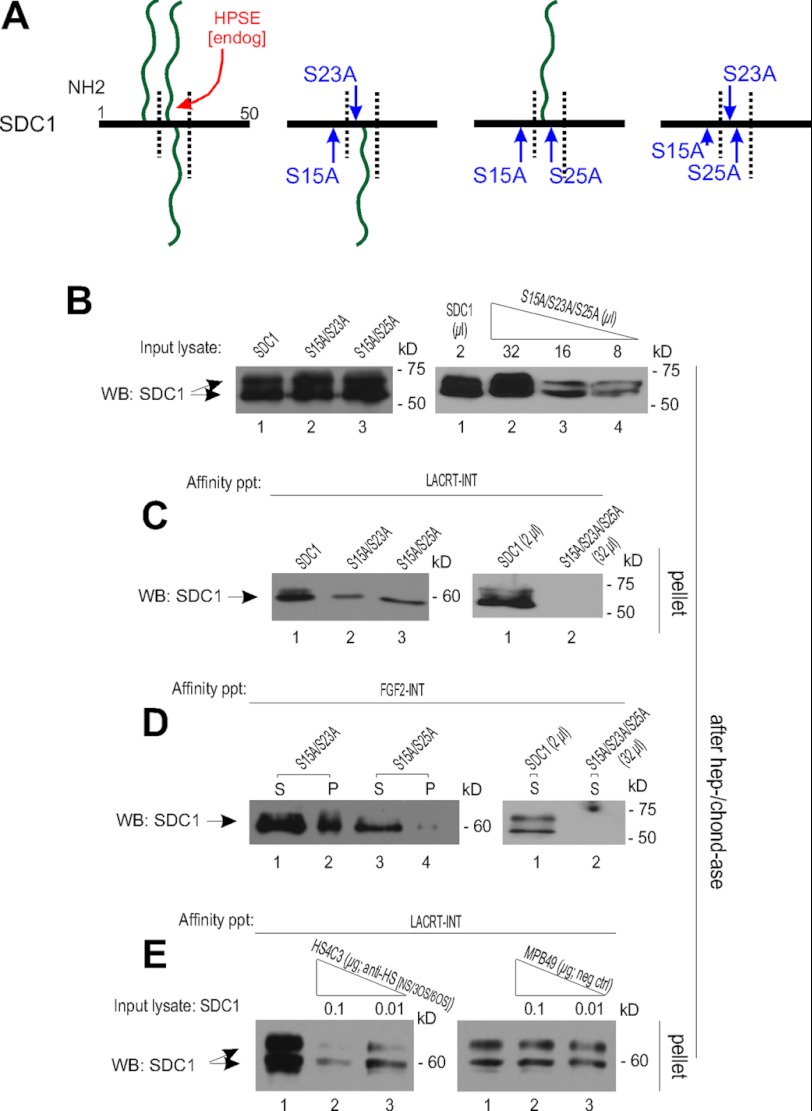
To address this observation more selectively, predicted SDC1 heparan sulfate attachment sites on serine were each mutated to alanine (Fig. 6A) and then subjected to lacritin affinity precipitation, after first checking relative levels of synthesis in which lysates were subjected to heparitinase and chondroitinase digestion, followed by blotting for SDC1 core protein (Fig. 6B). Synthesis of S15A/S23A and S15A/S25A were equivalent to that of SDC1, but not S15A/S23A/S25A, which required a 16-fold increase in lysate load (Fig. 6B). Ser-15 is most distal and Ser-25 most proximal to GAGAL (Fig. 2A). Binding to S15A/S23A and S15A/S25A were both slightly diminished (Fig. 6C). In contrast, lacritin binding to triple mutant S15A/S23A/S25A was not detectable (Fig. 6C).
FIGURE 6.
Sulfation of the N-terminal HS chain(s) of SDC1 is essential for lacritin binding. A, linear diagrams of the N-terminal 50 amino acids of SDC1 with HS(/CS) substitution sites at serines 15, 23, and 25 mutated to alanine. B, lysates from HEK293E cells transiently expressing SDC1, SDC1 S15A/S23A, SDC1 S15A/S25A, or SDC1 S15A/S23A/S25A were digested with heparitinase I/chondroitinase ABC (hep-/chond-ase) to control for equal loading by core protein and adjusted if necessary (right). Lysates were blotted (WB) with mAb B-B4 for SDC1 core protein. C, lacritin-intein beads were incubated with lysates from HEK293E cells transiently expressing SDC1 S15A/S23A, SDC1 S15A/S25A, or SDC1 S15A/S23A/S25A. After incubation, beads were washed extensively and treated with heparitinase I/chondroitinase ABC. Digests were centrifuged, and the pellets were blotted with mAb B-B4 for SDC1 core protein. D, FGF-2-intein beads were incubated with the same lysates. After incubation, washing, and treatment with heparitinase I/chondroitinase ABC, digests were centrifuged, and both the pellets and supernatants were blotted with mAb B-B4 for SDC1 core protein. Partially incomplete digestion gave rise to some FGF2-detectable SDC1 S15A/S23A in the pellet. E, lacritin-intein beads were incubated with lysates from HEK293E cells transiently expressing SDC1 in the presence of increasing amounts of purified HS4C3 or MPB49 antibody. HS4C3 is directed to HS with N-sulfation, 2-O-sulfation, 3-O-sulfation, or 6-O-sulfation, with highest affinity to 3-O-sulfation. MPB49 does not bind HS or CS. After incubation, beads were thoroughly washed and treated with heparitinase I/chondroitinase ABC. Digests were centrifuged, and the pellets were blotted with mAb B-B4 for SDC1 core protein.
As a positive control for HS chain assembly, parallel FGF2 affinity precipitations were performed (Fig. 6D). FGF2 precipitates were treated with heparitinase and chondroitinase, and then both the supernatant and pellet of the digest were blotted for SDC1 core protein. FGF2 bound S15A/S23A and S15A/S25A but not S15A/S23A/S25A (Fig. 6D). Bound S15A/S23A and S15A/S25A were detected in the supernatant (and some in the pellet from incomplete digestion).
Because FGF binds only sulfated HS (28), we wondered whether sulfation contributed to lacritin binding. Chlorate suppresses sulfation by competitively inhibiting 3′-phosphoadenosine 5′-phosphosulfate synthase (2ATP-sulfurylase) (28). After culturing cells overnight in 25, 50, or 100 mm sodium chlorate in low sulfur medium, the heterogeneity of SDC1 progressively decreased (Fig. 5A), but protein synthesis of SDC1 was unaffected (Fig. 5B). No lacritin-bound SDC1 (Fig. 5C) nor chondroitin-4-sulfated band of ∼60 kDa was detected (Fig. 5D). Some ∼60-kDa 3G10 epitope was detected, although less (Fig. 5C).
2-, 3-, or 6-O-sulfation or N-sulfation of HS dictates ligand binding specificity. N- and 2-O-sulfation are, for example, necessary for interaction with FGF2 (43). We therefore challenged lacritin affinity precipitations with several modification-selective antibodies (Figs. 6 and 7). The anti-HS single chain variable fragment antibody HS4C3 preferably binds HS with 3-O-sulfated glucosamine and interacts weakly with N-, 2-O-, and 6-O-sulfated saccharides (43). Purified HS4C3, but not HS4E4 (N-sulfation; not shown) or negative control MPB49, inhibited SDC1 binding, suggesting by elimination that 3-O-, 2-O-, or 6-O-sulfated HS may help specify lacritin targeting.
FIGURE 7.
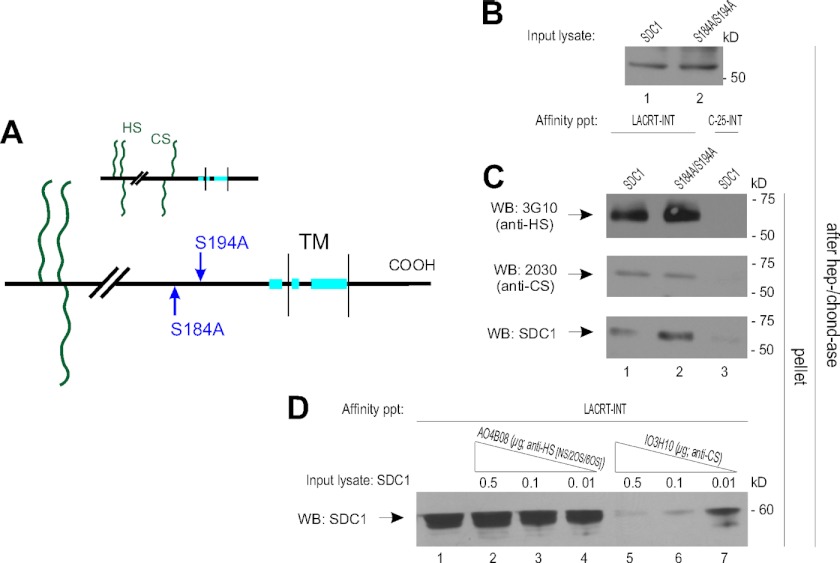
Substitution of a sulfated N-terminal CS chain(s) is necessary for lacritin binding. A, linear diagram of SDC1 with putative CS substitution sites at serines 184 and 194 mutated to alanine. TM, transmembrane domain. Inset, intact SDC1. B, lysates from HEK293E cells transiently expressing SDC1 or SDC1 S184A/S194A were digested with heparitinase I/chondroitinase ABC (hep-/chond-ase) to control for equal loading. Lysates were blotted with mAb B-B4 for SDC1 core protein. C, lacritin-intein beads were incubated with lysates from HEK293E cells transiently expressing SDC1 or SDC1 S184A/S194A. C-25-intein beads were incubated with SDC1 lysates. After incubation, beads were washed several times and treated with heparitinase I/chondroitinase ABC. Digests were centrifuged, and the pellets were blotted (WB) with mAb 3G10 for desaturated uronates in heparitinase-cleaved HS, mAb 2030 for chondroitin-4 sulfate in chondroitinase ABC-cleaved CS and with mAb B-B4 for SDC1 core protein. D, lacritin-intein beads were incubated with lysates from HEK293E cells transiently expressing SDC1 in the presence of increasing amounts of purified AO4B08 or IO3H10 antibody. AO4B08 is directed to HS with N-sulfation, 2-O-sulfation, or 6-O-sulfation. IO3H10 is directed to CS. After incubation, beads were thoroughly washed and treated with heparitinase I/chondroitinase ABC. Digests were centrifuged, and the pellets were blotted with mAb B-B4 for SDC1 core protein.
Curious about the loss of lacritin binding coincident with a complete absence of chondroitin-4 sulfation with chlorate or xyloside treatment, we wondered whether serine 15, 23, or 25 might be substituted with CS (34). Membrane-proximal serines 184 and 194 are well established substitution sites for CS (34). We therefore mutated both to alanine (Fig. 7A) as a double mutant in otherwise intact SDC1, thereby focusing anti-CS blotting on serines 15, 23, and 25. SDC1 S184A/S194A expression was equivalent to that of native SDC1, as indicated by core protein blotting following heparitinase and chondroitinase digestion (Fig. 7B). Affinity for lacritin was slightly enhanced as a mAb 2030 anti-chondroitin-4 sulfate- and mAb 3G10-blottable band (Fig. 7C). Negative control C-25 showed no SDC1 binding or background anti-CS or -HS staining (Fig. 7C).
To probe whether CS substitution of serine 15, 23, or 25 contributed, with HS, to the specification of lacritin-GAGAL binding, we challenged lacritin affinity precipitations of SDC1 lysates with the CS-directed single chain variable fragment antibody IO3H10. Also assayed was AO4B08 with specificity for N-, 2-O-, and 6-O-sulfated HS (43). Purified IO3H10 inhibited binding in a dose-dependent manner, whereas purified AO4B08 had no effect. Thus, N-terminal CS, together with 3-O-sulfated HS, appears to be required for GAGAL-dependent lacritin binding.
With lacritin-bound SDC1 predominated by 35S-labeled HS of ∼4–5 kDa in a heparanase-dependent manner (versus ∼40 kDa for FGF2-bound SDC1) (18), and with exogenous heparanase or heparitinase capable of rescuing lacritin-dependent mitogenesis in cells depleted of heparanase by siRNA (18), 3-O-sulfated glucosamine must reside on HS cleavage stubs. How then might lacritin bind to sulfated HS stubs and sulfated CS? Anionic stubs might electrostatically interact with Lys-107, Lys-110, and/or Lys-111 that contribute to the C-terminal hydrophilic face of lacritin. As an example, Arg-20 and Lys-27 of the hydrophilic face of glucagon bind the glucagon-like peptide 1 receptor (38). Similarly, His-335, Glu-339, and Arg-343 of the hydrophilic face of the oxytocin receptor are required for phopholipase C targeting (44).
To address whether binding is via the hydrophilic face of the lacritin C-terminal amphipathic α-helix, lacritin point mutants E103S/K107S, K110S, and K111S were developed. Although bacterially expressed K110S lacritin was insoluble (as was K95S and E99S outside of the helix), E103S/K107S and K111S lacritins were both soluble, and expression of E103S/K107S exceeded that of lacritin by 3-fold (40) (no quantitative data available for K111S). Interestingly, neither bound SDC1 (Fig. 4C, right; longer exposures show slight K111S binding (not shown)), suggesting that the cationic lacritin face was essential. Thus, a mixture of hydrophobic, electrostatic, and enzymatic elements combine to anchor lacritin on SDC1.
DISCUSSION
Morphogenetic, secretory, and homeostatic events are regulated in part by local interaction of growth factors with cell surface proteoglycans. Previously, we described a novel mechanism in which heparanase deglycanation of the HS-rich SDC1 N terminus, and not of SDC2 or -4 or long HS chains, is required for ligation of the epithelial prosecretory mitogen lacritin (18). We now discover that ligation requires the N-terminal GAGAL of SDC1 that enhances the α-helicity of the C-terminal amphipathic α-helix of lacritin, to which 3-O-sulfated HS (presumably as heparanase-shortened stubs) and 4-O-sulfated CS appear to gain anchorage, probably through lacritin cationic face residues Lys-107 and Lys-111. Thus, lacritin cell targeting of SDC1 is dependent on a mixture of protein-protein and protein-HS and -CS interactions.
Such hybrid binding has not been described previously, but it may not be uncommon. Ligation of cell surface proteoglycans was originally thought to be restricted to the anionic HS chains (45, 46). However, an increasing number of ligands are now known to be captured by core proteins, several of which also bind heparin sulfate. Bone morphogenetic protein, Hh/Shh, and Wnts were originally known only for their heparin sulfate affinity (47–51). However, new studies reveal how ligation of glypican-4 core protein by hedgehog and wingless (4, 5) and of glypican-5 core protein by bone morphogenetic protein in the fly (6), as well as of glypican-3 core protein in mice by sonic hedgehog, Wnt 3A, and Wnt 7B (8, 9), play important roles in wing morphogenesis and body size, respectively. In some cases, the relative importance of each core protein to HS or CS may shift with time. In cerebeller development, Shh-dependent granule cell proliferation displays no or substantial heparinase sensitivity, respectively, at 3 and 6 days postnatal (51). Similar examples from the syndecan family are unavailable, although core proteins of syndecan-1 (10 - 12), -2 (13, 14), and -4 (15) are cell-adhesive. Core protein involvement expands morphogenic complexity, seemingly with an HS or CS rheostat to modulate different cellular processes. However, the involvement of heparanase in this process may be rare and perhaps restricted to lacritin and SDC1.
Probing the GAGAL docking site revealed an interesting chain requirement in which S15A/S23A/S25A displayed no affinity, whereas S15A/S23A and S15A/S25A bound lacritin at levels slightly less than wild type. Because S15A does not affect binding (not shown), substitution at Ser-23 and Ser-25 may be key. HS chains in mouse Sdc1 are hierarchical. Alanine mutation of serine 25 (serine 47 with signal peptide) < S23A < S15A as stimulators of collagen gel invasion by human B lymphoid cells (12). Also, Sdc1 mutated at either serine 23 or 25, but not 15 (or wild type), failed to support spreading of monkey fibroblastic cells on thrombospondin-1 (52). What might be the spatial arrangement of HS stub(s) and CS? A location immediately proximal to GAGAL may be optimal because N-terminal CS is short (34), as is the HS stub. HS stub length can be estimated, although with some variability. Heparanase cleaves glucuronic acid-N-sulfoglucosamine linkages when N-sulfoglucosamine is 3-O- or 6-O-sulfated or when a 2-O-sulfated glucuronic acid residue is nearby (53). Single chain antibody HS4C3 was inhibitory, suggesting that the HS stub was 3-O-sulfated. From the expected HS structure (54), cleavage should generate a 19–29 monosaccharide stub of Xyl-Gal-Gal-GlcA-GlcNAc-GlcA5–10-GlcNAc-GlcA-GlcNAc (N-,6-sulfated)-GlcA (2-sulfated)-GlcNAc(3-sulfated)) that has a calculated molecular mass of 4.3–6.1 kDa. This compares favorably with HS stubs on lacritin-bound SDC1 that by gel filtration had a relative mass of ∼4–5 kDa, about one-tenth that of uncleaved HS (18). Recent x-ray solution scattering of 6–24 monosaccharide HS (3–10 nm, respectively) (55) makes feasible an HS stub estimate of 8–12 nm. By further approximating the distance between serine 15 and glycine 26 of GAGAL as 11.3 nm (assuming 0.8 nm per amino acid separated by 0.3 nm), it would appear that a stub at serine 23 or 25 would be preferred. However, core flexibility may obviate the need for proximity.
Recent studies suggest that lacritin, HS, and heparanase may be key elements in the “lacrimal functional unit,” whose dysfunction is manifested as “dry eye,” the most common eye disease (24, 25). The lacrimal functional unit consists of all ocular surface epithelia (cornea, conjunctival, lid), the lacrimal and meibomian glands, and lacrimal functional unit-coordinating afferent and efferent neural fibers (56). Lacritin stimulates basal tear secretion by rat lacrimal acinar cells (16) and basal tearing when topically added to rabbit eyes, an effect sustained for several h (22). Rather than densitization after 2 weeks of three times daily treatment, basal tearing gradually increases and remains elevated 1 week after treatment has ceased (22). Selective tear lacritin deficiency of “about 50%” (57) to “∼7-fold” (58) or to 0.24–0.56 of normal (23) has been noted in several proteomic studies of dry eye (23, 58) or of “blepharitis” (57), an inflammation of the eyelid often associated with dry eye. One study indicated no difference (59) but did not differentiate between monomeric (active) and multimeric lacritin (largely inactive) (60). Multimers form by tissue transglutaminase-catalyzed cross-linking involving Gln-106 of the SDC1 binding region (60).
Conjunctival cells from dry eye patients express less EXTL2 (exostoses (multiple)-like 2), HS2ST1 (heparan sulfate 2-O-sulfotransferase 1), and HS3ST6 (heparan sulfate (glucosamine) 3-O-sulfotransferase 6) (61). This could diminish the number of HS chains capable of heparanase cleavage, one rationale for decreased lacritin binding of SDC1 from cells cultured in low sulfate medium containing chlorate. Heparanase is readily detectable in normal tears but less so in a small sample of dry eye tears.4 Regulation of active heparanase secretion in cell culture is in part by ADP or ATP (62). In unrelated studies with other end points, the UTP analog diquafosol tetrasodium was recently approved for the treatment of dry eye. The implied interrelationship between lacritin, SDC1, and heparanase in normal and dry eye is intriguing. Taken together, we describe a hybrid core protein/HS/CS binding process as a novel mechanism in physiology/dysfunction and potentially in metazoan development.
Acknowledgments
We thank Jeffrey Romano for aid in blotting and Jian Liu (University of North Carolina) and Alan Rapraeger (University of Wisconsin) for critically reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 EY013143 and EY018222 (to G. W. L.).

This article contains supplemental Tables 1–4.
P. Ma and G. W. Laurie, unpublished observations.
- HS
- heparan sulfate
- CS
- chondroitin sulfate
- xyloside
- 4-methylumbelliferyl-β-d-xylopyranoside
- 293E
- HEK293-EBNA1.
REFERENCES
- 1. Ori A., Wilkinson M. C., Fernig D. G. (2011) A systems biology approach for the investigation of the heparin/heparan sulfate interactome. J. Biol. Chem. 286, 19892–19904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pinhal M. A., Smith B., Olson S., Aikawa J., Kimata K., Esko J. D. (2001) Enzyme interactions in heparan sulfate biosynthesis. Uronosyl 5-epimerase and -O-sulfotransferase interact in vivo. Proc. Natl. Acad. Sci. U.S.A. 98, 12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Presto J., Thuveson M., Carlsson P., Busse M., Wilén M., Eriksson I., Kusche-Gullberg M., Kjellén L. (2008) Heparan sulfate biosynthesis enzymes EXT1 and EXT2 affect NDST1 expression and heparan sulfate sulfation. Proc. Natl. Acad. Sci. U.S.A. 105, 4751–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yan D., Wu Y., Yang Y., Belenkaya T. Y., Tang X., Lin X. (2010) The core protein of glypican Dally-like determines its biphasic activity in wingless morphogen signaling. Development 137, 2033–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yan D., Wu Y., Feng Y., Lin S. C., Lin X. (2009) The core protein of glypican Dally-like determines its biphasic activity in wingless morphogen signaling. Dev. Cell 17, 470–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kirkpatrick C. A., Knox S. M., Staatz W. D., Fox B., Lercher D. M., Selleck S. B. (2006) The function of a Drosophila glypican does not depend entirely on heparan sulfate modification. Dev. Biol. 300, 570–582 [DOI] [PubMed] [Google Scholar]
- 7. Rawson J. M., Dimitroff B., Johnson K. G., Rawson J. M., Ge X., Van Vactor D., Selleck S. B. (2005) The heparan sulfate proteoglycans Dally-like and Syndecan have distinct functions in axon guidance and visual-system assembly in Drosophila. Curr. Biol. 15, 833–838 [DOI] [PubMed] [Google Scholar]
- 8. Capurro M. I., Xu P., Shi W., Li F., Jia A., Filmus J. (2008) Glypican-3 inhibits Hedgehog signaling during development by competing with patched for Hedgehog binding. Dev. Cell 14, 700–711 [DOI] [PubMed] [Google Scholar]
- 9. Capurro M. I., Xiang Y. Y., Lobe C., Filmus J. (2005) Glypican-3 promotes the growth of hepatocellular carcinoma by stimulating canonical Wnt signaling. Cancer Res. 65, 6245–6254 [DOI] [PubMed] [Google Scholar]
- 10. Beauvais D. M., Ell B. J., McWhorter A. R., Rapraeger A. C. (2009) Syndecan-1 regulates αvβ3 and αvβ5 integrin activation during angiogenesis and is blocked by synstatin, a novel peptide inhibitor. J. Exp. Med. 206, 691–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Beauvais D. M., Rapraeger A. C. (2010) Syndecan-1 couples the insulin-like growth factor-1 receptor to inside-out integrin activation. J. Cell Sci. 123, 3796–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Langford J. K., Yang Y., Kieber-Emmons T., Sanderson R. D. (2005) Identification of an invasion regulatory domain within the core protein of syndecan-1. J. Biol. Chem. 280, 3467–3473 [DOI] [PubMed] [Google Scholar]
- 13. Park H., Kim Y., Lim Y., Han I., Oh E. S. (2002) Syndecan-2 mediates adhesion and proliferation of colon carcinoma cells. J. Biol. Chem. 277, 29730–29736 [DOI] [PubMed] [Google Scholar]
- 14. Whiteford J. R., Behrends V., Kirby H., Kusche-Gullberg M., Muramatsu T., Couchman J. R. (2007) Syndecans promote integrin-mediated adhesion of mesenchymal cells in two distinct pathways. Exp. Cell Res. 313, 3902–3913 [DOI] [PubMed] [Google Scholar]
- 15. McFall A. J., Rapraeger A. C. (1998) Characterization of the high affinity cell-binding domain in the cell surface proteoglycan syndecan-4. J. Biol. Chem. 273, 28270–28276 [DOI] [PubMed] [Google Scholar]
- 16. Sanghi S., Kumar R., Lumsden A., Dickinson D., Klepeis V., Trinkaus-Randall V., Frierson H. F., Jr., Laurie G. W. (2001) cDNA and genomic cloning of lacritin, a novel secretion enhancing factor from the human lacrimal gland. J. Mol. Biol. 310, 127–139 [DOI] [PubMed] [Google Scholar]
- 17. Wang J., Wang N., Xie J., Walton S. C., McKown R. L., Raab R. W., Ma P., Beck S. L., Coffman G. L., Hussaini I. M., Laurie G. W. (2006) Restricted epithelial proliferation by lacritin via PKCα-dependent NFAT and mTOR pathways. J. Cell Biol. 174, 689–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ma P., Beck S. L., Raab R. W., McKown R. L., Coffman G. L., Utani A., Chirico W. J., Rapraeger A. C., Laurie G. W. (2006) Heparanase deglycanation of syndecan-1 is required for binding of the epithelial-restricted prosecretory mitogen lacritin. J. Cell Biol. 174, 1097–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weigelt B., Bosma A. J., van 't Veer L. J. (2003) Expression of a novel lacrimal gland gene lacritin in human breast tissues. J. Cancer Res. Clin. Oncol. 129, 735–736 [DOI] [PubMed] [Google Scholar]
- 20. Porter D., Weremowicz S., Chin K., Seth P., Keshaviah A., Lahti-Domenici J., Bae Y. K., Monitto C. L., Merlos-Suarez A., Chan J., Hulette C. M., Richardson A., Morton C. C., Marks J., Duyao M., Hruban R., Gabrielson E., Gelman R., Polyak K. (2003) A neural survival factor is a candidate oncogene in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 100, 10931–10936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Landgraf P., Wahle P., Pape H. C., Gundelfinger E. D., Kreutz M. R. (2008) The survival-promoting peptide Y-P30 enhances binding of pleiotrophin to syndecan-2 and -3 and supports its neuritogenic activity. J. Biol. Chem. 283, 25036–25045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Samudre S., Lattanzio F. A., Jr., Lossen V., Hosseini A., Sheppard J. D., Jr., McKown R. L., Laurie G. W., Williams P. B. (2011) Lacritin, a novel human tear glycoprotein, promotes sustained basal tearing and is well tolerated. Invest. Ophthalmol. Vis. Sci. 52, 6265–6270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Srinivasan S., Thangavelu M., Zhang L., Green K. B., Nichols K. K. (2012) iTRAQ quantitative proteomics in the analysis of tears in dry eye patients. Invest. Ophthalmol. Vis. Sci. 53, 5052–5059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schaumberg D. A., Sullivan D. A., Buring J. E., Dana M. R. (2003) Prevalence of dry eye syndrome among US women. Am. J. Ophthalmol. 136, 318–326 [DOI] [PubMed] [Google Scholar]
- 25. Lin P. Y., Cheng C. Y., Hsu W. M., Tsai S. Y., Lin M. W., Liu J. H., Chou P. (2005) Association between symptoms and signs of dry eye among an elderly Chinese population in Taiwan. The Shihpai Eye Study. Invest. Ophthalmol. Vis. Sci. 46, 1593–1598 [DOI] [PubMed] [Google Scholar]
- 26. Durocher Y, Perret S., Kamen A. (2002) High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 30, E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lohmander L.S., Hascall V.C., Caplan A.I. (1979) Effects of 4-methylumbelliferyl-β-d-xylopyranoside on chondrogenesis and proteoglycan synthesis in chick limb bud mesenchymal cell cultures. J. Biol. Chem. 254, 10551–10561 [PubMed] [Google Scholar]
- 28. Rapraeger A. C., Krufka A., Olwin B. B. (1991) Requirement of heparan sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science 252, 1705–1708 [DOI] [PubMed] [Google Scholar]
- 29. van Kuppevelt T. H., Dennissen M. A., van Venrooij W. J., Hoet R. M., Veerkamp J. H. (1998) Generation and application of type-specific anti-heparan sulfate antibodies using phage display technology. Further evidence for heparan sulfate heterogeneity in the kidney. J. Biol. Chem. 273, 12960–12966 [DOI] [PubMed] [Google Scholar]
- 30. Dore J.M., Morard F., Vita N., Wijdenes J. (1998) Identification and location on syndecan-1 core protein of the epitopes of B-B2 and B-B4 monoclonal antibodies. FEBS Lett. 426, 67–70 [DOI] [PubMed] [Google Scholar]
- 31. David G., Bai X. M., Van der Schueren B., Cassiman J. J., Van den Berghe H. (1992) Developmental changes in heparan sulfate expression. In situ detection with mAbs. J. Cell Biol. 119, 961–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Langford J. K., Stanley M. J., Cao D., Sanderson R. D. (1998) Multiple heparan sulfate chains are required for optimal syndecan-1 function. J. Biol. Chem. 273, 29965–29971 [DOI] [PubMed] [Google Scholar]
- 33. Deepa S. S., Umehara Y., Higashiyama S., Itoh N., Sugahara K. (2002) Specific molecular interactions of oversulfated chondroitin sulfate E with various heparin-binding growth factors. Implications as a physiological binding partner in the brain and other tissues. J. Biol. Chem. 277, 43707–43716 [DOI] [PubMed] [Google Scholar]
- 34. Kokenyesi R., Bernfield M. (1994) Core protein structure and sequence determine the site and presence of heparan sulfate and chondroitin sulfate on syndecan-1. J. Biol. Chem. 269, 12304–12309 [PubMed] [Google Scholar]
- 35. Barden J. A., Cuthbertson R. M., Jia-Zhen W., Moseley J. M., Kemp B. E. (1997) Solution structure of parathyroid hormone related protein (residues 1–34) containing an Ala substituted for an Ile in position 15 (PTHrP[Ala15]-(1–34)). J. Biol. Chem. 272, 29572–29578 [DOI] [PubMed] [Google Scholar]
- 36. Pioszak A. A., Parker N. R., Gardella T. J., Xu H. E. (2009) Structural basis for parathyroid hormone-related protein binding to the parathyroid hormone receptor and design of conformation-selective peptides. J. Biol. Chem. 284, 28382–28391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pioszak A. A., Parker N. R., Suino-Powell K., Xu H. E. (2008) Molecular recognition of corticotropin-releasing factor by its G-protein-coupled receptor CRFR1. J. Biol. Chem. 283, 32900–32912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Runge S., Thøgersen H., Madsen K., Lau J., Rudolph R. (2008) Crystal structure of the ligand-bound glucagon-like peptide-1 receptor extracellular domain. J. Biol. Chem. 283, 11340–11347 [DOI] [PubMed] [Google Scholar]
- 39. Parthier C., Kleinschmidt M., Neumann P., Rudolph R., Manhart S., Schlenzig D., Fanghänel J., Rahfeld J. U., Demuth H. U., Stubbs M. T. (2007) Crystal structure of the incretin-bound extracellular domain of a G protein-coupled receptor. Proc. Natl. Acad. Sci. U.S.A. 104, 13942–13947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McKown R. L., Raab R. W., Kachelries P., Caldwell S., Laurie G. W. (2013) Conserved regional 3′ grouping of rare codons in the coding sequence of ocular prosecretory mitogen lacritin. Invest. Ophthalmol. Vis. Sci. 54, 1979–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lugemwa F. N., Esko J. D. (1991) Estradiol β-d-xyloside, an efficient primer for heparan sulfate biosynthesis. J. Biol. Chem. 266, 6674–6677 [PubMed] [Google Scholar]
- 42. Dahlbäck M., Jørgensen L. M., Nielsen M. A., Clausen T. M., Ditlev S. B., Resende M., Pinto V. V., Arnot D. E., Theander T. G., Salanti A. (2011) The chondroitin sulfate A-binding site of the VAR2CSA protein involves multiple N-terminal domains. J. Biol. Chem. 286, 15908–15917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ten Dam G. B., Kurup S., van de Westerlo E. M., Versteeg E. M., Lindahl U., Spillmann D., van Kuppevelt T. H. (2006) 3-O-Sulfated oligosaccharide structures are recognized by anti-heparan sulfate antibody HS4C3. J. Biol. Chem. 281, 4654–4662 [DOI] [PubMed] [Google Scholar]
- 44. Zhong M., Navratil A. M., Clay C., Sanborn B. M. (2004) Residues in the hydrophilic face of putative helix 8 of oxytocin receptor are important for receptor function. Biochemistry 43, 3490–3498 [DOI] [PubMed] [Google Scholar]
- 45. Couchman J. R. (2003) Syndecans. Proteoglycan regulators of cell-surface microdomains? Nat. Rev. Mol. Cell Biol. 4, 926–937 [DOI] [PubMed] [Google Scholar]
- 46. Häcker U., Nybakken K., Perrimon N. (2005) Heparan sulfate proteoglycans. The sweet side of development. Nat. Rev. Mol. Cell Biol. 6, 530–541 [DOI] [PubMed] [Google Scholar]
- 47. Bradley R. S., Brown A. M. (1990) The proto-oncogene int-1 encodes a secreted protein associated with the extracellular matrix. EMBO J. 9, 1569–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li Y. S., Milner P. G., Chauhan A. K., Watson M. A., Hoffman R. M., Kodner C. M., Milbrandt J., Deuel T. F. (1990) Cloning and expression of a developmentally regulated protein that induces mitogenic and neurite outgrowth activity. Science 250, 1690–1694 [DOI] [PubMed] [Google Scholar]
- 49. Paralkar V. M., Hammonds R. G., Reddi A. H. (1991) Identification and characterization of cellular binding proteins (receptors) for recombinant human bone morphogenetic protein 2B, an initiator of bone differentiation cascade. Proc. Natl. Acad. Sci. U.S.A. 88, 3397–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tsutsui J., Uehara K., Kadomatsu K., Matsubara S., Muramatsu T. (1991) A new family of heparin-binding factors. Strong conservation of midkine (MK) sequences between the human and the mouse. Biochem. Biophys. Res. Commun. 176, 792–797 [DOI] [PubMed] [Google Scholar]
- 51. Rubin J. B., Choi Y., Segal R. A. (2002) Cerebellar proteoglycans regulate sonic hedgehog responses during development. Development 129, 2223–2232 [DOI] [PubMed] [Google Scholar]
- 52. Adams J. C., Kureishy N., Taylor A. L. (2001) A role for syndecan-1 in coupling fascin spike formation by thrombospondin-1. J. Cell Biol. 152, 1169–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Peterson S. B., Liu J. (2010) Unraveling the specificity of heparanase utilizing synthetic substrates. J. Biol. Chem. 285, 14504–14513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Esko J. D., Selleck S. B. (2002) Order out of chaos. Assembly of ligand binding sites in heparan sulfate. Annu. Rev. Biochem. 71, 435–471 [DOI] [PubMed] [Google Scholar]
- 55. Khan S., Rodriguez E., Patel R., Gor J., Mulloy B., Perkins S. J. (2011) The solution structure of heparan sulfate differs from that of heparin. Implications for function. J. Biol. Chem. 286, 24842–24854 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 56. McKown R. L., Wang N., Raab R. W., Karnati R., Zhang Y., Williams P. B., Laurie G. W. (2009) Lacritin and other new proteins of the lacrimal functional unit. Exp. Eye Res. 88, 848–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Koo B. S., Lee D. Y., Ha H. S., Kim J. C., Kim C. W. (2005) Comparative analysis of the tear protein expression in blepharitis patients using two-dimensional electrophoresis. J. Proteome Res. 4, 719–724 [DOI] [PubMed] [Google Scholar]
- 58. Nichols J. J., Green-Church K. B. (2009) Mass spectrometry-based proteomic analyses in contact lens-related dry eye. Cornea 28, 1109–1117 [DOI] [PubMed] [Google Scholar]
- 59. Zhou L., Beuerman R. W., Chan C. M., Zhao S. Z., Li X. R., Yang H., Tong L., Liu S., Stern M. E., Tan D. (2009) Identification of tear fluid biomarkers in dry eye syndrome using iTRAQ quantitative proteomics. J. Proteome Res. 8, 4889–4905 [DOI] [PubMed] [Google Scholar]
- 60. Velez V. F., Romano J. A., McKown R. L., Green K., Zhang L., Raab R. W., Ryan D. S., Hutnik C. M., Frierson H. F., Jr., Laurie G. W. (2013) Tissue transglutaminase is a negative regulator of monomeric lacritin bioactivity. Invest. Ophthal. Vis. Sci. 54, 2123–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mantelli F., Schaffer L., Dana R., Head S. R., Argüeso P. (2009) Glycogene expression in conjunctiva of patients with dry eye. Down-regulation of Notch signaling. Invest. Ophthalmol. Vis. Sci. 50, 2666–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shafat I., Vlodavsky I., Ilan N. (2006) Characterization of mechanisms involved in secretion of active heparanase. J. Biol. Chem. 281, 23804–23811 [DOI] [PubMed] [Google Scholar]